Abstract
Rad23 was identified as a DNA repair protein; although a role in protein degradation has been described. The protein degradation function of Rad23 contributes to cell cycle progression, stress response, ER proteolysis, DNA repair. Rad23 binds the proteasome through a ubiquitin-like (UbL) domain, and contains ubiquitin-associated (UBA) motifs that bind multiubiquitin chains. These domains allow Rad23 to function as a substrate shuttle-factor. This property is shared by structurally similar proteins (Dsk2 and Ddi1), and is conserved among the human and mouse counterparts of Rad23. Despite much effort, the regulation of Rad23 interactions with ubiquitinated substrates and the proteasome is unknown. We report here that Rad23 is extensively phosphorylated in vivo and in vitro. Serine residues in UbL are phosphorylated, and influence Rad23 interaction with proteasomes. Replacement of these serine residues with acidic residues, to mimic phosphorylation, reduced proteasome binding. We reported that when_UbL is overexpressed, it can compete with Rad23 for proteasome interaction and inhibit substrate turnover. This effect is not observed with UbL containing acidic substitutions, consistent with results that phosphorylation inhibits interaction with the proteasome. Loss of both Rad23 and Rpn10 caused pleiotropic defects that were suppressed by overexpressing either Rad23 or Rpn10. Rad23 bearing a UbL domain with acidic substitutions failed to suppress rad23Δ rpn10Δ, confirming the importance of regulated Rad23/proteasome binding. Strikingly, Threonine-75 in human HR23B also regulates interaction with the proteasome, suggesting that phosphorylation is a conserved mechanism for controlling Rad23/proteasome interaction.
Keywords: Rad23, proteasome, UbL, degradation, phosphorylation
Introduction
Rad23 was first characterized as a DNA repair factor that is required for nucleotide excision repair (NER: reviewed in references 1, 2). The NER mechanism is required for the removal of bulky DNA adducts, and mutations in multiple complementation groups can lead to xeroderma pigmentosum in humans (reviewed in 1). A complex consisting of Rad23 and Rad4 performs a key role in recognizing bulky lesions in DNA 2. The loss of Rad4 (XPC in human) prevents DNA incision, which leads to a complete NER defect. In contrast, loss of yeast Rad23 causes a partial decrease in UV survival. However, DNA incision occurs in rad23Δ, suggesting that the defect in this mutant occurs at a late step in the NER mechanism. We discovered that Rad23 could interact with the proteasome. This finding revealed a novel role for proteolysis in DNA repair 3. In agreement, proteasome mutant's show reduced survival after UV light, although the specific requirement for proteolysis in NER has not been determined.
Distinct methods have been utilized to assess the role of Rad23 and the proteasome in DNA repair 3; 4. However, these studies have not yielded concordant results. We note that DNA incision is an early step in NER that does not reflect the efficiency of subsequent events including DNA patch filling, ligation, chromatin remodeling and recovery from growth arrest. In contrast, yeast cell survival is an end-point assay that is determined several days after DNA injury, and does not take into account defects in earlier biochemical steps.
Rad4 protein is constitutively degraded by the ubiquitin/proteasome pathway, but is stabilized through its interaction with Rad23 5. The intermediate UV sensitivity of rad23Δ is partly due to accelerated turnover of Rad4. However, the UV sensitivity of rad23Δ is not overcome by stabilizing Rad4, suggesting that Rad23 has additional functions that cannot be bypassed by solely providing more Rad4. Because DNA incision occurs efficiently in rad23Δ, Rad23 appears to function later in the NER pathway. The Rad23 requirement for efficient cell-cycle progression 6, cellular response to environmental toxins 6, and ER-associated degradation 7, are all linked to its role as a shuttle-factor. The UbL domain in yeast Rad23 binds the Rpn1 subunit in the proteasome 8, and a substrate ubiquitination factor, Ufd2. In contrast, the UbL domains in two human counterparts (hHR23A; hHR23B) bind the proteasome subunit S5a 9 and Ataxin-3 10;11 A distinct binding domain in Rad23 binds Rad4 12, and similarly human Rad23 proteins contain a unique motif that binds XPC 13. The two UBA domains (ubiquitin-associated) in Rad23 bind multiUb chains. The carboxy-terminal UBA2 domain can also binds Png1 14. The human UBA2 domain interacts with other regulatory factors including HIV-1 encoded Vpr 15, p300/CREB 16 and Mpg1 17. The significance of these interactions is not clear.
The proteolytic role of Rad23 explains its widespread involvement in stress response, transcription, ER associated protein degradation, and NER 18. Removal of the UbL prevents interaction with the proteasome and causes UV sensitivity, underscoring a role for the proteasome in NER. An important property of a ‘shuttle-factor’ function (such as Rad23) is to interact with the proteasome only when it is bound to cargo (multiUb proteolytic substrates). In agreement, overexpression of the UbL domain caused unregulated interaction with the proteasome, and stabilization of proteolytic substrates. Significantly, it is not known how Rad23/proteasome interaction is regulated, to prevent such an unregulated interaction. Similarly, the regulation of Rad23 interactions with multiubiquitinated substrates has not been investigated. Regulation of Rad23/proteasome interaction provides a way to repeatedly deliver multiubiquitinated proteins to the proteasome 19. Although there is some evidence that monomeric and dimeric states of Rad23 might regulate its activity 20; 21, it is unclear how it oscillates between these two forms. Rad23 interaction with the proteasome is independent of its interactions with other factors including MPG 17, Png1 14, Vpr 15 and p300 16, because the isolated UbL domain can bind proteasomes efficiently. It is also unclear how Rad23 interaction with Rad4 is distinct from its interaction with multiubiquitinated proteolytic substrates.
We determined that Rad23 is phosphorylated in vivo. This post-translational modification provides a promising avenue for understanding its regulation and function. Mass spectrometry analysis showed that several residues in Rad23 are phosphorylated in vivo, and in vitro. In this report we examine the phosphorylation of serine residues in the UbL domain, and test their effect on proteasome binding and substrate turnover. We determined that UbL-phosphorylation inhibited Rad23/proteasome binding and reduced protein degradation by the proteasome. We speculate that Rad23 phosphorylation controls its association with the proteasome, thereby accomplishing the repeated delivery of ubiquitinated proteolytic substrates.
Results
Multiple residues in Rad23 are phosphorylated in vivo
Yeast cells were grown in medium containing [32P]-orthophosphate and protein extracts were prepared. Flag-Rad23 was immunoprecipitated and separated in a denaturing polyacrylamide gel. 32P-Flag-Rad23 was excised and subjected to acid hydrolysis, and the hydrolysate was resolved by thin-layer chromatography (TLC) (Fig. 1a). Unlabeled serine, threonine and tyrosine were combined and two different loadings were separated (lanes 4 and 5). The positions of these unlabeled amino acid residues were determined by staining with ninhydrin. The TLC plate was subsequently exposed to X-ray film and 32P-serine (S), 32P-threonine (T) and 32P-tyrosine (Y) were detected. To confirm these results we immunoprecipitated Flag-Rad23 from unlabeled cells and treated one half of the sample with lambda phosphatase (+ λPPase; Fig. 1b). The proteins were separated by 2-dimensional gel electrophoresis, and an antibody against Rad23 detected five spots. Significantly, most of these spots were lost following exposure to λPPase, consistent with their dephosphorylation. Two spots remained after dephosphorylation, representing the unphosphorylated form of Rad23, and one containing a single phosphate. We speculate this could arise if this residue were inaccessible to λPPase.
Fig. 1.
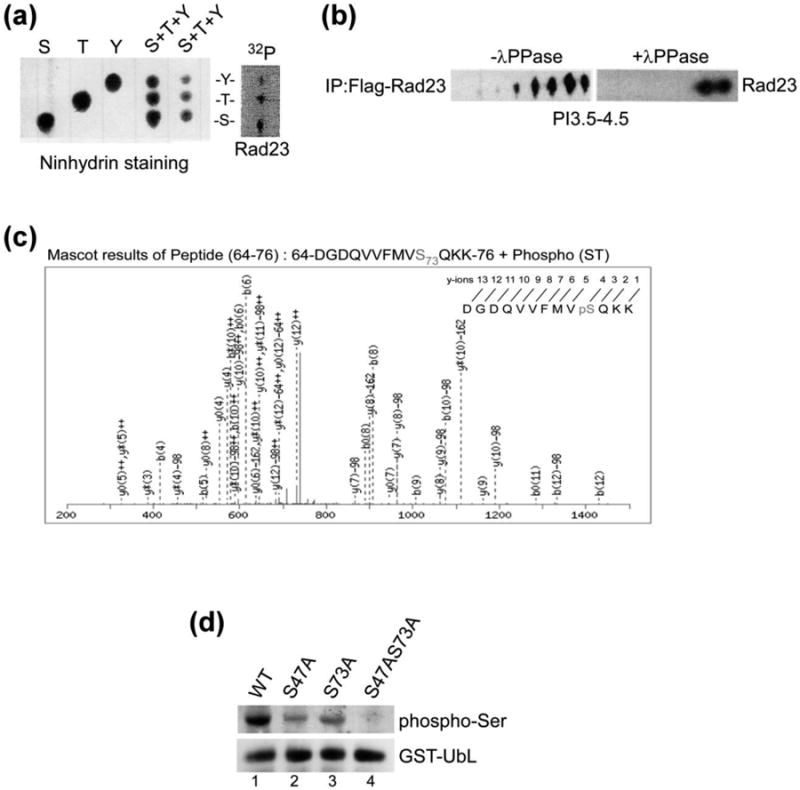
Rad23 are phosphorylated at multiple residues in vivo. (a) Yeast cells expressing Flag-Rad23 was affinity purified from exponential-phase yeast cells that were incubated with [32P]-orthophosphate for 1 h. The in vivo phosphorylated Flag-Rad23 was separated by SDS-PAGE, and transferred to PVDF membrane. 32P-labeled Flag-Rad23 was identified by autoradiography, excised and digested in acid. The hydrolysate was separated by one-dimensional thin-layer chromatography (TLC). The positions of non-radioactive phosphoamino acid standards were detected by staining with ninhydrin (left panel). The independent standards and two different amounts of a mixture of the three combined standards are shown. The pattern of radioactive spots generated from the hydrolyzed of 32P-Flag-Rad23 is shown on the right (Flag-Rad23), and the positions of phosphorylated tyrosine (Y), threonine (T), and serine (S) residues are indicated. (b) Flag-Rad23 was immunoprecipitated from yeast cells, and incubated with or without λ-phosphatase for 1 h. Proteins were released from the affinity beads and resolved by isoelectric focusing (IEF). The separated proteins were resolved in the second dimension in SDS-PAGE, transferred to nitrocellulose and incubated with anti-Flag antibody. (c) Recombinant GST-Rad23 was purified from E. coli BL21 (DE3) cells, extensively washed by lysis buffer and subjected to in vitro kinase assay followed by LC-MS/MS analysis. (d) GST-UbL and mutants with changes in candidate phosphorylation sites were isolated from rad23Δ strain and in vivo phosphorylation was investigated by immunoblotting, using antibodies that recognize phospho-serine residues.
We purified GST-Rad23 from E. coli and incubated the immobilized protein with extract prepared from wild type yeast. GST-Rad23 was then subjected to mass spectrometry analysis (LC-MS/MS), and a number of phosphorylated residues were identified. We were intrigued by the phosphorylation of residues in the UbL domain, because this structure has a well-characterized role in binding the Rpn2 protein in yeast proteasomes. In contrast, the UBA domains in Rad23 have multiple binding partners that could confound the characterization of their phosphorylation. Because UbL/proteasome interaction is essential for all Rad23 activities, the regulation of this function is important. To strengthen our in vitro studies we isolated GST-UbL from yeast cells and characterized the protein by mass spectrometry. These studies confirmed that Ser-73 in the UbL domain is also phosphorylated in vivo. A representative mass spectrometry profile of peptide 64-76 shows the in vitro phosphorylation of Ser-73 (Fig. 1c). However, we also identified residues that were differentially phosphorylated in vitro and in vivo. For instance, Ser-47 was phosphorylated in vitro, whereas Ser-57 and Ser-59 were phosphorylated in vivo. Since there are seven Ser/Thr residues in this general region, there may be flexibility in which residues are targeted for phosphorylation. We also note that the phosphorylation of specific residues could be regulated in vivo, but not in vitro. Further study will be required to verify the physiological relevance of other phosphorylated residues. Full-length Rad23 that was characterized in vivo showed phosphorylation of Thr-94 and Thr-139. Both residues lie outside the UbL domain. Intriguingly the polypeptide sequence flanking these residues are highly similar (90-ESASTPG-96 and 135-ESATTPG-141, respectively), suggesting that they may be targeted by the same kinase. We note that ∼ 70 amino acid sequence between UbL and UBA1 is highly enriched in Ser/Thr residues (> ∼ 1/3rd), and many conform to potential phosphorylation sites.
Serine-47 and Serine-73 in the UbL domain are important sites for phosphorylation
The structure of the yeast ubiquitin-like (UbL) domain was determined at the atomic level, and strong similarity to ubiquitin was observed 22. However, unlike ubiquitin and other ubiquitin-like modifiers, the UbL domain in Rad23 protein is not excised 23 and conjugated to other proteins. The yeast UbL domain binds the proteasome subunit Rpn1 8, whereas the human counterparts of Rad23 bind the S5a subunit in the proteasome 9. The Rad23 UbL domain also interacts with Ufd2 11; 24 and ataxin-3 10, which are also associated with the protein degradation pathway. The absolute requirement for UbL in binding the proteasome 3 led us to focus on the effect of phosphorylation on its function. Human and mouse Rad23 counterparts contain a threonine residue at the position corresponding to Ser-73 in yeast Rad23. Although serine and threonine residues are not necessarily interchangeable, as illustrated by the fact that only threonine can function as a nucleophile in the proteasome peptidases 25, both residues are structurally similar and can be phosphorylated. In addition to Ser-73, mass spectrometry of UbL purified from yeast showed that three additional Ser/Thr residues were phosphorylated in vivo.
UbL was expressed as a fusion to glutathione S-transferase (GST) and Ser-47 and Ser-73 were converted to alanine. GST-UbL was affinity purified, separated by SDS-PAGE, and an immunoblot was incubated with antibody against phospho-serine (Fig. 1d). A strong cross-reaction was observed with the anti-phosphoserine antibody, whereas the interaction with GST-UbLS47A and GST-UbLS73A was reduced significantly. We characterized GST-UbL to specifically focus on residues in this domain that might be phosphorylated. As expected, the phosphorylation of a double mutant (GST-UbLS47A S73A) was reduced further.
We reasoned that conversion of a phosphorylated residue to an acidic amino acid might mimic the effect of a phosphorylated residue, as has been described by others 26; 27. Ser-47 and Ser-73 were converted to acidic residues (S47E; S73D). GST-UbL and mutant derivatives were expressed in yeast containing an epitope-tagged proteasome subunit (Pre1-FLAG). GST proteins modified at Ser-47 were affinity purified on glutathione-Sepharose and immunoblots were reacted with antibodies against Flag and GST (Fig. 2a). Antibody reaction against the GST and GST-UbL proteins in whole cell extracts (WCE) showed that they were expressed at similar levels. The level of Pre1-FLAG was also equivalent in all strains. After affinity purification, similar amounts of GST proteins were isolated. GST alone did not co-purify Pre1-Flag. (A higher-molecular weight band represents a cross-reaction against GST). Both GST-UbL (WT) and GST-UbLS47A co-precipitated Pre1-Flag (lanes 2 and 3). In contrast, GST-UbLS47E showed significantly reduced interaction with the proteasome, as indicated by the lower co-purification of Pre1-Flag, despite high levels in the extract (lane 4). Similarly, the interaction between the proteasome and GST-UbLS73A and GST-UbLS73D was examined (Fig. 2b). In agreement with the findings in Fig. 2a, we observed reduced proteasome (Pre1-FLAG) interaction with GST-UbLS73D (lane 4). (The FLAG antibody reaction showed non-specific reaction against GST, seen at the top of Lane 1). We also tested the co-purification of another proteasome subunit and similar findings were observed (Fig. S1).
Fig. 2.
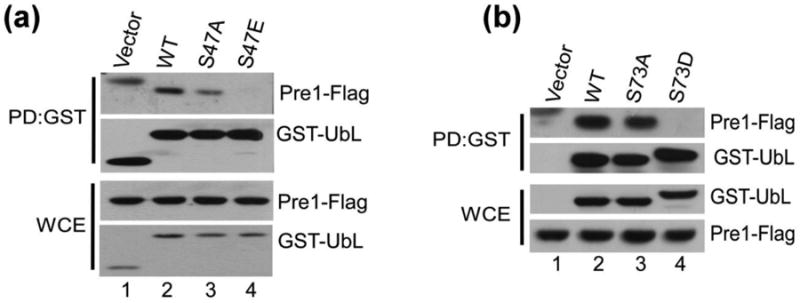
Phospho-mimetic mutations of Ser47 and Ser73 in UbL domain prevent Rad23/proteasome interaction in vivo. (a)(b) GST-UbL and variants with mutations in this domain were expressed in rad23Δ strain that expressed Pre1-Flag, and the interaction between the proteasome and the UbL domain was examined.
We investigated if the reduced proteasome-interaction displayed by specific UbL mutants also occurred in the context of the full-length protein. A yeast strain expressing the 20S proteasome subunit Pre2-HA was transformed with an empty vector, or plasmids expressing wildtype Flag-Rad23, Flag-rad23S47A, and Flag-rad23S47E. Protein extracts were prepared and applied to Flag-agarose, and immunoblots were incubated with antibodies against HA (Fig. 3a). The Flag-tagged Rad23/rad23 proteins were recovered efficiently on the affinity beads. However, the co-purification of Pre2-HA was reduced with Flag-rad23S47E (lane 4), but not Flag-rad23S47A. The filter was incubated next with antibody against Rpt1 and reduced binding to this 19S proteasome subunit was observed. In contrast, the co-purification of Rpt1 with Flag-rad23S47A was not affected. There were no detectable non-specific interactions associated with extracts containing vector and the Flag-agarose matrix (lane 1).
Fig. 3.
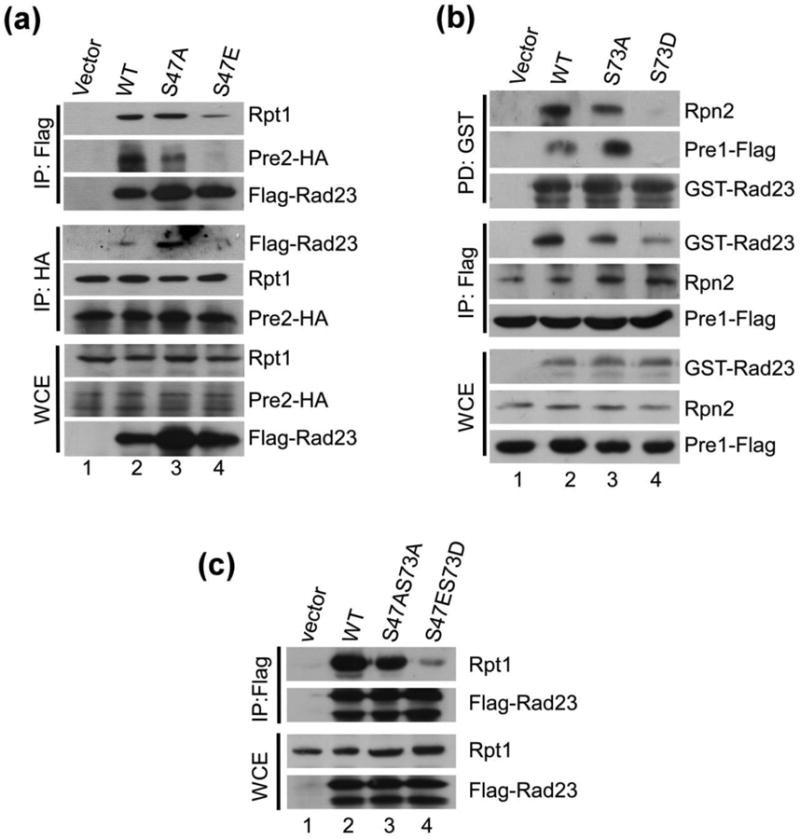
Phospho-mimetic mutations of Ser47 and Ser73 in Rad23 prevent Rad23/proteasome interaction in vivo. (a) Flag-Rad23, Flag-Rad23S47A and Flag-Rad23S47E were expressed in rad23Δ that also expressed proteasome subunit Pre2-HA. Whole cell extracts (WCE) were prepared and equal amount of protein was incubated with Flag-agarose. The co-purification of the proteasome, with Rad23 and mutant derivatives, was determined by immunoblotting. (b) GST-Rad23, GST-Rad23S73A and GST-Rad23S73D were expressed in rad23Δ expressing a different epitope-tagged proteasome subunit, Pre1-Flag. As described in (a), the interaction between 26S proteasome and Rad23 was investigated by either purifying the GST-tagged Rad23 proteins, or the proteasome subunit Pre1-Flag. Rad23/proteasome interaction was gauged by immunoblotting. (c) The consequence of a UbL mutant bearing both Rad23S47AS73A and Rad23S47ES73D mutations was examined by isolating the full-length proteins from yeast cells and examining interaction with the proteasome. A vector control (lane 1) showed that nonspecific precipitation of the proteasome and Rad23 was negligible.
In a reciprocal study Pre2-HA was immunoprecipitated and the co-purification of Flag-Rad23 was investigated. Immunoblotting showed that a lower amount of Flag-rad23S47E was co-purified with Pre2-HA (lane 4). In contrast, Pre2-HA interaction with the 19S subunit Rpt1 was not affected, indicating that the interaction between the 19S and 20S proteasome particles was unaffected. The Flag-tagged Rad23 proteins were expressed efficiently in all strains (WCE: lower panels). We also tested the effect of mutations of Ser-73 (Fig. 3b). These Rad23 derivatives were expressed as fusions to glutathione S-transferase (GST). GST, GST-Rad23, GST-rad23S73A and GST-rad23S73D were expressed in a yeast strain containing Pre1-FLAG. Protein extracts were applied to glutathione-Sepharose and immunoblotting showed significantly reduced interaction between GST-rad23S73D and the proteasome (lane 4). Neither Rpn2, nor Pre1-FLAG was isolated with GST-rad23S73D, whereas GST-Rad23 and GST-rad23S73A showed similar interactions with the proteasome. In agreement, Pre1-FLAG was purified with reduced levels of GST-rad23S73D, but GST-Rad23 and GST-rad23S73A were efficiently co-purified.
We investigated if a double mutant would have a more severe defect in binding the proteasome. Flag-Rad23, Flag-rad23S47A S73A and Flag-rad23S47E S73D were expressed in wildtype cells. Protein extracts were applied to Flag-agarose and the bound proteins were characterized by immunoblotting (Fig. 3c). The interaction between Flag-rad23S47E,S73D and the proteasome was significantly reduced, whereas Flag-rad23S47A,S73A interaction with the proteasome was similar to that observed with Flag-Rad23.
Unregulated UbL/proteasome binding can inhibit protein turnover
We showed previously that overexpression of the UbL domain can stabilize proteolytic substrates 19. This effect is caused by a non-productive interaction between the yeast UbL domain and the proteasome. We therefore surmised that if UbL phosphorylation reduced proteasome binding then a mutation such as S73D should not affect the turnover of proteasome substrates. We expressed GST-UbL and GST-UbLS73D in yeast cells that contained Ub-Pro-β-gal, which is a well characterized test substrate of the proteasome 28. High-levels of GST-UbL caused stabilization of Ub-Pro-βgal (Fig. 4a), which is degraded rapidly by the proteasome. The expression levels of native Rad23, GST-UbL and a stable protein (Pab1) were unaffected during the cycloheximide chase. In contrast, expression of GST-UbLS73D failed to affect the rapid turnover of Ub-Pro-β-gal, consistent with our findings reported here that phosphorylation of the UbL domain reduces interaction with the proteasome. Expression of a double mutant GST-UbLS47E S73D also failed to inhibit the rapid degradation of Ub-Pro-β-gal, whereas the expression of GST-UbL caused strong stabilization (Fig. 4b). The double mutant exerted a stronger effect than the Ser-73 → Asp-73 substitution mutation, indicating that phosphorylation of both Ser-47 and Ser-73 affect proteasome binding. Next, we examined the effect of the Rad23S47E,S73D mutation on proteolysis by the ubiquitin-proteasome pathway. Flag-Rad23 and its mutant variants were co-expressed in rad23Δ cells with the test substrate Ub-Pro- β-galactosidase (Ub-Pro-β-gal). An expression shut-off assay was used to measure steady-state β-gal enzymatic activity. We detected ∼ 3-fold higher β-gal activity in rad23Δ cells expressing the double mutant Flag-Rad23S47E,S73D (Fig. 4c). In contrast, similar β-gal activity was measured in extracts containing Flag-Rad23 and Flag-Rad23S47A,S73A. We conclude that the lower β-gal activity was the result of successful delivery of Ub-Pro-β-gal to the proteasome by Flag-Rad23 and Flag-Rad23S47A,S73A. These results were confirmed by immunoblotting analysis (Fig. S2).
Fig. 4.
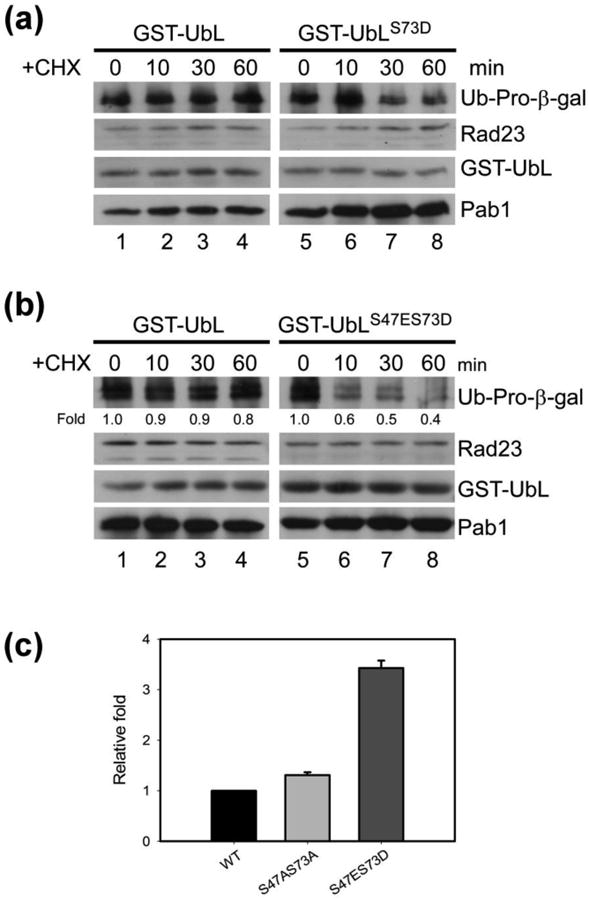
Stabilization of proteolytic substrates by Rad23 phospho-mimetic mutants. (a) overexpression of the Rad23 UbL domain can interfere with protein degradation. Ub-Pro-β-gal was co-expressed with either GST-UbL or GST-UbLS73D. Ub-Pro-β-gal was detected in total extract during a cycloheximide chase, by immunoblotting, and moderate stabilization was observed. (b) Ub-Pro-β-gal was co-expressed with GST-UbL or the double mutant GST-UbLS47ES73D in wildtype yeast Pab1 levels were determined as a loading control. These results are representative of three independent experiments. (c) The reporter substrate Ub-Pro-β-gal was co-expressed with Flag-Rad23, Flag-Rad23S47AS73A, or Flag-Rad23 S47ES73D in rad23Δ. β-galactosidase activity was measured in whole cell extracts to estimate Ub-Pro-β-gal stability. β-gal activity representing the average of four independent experiments were normalized to the value detected in the wildtype strain.
Phosphorylation of the UbL domain is required for efficient growth and response to stress
We reported previously that the Rad23 and Rpn10 proteins have overlapping functions 6. Loss of both proteins (rad23Δ rpn10Δ) causes pleiotropic defects that are not observed in either single mutant (rad23Δ; rpn10Δ). As expected, expression of either Rad23 or Rpn10 fully rescued the pleiotropic defects of rad23Δ rpn10Δ. This finding offered an opportunity to test the importance of UbL/proteasome interaction using specific phosphorylation mutants. Removal of the UbL domain from yeast Rad23 causes intermediate UV sensitivity 5; 6. However, the removal of this domain (77 aa residues) could cause unforeseen structural changes. The ability to regulate Rad23/proteasome binding through single amino acid substitutions (rad23S47A,S73A) offered a unique way to characterize the significance of this interaction without inducing major steric and structural perturbations. We expressed Rad23, rad23S47A,S73A and rad23S47ES73D in rad23Δ rpn10Δ, and spotted ten-fold dilutions of actively growing cultures on synthetic medium (Fig. 5a). The agar plates were incubated at the permissive (30°C; left panel) and non-permissive temperatures (13°C; right panel). The poor growth of rad23Δ rpn10Δ at 13°C was suppressed by expression of Rad23, and rad23S47A,S73A. In contrast, rad23S47E,S73D was unable to confer growth to rad23Δ rpn10Δ at 13°C.
Fig. 5.
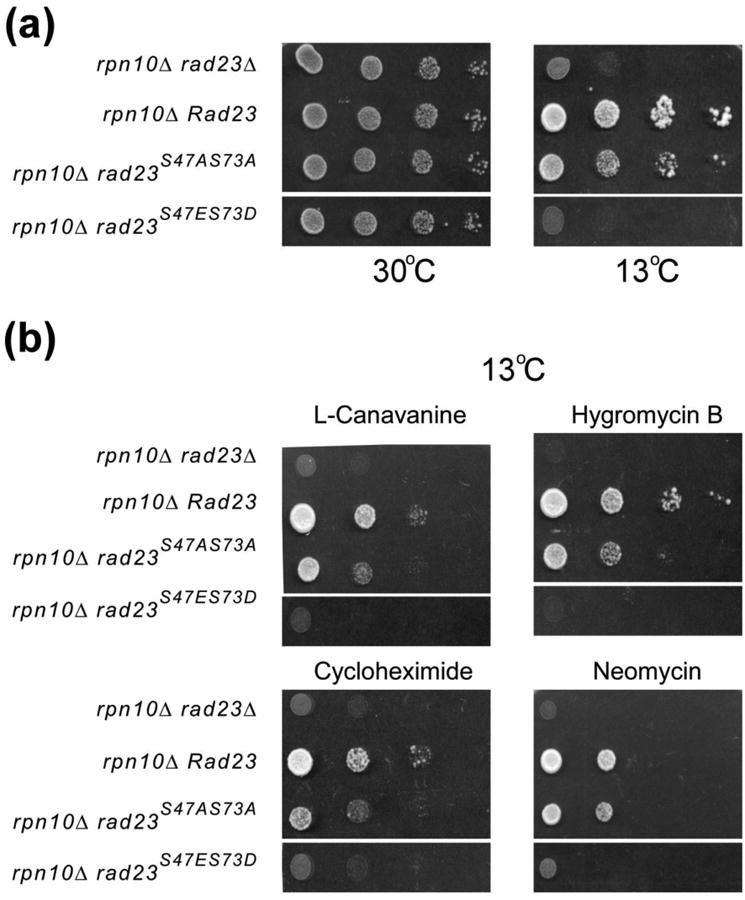
Altered stress response by yeast cells expressing Rad23 phospho-mimetic mutations. Wildtype Rad23 or derivatives with mutations in Ser47 and Ser73 were expressed in rad23Δ rpn10Δ. Mid-log-phase cultures were normalized to a density of A600 ∼ 1.0, and ten-fold dilutions were spotted on selective synthetic medium plates. (a) The plates were incubated at 30°C or at 13°C until growth was observed. (b) Mid-log-phase cultures were spotted on medium containing 0.5 μg/ml L-canavanine, 0.1 μg/ml cycloheximide, 2 μM Neomycin, or 0.2 μM Hygromycin B. These results are representative of three independent experiments.
We also examined growth in the presence of drugs that severely impede the growth of rad23Δ rpn10Δ. We confirmed that expression of Rad23 suppressed all the drug-specific effects in rad23Δ rpn10Δ (Fig. 5b). Expression of rad23S47A,S73A partially suppressed the poor growth of in rad23Δ rpn10Δ, caused by L-canavanine, hygromycin-B, cycloheximide and neomycin. The proteasome is required for the elimination of damaged proteins that are induced by exposure to these drugs 29.
Regulated proteasome interaction by Rad23 is required for efficient cell-cycle progression
The rad23Δ rpn10Δ mutant displays a strong G2-specific delay during cell cycle progression 6. This growth defect is likely to be caused by a failure to degrade cell-cycle specific regulatory factors. Regulated Rad23/proteasome interaction might contribute directly to the turnover of these key proteins. We therefore investigated if the absence of Rad23/proteasome interaction contributed to the G2-phase delay in rad23Δ rpn10Δ. We used a cell sorter to monitor cell cycle progression in rad23Δ rpn10Δ expressing Rad23 or phosphorylation defective mutants (Fig. 6). At 30°C the distribution of G1, G2 and M phase cells was similar in all four strains (panel a), although a higher proportion of G2 cells was evident in rad23Δ rpn10Δ. After transfer to 13°C cells expressing Rad23 had an equivalent proportion of G1 and G2 phase cells (38.4% and 34.8% respectively). However, the level of G2-phase cells increased dramatically in rad23Δ rpn10Δ (G1 = 13.4%; G2 = 62.7%). Expression of either Rad23 or rad23S47A S73A in the double mutant restored normal distribution of cells at 13°C. In contrast, rad23S47E S73D failed to alleviate the G2-phase growth delay in rad23Δ rpn10Δ, demonstrating that Rad23/proteasome interaction is required for efficient cell-cycle progression.
Fig. 6.
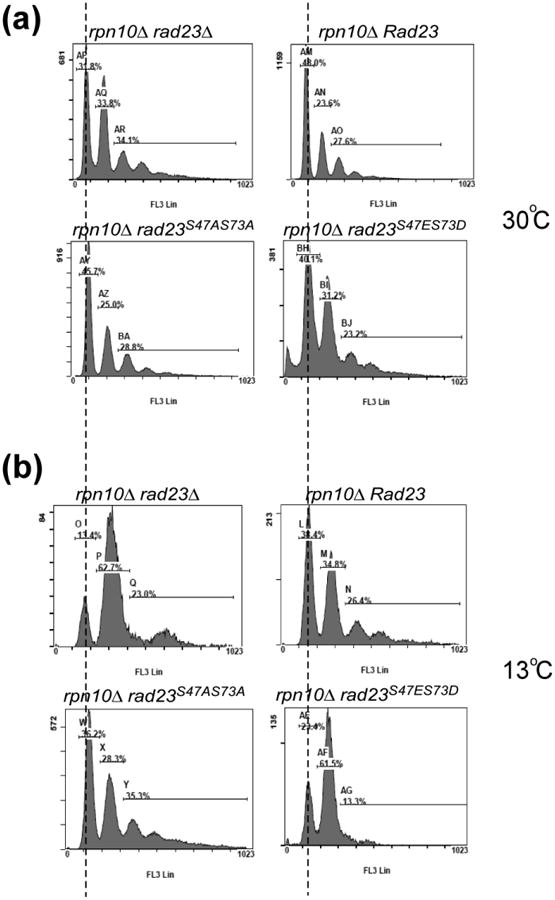
The G2/M-phase delay in rad23Δ rpn10Δ is not suppressed by Rad23S47ES73D Wildtype Rad23 or mutations in various phosphorylation sites were expressed in rad23Δ rpn10Δ. Yeast cells were grown to the mid exponential-phase at (a) 30°C or (b) 13°C, fixed in 70% ethanol at -20°C, and examined by flow cytometry.
Phosphorylation of the UbL domain in human hHR23B inhibits interaction with the proteasome
To determine if the mechanism for regulating yeast Rad23/proteasome interaction is conserved, we examined the human Rad23 counterpart hHR23B. Sequence analysis showed that Thr-75 in hHR23B corresponds to the Ser-73 residue in Rad23 that we examined here. Thr-75 was converted to either alanine (T75A) or aspartate (T75D), and the protein derivatives were expressed with a Myc epitope in human lung carcinoma cell line A549. A549 contained a hHR23B-specific lentiviral expressed knock-down construct. Protein extracts were incubated with anti-Myc antibody to immunoprecipitate Myc-hHR23B and mutant derivatives. We found that both wildtype (WT) and T75A mutants were able to co-precipitate the Rpn2 proteasome subunit (Fig. 7). In contrast, the T75D mutant showed significantly reduced interaction with Rpn2. The expression of Rpn2 and the hHR23B proteins was similar in all strains (WCE). These findings demonstrate that our characterization of the Rad23 protein in yeast is applicable to the human Rad23 protein, implying a common regulatory mechanism.
Fig. 7.
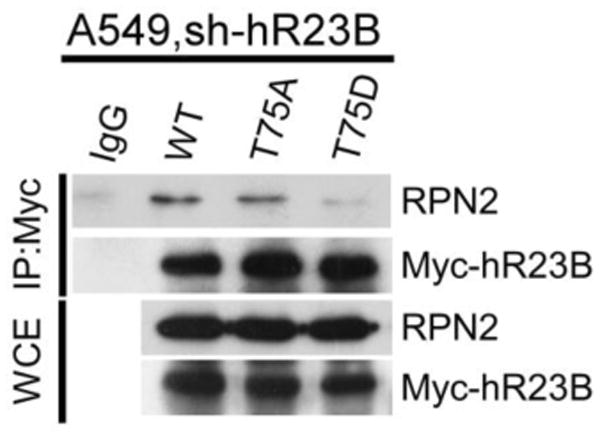
Phosphorylation of the UbL domain in human hHR23B inhibits interaction with the proteasome. Thr-75 in hHR23B was converted to either alanine (T75A) or aspartic acid (T75D), and the protein derivatives were expressed with a Myc epitope in human lung carcinoma cell line A549. A549 contained a hHR23B-specific lentiviral expressed knock-down construct. Protein extracts were incubated with anti-Myc antibody to immunoprecipitate Myc-hHR23B and mutant derivatives. The co-precipitated proteasome was determined by immunoblot analysis using antibody against Rpn2.
Discussion
Three distinct structural motifs have been identified in the family of Rad23 proteins. The yeast Rad23 protein is the paradigm for this class of proteins, and is generally believed to functions as a shuttle-factor that can deliver proteolytic substrates to the proteasome 19. This action requires two structures; an amino terminal ubiquitin-like (UbL) domain that binds the proteasome 3, and two ubiquitin-associated (UBA) domains that bind multiubiquitin chains 30; 31. In addition, Rad23 contains a Rad4-binding sequence that can bind and control Rad4 stability 32. The UbL domain has been reported to bind the Rpn1 subunit in the proteasome 8, the Ufd2 factor 11, and Ataxin-3 10. The UBA domains are believed to bind many multiubiquitinated cellular proteins, as well as numerous specific factors, including HIV-1 encoded Vpr protein 33, Png1 14, and p300 16. The significance of these interactions is not understood.
Rad23/proteasome interaction has been examined extensively, and its link to the Rpn10 proteasome receptor and other factors has been investigated 6; 34; 35. This interaction can be easily monitored, and its effect on protein turnover has been investigated. Previous studies that characterized a rad23 mutant protein lacking the entire UbL domain revealed proteolytic defects. Therefore, the discovery that phosphorylation of specific residues in the UbL domain affect Rad23/proteasome interaction allowed us to focus on investigating the significance of this association in protein degradation. Proteomic analysis of human Rad23 (hHR23B) that was purified in association with the proteasome showed phosphorylation of Serine-160.
We show here that residues in the UbL domain are phosphorylated in vivo and in vitro. Conversion of these residues to alanine did not have an appreciable effect on growth, or response to environmental stresses. However, conversion to an acidic residue caused strong inhibition of binding to the proteasome. This deficiency could be assessed directly by measuring the co-purification of proteasome subunits, and also using recombinant protein that was incubated with yeast extracts. The biological relevance of blocking proteasome interaction using either the full-length protein (rad23S47E,S73D), or just the UbL domain (UbLS47E,S73D), was tested. Unregulated interaction between the UbL domain and the proteasome can strongly interfere with intracellular protein breakdown. However, a mutant version that is unable to bind the proteasome (UbLS47E,S73D) did not interfere with the turnover of a test substrate of the proteasome. We suggest that regulated Rad23/proteasome interaction is physiologically important because rad23Δ rpn10Δ cells expressing rad23S47E,S73D were sensitive to drugs that cause protein unfolding, and showed a strong delay in the G2-phase of the cell cycle.
Our findings raise interesting questions about the nature of Rad23/proteasome binding. Based on the hypothesis that Rad23 can deliver multiubiquitinated proteins to the proteasome, it is important to determine if this interaction occurs only after Rad23 has bound cargo. Specifically, an unregulated interaction between UbL and the proteasome can inhibit protein degradation 19. We speculate that unphosphorylated Rad23 binds multiubiquitinated proteins and traffics them to the proteasome. Following the delivery of a proteolytic substrate Rad23 might become phosphorylated to trigger its release from the proteasome. This mechanism would allow Rad23 to renew another cycle of substrate translocation to the proteasome. This mechanism might require a proteasome-associated kinase that specifically phosphorylated Rad23, and other shuttle-factors, after the delivery of substrates. Other scenarios are also possible, although we believe this model provides a straightforward interpretation of the data. Many kinases are associated with the proteasome, although further study will be required to identify one that can target Rad23. It is intriguing in this regard that the Snf1 kinase has been linked to Rad23 function in DNA repair, although its site of action has not been described.
Materials and Methods
Yeast strains and plasmids
Yeast cultures were grown in rich (YPD) or synthetic media containing 2% glucose or galactose. The Saccharomyces cerevisiae rad23Δ and rad23Δrpn10Δ strains, and strains expressing chromosomal hemagglutinin (HA)-tagged Rad4 were as previously described 5. Similarly, plasmids expressing Flag-tagged Rad23, rad23ΔUbL, Pre2-HA, Pre1-Flag, Ub-Arg-β-galactosidase (Arg-β-gal), Ub-Pro-β-galactosidase (Ub-Pro-β-gal), Rpn8-V5, GST-tagged Rad23, and UbL were as described 3; 19; 32,36. Full-length human Rad23B (hHR23B) cDNA was amplified from A549 cells and cloned into pCMV-Myc with XhoI and EcoRI sites. The substitutions of the serine-47 and serine-73 residues to alanine or aspartic acid/glutamic acid were achieved using a QuickChange II Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) and the following DNA oligonucleotides:
S47A forward: 5′-CAAATAAAACTGATCTACGCGGGTAAAGTGCTAC-3′
S47A reverse: 5′-GTAGCACTTTACCCGCGTAGTACAGTTTTATTTTG-3′
S47E forward: 5′-CAAATAAAACTGATCTACGAGGGTAAAGTGCTAC-3′
S47E reverse: 5′-GTAGCACTTTACCCTCGTAGTACAGTTTTATTTTG-3′
S73A forward: 5′-GTCTTCATGGTTGCTCAAAAAAAG-3′
S73A reverse: 5′-CTTTTTTTGAGCAACCATGAAGAC-3′
S73D forward: 5′-GTCTTCATGGTTGATCAAAAAAAG-3′
S73D reverse: 5′-CTTTTTTTGATCAACCATGAAGAC-3′.
T75A (hHR23B) forward: 5′-GTGGTTATGGTGGCCAAACCCAAAG-3′
T75A (hHR23B) reverse: 5′-CTTTGGGTTTGGCCACCATAACCAC-3′
T75D (hHR23B) forward: 5′-GTGGTTATGGTGGACAAACCCAAAG-3′
T75D (hHR23B) reverse: 5′-CTTTGGGTTTGTCCACCATAACCAC-3′
In all cases, multiple isolates were confirmed by DNA sequencing.
Chemicals and antibodies
The antibody against Pab1 was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The polyclonal antibodies against β-galactosidase, Rpn2, HA, and V5 were from Abcam, Inc. (Cambridge, MA, USA). Anti-Flag M2-agarose beads, iodoacetamide, hydroxyurea (HU), L-canavanine, neomycin, hygromycin B and cycloheximide were purchased from Sigma Chemical Co. (St. Louis, MO, USA). The polyclonal antibodies against Rpt1 and Rad23 were gifted from Dr. Madura (UMDNJ, Piscataway, NJ, USA). The Glutathione-Sepharose 4B beads were purchased from GE Healthcare (Piscataway, NJ, USA). Ortho-nitrophenyl-β-galactoside (ONPG) was purchased from MP Biomedicals, Inc. (Solon, Ohio, USA). Dithiothreitol (DTT) was purchased from Amresco, Inc. (Solon, Ohio, USA). λ-Phosphatase was purchased from Cell Signaling Technology (Beverly, MA, USA). 4-Nitroquinoline-1-oxide (4-NQO) was from Lancaster (Morecambe, UK). All other chemicals were purchased from Sigma, unless otherwise specified.
Protein expression shut-off assay
Yeast cells expressing β-galactosidase from the GAL1 promoter were grown at 30°C to an OD600 of ∼1 in a synthetic 2% raffinose medium, lacking uracil. Protein expression was induced by the addition of 2% galactose for 2 h and then repressed by the addition of 2% glucose. Cycloheximide (0.5 mg/ml) was then added to stop protein synthesis. Samples were withdrawn at the indicated time points and cells harvested by centrifugation, and protein extracts were prepared for immunoblotting or immunoprecipitation, as indicated.
Immunoprecipitation and Western blotting
Yeast strains containing plasmids were grown in synthetic medium, pelleted, and frozen at -20°C. For analysis, the cells were suspended in buffer A [50 mM HEPES, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 50 mM NaF, 1 mM Na3VO4, 10% glycerol, and protease inhibitors (Roche, Mannheim, Germany)] and lysed by disruption with glass beads. The extracts were centrifuged at 12,000 rpm for 5 min at 4°C, and the protein concentrations were determined using the Bradford assay (Bio-Rad, Hercules, CA, USA). Equal amounts of total protein were adjusted to 500 μl with buffer A containing 20 μl anti-Flag M2-agarose beads (for Flag-tagged proteins) or Glutathione- Sepharose 4B beads (for GST-fusion proteins). The samples were incubated at 4°C for 2-3 h, and the beads were washed three times with l ml buffer A. The bound proteins were boiled for 5 min, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and incubated with the appropriate primary antibodies. For the detection of ubiquitin conjugates, the nitrocellulose membranes were boiled for 5 min prior to incubation with appropriate secondary antibodies. The signal was developed using an enhanced chemiluminescence (ECL) kit (Perkin Elmer, Boston, Massachusetts, USA).
One-dimensional thin-layer chromatography (TLC)
Yeast cells expressing Flag-Rad23 were grown to the logarithmic (Log) phase in the presence of [32P]-orthophosphate (GE Healthcare) for 1 h. In vivo phosphorylated Flag-Rad23 was recovered by immunoprecipitation, resolved by SDS-PAGE, and transferred to a PVDF membrane. 32P-labeled Flag-Rad23 was excised and digested in acid, and the hydrolysate was separated by one-dimensional thin-layer chromatography. The positions of non-radioactive phosphoamino acid standards were detected by staining with ninhydrin.
Drug sensitivity assay
Yeast cells were grown to mid-log-phase in selective medium and normalized to a density of A600 nm ∼ 1. Ten-fold serial dilutions were spotted on selective synthetic medium plates containing different chemicals. The plates were then wrapped in aluminum foil and incubated at 30°C or 13°C until colonies could be imaged.
Yeast cell cycle phase analysis
Yeast cells were grown to mid-log-phase in selective synthetic medium, washed with phosphate-buffered saline (PBS), and fixed in 70% ethanol at -20°C. The cells were washed with 50 mM Tris (pH7.5), suspended in PBS containing 1 mg/ml RNase A, and incubated at 37°C for 2 h. The cells were centrifuged and washed with PBS, proteinase K (40 μg/ml) was added, and the cells were incubated at 55°C for an additional hour. Propidium iodide (PI) was added at a final concentration of 20 μg/ml. For analysis, the cells were sonicated twice to disrupt aggregates, and then immediately subjected to flow cytometry using a Beckman Coulter FC500 (Beckman, Brea, CA, USA). Cell cycle phases were identified and plotted.
Supplementary Material
Highlights.
Rad23 is a shuttle-factor that delivers proteolytic substrates to the proteasome
Rad23 is phosphorylated in vivo.
Ser residues in the UbL domain, which binds the proteasome, are phosphorylated.
The phosphorylation of UbL in Rad23 prevents interaction with the proteasome.
Phosphorylation provides a mechanism to regulate Rad23/proteasome interaction.
Acknowledgments
This work was supported in part by grants to S.-M. Chuang (NSC94-2312-B-005-003; NSC95-2311-B-005-014-MY3 from the National Science Council, Taiwan) and to KM (CA083875 from the National Cancer Institute and GM104968 from the National Institutes of Health). We thank Irving Vega for conducting preliminary studies. Members of the laboratory are thanked for critical review of the manuscript.
The mass spectrometry data were obtained from an Orbitrap instrument funded in part by an NIH grant NS046593 (H. Li), for the support of the Neuroproteomics Core Facility-New Jersey Medical School.
Abbreviations used
- Ub
ubiquitin
- UbL
ubiquitin-like
- UBA
ubiquitin-associated
- NER
nucleotide excision repair
- TLC
thin-layer chromatography
- λPPase
lambda phosphatase
- GST
glutathione S-transferase
- WCE
whole cell extracts
Footnotes
Authors Statement: The authors have no competing financial interests in this work
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guzder SN, Sung P, Prakash L, Prakash S. Affinity of yeast nucleotide excision repair factor 2, consisting of the Rad4 and Rad23 proteins, for ultraviolet damaged DNA. J Biol Chem. 1998;273:31541–6. doi: 10.1074/jbc.273.47.31541. [DOI] [PubMed] [Google Scholar]
- 3.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–8. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 4.Russell SJ, Reed SH, Huang W, Friedberg EC, Johnston SA. The 19S regulatory complex of the proteasome functions independently of proteolysis in nucleotide excision repair. Mol Cell. 1999;3:687–95. doi: 10.1016/s1097-2765(01)80001-0. [DOI] [PubMed] [Google Scholar]
- 5.Ortolan TG, Chen L, Tongaonkar P, Madura K. Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Res. 2004;32:6490–500. doi: 10.1093/nar/gkh987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–7. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–30. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 9.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–25. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 10.Doss-Pepe EW, Stenroos ES, Johnson WG, Madura K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol Cell Biol. 2003;23:6469–83. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–65. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen LE, Verhage RA, Brouwer J. Preferential binding of yeast Rad4.Rad23 complex to damaged DNA. J Biol Chem. 1998;273:33111–4. doi: 10.1074/jbc.273.50.33111. [DOI] [PubMed] [Google Scholar]
- 13.van der Spek PJ, Eker A, Rademakers S, Visser C, Sugasawa K, Masutani C, Hanaoka F, Bootsma D, Hoeijmakers JH. XPC and human homologs of RAD23: intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res. 1996;24:2551–9. doi: 10.1093/nar/24.13.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki T, Park H, Kwofie MA, Lennarz WJ. Rad23 provides a link between the Png1 deglycosylating enzyme and the 26 S proteasome in yeast. J Biol Chem. 2001;276:21601–7. doi: 10.1074/jbc.M100826200. [DOI] [PubMed] [Google Scholar]
- 15.Withers-Ward ES, Jowett JB, Stewart SA, Xie YM, Garfinkel A, Shibagaki Y, Chow SA, Shah N, Hanaoka F, Sawitz DG, Armstrong RW, Souza LM, Chen IS. Human immunodeficiency virus type 1 Vpr interacts with HHR23A, a cellular protein implicated in nucleotide excision DNA repair. J Virol. 1997;71:9732–42. doi: 10.1128/jvi.71.12.9732-9742.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu Q, Wani G, Wani MA, Wani AA. Human homologue of yeast Rad23 protein A interacts with p300/cyclic AMP-responsive element binding (CREB)-binding protein to down-regulate transcriptional activity of p53. Cancer Res. 2001;61:64–70. [PubMed] [Google Scholar]
- 17.Miao F, Bouziane M, Dammann R, Masutani C, Hanaoka F, Pfeifer G, O'Connor TR. 3-Methyladenine-DNA glycosylase (MPG protein) interacts with human RAD23 proteins. J Biol Chem. 2000;275:28433–8. doi: 10.1074/jbc.M001064200. [DOI] [PubMed] [Google Scholar]
- 18.Dantuma NP, Heinen C, Hoogstraten D. The ubiquitin receptor Rad23: at the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 2009;8:449–60. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–13. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diaz-Martinez LA, Kang Y, Walters KJ, Clarke DJ. Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div. 2006;1:28. doi: 10.1186/1747-1028-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walters KJ, Lech PJ, Goh AM, Wang Q, Howley PM. DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc Natl Acad Sci U S A. 2003;100:12694–9. doi: 10.1073/pnas.1634989100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen YW, Tajima T, Agrawal S. The crystal structure of the ubiquitin-like (UbL) domain of human homologue A of Rad23 (hHR23A) protein. Protein Eng Des Sel. 2011;24:131–8. doi: 10.1093/protein/gzq084. [DOI] [PubMed] [Google Scholar]
- 23.Watkins JF, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13:7757–65. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baek GH, Kim I, Rao H. The Cdc48 ATPase modulates the interaction between two proteolytic factors Ufd2 and Rad23. Proc Natl Acad Sci U S A. 2011;108:13558–63. doi: 10.1073/pnas.1104051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kisselev AF, Songyang Z, Goldberg AL. Why does threonine, and not serine, function as the active site nucleophile in proteasomes? J Biol Chem. 2000;275:14831–7. doi: 10.1074/jbc.275.20.14831. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhary A, King WG, Mattaliano MD, Frost JA, Diaz B, Morrison DK, Cobb MH, Marshall MS, Brugge JS. Phosphatidylinositol 3-kinase regulates Raf1 through Pak phosphorylation of serine 338. Curr Biol. 2000;10:551–4. doi: 10.1016/s0960-9822(00)00475-9. [DOI] [PubMed] [Google Scholar]
- 27.Sweeney HL, Yang Z, Zhi G, Stull JT, Trybus KM. Charge replacement near the phosphorylatable serine of the myosin regulatory light chain mimics aspects of phosphorylation. Proc Natl Acad Sci U S A. 1994;91:1490–4. doi: 10.1073/pnas.91.4.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 29.Chuang SM, Chen L, Lambertson D, Anand M, Kinzy TG, Madura K. Proteasome-mediated degradation of cotranslationally damaged proteins involves translation elongation factor 1A. Mol Cell Biol. 2005;25:403–13. doi: 10.1128/MCB.25.1.403-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat Struct Biol. 2001;8:417–22. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Shinde U, Ortolan TG, Madura K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2001;2:933–8. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ortolan TG, Tongaonkar P, Lambertson D, Chen L, Schauber C, Madura K. The DNA repair protein rad23 is a negative regulator of multi-ubiquitin chain assembly. Nat Cell Biol. 2000;2:601–8. doi: 10.1038/35023547. [DOI] [PubMed] [Google Scholar]
- 33.Dieckmann T, Withers-Ward ES, Jarosinski MA, Liu CF, Chen IS, Feigon J. Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat Struct Biol. 1998;5:1042–7. doi: 10.1038/4220. [DOI] [PubMed] [Google Scholar]
- 34.Elsasser S, Chandler-Militello D, Muller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–22. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 35.Goh AM, Walters KJ, Elsasser S, Verma R, Deshaies RJ, Finley D, Howley PM. Components of the ubiquitin-proteasome pathway compete for surfaces on Rad23 family proteins. BMC Biochem. 2008;9:4. doi: 10.1186/1471-2091-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L, Madura K. Centrin/Cdc31 is a novel regulator of protein degradation. Mol Cell Biol. 2008;28:1829–40. doi: 10.1128/MCB.01256-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


