Abstract
BRAF inhibitors (BRAFi) have led to clinical benefit in patients with melanoma. The development of a blood-based assay to detect and quantify BRAF levels in these patients has diagnostic, prognostic, and predictive capabilities that could guide treatment decisions. Blood BRAFV600E detection and quantification was performed on samples from 128 patients with Stage II (19), III (67), and IV (42) melanoma. Tissue BRAF analysis was performed in all patients with Stage IV disease and in selected patients with Stage II and III disease. Clinical outcomes were correlated to initial BRAF levels as well as BRAF level dynamics. Serial analysis was performed on 17 Stage IV melanoma patients treated with BRAFi and compared to tumor measurements by RECIST. The assay was highly sensitive (96%) and specific (95%) in the Stage IV setting, using a blood level of 4.8 pg as “positive”. BRAF levels typically decreased following BRAFi. A subset of these patients (5) had an increase in BRAF V600E values 42-112 days prior to clinical or radiographic disease progression (PD). From 86 patients with resected, stage II or III melanoma, 39 had evidence of disease relapse (45.3%). Furthermore, BRAF mutation in the blood after surgical resection in these patients was not associated with a difference in relapse risk, though tissue BRAF status was only available for a subset of patients. In summary we have developed a highly sensitive and specific, blood-based assay to detect BRAFV600 mutation in patients with melanoma.
Keywords: BRAF V600E, biomarker, melanoma test, TspR1, vemurafenib, daBRAFenib, trametanib
Introduction
Metastatic melanoma is currently the 5th and 7th most common cancer in American men and women, respectively, and remains one of the few cancers with a rising incidence.(1) Over 9000 people are expected to die in the United States in 2013 from this disease.(1) Recent treatment advances have led to the FDA approval of two BRAF inhibitors, vemurafenib (Zelboraf) and dabrafenib (Tafinlar), a MEK inhibitor, trametinib (Mekinist), and the immunotherapy ipilimumab (Yervoy) for the treatment of patients with advanced melanoma.(2-6) Unfortunately, resistance to BRAF and MEK inhibitor therapy is common, response to ipilimumab uncommon, and durable response to any therapy infrequent; as such, the overwhelming majority of these patients eventually will die of their disease.(7, 8) Most patients with BRAF mutant disease will be candidates for multiple lines of therapy, but conventional radiographic monitoring to track response and progression fails to identify patients at a point when they can receive benefit from follow-on therapy. There is a critical need to develop highly sensitive blood-based biomarkers that could enable better treatment selection and improved monitoring of patients with advanced and high-risk melanoma.
Current, standard BRAF testing methods are tissue-based and provide only qualitative data, i.e. positive or negative.(9-14) The major limitations to these approaches are lack of sensitivity and the need to acquire tissue (either via location of an archived tumor block or fresh biopsy). Most tissue-based assays have the ability to identify one mutant allele in ten or twenty wild-type alleles and thus require tumor specimens that contain approximately 40-50% tumor cellularity to account for heterozygosity and stromal and lymphoid elements typically present in melanoma metastases.(9-15) While most metastatic tumor biopsies have little trouble meeting this benchmark, analysis of primary melanomas and microscopically involved sentinel nodes are less reliable due to tumor heterogeneity (primary tumors) and/or relative infrequency of tumor cells (sentinel lymph nodes).(16, 17) Further, the identification of an appropriate block or the coordination of biopsy and subsequent analysis delays the start of systemic therapy. In these circumstances, a highly sensitive blood-based assay would provide a superior diagnostic tool.
A blood-based assay also would provide serial data about the state of the disease. For example, patients with resected melanoma have a risk of recurrence and death that ranges from 7-80%. While clinical and pathological staging can narrow the range, it is still broad for each stage of cancer and serial blood testing and imaging is of little value in improving prognostic accuracy.(18) An assay that rises in the setting of disease recurrence would likely enhance the predictive value of imaging and allow for timely diagnosis and treatment of recurrent melanoma. During the treatment of metastatic disease, blood tests that can serve as a surrogate marker of disease status and substitute for more expensive and difficult radiographic imaging, would offer a cost effective option to imaging and allow earlier transition to next line therapy for patients with emerging resistant disease.
We previously described the development of a highly sensitive and inexpensive, blood-based BRAF assay that took advantage of a unique restriction enzyme site in wild-type BRAF at the V600 position.(19) Our current assay continues to target this restriction digest site and also adds a Real-Time PCR step that allows for precise quantification of BRAF levels. In this report, we describe this enhanced assay and present testing results from patients with stage II, III, and IV melanoma.
Materials and Methods
Cell lines, tissue acquisition and oligonucleotides
The melanoma cell line A375, kidney cancer cell line 786-0, colon adenocarcinoma HT29 and prostate carcinoma DU145 were purchased in 2013 from ATCC (Manassas, Virginia, USA). All four cell lines were authenticated by isoenzymology and the Cytochrome C subunit I (COI) PCR assay was performed for confirmation of species. In addition, the cell lines had their identity confirmed by STR analyses. Oligonucleotides were custom synthesized from Invitrogen (Carlsbad, California, USA) and Sigma (St Louis, Mo).
The protocol in brief
The protocol is a quantitative modification of a previous assay. As illustrated in Figure 1A, an initial RT-PCR is followed by digestion with TspR1 (restriction site=NNCASTGNN), which preferentially digests the wild-type (TACAGTGAA) product but not the V600E mutated (TACAGAGAA) PCR product. In addition, none of the other less frequently reported V600 mutations (V600D, V600M, V600G, V600A, V600R, V600K, or V600G) are substrates for TspR1. A second, nested PCR using the digested material follows. After a second digestion with TspR1, the product is subjected to a real time PCR specific for the V600E mutation and not wild type sequences.
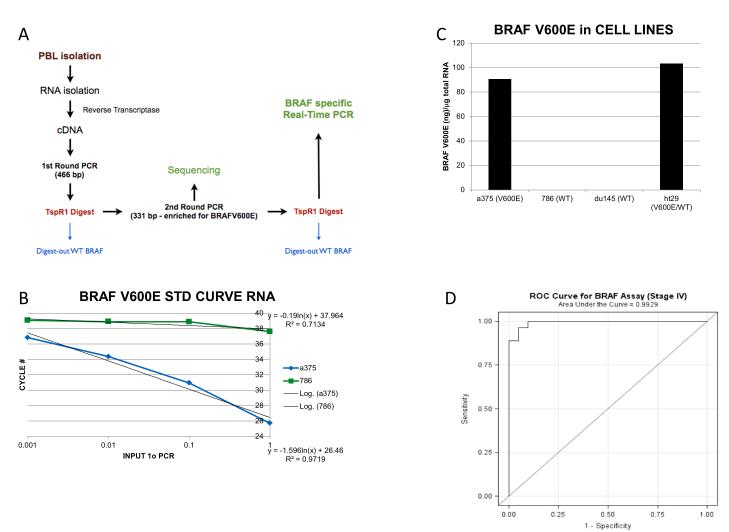
Figure 1.
(A) Schematic of the BRAFV600 assay; (B) Standard curve of the BRAF assay. The equation representative of the best fit line for the BRAFV600E (lower equation) and wild type BRAF (upper equation) are shown. (C) BRAF expression level in individual cell lines including a homozygous BRAFV600E mutant line (A375), two BRAF wild-type lines (786-0, Du145), and a heterozygous line (HT29). (D) Receiver operator curve for stage IV BRAF V600E patients. With AUC of 0.9929, the assay has excellent classification ability for Stage IV melanoma.
The protocol in detail
RNA from Ficoll purified PBLs or cell lines was isolated by the trizol method (Invitrogen) and (3 g) reverse transcribed to cDNA by standard methods using M-MLV reverse transcriptase (Invitrogen) and oligo (dt)15 (Promega).(20) The cDNA was subjected to real time PCR for 18S RNA in order to normalize the quantity, as well as quality of the input RNA prior to the next step (ABI for oligo/probe set). The equilibrated cDNA was PCR amplified using PCR master mix (Promega) and oligonucleotides [5 (CCATATCATTGAGACCAAATTTGAGATG)3 and 5 (GGCACTCTGCCATTAATCTCTTCATGG)3] that produced a product of 466 bp including the mutation site at position 600. The PCR conditions were 94 ° for 2 min followed by 40 cycles of 94 ° for 1 min, 60 ° for 2 min and 72 ° for 2 min with a final incubation of 72 ° for 7 min. After cleanup using a nucleospin extract column (Clontech), a portion of the PCR product was digested with TSPR1 (restriction site=NNCASTGNN, New England Biolabs, Beverly, Massachusetts, USA) at 65 ° for 16h. Only wild-type BRAF and not V600E mutant BRAF PCR product was digested by this enzyme. This digestion was added to reduce the amount of contaminating normal BRAF from surrounding and infiltrating normal tissue in the blood samples. A 1/100 dilution of the TSPR1 digested material was then PCR amplified a second time using nested oligonucleotides 5 (ACGCCAAGTCAATCATCCACAGAG)3 and 5 (CCGTACCTTACTGAGATCTGGAGACAGG)3 producing a product of 331 bp, which was enriched in PCR products containing the position 600 mutation. The conditions of the PCR were the same as the first PCR except the amplification was 45 cycles for PBLs instead of 40 cycles. After a second cleanup using a nucleo-spin extract column, the DNA (1/1000 dilution) was digested again with TspR1 and then subjected to a BRAFV600E real time PCR as described(21) The annealing and extension temperature was adjusted to 64° resulting in a more favorable amplification of the mutant as compared to the wild type templates (Figure 1B) than was reported (21). To further favor the mutant over the wild type product, a 33-fold excess of the reverse (common sequence in mutant and wild type) to forward (exact match for mutant and 1 base mismatch for wild type sequences) primers were used in the real time PCR assay. Therefore, after two rounds of TspR1 digestion it is highly unlikely that any remaining wild type product would have a significant impact on the assay. Purified BRAFV600E first round PCR product with a known concentration was also run through the assay and was used to create a standard curve. Using the standard curve the amount of end product was determined.
Peripheral blood isolation
Peripheral blood lymphocytes (PBLs) were obtained from 128 patients with advanced or cutaneous melanoma as part of IRB approved tissue-banking protocols (DFHCC 02-017 and 11-181). PBLs were isolated by Ficoll density centrifugation.(22) Of these 128 patients, 42 had stage IV disease and had blood collected specifically for this analysis between 2009 and 2012. PBL from the 19 patients with stage II melanoma and 67 patients with stage III disease were collected and isolated approximately 4-8 weeks following completion of surgical management as part of the Harvard Skin SPORE between 2002 and 2006. These samples were stored in freezing medium (95% fetal calf serum with 5% DMSO) at −80 deg. C (stage IV samples) or in liquid nitrogen (stage II and III samples). Furthermore, blood was drawn pre- and post-resection from 8 stage III patients previously determined to be BRAFV600E by tissue analysis. Only one sample per patient was available for most patients involved in this study and in all of the patients with Stage II or III disease (except for the aforementioned 8 patients with stage III disease with pre- and post-operative samples).
Serial blood samples were collected and assayed from twelve patients receiving the BRAF inhibitor vemurafenib and six patients receiving the combination of dabrafenib and trametinib. Samples were collected until RECIST-determined disease progression was documented in 17 of these 18 patients. In a subset of patients, different tubes were utilized after initiation of BRAF-directed therapy. Specifically, CPT tubes were utilized and the cellular component was removed and analyzed. Analyses comparing the two techniques (Ficoll isolation and CPT tube isolation) revealed a 7.5-fold greater BRAF level when Ficoll was used compared with CPT tube isolation (Supplemental Figure 1). As a result, only samples isolated using Ficoll isolation were analyzed in this study.
Tissue-based BRAF analysis
Patients with stage IV melanoma had BRAF mutational analysis on tumor tissue as part of standard of care either via the Cobas assay (Clarient Labs) or via SNaPshot (Massachusetts General Hospital Cancer Center Translational Research Laboratory). (23, 24)
Statistical analysis
Summaries of BRAF expression levels over time are presented using descriptive methods. Comparisons of BRAF expression according to mutational status or stage of disease were based linear regression models of natural log(BRAF) with mutational status or stage as the single predictor. Bonferroni corrections were used for pairwise comparisons to adjust for multiplicity. The distributions of relapse-free survival (RFS) and overall survival (OS) are described using the method of Kaplan-Meier. Five-year estimates of RFS and OS are presented with 95% confidence intervals calculated using log(-log(RFS or OS)) methodology. Statistical significance is defined as p<0.05; there are no adjustments for multiple comparisons.
Results
Generation of standard curves
Standard curves were generated using known amounts of purified 1° PCR products of wild type and V600E BRAF as templates. The assay can reliably detect as low as 1 pg. of BRAFV600E and exhibits a nearly 1000-fold difference in sensitivity for the V600E as compared to wild type BRAF PCR product (Figure 1B).
Sensitivity and specificity of assay
To examine the specificity of the assay, the protocol was used in four cell lines: A375, a melanoma line with a homozygous BRAFV600E mutation, HT29, an adenocarcinoma which is heterozygous for the BRAFV600E and 786-0 and Du145, renal cell carcinoma (RCC) and prostate cell lines, both wild type for BRAF. As shown in Figure 1C, using equal amounts of input RNA (3 ug.), the A375 and HT29 cells expressed nearly 10,000 fold greater BRAFV600E than either wild type cell line. Using a cut-off value of 4.8 pg. in blood samples, the sensitivity of the assay for stage IV patients is 96% and the specificity 95%. The area under the receiver operator curve (ROC) was 0.9929, demonstrating an excellent ability to discriminate patients with and without BRAF-mutant melanoma (Figure 1D).
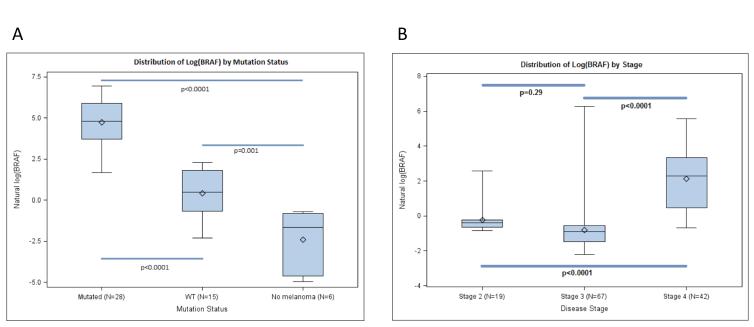
Samples obtained from 128 (42 Stage IV, 67 Stage III, and 19 Stage II) patients with melanoma were analyzed. A comparison of BRAF levels from 42 stage IV melanoma patients whose tumor biopsies had been previously determined to contain a V600E mutant (27 patients) or wild type BRAF (15 patients), 4 RCC patients (known to be BRAF WT), and two normal controls was performed (Figure 2A). Mean BRAFV600E levels were 50.3 pg., 1.7 pg., and 1.2 pg. for patients with mutant disease, wild type melanoma, or RCC/non-melanoma, respectively (p<0.0001).
Figure 2.
Distributions of BRAF levels by (A) disease stage or (B) mutation status. BRAF levels were transformed using natural logarithms (base e) (A) BRAFV600E level is higher in patients with tissue-detected BRAF mutation than in patients with Stage IV BRAF WT melanoma (p< 0.0001), and patients without melanoma (normal)(P<0.0001); (B) BRAFV600E level is higher among patients with Stage IV, BRAF mutant melanoma than in patients with Stage II and III melanoma with a “positive” BRAF level (> 4.8 pg; p<0.0001 for each). Statistical comparisons based on linear models with disease stage or mutation status as single predictor. Bonferroni corrections were used to adjust for multiple comparisons.
In the 86 patients with Stage II and III melanoma, the median BRAF value from the post resection blood draws was 0.48 pg, regardless of the BRAF status by tissue analysis.. A comparison of BRAF values in patients across stage II, III, and IV show that patients with stage IV melanoma had significantly higher blood BRAFV600E values (Figure 2B).
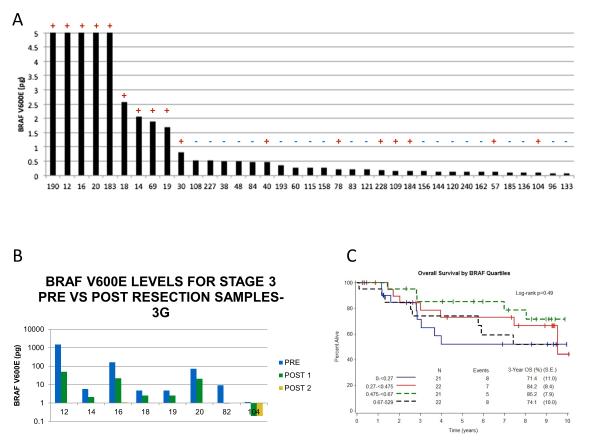
Thirty-seven of the 67 Stage III patients had tumor blocks suitable for tissue BRAF mutation analysis. In these patients, tissue-based analysis detected a BRAFV600E mutation in 17 patients. Using a cut-off value of 4.8 pg (determined from the Stage IV ROC curve) in blood samples, 5 of the 17 patients with tissue BRAF detection had an elevated blood value. Further, the ten highest post-operative BRAFV600E levels in these patients were all patients with known BRAF mutations based on tissue analysis and none of the 20 patients without BRAF V600E detection in tissue had an elevated V600E value (Figure 3A). Of note, the mean BRAF value in the 17 patients with known BRAF mutation in tissue was 21pg compared with 0.26 pg in the 20 patients without identified mutation in tissue, though this difference did not meet statistical significance (p< 0.128). In order to potentially explain the high number of post resection blood samples that had negative BRAFV600E levels despite being tissue positive, pre- vs. post-resection samples were compared. All samples were previously determined to be BRAFV600E by tissue analysis. As shown in Figure 3B, all 8 patients showed a marked decreased in BRAFV600E levels post surgery. Furthermore five patients had post-operative levels below the level of detection (4.8 pg) for this assay. The reduced tissue burden as a result of surgical resection provides a plausible explanation for the approximate 70% discordance between tissue and post surgical blood analysis by this assay. Additionally, comparisons in all 86 stage 2 and 3 patients of blood BRAF values post surgery were made according to stage (Stage II vs. Stage III), sub-stage (Stages II and III A, B, and C), and intermediate versus high risk, defined as risk of death < 50% (AJCC Stage IIA, IIB, or IIIA) or ≥ 50% (Stage IIC, IIIB, IIIC), and showed no significant differences.
Figure 3.
(A) BRAF V600E blood levels from stage 3 patients post resection compared to separate tissue analysis. The bar graph represents the blood BRAF V600E level in pg to a maximum level of 5 pg. The tissue BRAF V600E status is indicate by a + or - above each bar. (B) Blood BRAF V600E levels from eight patients pre- and post-surgical resection. All patients were previously determined to be BRAF V600E positive by tissue analysis. (C) Kaplan-Meier estimates for overall survival according to quartiles of the distribution of BRAFV600E levels (0-.27 pg; 0.27-0.45 pg; 0.45-0.67 pg and > 0.67 pg).
Predictive value of V600E BRAF blood in Stage II and III melanoma
In the 86 patients with resected, stage II or III melanoma, 39 had evidence of disease relapse (45.3%). In all these patients, detectable oncogenic BRAF mutation in the blood was not associated with a difference in the risk of relapse (5-year RFS: 52% vs. 57%, log-rank p=0.98) or death (5-year OS: 73% vs. 75%, log-rank p=0.88). Analysis of BRAF levels quartiles similarly showed no evidence of OS difference (Figure 3C). These findings show that, although not predictive of outcome at this time, this assay can detect the BRAFV600E mutation in resected stage II and III patients, regardless of the tumor BRAF status.
Analysis of serial blood samples from patients receiving BRAF targeted therapy
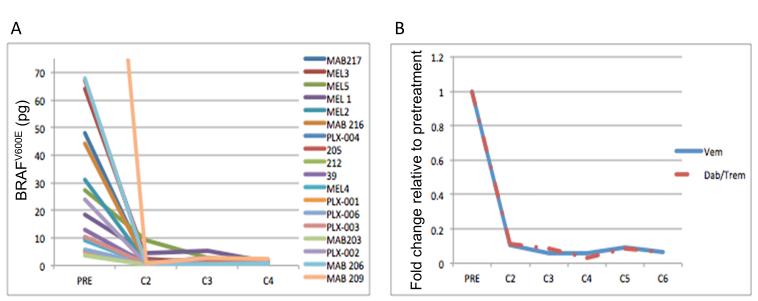
Blood BRAFV600E levels in 18 patients with BRAFV600E melanoma treated with either vemurafenib (12 patients) or the combination of dabrafenib and trametinib (6 patients) dramatically reduce following the commencement of therapy (Figure 4), and this reduction is similar in patients treated with single-agent BRAF or combination BRAF-MEK inhibitor therapy. Supplemental figure 2A-C shows the serial values of the BRAFV600E level in the blood of eleven patients treated with single agent vemurafenib and all six patients treated with dabrafenib and trametinib in whom tumor measurements from serial CAT scans acquired by patients until disease progression were also plotted alongside the blood BRAFV600E data. In the majority (15/17) of patients a reduction in blood BRAFV600E level correlated with disease response on imaging. After the decrease in BRAFV600E, five of the seventeen patients showed an increase in BRAFV600E in the blood 42-112 days prior to having treatment stopped due to disease progression (supplemental figure 2C). Of note in all 17 patients treatment was halted due to the occurrence of new lesions or non-target lesion progression and not due to target lesions progression.
Figure 4.
Blood BRAFV600E levels in 18 patients with BRAFV600E melanoma treated with either vemurafenib (12 patients) or the combination of dabrafenib and trametinib (6 patients) dramatically reduce following the commencement of therapy. The data in (A) show the response of individual patients to the drug regimen. (B) Mean BRAFV600E levels in response to either vemurafenib or the dabrafenib and trametinib combination. The values in (B) are relative to pretreatment levels (set as 1.0).
Discussion
Blood-based detection of oncogenic mutations has become increasingly widespread. Alternative methods make use of real time PCR, mass spectrometry, allelic specific PCR, PCR using locked oligonucleotides to suppress wild type sequences, direct sequencing of RNA or DNA to preferentially distinguish the mutant V600E from wild type BRAF, as well as a combination of emulsion-based digital PCR and flow cytometry (so-called Beads, Emulsion, Amplification, and Magnetics or BEAMing).(9, 15, 25-32) Our assay is unique due to our approach that leads to its high sensitivity and specificity. This is achieved by both the use of RNA from PBLs isolated from Ficoll, as it is postulated these cells contain either circulating tumor cells (CTCs) or other encapsulated, circulating RNA-containing factors released by tumors, and the reduction of background from wild type BRAF with the use of TspR1, a restriction enzyme that preferentially digests only the wild type sequence from the first PCR product.(19, 21) As a result, we have a highly sensitive assay (96% in patients with active, metastatic disease) that has exquisite specificity (95% in these same patients) and is able to provide quantitative information due to the use of V600E-specific Real-Time PCR.
An initial application of this assay would be to diagnose BRAF-mutant disease. Current tissue-based BRAF mutational methods can be challenging in archived primary melanomas (tumor heterogeneity) and microscopic nodal metastases (lack of sensitivity), and patients with newly metastatic melanoma often have rapidly progressive disease that requires urgent identification of mutational analysis. Analyzing blood for the BRAF mutation would prove to be a more efficient and possibly more reliable method of determining a patient’s BRAF status. The sensitivity and specificity seen with our assay is encouraging, though is limited by a relatively small sample size. Confirmation of this degree of sensitivity and specificity is required before broad-scale, blood-based analysis as the sole determinant of whether a melanoma harbors a BRAF-mutation can be adopted.
A second use of this assay would be in the Stage II and III setting, as the optimal follow-up of patients who are rendered disease-free with surgery for their melanoma is unknown.(33) The development and validation of a blood-based prognostic biomarker would offer the potential to improve the NCCN guidelines and direct radiographic imaging. Our data show that while blood BRAFV600E levels reduce following surgery, post-operative blood-based analysis may be useful in diagnosing BRAFV600E status. The major weakness with this data, however, is that in the majority of cases, tissue analysis was not performed due to either a lack of access to tumor blocks or insufficient disease in the blocks. Still, we demonstrate proof-of-concept that blood BRAFV600E levels can be detected in the blood of patients with resected melanoma. A second weakness is that we only have one post-operative blood value in the majority of these patients. It is conceivable that the BRAF level would change over time and that this change in level over time might be more predictive of outcome than a one-time value. Investigation into this specific application of this assay is underway (NCT01840527).
A third potential use of our assay would be to help guide the use of adjuvant therapy in patients with BRAF-mutant melanoma. There are two open phase III trials (NCT01667419, NCT01682083) testing the effectiveness of BRAF-directed therapy in patients with resected, high-risk disease; it is conceivable that adjuvant BRAF-directed therapy will become standard of care. If so, it is clear that the presence of a BRAF mutation in tissue will be requisite to qualify for this type of therapy. However, assessment of BRAF mutations in primary melanoma samples may be complicated by substantial tumor heterogeneity among cells in the primary tumor and between primary and metastatic tumors.(16, 17) This may lead to both false negatives and false positives, as it may not be clear which clones will ultimately establish metastasis. Our analysis of 37 patients with either Stage II or III disease who had both tissue and blood analysis performed showed congruous findings (i.e., both positive or both negative) in 25 (67%). Importantly, all 20 wild-type patients by tissue analysis were also wild type by blood analysis. Of the 17 patients with BRAFV600E by tissue analysis, only 5 were also V600E by blood. A plausible explanation for the 12 patients with a BRAF-mutation identified in tissue without detectable mutation in the blood may have had such low tumor volume following surgery that the circulating BRAF levels were below the limits of detection; thus a scenario where both the tissue and blood analysis may be true yet discordant. An alternative explanation would be that either the tissue analysis was falsely positive or the blood assay falsely negative. Prospective collection and analysis is underway in both the stage II and III setting to better define the utility of this assay in this setting (NCT01840527).
Finally, an assay that has the potential to identify tumor resistance to BRAF-directed therapy at an earlier time point is desperately needed. BRAFi resistance typically develops within 6-8 months following initial tumor regression, but with a range of 2 months to 2 years.(2, 4, 34, 35) Importantly, each described mechanism involves the retention of the initiating BRAF mutation.(36-44) As the mechanisms of resistance are now being elucidated, we feel that diagnostic assays, which may identify emerging resistance at an earlier time-point than standard clinical or radiographic assessments, will enable more prompt switching to another therapy. This is particularly important due to the fact that a number of patients treated with BRAF inhibitors progress quite quickly following initial disease regression.(45, 46) While there are no current data which specifically support such a strategy, it is thought that more advanced notice of disease progression, when disease growth is more modest, would allow for a more timely change in treatment and improved benefit of next line therapy. To date, only a small number of patients have been followed serially (17 presented here) with our assay in the context of BRAF-directed therapy. Still, this is the largest number presented to date followed with serial testing with a quantitative BRAF assay. Our findings - that BRAF level reduces with the initiation of BRAF inhibitor therapy and typically increases at the time of or in advance of radiographically-defined disease progression - are compelling. While these findings require confirmation, they also serve as a proof of concept that this type of assay may have value in this patient population and treatment setting.
There are a number of limitations of this study that serve to temper enthusiasm for the widespread adoption of this assay. First, our assay only measures BRAFV600E levels and is likely not useful for patients with BRAFV600K, or another non-V600E, V600 BRAF mutation. Second, all the samples used in the Stage II and III analysis were obtained between 2001-2006 while all the samples used for the Stage IV analysis were collected prospectively from 2010-2012. It is very possible that long-term storage (in this case stored frozen in liquid nitrogen) might affect the levels of BRAF expression. Third, it is difficult to interpret the Stage II and III data in the absence of corresponding tissue BRAF mutational analysis. Given the change of accurate testing of primary tumors and microscopically involved lymph nodes, this is a problem that may never be completely solvable, though prospective collection will likely lead to a higher percentage of patients with available tissue amenable to tissue-based analysis. Fourth, the fact that we were analyzing samples from a retrospective cohort meant that we only had one time point available for analysis in the majority of these Stage II and III patients. Depending on just one time-point in the clinical care continuum of these patients reduces the chances of showing the prognostic value of this assay. Fifth, the serial analyses of the BRAF mutant melanoma patients treated with BRAF-directed therapy involves a small number of patients with variable time points obtained. In particular, the on-treatment blood draws were initially going to be obtained at 2 weeks, 4 weeks, and then every 4 weeks; but soon after we changed this to every 4 weeks. Still, the inclusion of all patients allows for a more comprehensive look at this assay.
In conclusion, we have reported the clinical utility of the first blood-based, BRAF detection and quantification assay in a number of clinical settings. While our findings require confirmation prior to more extended adoption of this or similar clinical assays, the data provide proof of concept that circulating blood-BRAF can be collected, quantified, flowed, and utilized in patients with Stage II, III, and IV melanoma.
Supplementary Material
Acknowledgements
This study was funded by NCI SPORE in Skin Cancer 2P50CA93683, the Egan Memorial Research Laboratory for Melanoma Translational Research, Conquer Cancer Foundation Career Development Award, and Clinical Investigator Training Program.
Financial Support
JW Mier NCI SPORE in Skin Cancer 2P50CA93683.
JW Mier and RJ Sullivan the Egan Memorial Research Laboratory for Melanoma Translational Research
RJ Sullivan: Conquer Cancer Foundation Career Development Award, K12, and Clinical Investigator Training Program.
Footnotes
There are the following conflicts of interest to declare with regard to this manuscript:
Michael B. Atkins-Consultant for Genentech, Bristol Myers Squibb and on an Advisory Board for Glaxo Smith Kline;
F. Stephen Hodi-Nonpaid consultant Genentech;
Keith T. Flaherty-Consultant for Glaxo Smith Kline, Roche Genentech and Novartis;
David F. McDermott-Consultant for Genentech.
Jennifer A Wargo-received an honorarium from Dava Oncology and consultant for GlaxoSmithKline and Genentech.
There are no other conflicts of interests to declare among the remaining authors.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. The New England journal of medicine. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved Survival with MEK Inhibition in BRAF-Mutated Melanoma. The New England journal of medicine. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 4.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 5.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. The New England journal of medicine. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robert C, Thomas L, Bondarenko I, O’Day S, M DJ, Garbe C, et al. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. The New England journal of medicine. 2011;364:2517–26. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 7.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49:1297–304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Weber JS, Kahler KC, Hauschild A. Management of immune-related adverse events and kinetics of response with ipilimumab. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2012;30:2691–7. doi: 10.1200/JCO.2012.41.6750. [DOI] [PubMed] [Google Scholar]
- 9.Anderson S, Bloom KJ, Vallera DU, Rueschoff J, Meldrum C, Schilling R, et al. Multisite analytic performance studies of a real-time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed, paraffin-embedded tissue specimens of malignant melanoma. Arch Pathol Lab Med. 2012;136:1385–91. doi: 10.5858/arpa.2011-0505-OA. [DOI] [PubMed] [Google Scholar]
- 10.Su Z, Dias-Santagata D, Duke M, Hutchinson K, Lin YL, Borger DR, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mancini I, Santucci C, Sestini R, Simi L, Pratesi N, Cianchi F, et al. The use of COLD-PCR and high-resolution melting analysis improves the limit of detection of KRAS and BRAF mutations in colorectal cancer. J Mol Diagn. 2010;12:705–11. doi: 10.2353/jmoldx.2010.100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fadhil W, Ibrahem S, Seth R, Ilyas M. Quick-multiplex-consensus (QMC)-PCR followed by high-resolution melting: a simple and robust method for mutation detection in formalin-fixed paraffin-embedded tissue. J Clin Pathol. 2010;63:134–40. doi: 10.1136/jcp.2009.070508. [DOI] [PubMed] [Google Scholar]
- 13.MacConaill LE, Campbell CD, Kehoe SM, Bass AJ, Hatton C, Niu L, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan YH, Liu Y, Eu KW, Ang PW, Li WQ, Salto-Tellez M, et al. Detection of BRAF V600E mutation by pyrosequencing. Pathology. 2008;40:295–8. doi: 10.1080/00313020801911512. [DOI] [PubMed] [Google Scholar]
- 15.Miller CJ, Cheung M, Sharma A, Clarke L, Helm K, Mauger D, et al. Method of mutation analysis may contribute to discrepancies in reports of (V599E)BRAF mutation frequencies in melanocytic neoplasms. The Journal of investigative dermatology. 2004;123:990–2. doi: 10.1111/j.0022-202X.2004.23468.x. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Goto Y, Murata H, Sakaizawa K, Uchiyama A, Saida T, et al. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. British journal of cancer. 2011;104:464–8. doi: 10.1038/sj.bjc.6606072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, et al. Intra- and inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PloS one. 2012;7:e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panka DJ, Sullivan RJ, Mier JW. An inexpensive, specific and highly sensitive protocol to detect the BrafV600E mutation in melanoma tumor biopsies and blood. Melanoma research. 2010;20:401–7. doi: 10.1097/CMR.0b013e32833d8d48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ju ST, Panka DJ, Cui H, Ettinger R, el-Khatib M, Sherr DH, et al. Fas(CD95)/FasL interactions required for programmed cell death after T-cell activation. Nature. 1995;373:444–8. doi: 10.1038/373444a0. [DOI] [PubMed] [Google Scholar]
- 21.Fusi A, Berdel R, Havemann S, Nonnenmacher A, Keilholz U. Enhanced detection of BRAF-mutants by pre-PCR cleavage of wild-type sequences revealed circulating melanoma cells heterogeneity. Eur J Cancer. 2011;47:1971–6. doi: 10.1016/j.ejca.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Wilson BJ, Kocvara H. A simple rapid method for layering blood on Ficoll-Isopaque gradients. J Immunol Methods. 1975;9:67–8. doi: 10.1016/0022-1759(75)90036-8. [DOI] [PubMed] [Google Scholar]
- 23.Halait H, Demartin K, Shah S, Soviero S, Langland R, Cheng S, et al. Analytical performance of a real-time PCR-based assay for V600 mutations in the BRAF gene, used as the companion diagnostic test for the novel BRAF inhibitor vemurafenib in metastatic melanoma. Diagnostic molecular pathology: the American journal of surgical pathology, part B. 2012;21:1–8. doi: 10.1097/PDM.0b013e31823b216f. [DOI] [PubMed] [Google Scholar]
- 24.Su Z, Dias-Santagata D, Duke M, Hutchinson K, Lin YL, Borger DR, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. The Journal of molecular diagnostics: JMD. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elvidge GP, Price TS, Glenny L, Ragoussis J. Development and evaluation of real competitive PCR for high-throughput quantitative applications. Anal Biochem. 2005;339:231–41. doi: 10.1016/j.ab.2005.01.040. [DOI] [PubMed] [Google Scholar]
- 26.Kitago M, Koyanagi K, Nakamura T, Goto Y, Faries M, O’Day SJ, et al. mRNA expression and BRAF mutation in circulating melanoma cells isolated from peripheral blood with high molecular weight melanoma-associated antigen-specific monoclonal antibody beads. Clinical chemistry. 2009;55:757–64. doi: 10.1373/clinchem.2008.116467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kwak JY, Kim EK, Kim JK, Han JH, Hong SW, Park TS, et al. Dual priming oligonucleotide-based multiplex PCR analysis for detection of BRAFV600E mutation in FNAB samples of thyroid nodules in BRAFV600E mutation-prevalent area. Head Neck. 2010;32:490–8. doi: 10.1002/hed.21210. [DOI] [PubMed] [Google Scholar]
- 28.Morlan J, Baker J, Sinicropi D. Mutation detection by real-time PCR: a simple, robust and highly selective method. PLoS One. 2009;4:e4584. doi: 10.1371/journal.pone.0004584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oldenburg RP, Liu MS, Kolodney MS. Selective amplification of rare mutations using locked nucleic acid oligonucleotides that competitively inhibit primer binding to wild-type DNA. J Invest Dermatol. 2008;128:398–402. doi: 10.1038/sj.jid.5700920. [DOI] [PubMed] [Google Scholar]
- 30.Dominguez PL, Kolodney MS. Wild-type blocking polymerase chain reaction for detection of single nucleotide minority mutations from clinical specimens. Oncogene. 2005;24:6830–4. doi: 10.1038/sj.onc.1208832. [DOI] [PubMed] [Google Scholar]
- 31.Zatelli MC, Trasforini G, Leoni S, Frigato G, Buratto M, Tagliati F, et al. BRAF V600E mutation analysis increases diagnostic accuracy for papillary thyroid carcinoma in fine-needle aspiration biopsies. Eur J Endocrinol. 2009;161:467–73. doi: 10.1530/EJE-09-0353. [DOI] [PubMed] [Google Scholar]
- 32.Higgins MJ, Jelovac D, Barnathan E, Blair B, Slater S, Powers P, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin Cancer Res. 2012;18:3462–9. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Coit DG, Andtbacka R, Anker CJ, Bichakjian CK, Carson WE, 3rd, Daud A, et al. Melanoma, version 2.2013: featured updates to the NCCN guidelines. J Natl Compr Canc Netw. 2013;11:395–407. doi: 10.6004/jnccn.2013.0055. [DOI] [PubMed] [Google Scholar]
- 34.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. The New England journal of medicine. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, et al. Identification of a mutation in the extracellular domain of the Epidermal Growth Factor Receptor conferring cetuximab resistance in colorectal cancer. Nature medicine. 2012;18:221–3. doi: 10.1038/nm.2609. [DOI] [PubMed] [Google Scholar]
- 39.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer research. 2008;68:4853–61. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nature communications. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ackerman A, McDermott DF, Lawrence DP, Gunturi A, Flaherty KT, Hodi FS, et al. Outcomes of patients with malignant melanoma treated with immunotherapy prior to or after vemurafenib. J Clin Oncol. 2012;30 doi: 10.1002/cncr.28620. [DOI] [PubMed] [Google Scholar]
- 46.Ascierto PA, Simeone E, Giannarelli D, Grimaldi AM, Romano A, Mozzillo N. Sequencing of BRAF inhibitors and ipilimumab in patients with metastatic melanoma: a possible algorithm for clinical use. Journal of translational medicine. 2012;10:107. doi: 10.1186/1479-5876-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.