Abstract
Cancer cells condition macrophages and other inflammatory cells in the tumor microenvironment so that these cells are more permissive for cancer growth and metastasis. Conditioning of inflammatory cells reflects, at least in part, soluble mediators (such as transforming growth factor β and IL-4) that are released by cancer cells and alter the phenotype of cells of the innate immune system. Signaling pathways in cancer cells that potentiate this activity are incompletely understood. The urokinase receptor (uPAR) is a cell-signaling receptor known to promote cancer cell survival, proliferation, metastasis, and cancer stem cell–like properties. The present findings show that uPAR expression in diverse cancer cells, including breast cancer, pancreatic cancer, and glioblastoma cells, promotes the ability of these cells to condition co-cultured bone marrow–derived macrophages so that the macrophages express significantly increased levels of arginase 1, a biomarker of the alternatively activated M2 macrophage phenotype. Expression of transforming growth factor β was substantially increased in uPAR-expressing cancer cells via a mechanism that requires uPA-initiated cell signaling. uPAR also controlled expression of IL-4 in cancer cells via a mechanism that involves activation of ERK1/2. The ability of uPAR to induce expression of factors that condition macrophages in the tumor microenvironment may constitute an important mechanism by which uPAR promotes cancer progression.
It is well established that certain chronic infections and inflammation predispose to the development of malignancy.1–3 Once cancer develops, inflammatory cells that infiltrate the tumor may promote disease progression.4–6 This process is mediated by bidirectional paracrine pathways involving cancer and inflammatory cells. Growth factors and cytokines released by cancer cells are immunosuppressive, and also condition inflammatory cells so that these cells release mediators that support cancer cell growth, survival, metastasis, and angiogenesis.7–10 Inflammatory cell conditioning is prevalent in breast cancer. These tumors include large numbers of macrophages, dendritic cells, mast cells, and T cells, and the extent to which the tumor is infiltrated by these inflammatory cells correlates with the incidence of metastasis.11–13 A high density of tumor-associated macrophages (TAMs) is also correlated with higher breast cancer tumor grade and decreased relapse-free and overall survival.14–17
Although macrophages express a wide spectrum of phenotypic properties, these cells are frequently categorized as classically activated (M1) or alternatively activated (M2).18–21 In response to pathogens, tissue damage, and Th1 cytokines such as IFN-γ and TNF-α, M1-polarized macrophages release cytotoxic compounds and proteins, including nitric oxide, reactive oxygen species, and proinflammatory cytokines (including IL-12, IL-23, and TNF-α). M2-polarized macrophage have been classified into a number of subcategories; in many contexts, these cells demonstrate enhanced activity in the resolution of inflammation, tissue remodeling, and healing.18–21 Arginase 1 (Arg1), which is expressed selectively by M2-polarized macrophages, diverts substrate from the enzyme systems that produce cytotoxic levels of nitric oxide.22,23 In general, it is thought that TAMs, which have been conditioned by cancer cells to express tumor-permissive gene products, demonstrate characteristics in common with M2-polarized macrophages, although a recent report highlights phenotypic differences.18,19,24 Cell-signaling systems in tumor cells that promote the ability of these cells to regulate macrophage phenotype remain incompletely understood.
In many forms of cancer, expression of the urokinase receptor [urokinase plasminogen activator receptor (uPAR)] correlates with poor prognosis and shortened survival.25–28 Originally, the activity of uPAR in cancer was attributed to its ability to bind the serine protease, urokinase-type plasminogen activator (uPA), and activate a cascade of extracellular proteases involved in matrix remodeling and cell migration through tissue boundaries. The current understanding, however, is that uPAR also is a cell-signaling receptor that activates diverse signaling pathways.29 Although uPAR may signal autonomously when expressed at high levels, uPA binding to uPAR robustly activates cell signaling even when the cell-surface abundance of uPAR is low.29–32 uPAR-initiated cell signaling promotes cancer cell survival, release from states of dormancy, migration, epithelial–mesenchymal transition, cancer stem cell–like properties, and metastasis independently of protease activation.33–38
Here, we show that in multiple forms of cancer, including breast cancer, pancreatic cancer, and glioblastoma (GBM), uPAR expression promotes the ability of the cancer cells to M2-polarize co-cultured macrophages. The mediators that are released selectively by uPAR-expressing cancer cells to regulate macrophage phenotype may vary across different cancer cells; however, we provide evidence that both TGF-β and IL-4 are involved. The ability of cancer-cell uPAR to promote conditioning of inflammatory cells in the tumor microenvironment is a novel mechanism by which uPAR may promote cancer progression.
Materials and Methods
Reagents
Anti–F4/80 (phycoerythrin PE conjugated) and isotype-matched IgG were from eBioscience (San Diego, CA). Monoclonal human uPAR-specific antibody (ATN658) for flow cytometry was provided by Dr. Andrew Mazar (Northwestern University). Rabbit monoclonal antibody that detects both TGF-β1 and TGF-β3 (56E4) was from Cell Signaling Technology (Danvers, MA). Monoclonal antibody that detects α-tubulin was from Sigma-Aldrich (St. Louis, MO). Mouse uPAR-specific antibody for flow cytometry was from R&D Systems (Minneapolis, MN). The MAP kinase (MEK) inhibitor PD98059 was from Calbiochem (San Diego, CA). qPCR reagents, including primers and probes for mouse Arg1, uPA, TGF-β, IL-4, and HPRT-1 were from Applied Biosystems–Life Technologies (Carlsbad, CA). The TGF-β primers and probe were selective for TGF-β1. The siRNA targeting human uPA is previously described.39 An siCONTROL nontargeting siRNA pool [non-targeting control (NTC)] was obtained from Dharmacon (GE Dharmacon, Lafayette, CO).
Cell Culture
MDA-MB 468 breast cancer cells were obtained from the ATCC (Manassas, VA). Low-passage MCF7 cells were originally provided by Dr. Sally Parsons (University of Virginia, Charlottesville, VA). Stable derivative cell lines were used in which MDA-MB 468 and MCF7 cells were transfected to overexpress human uPAR or with empty vector (EV), as described previously.32,36,38 Expression of uPAR was confirmed by flow cytometry (FACSCanto II system; BD Biosciences, San Jose, CA) and the results were analyzed using FlowJo software version 7.6.4 (TreeStar, Ashland, OR). MCF-7 and MDA-MB 468 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. siRNA (40 nmol/L) was introduced into control and uPAR-overexpressing MDA-MB 468 cells by incubation with Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer’s instructions. 4T1 and 168FARN mouse mammary cancer cells (generously provided by Dr. Fred R. Miller, Wayne State University, Detroit, MI) were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. PanO2 mouse pancreatic adenocarcinoma cells were obtained from the National Cancer Institute Developmental Therapeutics Program Tumor Repository and cultured as described previously.40 These cells were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum. uPAR was silenced in PanO2 and 4T1 cells by transducing these cells with a lentivirus that encodes shRNA targeting mouse uPAR (pLKO-shmuPAR; RNAi Consortium, Broad Institute, Cambridge, MA). The sequence 5′-ACCAACAGGACCATG-3′ in the shRNA corresponds to nucleotides 231 to 246 in the coding region of mouse uPAR. Control cells were transduced with viral particles that contain EV. Transduced cells were selected with 2 μg/mL puromycin for 1 week. uPAR protein expression was then assessed by flow cytometry or immunoblot analysis. ESC1 and ESC2 (Ludwig Institute for Cancer Research, University of California, San Diego) are GBM cell lines established by harvesting two separate xenografts formed by U373MG GBM cells.41 These cells, which are known to express high levels of uPA and uPAR, were cultured as described previously.39
Co-Culturing Experiments with BMDMs
Bone marrow–derived macrophages (BMDMs) were harvested from femurs and tibias of 8- to 10-week-old female C57BL/6 mice (Charles River Laboratories, San Diego, CA) and cultured in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 20% L929 cell–conditioned medium for 7 days.40,42 The BMDMs were released with trypsin. The purity of BMDMs was confirmed by performing flow cytometry for the macrophage marker F4/80. For co-culturing experiments, co-culture inserts (0.4-μm pores; BD Biosciences) were placed in 6-well tissue culture plates. BMDMs were added to the tissue culture wells (1 × 106 cells per well) and cancer cells (1.5 × 105) were added to the inserts. The cells were washed with serum-free medium and then cultured in serum-free medium for four additional days before harvesting the BMDMs for gene expression analysis or Arg1 activity measurements.
qPCR
For quantitative real-time PCR (qPCR), total RNA was isolated from BMDMs, which were co-cultured with cancer cells, or from the cancer cells using a NucleoSpin RNA II kit (Macherey-Nagel, Bethlehem, PA). cDNA was synthesized using an iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA). qPCR was performed using an Applied Biosystems StepOnePlus instrument (Life Technologies) and a one-step program: 95°C, 10 minutes; 95°C, 30 seconds, and 60°C, 1 minute for 40 cycles. HPRT1 gene expression was measured as a normalizer. Results were analyzed by the relative quantification method. All experiments were performed in triplicate with internal duplicate determinations.
Arginase Activity Assay
Arginase activity was measured using a QuantiChrom arginase assay kit (BioAssay Systems, Hayward, CA). BMDMs that had been co-cultured with breast cancer cells were washed with 20 mmol/L sodium phospate, 150 mmol/L NaCl, pH 7.4. Cell extracts were prepared in 10 mmol/L Tris–HCl, pH 7.4, 0.4% Triton X-100, 1 μmol/L pepstatin A, and 1 μmol/L leupeptin. Cell extracts were incubated with l-arginine for 2 hours, and then urea detection reagent was added for 2 hours. The absorbance (OD) at 520 nm was measured. As a control, we also measured the absorbance of 1 mmol of urea. Arginase activity was calculated as follows: arginase activity index (mmol urea/1 × 106 cells) = (ODsample − ODblank)/(ODstandard − ODwater) × 1 mmol × dilution factor.
Immunoblot Analysis
Cell extracts were prepared in radioimmunoprecipitation assay buffer containing complete protease inhibitor cocktail. Protein concentrations were determined by bicinchoninic acid assay (Sigma-Aldrich). Equal amounts of cell extract were resolved by SDS-PAGE, electrotransferred to polyvinylidene difluoride membranes, and probed to detect the protein of interest and tubulin as a loading control.
TGF-β ELISA
Breast cancer cells were plated in 60-mm dishes (1 × 106 cells per plate) for 24 hours and then washed extensively. The cells were incubated in serum-free medium for an additional 24 hours. Conditioned medium was harvested. TGF-β released from cells was measured using enzyme-linked immunosorbent assay (TGF-β Quantikine ELISA; R&D Systems). This assay is selective for TGF-β1.
Results
uPAR Expression in Breast Cancer Cells Induces Arg1 in Co-Cultured Macrophages
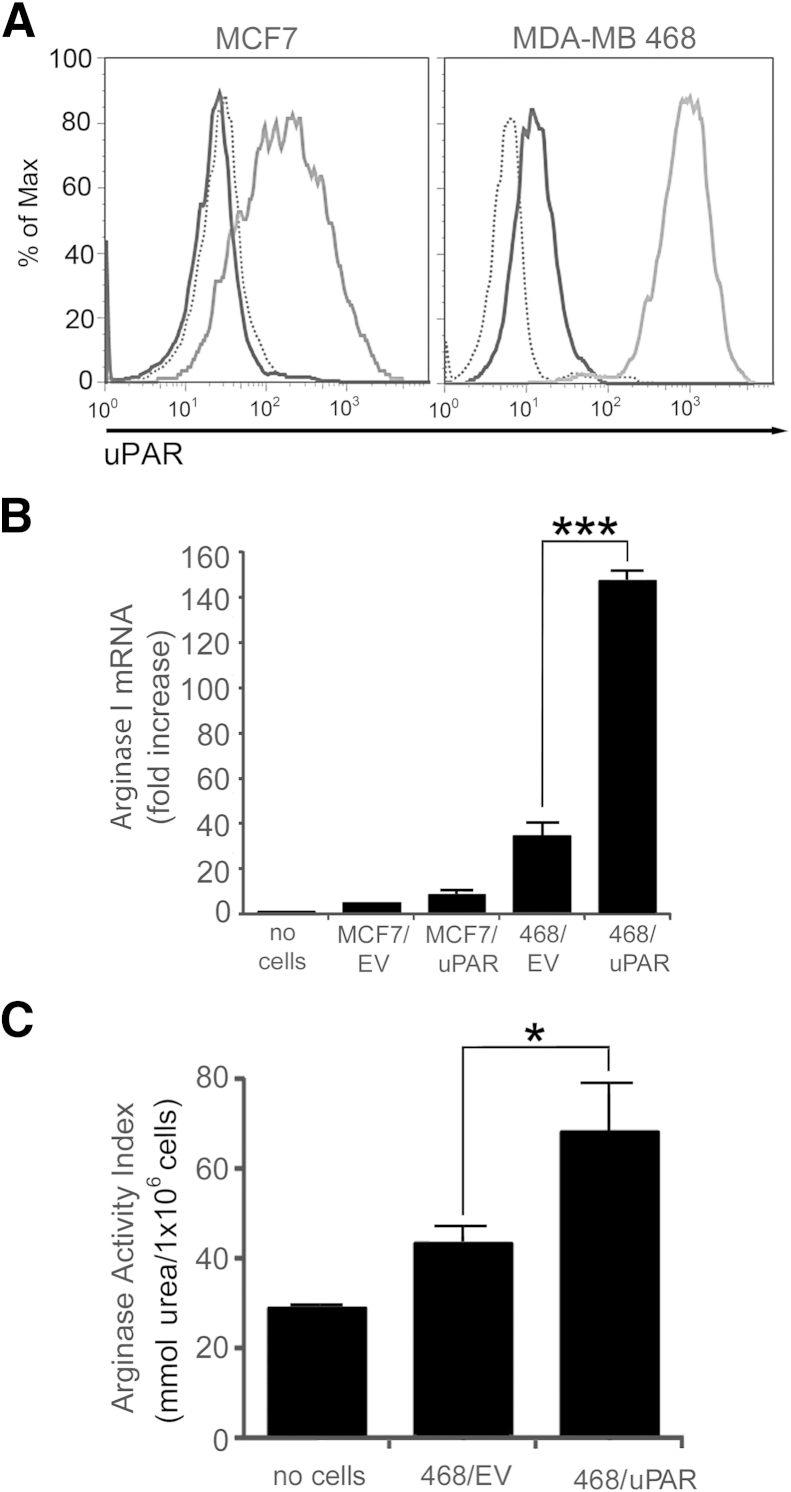
MCF-7 cells are estrogen receptor-α–expressing breast cancer cells that express low levels of uPAR and undetectable levels of uPA.30 MDA-MB 468 breast cancer cells express higher levels of uPAR and also express uPA, allowing autocrine uPAR signaling.36,38 We overexpressed uPAR in both cell types to generate stable cell lines, as described previously.32,33,38 Control cells were transfected with EV. Cell-surface uPAR expression was compared by flow cytometry. In control EV-transfected MCF-7 cells, uPAR expression was insufficient for detection by flow cytometry (Figure 1A). uPAR was readily detected in control MDA-MB 468 cells. A marked increased in cell-surface uPAR was observed when MCF-7 or MDA-MB 468 cells were transfected to overexpress uPAR.
Figure 1.
Urokinase receptor (uPAR) overexpression in MDA-MB 468 breast cancer cells increases arginase 1 (Arg1) expression in co-cultured bone marrow–derived macrophages (BMDMs). A: Flow cytometry was performed to detect uPAR in MCF7 and MDA-MB 468 cells. uPAR-overexpressing cells (gray curve); empty vector (EV)–transfected control cells (black curve); and isotype-matched IgG (dotted curve). B: BMDM Arg1 mRNA was analyzed by quantitative real-time PCR (qPCR) after co-culturing with control cancer cells, transfected with EV, or with cells transfected to overexpress human uPAR (uPAR). C: After co-culturing with cancer cells, BMDM arginase activity was assessed as the arginase activity index, an indicator of the amount of urea produced by BMDMs. Data are expressed as means ± SEM. n = 3, with internal duplicate replicates. ∗P < 0.05; ∗∗∗P < 0.001, one-way analysis of variance. Max, maximum; no cells, no cancer cells were added to the co-culture inserts.
To investigate whether uPAR expression in breast cancer cells promotes conditioning of TAMs, an in vitro co-culture model system was established. Arg1 mRNA expression was measured as a biomarker of the M2-polarized macrophage phenotype in BMDMs that were co-cultured with cancer cells. Arg1 mRNA was increased in BMDMs co-cultured with MDA-MB 468 cells, compared with MCF-7 cells (Figure 1B). uPAR overexpression in MDA-MB 468 cells further increased Arg1 mRNA expression (P < 0.001). Although uPAR overexpression in MCF-7 cells was associated with a trend toward increased Arg1 mRNA in co-cultured BMDMs, the difference was not statistically significant.
To confirm the results of the qPCR experiments, we examined arginase activity using a spectrophotometric assay.43 A significant increase in arginase activity was detected in BMDMs that were co-cultured with uPAR-overexpressing MDA-MB 468 breast cancer cells compared with control, EV-transfected MDA-MB 468 cells (P < 0.05) (Figure 1C). The increase in arginase activity was lower in magnitude than the increase in Arg1 mRNA. This may be explained by the fact that the arginase activity assay detects two separate arginase isozymes and is affected by interfering compounds such as ornithine and urea.43
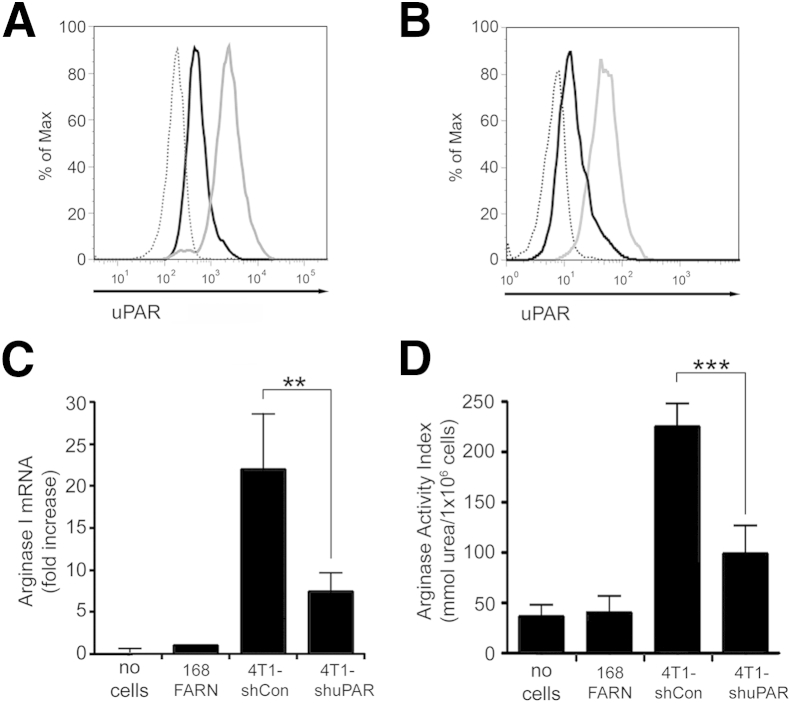
Because the BMDMs were isolated from mice, we performed co-culturing experiments with mouse breast cancer cell lines. 4T1 cells are highly metastatic breast cancer cells isolated from a spontaneously developed tumor in BALB/c mice. 4T1 cells are known to express high levels of uPA and uPAR.44,45 168FARN cells also were isolated from a BALB/c mammary tumor. These cells are tumorigenic, but rarely metastasize. We confirmed that 4T1 cells express uPAR by flow cytometry (Figure 2A). The level of cell-surface uPAR in 4T1 cells was approximately fourfold higher than in 168FARN cells. To derive stable cell lines in which uPAR expression is silenced, 4T1 cells were transduced with the lentiviral vector pLKO-shmuPAR, which encodes uPAR-specific shRNA. Flow cytometry indicated that gene silencing decreased cell-surface uPAR in 4T1 cells by >80% (Figure 2B).
Figure 2.
Urokinase receptor (uPAR) controls the ability of 4T1 metastatic breast cancer cells to regulate expression of arginase 1 (Arg 1) in co-cultured bone marrow-derived macrophages (BMDMs). A: Flow cytometry was performed to detect mouse uPAR in 168FARN (black curve) and 4T1 (gray curve) cells and in isotype-matched IgG (dotted curve). B: Flow cytometry was performed to detect mouse uPAR in 4T1 cells transduced with a control lentiviral vector, 4T1-shCon cells (gray curve), or with a lentiviral vector encoding shRNA against uPAR, 4T1-shuPAR cells (black curve). An isotype-matched IgG control study is also shown (dotted curve). C and D: After co-culturing with cancer cells, BMDM Arg1 mRNA was analyzed by qPCR (C) and BMDM arginase activity was assessed as the arginase activity index (D). Data are expressed as means ± SEM. n = 3. ∗∗P < 0.01, ∗∗∗P < 0.001, one-way analysis of variance.
In co-culturing experiments, 4T1 cells induced Arg1 expression in BMDMs more effectively than did 168FARN cells, consistent with our results showing higher uPAR expression in 4T1 cells (Figure 2C). uPAR gene silencing in 4T1 cells significantly decreased the ability of these cells to induce Arg1 expression (P < 0.005). These results were confirmed by measuring BMDM arginase activity (Figure 2D).
uPAR Controls the Ability of Diverse Cancer Cells to Condition Macrophages
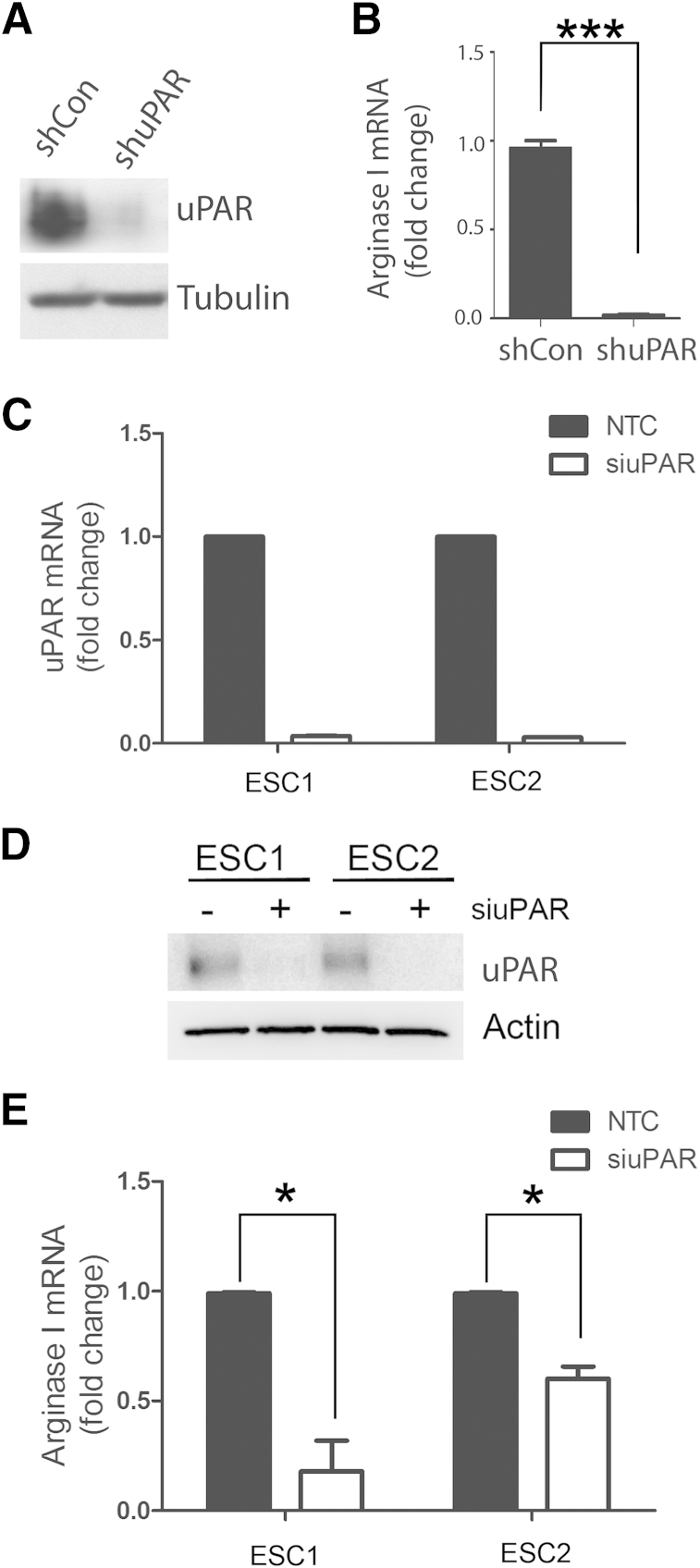
PanO2 mouse pancreatic adenocarcinoma cells express substantial levels of uPAR, as determined by immunoblot analysis (Figure 3A). uPAR gene silencing with pLKO-shmuPAR essentially blocked expression of uPAR at the protein level in these cells. Control uPAR-expressing PanO2 cells were substantially more effective at inducing Arg1 mRNA expression in BMDMs, compared with cells in which uPAR was silenced (Figure 3B).
Figure 3.
Urokinase receptor (uPAR) gene silencing in mouse pancreatic adenocarcinoma PanO2 and glioblastoma (GBM) cells decreases Arg1 expression in co-cultured bone marrow-derived macrophages (BMDMs). A: Immunoblotting was performed to detect mouse uPAR and tubulin in PanO2 cells transfected with a control lentiviral vector (shCon) or with a lentiviral vector encoding shRNA against uPAR (shuPAR). B: BMDM Arg1 mRNA was analyzed by qPCR after co-culturing with PanO2 cells that were transduced with shCon or shuPAR. C: ESC1 and ESC2 GBM cells were transfected with non-targeting control (NTC) or uPAR-specific siRNA (siuPAR), and uPAR mRNA levels were determined by qPCR, standardized against levels present in cells treated with NTC siRNA. D: uPAR protein expression was determined by immunoblot analysis in cells transfected with NTC or uPAR-specific siRNA. E: ESC1 and ESC2 GBM cells were transfected with NTC or uPAR-specific siRNA. mRNA levels of BMDM Arg1 were determined by qPCR after co-culturing with ESC cells, standardized against levels present in cells co-cultured with ESC cells transfected with NTC siRNA. Data are expressed as means ± SEM. n = 3. ∗P < 0.05, Student’s t-test; ∗∗∗P < 0.001, one-way analysis of variance.
We also studied ESC1 and ESC2 GBM cells. These cells are derived from different tumors formed by U373MG GBM cells in mice.39,41 When initially inoculated, the U373MG cells expressed a constitutively active form of the EGF receptor; however, expression of this receptor was blocked in vivo after the tumors were established forcing the tumors into a state of dormancy.41 ESC cell lines were prepared from tumors that regained the capacity to grow in vivo and are known to express high levels of uPA and uPAR.39 For the studies reported here, uPAR was silenced in ESC1 and ESC2 cells using siRNA. uPAR gene silencing was highly effective at the mRNA level, as determined by qPCR (Figure 3C), and at the protein level, as determined by immunoblot analysis (Figure 3D). uPAR gene silencing in ESC1 and ESC2 cells significantly decreased the ability of these cells to induce Arg1 mRNA expression in co-cultured BMDMs (Figure 3E).
uPAR Induces Expression of TGF-β in Cancer Cells
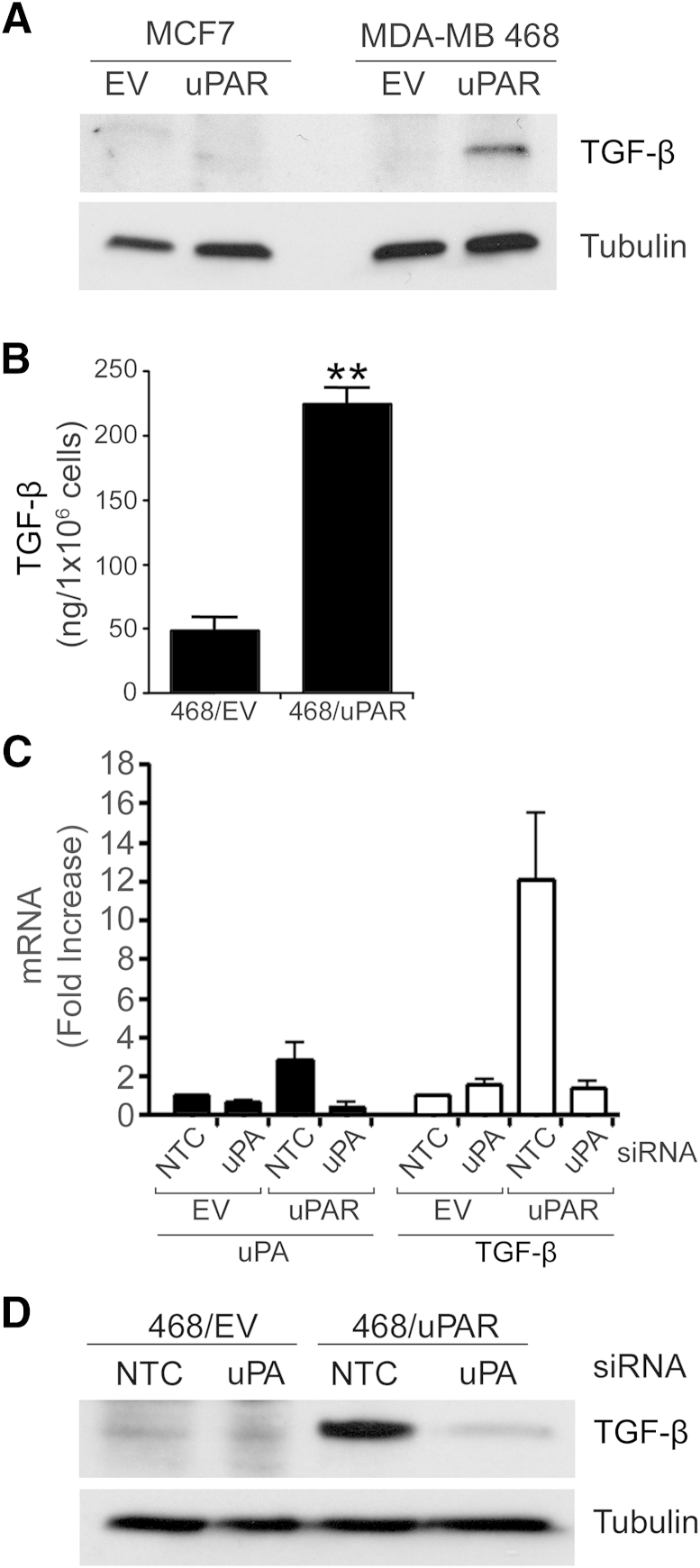
TGF-β is well recognized as an important growth factor released by cancer cells, that expresses immunosuppressive activities and that conditions TAMs so that they express M2-polarized phenotypic properties.46–50 We examined TGF-β in cell extracts from the control and uPAR-overexpressing MCF7 and MDA-MB 468 cells described previously (Figure 1A). TGF-β was readily detected in extracts of MDA-MB 468 cells only after uPAR overexpression (Figure 4A). uPAR overexpression also increased the level of TGF-β detected in serum-free medium conditioned by MDA-MB 468 cells, as determined by ELISA selective for TGF-β1 (Figure 4B).
Figure 4.
Urokinase receptor (uPAR) induces transforming growth factor β (TGF-β) expression in MDA-MB breast cancer 468 cells. A: Immunoblot analysis to detect TGF-β and tubulin was performed on cell extracts from empty vector-transfected control (EV) and uPAR-overexpressing MCF7 and MDA-MB 468 cells. B: Conditioned medium from control MDA-MB 468 cells (468/EV) and uPAR-overexpressing MDA-MB 468 cells (468/uPAR) was subjected to enzyme-linked immunosorbent assay (ELISA) to detect TGF-β. C: Control MDA-MB 468 cells (EV) and uPAR-overexpressing MDA-MB 468 cells (uPAR) were transfected with uPA-specific siRNA (40 nmol/L) or NTC siRNA pool (40 nmol/L). After 24 hours, the cells were cultured in serum-free medium for an additional 24 hours. mRNA levels were determined by qPCR for urokinase-type plasminogen activator (uPA) and TGF-β, standardized against levels present in control EV cells transfected with control siRNA. D: Immunoblot analysis to detect TGF-β and tubulin was performed on cell extracts from control and uPAR-overexpressing MDA-MB 468 cells that were transfected with uPA-specific or NTC siRNA. Data are expressed as means ± SEM. n = 3. ∗∗P < 0.01, Student’s t-test.
We hypothesized that activation of uPAR signaling secondary to uPAR overexpression increases expression of TGF-β in MDA-MB 468 cells. To test this hypothesis, we silenced uPA in control and uPAR-overexpressing MDA-MB 468 cells, precluding the possibility for autocrine uPA-initiated uPAR cell signaling. In cells transfected with NTC siRNA, uPA mRNA was slightly increased by uPAR overexpression; TGF-β mRNA was increased approximately 10-fold, consistent with the demonstrated increases in TGF-β protein (Figure 4C). uPA gene silencing in uPAR-overexpressing MDA-MB 468 cells was greater than 80% effective, as determined by qPCR, and was accompanied by a 90% decrease in TGF-β mRNA expression. We confirmed that uPA gene silencing inhibits TGF-β expression in uPAR-overexpressing MDA-MB 468 cells by performing immunoblot analysis (Figure 4D). These results support a model in which uPA-initiated uPAR signaling induces TGF-β expression.
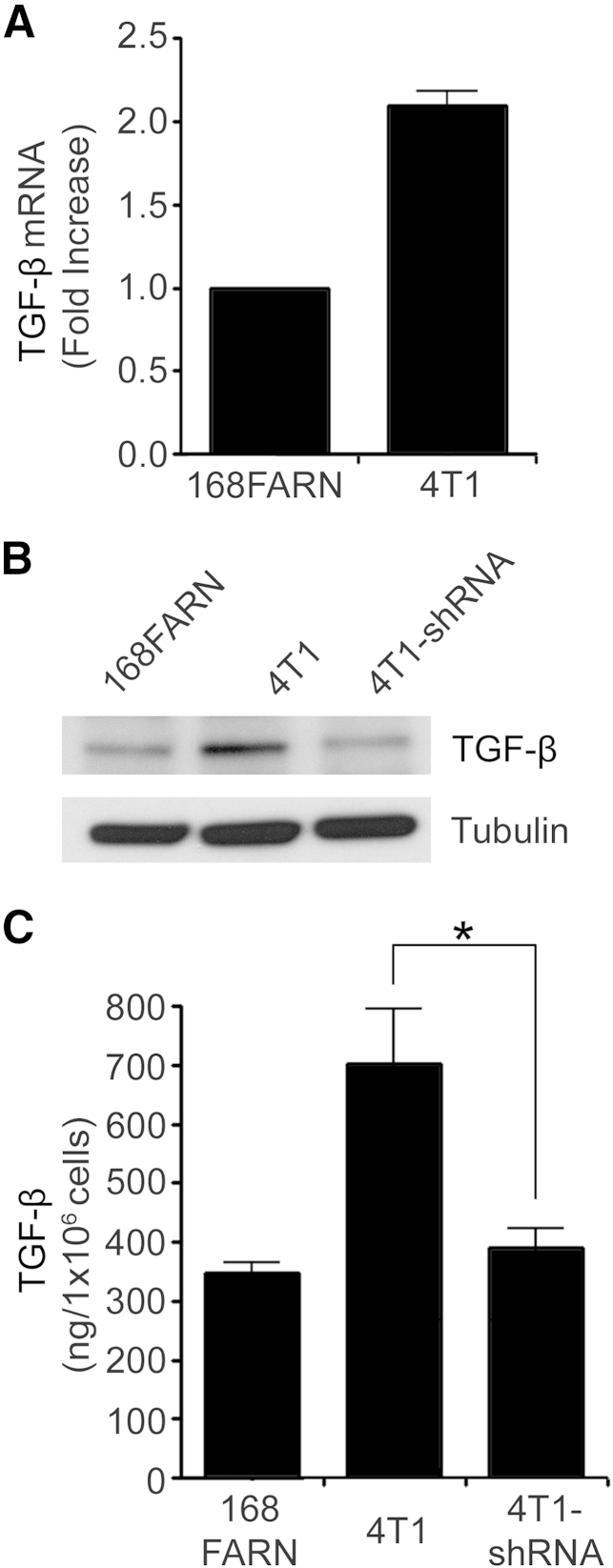
Next, we examined TGF-β expression in 168FARN cells, in wild-type 4T1 cells, and in 4T1 cells in which uPAR was silenced (Figure 2B). The level of TGF-β mRNA was approximately twofold higher in 4T1 cells, compared with 168FARN cells (Figure 5A). This difference was confirmed at the protein level by immunoblot analysis (Figure 5B) and ELISA (Figure 5C). uPAR gene silencing in 4T1 cells significantly decreased TGF-β protein production and secretion into the medium (P < 0.05).
Figure 5.
Urokinase receptor (uPAR) induces transforming growth factor-beta (TGF-β) expression in metastatic breast cancer 4T1 cells. A: mRNA levels of TGF-β were determined by qPCR, standardized against levels present in 168FARN mouse mammary cancer cells. B: Immunoblot analysis to detect TGF-β and tubulin was performed on cell extracts from 168FARN, control 4T1, and 4T1 cells in which uPAR was silenced (4T1-shRNA). C: ELISA to detect TGF-β was performed on conditioned medium from 168FARN, control 4T1, and 4T1 cells in which uPAR was silenced. Data are expressed as means ± SEM. n = 3. ∗P < 0.05, one-way analysis of variance.
IL-4 Is Regulated Downstream of ERK1/2 in GBM Cells
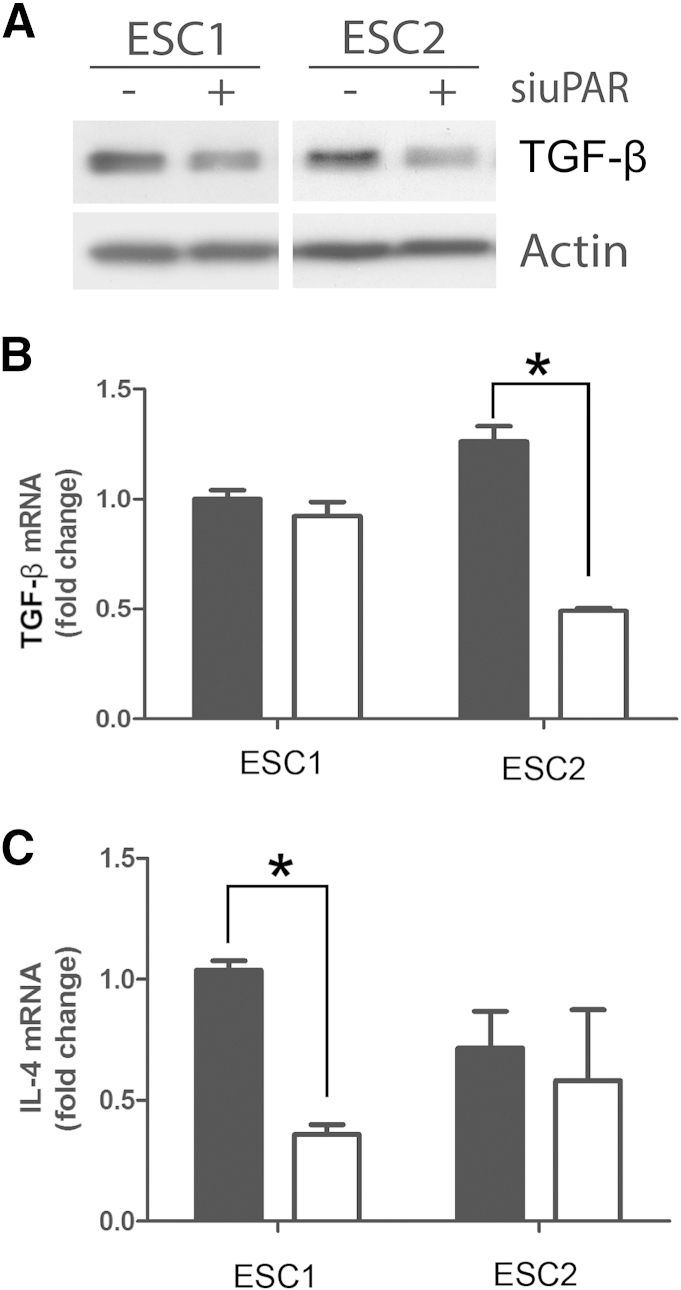
Next, we examined the effects of uPAR gene silencing on TGF-β expression in GBM cells. Analysis of cell extracts demonstrated a decrease in TGF-β protein in ESC1 and ESC2 cells in which uPAR was silenced (Figure 6A). The decrease in TGF-β protein was confirmed at the mRNA level in ESC2 cells (Figure 6B). Interestingly, uPAR gene silencing did not cause a significant decrease in TGF-β mRNA in ESC1 cells. We therefore decided to analyze IL-4 levels in control and uPAR gene-silenced ESC1 and ESC2 cells. uPAR gene silencing induced a significant decrease in IL-4 mRNA expression in ESC1 cells (P < 0.05) (Figure 6C). In ESC2 cells, IL-4 mRNA expression trended downward, but the decrease was not significant. These results suggest that uPAR expression in cancer cells may affect expression of different mediators that have the potential to condition macrophages.
Figure 6.
Urokinase receptor (uPAR) regulates transforming growth factor-beta (TGF-β) and IL-4 expression in glioblastoma (GBM) cells. A: Cell extracts from ESC1 and ESC2 cells that were transfected with uPAR-specific (+) or NTC siRNA (−) were subjected to immunoblot analysis to detect TGF-β and β-actin. B and C: ESC1 and ESC2 cells were transfected with uPAR-specific siRNA (white bars) or NTC siRNA (black bars). After 24 hours, the cells were cultured in serum-free medium for an additional 24 hours. mRNA levels of TGF-β (B) and IL-4 (C) were determined by qPCR, standardized against levels present in ESC1 cells transfected with NTC siRNA. Data are expressed as means ± SEM. n = 3. ∗P < 0.05, Student’s t-test.
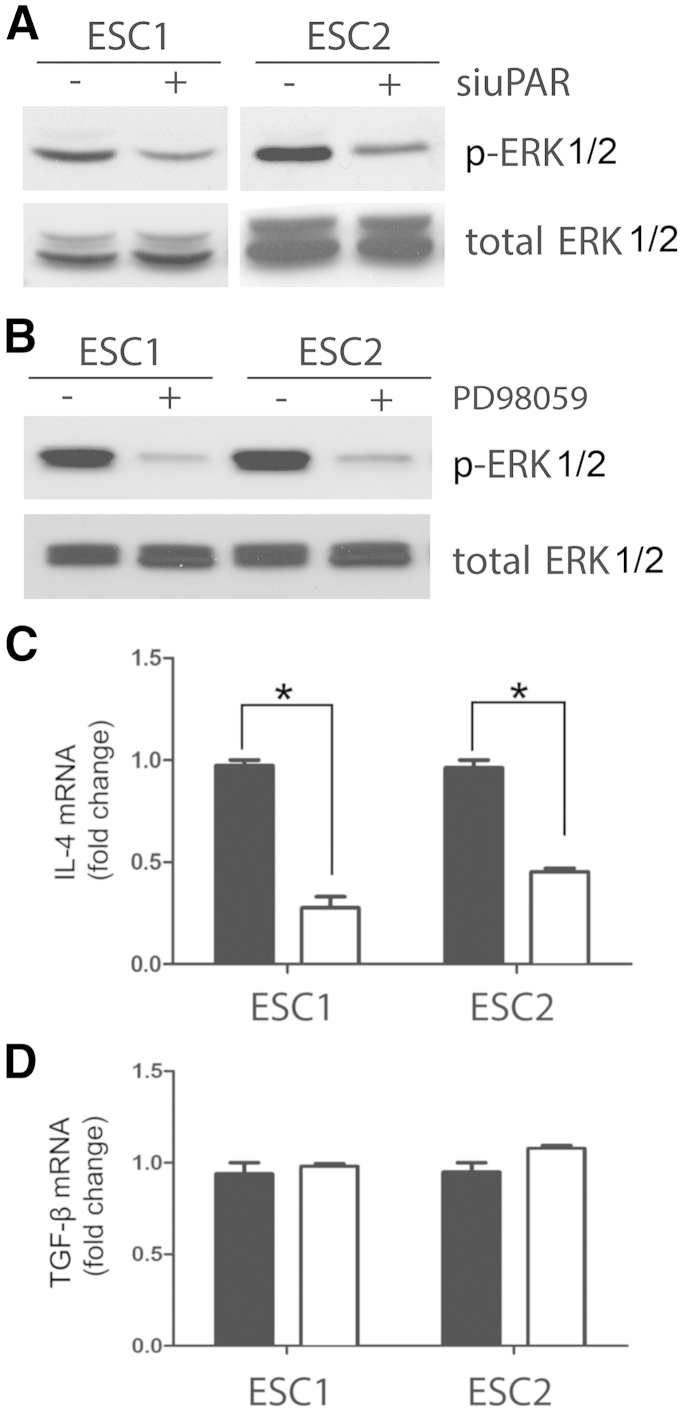
uPA binding to uPAR activates a network of interconnected signaling pathways,29 providing many mechanisms by which expression of factors such as TGF-β and IL-4 may be regulated. In ESC1 and ESC2 cells, uPAR is a significant regulator of ERK1/2 activation.39 ERK1/2 is also activated downstream of uPAR in other cancer cells, including MCF-7 and MDA-MB 468 breast cancer cells.30,32 When uPAR was silenced in ESC1 and ESC2 cells, phosphorylation of ERK1/2 was decreased (Figure 7A). We therefore examined whether inhibiting ERK1/2 activation blocks expression of TGF-β or IL-4 in ESC1 and ESC2 cells. In both cell lines, PD98059 (50 μmol/L) almost entirely blocked phosphorylation of ERK1/2 (Figure 7B). PD98059 also significantly decreased IL-4 mRNA expression (Figure 7C). TGF-β mRNA expression was unaffected (Figure 7D). These results suggest that, in cancer cells, expression of factors that condition TAMs may be regulated by uPAR through its effects on activation of ERK1/2 or other signaling factors controlled in concert.
Figure 7.
IL-4 is regulated downstream of ERK1/2 in ESC cells. A: Immunoblot analysis to detect detected phosphorylated (p-) and total ERK1/2 on cell extracts from ESC1 and ESC2 cells transfected with uPAR-specific (+) or NTC siRNA (−). B: ESC1 and ESC2 cells were treated with PD98059 (50 μmol/L) or with vehicle in serum-free medium for 30 minutes. p-ERK1/2 and total ERK1/2 were determined by immunoblot analysis. C and D: ESC1 and ESC2 cells were treated with PD98059 (50 μmol/L) (white bars) or vehicle (black bars) in serum-free medium for 4 hours. mRNA levels of IL-4 (D) and TGF-β (E) were determined by quantitative real-time PCR, standardized against levels present in vehicle-treated cells. Data are expressed as means ± SEM. n = 3. ∗P < 0.05, Student’s t-test.
Discussion
Mechanisms by which uPAR promotes cancer progression remain incompletely understood. Cell-signaling pathways activated downstream of uPAR promote cancer cell survival, proliferation, migration, invasion, and epithelial–mesenchymal transition.29–37 uPAR also induces stem cell–like properties in cancer cells.38 The role of cancer-cell uPAR in the conditioning of inflammatory cells in the tumor microenvironment had not been explored previously. In the present study, we investigated whether uPAR expression by cancer cells regulates the ability of these cells to control the phenotype of macrophages. Because we performed co-culturing experiments with cell-culture inserts, there was no direct contact between the cancer cells and the BMDMs and therefore only soluble mediators had an opportunity to affect the results.
In these studies with human and mouse breast cancer cells, we showed that uPAR expression correlates with the ability of these cells to secrete mediators that condition BMDMs so that the BMDMs express the M2 macrophage biomarker Arg1. Macrophage Arg1 may promote cancer progression by suppressing production of cytotoxic levels of nitric oxide.22,23 Our observation regarding uPAR and the ability of cancer cells to induce Arg1 expression in macrophages was reproduced when we examined pancreatic adenocarcinoma cells and GBM cells. Thus, the ability of cancer-cell uPAR to promote tumor-permissive conditioning of macrophages appears to be conserved across diverse neoplasms.
We propose that uPAR-activated cell signaling in cancer cells increases expression of soluble mediators that condition TAMs in the tumor microenvironment to allow cancer progression. The complete spectrum of soluble mediators that may be induced by uPAR remains to be determined. In MDA-MB 468, 4T1, and ESC GBM cells, uPAR increased expression of TGF-β, a known immunosuppressant in the tumor microenvironment.46 TGF-β targets multiple immune system cell types and, in macrophages, induces M2-polarized characteristics.46–50 In addition to regulating TGF-β expression, as demonstrated here, uPAR also may be involved in TGF-β activation. Plasminogen, which is activated by uPAR-associated uPA to form plasmin, converts biologically inactive, latent TGF-β into the active growth factor.51 Active TGF-β increases expression not only of uPA, but also of other proteins that may participate in uPAR-initiated cell signaling, such as plasminogen activator inhibitor-1.52,53 These synergistic pathways may combine to form positive feedback loops, which further activate uPAR signaling in cancer cells and further enable cancer cells to condition local TAMs.
To determine the role of uPAR-initiated cell signaling in regulation of TGF-β expression, we silenced uPA in uPAR-overexpressing MDA-MB 468 cells. Although uPAR signals independently of uPA when expressed at high levels,32 uPA binding to uPAR substantially increases the amplitude of uPAR signaling and also triggers cell signaling when the uPAR copy number is low.30 uPA gene silencing completely blocked the increase in TGF-β expression associated with uPAR overexpression. Next, we studied ESC1 and ESC2 GBM cells. We previously demonstrated that uPAR is a major regulator of ERK1/2 activation in these cells.39 uPAR gene silencing decreased phosphorylated ERK1/2 in both ESC1 and ESC2 cells. When ERK1/2 activation was pharmacologically blocked with PD98059, IL-4 expression was significantly decreased in the ESC cells. IL-4 expression also was significantly decreased by uPAR gene silencing in one of the two GBM cell lines (ESC1). IL-4 is expressed by cancer cells and involved in M2 polarization of macrophages.18–21 Importantly, TGF-β expression was not regulated by PD98059, suggesting that the effects of uPAR on TGF-β expression are unrelated to its activity in controlling ERK1/2 activation in these cells.
ESC1 and ESC2 cells were derived from the same parental cells, which were propagated as tumors in separate mice.41 Expression of a constitutively active form of the EGF receptor in the parental cells was silenced in vivo. This silencing placed pressure on the cancer cells, which subsequently entered a state of dormancy. Multiple molecular changes may occur in cancer cells when they emerge from dormancy.54 In ESC1 and ESC2 cells, increased expression of uPA and activation of uPAR signaling were observed.39 The difference in regulation of IL-4 and TGF-β observed when uPAR was silenced in ESC1 and ESC2 cells probably reflects other molecular changes that allowed re-establishment of growth in vivo. Nevertheless, uPAR-driven soluble mediators controlled macrophage Arg1 similarly in both GBM cell lines. Taken together, our results suggest that cancer-permissive conditioning of macrophages may be a general activity of cancer-cell uPAR, mediated by a set of secreted immunomodulatory factors in distinct proportions in different cancer cells.
Acknowledgments
We thank Dr. Fred R. Miller for 4T1 and 168FARN cells and Drs. Frank Furnari and Webster Cavenee for ESC1 and ESC2 cells.
Footnotes
Supported by NIH grant R01-CA169096 (S.L.G.).
J.H. and M.J. contributed equally to this work.
Disclosures: M.J. has no conflicts of interest or financial interests and no disclosures relative to her new affiliation with Isis Pharmaceuticals.
Current address of M.J., Isis Pharmaceuticals, Carlsbad, CA.
References
- 1.Houghton J., Stoicov C., Nomura S., Rogers A.B., Carlson J., Li H., Cai X., Fox J.G., Goldenring J.R., Wang T.C. Gastric cancer originating from bone marrow-derived cells. Science. 2004;306:1568–1571. doi: 10.1126/science.1099513. [DOI] [PubMed] [Google Scholar]
- 2.Karin M., Lawrence T., Nizet V. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell. 2006;124:823–835. doi: 10.1016/j.cell.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 3.Kuper H., Adami H.O., Trichopoulos D. Infections as a major preventable cause of human cancer. J Intern Med. 2000;248:171–183. doi: 10.1046/j.1365-2796.2000.00742.x. [DOI] [PubMed] [Google Scholar]
- 4.Vakkila J., Lotze M.T. Inflammation and necrosis promote tumour growth. Nat Rev Immunol. 2004;4:641–648. doi: 10.1038/nri1415. [DOI] [PubMed] [Google Scholar]
- 5.Coussens L.M., Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Condeelis J., Pollard J.W. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Wyckoff J., Wang W., Lin E.Y., Wang Y., Pixley F., Stanley E.R., Graf T., Pollard J.W., Segall J., Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 8.Lin E.Y., Li J.F., Gnatovskiy L., Deng Y., Zhu L., Grzesik D.A., Qian H., Xue X.N., Pollard J.W. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006;66:11238–11246. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- 9.Lin E.Y., Pollard J.W. Tumor-associated macrophages press the angiogenic switch in breast cancer. Cancer Res. 2007;67:5064–5066. doi: 10.1158/0008-5472.CAN-07-0912. [DOI] [PubMed] [Google Scholar]
- 10.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–550. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 11.Bates G.J., Fox S.B., Han C., Leek R.D., Garcia J.F., Harris A.L., Banham A.H. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24:5373–5380. doi: 10.1200/JCO.2006.05.9584. [DOI] [PubMed] [Google Scholar]
- 12.DeNardo D.G., Barreto J.B., Andreu P., Vasquez L., Tawfik D., Kolhatkar N., Coussens L.M. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16:91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeNardo D.G., Coussens L.M. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee A.H., Happerfield L.C., Bobrow L.G., Millis R.R. Angiogenesis and inflammation in invasive carcinoma of the breast. J Clin Pathol. 1997;50:669–673. doi: 10.1136/jcp.50.8.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leek R.D., Harris A.L. Tumor-associated macrophages in breast cancer. J Mammary Gland Biol Neoplasia. 2002;7:177–189. doi: 10.1023/a:1020304003704. [DOI] [PubMed] [Google Scholar]
- 16.Leek R.D., Lewis C.E., Whitehouse R., Greenall M., Clarke J., Harris A.L. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 1996;56:4625–4629. [PubMed] [Google Scholar]
- 17.Volodko N., Reiner A., Rudas M., Jakesz R. Tumour-associated macrophages in breast cancer and their prognostic correlations. Breast. 1998;7:99–105. [Google Scholar]
- 18.Mantovani A., Allavena P., Sica A. Tumour-associated macrophages as a prototypic type II polarised phagocyte population: role in tumour progression. Eur J Cancer. 2004;40:1660–1667. doi: 10.1016/j.ejca.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Mantovani A., Sozzani S., Locati M., Allavena P., Sica A. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002;23:549–555. doi: 10.1016/s1471-4906(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 20.Van Ginderachter J.A., Movahedi K., Hassanzadeh Ghassabeh G., Meerschaut S., Beschin A., Raes G., De Baetselier P. Classical and alternative activation of mononuclear phagocytes: picking the best of both worlds for tumor promotion. Immunobiology. 2006;211:487–501. doi: 10.1016/j.imbio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Martinez F.O., Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bronte V., Zanovello P. Regulation of immune responses by L-arginine metabolism. Nat Rev Immunol. 2005;5:641–654. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 23.Chang C.I., Liao J.C., Kuo L. Macrophage arginase promotes tumor cell growth and suppresses nitric oxide-mediated tumor cytotoxicity. Cancer Res. 2001;61:1100–1106. [PubMed] [Google Scholar]
- 24.Franklin R.A., Liao W., Sarkar A., Kim M.V., Bivona M.R., Liu K., Pamer E.G., Li M.O. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344:921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bianchi E., Cohen R.L., Thor A.T., Todd R.F., 3rd, Mizukami I.F., Lawrence D.A., Ljung B.M., Shuman M.A., Smith H.S. The urokinase receptor is expressed in invasive breast cancer but not in normal breast tissue. Cancer Res. 1994;54:861–866. [PubMed] [Google Scholar]
- 26.Cozzi P.J., Wang J., Delprado W., Madigan M.C., Fairy S., Russell P.J., Li Y. Evaluation of urokinase plasminogen activator and its receptor in different grades of human prostate cancer. Hum Pathol. 2006;37:1442–1451. doi: 10.1016/j.humpath.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Andreasen P.A., Kjoller L., Christensen L., Duffy M.J. The urokinase-type plasminogen activator system in cancer metastasis: a review. Int J Cancer. 1997;72:1–22. doi: 10.1002/(sici)1097-0215(19970703)72:1<1::aid-ijc1>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 28.de Bock C.E., Wang Y. Clinical significance of urokinase-type plasminogen activator receptor (uPAR) expression in cancer. Med Res Rev. 2004;24:13–39. doi: 10.1002/med.10054. [DOI] [PubMed] [Google Scholar]
- 29.Blasi F., Carmeliet P. uPAR: a versatile signalling orchestrator. Nat Rev Mol Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen D.H., Hussaini I.M., Gonias S.L. Binding of urokinase-type plasminogen activator to its receptor in MCF-7 cells activates extracellular signal-regulated kinase 1 and 2 which is required for increased cellular motility. J Biol Chem. 1998;273:5802–8507. doi: 10.1074/jbc.273.14.8502. [DOI] [PubMed] [Google Scholar]
- 31.Madsen C.D., Ferraris G.M., Andolfo A., Cunningham O., Sidenius N. uPAR-induced cell adhesion and migration: vitronectin provides the key. J Cell Biol. 2007;177:927–939. doi: 10.1083/jcb.200612058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eastman B.M., Jo M., Webb D.L., Takimoto S., Gonias S.L. A transformation in the mechanism by which the urokinase receptor signals provides a selection advantage for estrogen receptor-expressing breast cancer cells in the absence of estrogen. Cell Signal. 2012;24:1847–1855. doi: 10.1016/j.cellsig.2012.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma Z., Webb D.J., Jo M., Gonias S.L. Endogenously produced urokinase-type plasminogen activator is a major determinant of the basal level of activated ERK/MAP kinase and prevents apoptosis in MDA-MB-231 breast cancer cells. J Cell Sci. 2001;114:3387–3396. doi: 10.1242/jcs.114.18.3387. [DOI] [PubMed] [Google Scholar]
- 34.Ossowski L., Aguirre-Ghiso J.A. Urokinase receptor and integrin partnership: coordination of signaling for cell adhesion, migration and growth. Curr Opin Cell Biol. 2000;12:613–620. doi: 10.1016/s0955-0674(00)00140-x. [DOI] [PubMed] [Google Scholar]
- 35.Kjøller L., Hall A. Rac mediates cytoskeletal rearrangements and increased cell motility induced by urokinase-type plasminogen activator receptor binding to vitronectin. J Cell Biol. 2001;152:1145–1157. doi: 10.1083/jcb.152.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lester R.D., Jo M., Montel V., Takimoto S., Gonias S.L. uPAR induces epithelial-mesenchymal transition in hypoxic breast cancer cells. J Cell Biol. 2007;178:425–436. doi: 10.1083/jcb.200701092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jo M., Takimoto S., Montel V., Gonias S.L. The urokinase receptor promotes cancer metastasis independently of urokinase-type plasminogen activator in mice. Am J Pathol. 2009;175:190–200. doi: 10.2353/ajpath.2009.081053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jo M., Eastman B.M., Webb D.L., Stoletov K., Klemke R., Gonias S.L. Cell signaling by urokinase-type plasminogen activator receptor induces stem cell-like properties in breast cancer cells. Cancer Res. 2010;70:8948–8958. doi: 10.1158/0008-5472.CAN-10-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu J., Jo M., Cavenee W.K., Furnari F., VandenBerg S.R., Gonias S.L. Crosstalk between the urokinase-type plasminogen activator receptor and EGF receptor variant III supports survival and growth of glioblastoma cells. Proc Natl Acad Sci USA. 2011;108:15984–15989. doi: 10.1073/pnas.1113416108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Staudt N.D., Jo M., Hu J., Bristow J.M., Pizzo D.P., Gaultier A., VandenBerg S.R., Gonias S.L. Myeloid cell receptor LRP1/CD91 regulates monocyte recruitment and angiogenesis in tumors. Cancer Res. 2013;73:3902–3912. doi: 10.1158/0008-5472.CAN-12-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukasa A., Wykosky J., Ligon K.L., Chin L., Cavenee W.K., Furnari F. Mutant EGFR is required for maintenance of glioma growth in vivo, and its ablation leads to escape from receptor dependence. Proc Natl Acad Sci USA. 2010;107:2616–2621. doi: 10.1073/pnas.0914356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang X., Goncalves R., Mosser D.M. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008 doi: 10.1002/0471142735.im1401s83. Chapter 14:Unit 14.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kepka-Lenhart D., Ash D.E., Morris S.M. Determination of mammalian arginase activity. Methods Enzymol. 2008;440:221–230. doi: 10.1016/S0076-6879(07)00813-0. [DOI] [PubMed] [Google Scholar]
- 44.Aslakson C.J., Miller F.R. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 45.Mi Z., Guo H., Wai P.Y., Gao C., Kuo P.C. Integrin-linked kinase regulates osteopontin-dependent MMP-2 and uPA expression to convey metastatic function in murine mammary epithelial cancer cells. Carcinogenesis. 2006;27:1134–1145. doi: 10.1093/carcin/bgi352. [DOI] [PubMed] [Google Scholar]
- 46.Flavell R.A., Sanjabi S., Wrzesinski S.H., Licona-Limón P. The polarization of immune cells in the tumour environment by TGFbeta. Nat Rev Immunol. 2010;10:554–567. doi: 10.1038/nri2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harthun N.L., Weaver A.M., Brinckerhoff L.H., Deacon D.H., Gonias S.L., Slingluff C.L., Jr. Activated alpha 2-macroglobulin reverses the immunosuppressive activity in human breast cancer cell-conditioned medium by selectively neutralizing transforming growth factor-beta in the presence of interleukin-2. J Immunother. 1998;21:85–94. [PubMed] [Google Scholar]
- 48.Standiford T.J., Kuick R., Bhan U., Chen J., Newstead M., Keshamouni V.G. TGF-beta-induced IRAK-M expression in tumor-associated macrophages regulates lung tumor growth. Oncogene. 2011;30:2475–2484. doi: 10.1038/onc.2010.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo H., Hao Y., Tang B., Zeng D., Shi Y., Yu P. Mouse forestomach carcinoma cells immunosuppress macrophages through transforming growth factor-β1. Mol Med Rep. 2012;5:988–992. doi: 10.3892/mmr.2012.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng J., Tsang J.Y., Li D., Niu N., Ho D.H., Lau K.F., Lui V.C., Lamb J.R., Chen Y., Tam P.K. Inhibition of TGF-β signaling in combination with TLR7 ligation re-programs a tumoricidal phenotype in tumor-associated macrophages. Cancer Lett. 2013;331:239–249. doi: 10.1016/j.canlet.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Odekon L.E., Blasi F., Rifkin D.B. Requirement for receptor-bound urokinase in plasmin-dependent cellular conversion of latent TGF-beta to TGF-beta. J Cell Physiol. 1994;158:398–407. doi: 10.1002/jcp.1041580303. [DOI] [PubMed] [Google Scholar]
- 52.Gerwin B.I., Keski-Oja J., Seddon M., Lechner J.F., Harris C.C. TGF-beta 1 modulation of urokinase and PAI-1 expression in human bronchial epithelial cells. Am J Physiol. 1990;259:L262–L269. doi: 10.1152/ajplung.1990.259.4.L262. [DOI] [PubMed] [Google Scholar]
- 53.Webb D.J., Thomas K.S., Gonias S.L. Plasminogen activator inhibitor 1 functions as a urokinase response modifier at the level of cell signaling and thereby promotes MCF-7 cell growth. J Cell Biol. 2001;152:741–752. doi: 10.1083/jcb.152.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor T.E., Furnari F.B., Cavenee W.K. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr Cancer Drug Targets. 2012;12:197–209. doi: 10.2174/156800912799277557. [DOI] [PMC free article] [PubMed] [Google Scholar]