Abstract
Obstructive nephropathy, the leading cause of kidney failure in children, can be anatomic or functional. The underlying causes of functional hydronephrosis are not well understood. miRNAs, which are small noncoding RNAs, regulate gene expression at the post-transcriptional level. We found that miR-145-5p, a member of the miR-143/145 cluster that is highly expressed in smooth muscle cells of the renal vasculature, was present in the pelvicalyceal system and the ureter. To evaluate whether the miR-143/145 cluster is involved in urinary tract function we performed morphologic, functional, and gene expression studies in mice carrying a whole-body deletion of miR-143/145. miR-143/145–deficient mice developed hydronephrosis, characterized by severe papillary atrophy and dilatation of the pelvicalyceal system without obvious physical obstruction. Moreover, mutant mice showed abnormal ureteral peristalsis. The number of ureter contractions was significantly higher in miR-143/145–deficient mice. Peristalsis was replaced by incomplete, short, and more frequent contractions that failed to completely propagate in a proximal-distal direction. Microarray analysis showed 108 differentially expressed genes in ureters of miR-143/145–deficient mice. Ninety genes were up-regulated and 18 genes were down-regulated, including genes with potential regulatory roles in smooth muscle contraction and extracellular matrix-receptor interaction. We show that miR-143/145 are important for the normal peristalsis of the ureter and report an association between the expression of these miRNAs and hydronephrosis.
Hydronephrosis is a common consequence of obstructive nephropathy and is one of the most frequent congenital defects diagnosed before birth, affecting 0.9% to 7.7% of human pregnancies.1 Although most cases of antenatal urinary dilation resolve spontaneously, approximately 20% to 30% of the cases become clinically significant after birth.2,3 An interesting feature of urinary tract dilatation is that approximately half of the cases are not accompanied by physical obstruction or major structural abnormalities of the pelvis or ureter. Instead, they are caused by a functional obstruction, often caused by a failure of the ureteral peristaltic mechanism.
miRNAs are a group of small, noncoding, evolutionarily conserved RNAs that bind to the mRNA of target genes and negatively regulate their expression. Given that more than 60% of human protein-coding genes have potential binding sites for miRNAs and single miRNAs can simultaneously target hundreds of genes,4,5 it is believed that miRNAs have a far-reaching effect on gene expression. miRNAs regulate diverse cellular processes including stem cell differentiation, apoptosis, proliferation, myogenesis, angiogenesis, hematopoiesis, neurogenesis, and epithelial morphogenesis.6–13 Moreover, altered expression of miRNAs has a causative effect in a wide variety of human diseases including cancer, as well as renal, cardiovascular, neurologic, and age-related pathologies.14
miR-143 and miR-145 are transcribed as a cluster from a single gene and regulate the fate and plasticity of vascular smooth muscle cells (SMCs).15 Mice deficient in miR-143/145 are viable and fertile, but present with low blood pressure as a result of reduced vascular tone.16 These animals show thinning of the smooth muscle layer of the aorta, accompanied by a thickening of the extracellular matrix (ECM). SMCs in the aorta show a reduced number of actin-stress fibers and a more abundant rough endoplasmic reticulum, which is consistent with a more synthetic phenotype. In addition, neointima formation in response to vascular injury is impeded profoundly in these mice. We previously reported that miR-145-5p is highly expressed in the juxtaglomerular (JG) cells and SMCs of renal blood vessels,17 but its role in the urinary system is not known.
Here, we report that miR-145-5p is expressed in the pelvicalyceal system and the ureter of normal mice. Based on these observations, we sought to determine the role of the miR-143/145 cluster in the urinary system. We found that miR-143/145–deficient mice develop hydronephrosis after birth as a result of abnormal ureter peristalsis. Our results indicate that miR-143/145 are essential for the normal contractility of the ureter and therefore for normal transport of urine from the kidney to the bladder.
Materials and Methods
Animals
Mice carrying a whole-body deletion of both miR-143 and miR-145 were kindly provided by Dr. Eric Olson (University of Texas, Dallas, TX).16 Mice heterozygous for the deletion were crossed to generate wild-type (WT) and miR-143/145 knockout (KO) study animals. All animal work was conducted in accordance with the guidelines approved by the Council of the American Physiological Society and with federal laws and regulations. The protocol was reviewed and approved by the Animal Care and Use Committee of the University of Virginia. Mice were maintained on a 12-hour light/dark cycle and given food and water ad libitum.
In Situ Hybridization
Ureters were removed from WT and miR-143/145 KO mice, immediately fixed in 4% paraformaldehyde at 4°C for 24 hours, and embedded in paraffin. In situ hybridization was performed on 5-μm–thick sections. Detection of miR-145-5p was performed as previously described,17 with modifications. Sections were deparaffinized, then fixed in 4% paraformaldehyde/phosphate-buffered saline, acetylated, and incubated in hybridization solution consisting of 50% formamide, 5× standard saline citrate, 50 μg/mL tRNA, 1% SDS, and 5 μg/mL heparin at room temperature for 1 hour. Hybridization was conducted at 37°C for 18 hours using 40 nmol/L digoxigenin-labeled locked nucleic acid probe (Exiqon, Woburn, MA) specific for mouse miR-145-5p (5′-AGGGATTCCTGGGAAAACTGGAC-3′) in hybridization solution. Sections were washed sequentially once with 5× standard saline citrate at 37°C, three times with 0.2× standard saline citrate at 40°C, and once with 0.2× standard saline citrate at room temperature. Sites of hybridization were detected using alkaline phosphatase–conjugated digoxigenin antibody (Roche Diagnostics Corp., Indianapolis, IN) at a 1:4000 dilution, followed by BM Purple AP substrate color development (Roche). Negative controls were performed by omitting the probe in the hybridization step and by using a commercial nontargeting miRNA probe.
Histologic Analysis and Immunostaining
Bouin's-fixed, paraffin-embedded ureter and kidney sections were stained with hematoxylin and eosin and Masson's trichrome as described.17 Immunostaining for α-smooth muscle actin (α-SMA) and renin was performed using mouse monoclonal anti–α-SMA (1:10,000 dilution), rabbit anti-mouse renin (1:500 dilution), and the appropriate Vectastain ABC kit (Vector Laboratories, Inc., Burlingame, CA) as previously described.18 Smooth muscle layer thickness was quantified in ureter sections stained with Masson's trichrome using ImageJ version 1.47v software (NIH, Bethesda, MD). Three ureter sections were measured from each animal.
Assessment of Peristaltic Activity
For in vivo analysis of peristaltic activity, WT and miR-143/145 KO mice between 3 and 10 months of age were anesthetized with 250 mg/kg tribromoethanol and placed on a heating pad at 37°C. Ureters were exposed and bathed continuously in saline prewarmed to 37°C. The number of peristaltic waves in a 3-minute period was recorded by direct visualization and filmed.
In vitro peristalsis was assessed in cultured embryo ureter explants. Ureters were isolated from WT and miR-143/145 KO-embryonic day (E) 16.5 embryos, placed on 6.5-mm polycarbonate membrane inserts with a pore size of 0.4 μm (Costar, Cambridge, MA), and cultured at the air-medium interface in DMEM/F12 media supplemented with 5 μg/mL transferrin, 100 μg/mL penicillin, and 100 U/mL streptomycin (all obtained from Gibco, Life Technologies, Grand Island, NY) at 37°C in a humidified 95% air/5% CO2 atmosphere. Culture medium was replaced every 48 hours. Under these conditions, ureters began to show spontaneous peristalsis after 3 days in culture. Time-lapse imaging of ureters was performed after 5 days in culture using a Leica DM IRE2 inverted microscope (Leica Microsystems Inc., Buffalo Grove, IL). Phase-contrast images were captured every second for 2 minutes and analyzed using SimplePCI imaging software version 5.3.1.081004 (Hamamatsu Corporation, Sewickley, PA).
Microarray Analysis
Ureters were dissected from two WT and miR-143/145 KO male mice at 2.5 to 4.5 months of age and immediately placed in RNAlater (Qiagen, Valencia, CA). RNA was prepared using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. RNA was tested for purity using the 260 nm/280 nm and 260 nm/230 nm absorbance ratios, and for integrity using an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA). Microarray gene expression analysis was performed by the University of Virginia Core Lab Microarray Facility using the Affymetrix mouse gene array ST1.0 (Affymetrix, Santa Clara, CA). All preprocessing and analysis were performed using R version 2.15.0. Affymetrix CEL files were imported using the Affymetrix package version 1.34.0. Expression intensities were summarized, normalized, and transformed using the robust multiarray average algorithm.19 Probe sets were annotated using the Bioconductor annotation packages mogene10sttranscriptcluster.db version 8.0.1, annotated version 1.34.0 (Bioconductor, Fred Hutchinson Cancer Research Center, Seattle, WA). Probe sets that did not map to an NCBI Entrez Gene Database were excluded. The dimension reduction and visualization technique of Principal Components Analysis was used to project the multivariate data vector of each array into a two-dimensional plot to reflect the overall data (dis)similarity between the arrays and to confirm clustering between samples of each genotype. The overall sample similarity was confirmed further by taking the 500 most differentially expressed genes and performing both hierarchical clustering and a heatmap representation. For differential gene expression analysis, linear models with empiric Bayes moderated standard errors were fit using the Bioconductor limma package version 3.12.1. The false discovery rate (FDR) was used to correct for multiple testing.20 Data can be accessed at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo; accession number GSE58274).
To determine pathways that could be altered in the miR-143/145 KO ureters we performed signaling pathway impact analysis (SPIA), which takes into account the directionality as well as the topology of the gene networks.21 For this analysis the 500 most differentially expressed genes were considered. Data were subjected to both Bonferroni and FDR corrections.
RNA Extraction and qPCR
Total RNA was extracted as described in the Microarray Analysis section. cDNA was prepared from 3 μg of RNA using M-MLV Reverse Transcriptase and an oligo(dT)15 primer mix (both from Promega, Madison, WI). Real-time quantitative PCR (qPCR) was performed in a CFX Connect system (Bio-Rad, Hercules, CA) using SYBR Green I (Life Technologies) and Taq DNA polymerase (Promega). Primer sequences are listed in Table 1. PCR conditions were run at 95°C, 58°C, and 72°C for 40 cycles.
Table 1.
Primer Sequences for RT-qPCR
| Gene | Forward | Reverse |
|---|---|---|
| Cacna1d | 5′-TTCAGAAATGTCTCCACTGTAAAAA-3′ | 5′-CATCCCATTCCCTGAAGATG-3′ |
| Kcnma1 | 5′-CGTCCACTGGCTTGAGAGTA-3′ | 5′-GTACCTGTGGACCGTTTGCT-3′ |
| Pla2g4a | 5′-TTGGTCCCAGTTGCAGAAAT-3′ | 5′-GAAGGCACAGAGAAGCCTGA-3′ |
| Ppp1r14a | 5′-GATTTTCCGGCATCTTTCCT-3′ | 5′-GGATGCTTGGAGGAGCTGTA-3′ |
| Itga4 | 5′-GTTATCCCTCTCCTCCAGGC-3′ | 5′-ACAGATGCGGGATCAGAAAG-3′ |
| Itgb6 | 5′-AGCTGAAGGATGTTCCCAGA-3′ | 5′-TGCAGTCACCCAAGAACAAG-3′ |
| Itga3 | 5′-TGAGGGGACACAGGTACACA-3′ | 5′-AGACTGAGCGACAACAGCG-3′ |
| Thbs2 | 5′-AGTGCACAGCTACAGCCTGA-3′ | 5′-ACAGAGTACTGGCGTCGGTC-3′ |
| Myocd | 5′-GAGCCAGTTTCTGGTGGAAA-3′ | 5′-GTCCTTCTCTGTTGAAGCCG-3′ |
| Acta2 | 5′-TTGCTGACAGGATGCAGAAGGAGA-3′ | 5′-ATCTGCTGGAAGGTAGACAGCGAA-3′ |
| GAPDH | 5′-TTGATGGCAACAATCTCCAC-3′ | 5′-CGTCCCGTAGACAAAATGGT-3′ |
Statistics
Results are expressed as means ± SEM. Significance in the difference of mean values was determined by the Student's t-test.
Results
miR-145-5p Is Expressed in the Smooth Muscle Layer of the Ureter
The miR-143/145 cluster has been studied extensively in the smooth muscle of the vasculature,15,16 however, its expression in the ureter has not been reported. To this end, we performed in situ hybridization in ureter sections of WT mice using digoxigenin-labeled oligonucleotide locked nucleic acid probes. We found that miR-145-5p was expressed exclusively in the SMC layer of the ureter and the pelvicalyceal system (Figure 1). The specificity of the staining was shown by the absence of signal in ureters of miR-143/145–deficient mice (Figure 1A), or when the probe was omitted from the hybridization mix (not shown).
Figure 1.
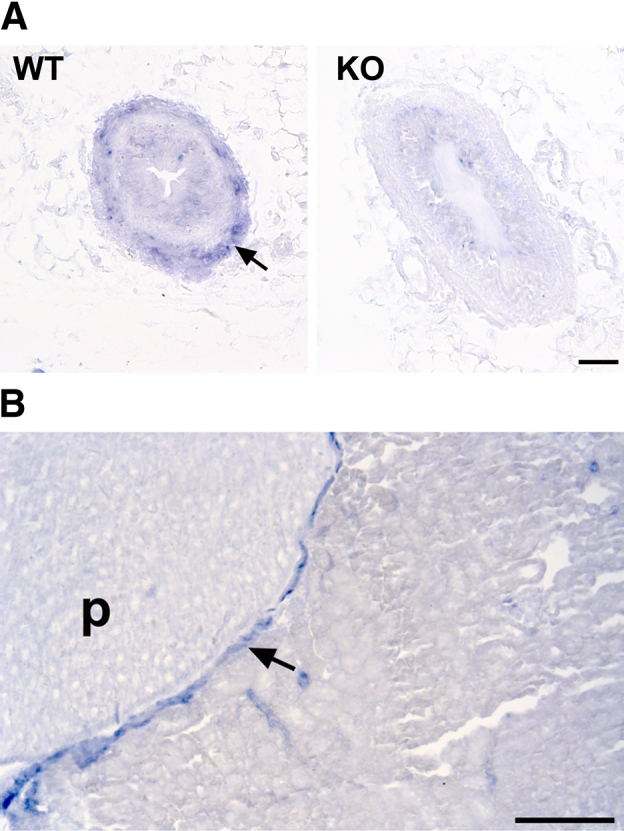
miR-145-5p is expressed in the smooth muscle layer of the ureter. A:In situ hybridization for miR-145-5p in ureter sections from WT and miR-143/145 KO mice. miR-145-5p was expressed in the smooth muscle layer of the ureter of WT mice (arrow). The specificity of the staining was confirmed by the lack of signal in ureters from the miR-143/145 KO mice. B: In situ hybridization for miR-145-5p at the level of the pelvicalyceal system in WT mice. miR-145-5p expression was localized to the smooth muscle layer (arrow). Scale bars: 50 μm (A); 200 μm (B). p, papilla.
miR-143/145–Deficient Mice Develop Hydronephrosis
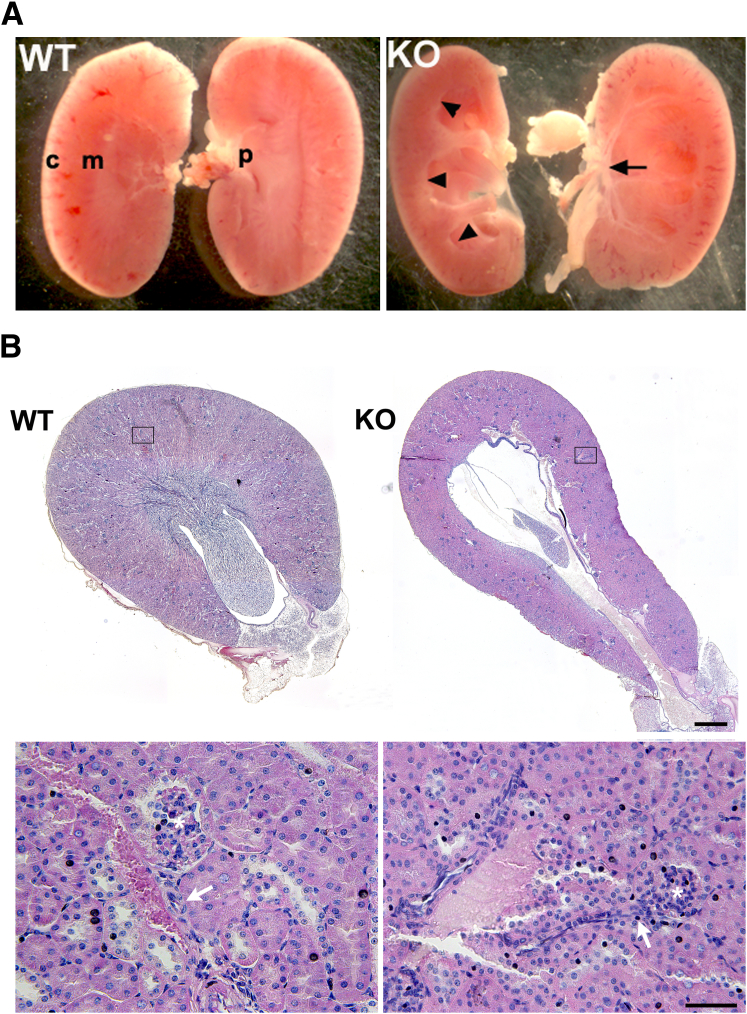
To determine whether the miR-143/145 cluster plays a role in maintaining the structure and function of the urinary tract, we conducted morphologic and functional studies in kidneys and ureters of miR-143/145–deficient mice. We found that mutant mice developed hydronephrosis characterized by marked dilatation of the pelvis and calyces accompanied by severe atrophy of the papilla (Figure 2). Histologic analysis by hematoxylin and eosin staining did not show morphologic changes at the level of the renal cortex in mice lacking miR-143/145. The structure of glomeruli, blood vessels, and proximal and distal tubules appeared normal in the mutant mice (Figure 2B). Interestingly, immunostaining experiments showed normal expression of the smooth muscle marker α-SMA in JG cells and renal arterioles of miR-143/145 KO mice (Supplemental Figure S1). This was an unexpected result owing to the important role of miR-143/145 in vascular smooth muscle development15 and the high expression of miR145-5p in JG cells and in the kidney vasculature.17 Immunostaining for renin showed that JG cells in miR-143/145 KO mice are plump compared with controls. This difference is consistent with the low blood pressure in miR-143/145 KO mice.
Figure 2.
miR-143/145–deficient mice develop hydronephrosis. A: Kidneys from 2-month-old WT and miR-143/145 KO mice. miR-143/145 KO mice develop hydronephrosis, which is characterized by marked dilatation of the pelvis and calyces (arrowheads) and accompanied by severe atrophy of the papilla (arrow). B: Hematoxylin and eosin staining of transversal kidney sections of 3-month-old male mice. miR-143/145 KO kidneys show severe atrophy of the papilla and dilatation of the pelvis. Lower panels show in more detail the boxed areas indicated in the upper panels. Hematoxylin and eosin staining does not show morphologic changes at the level of the renal cortex in mice lacking miR-143/145. The structure of glomeruli, blood vessels, and tubules is conserved in mutant mice. Asterisk, glomerulus; arrows, afferent arteriole. Scale bars: 500 μm (B, upper panel); 50 μm (B, lower panel). c, cortex; m, medulla; p, papilla.
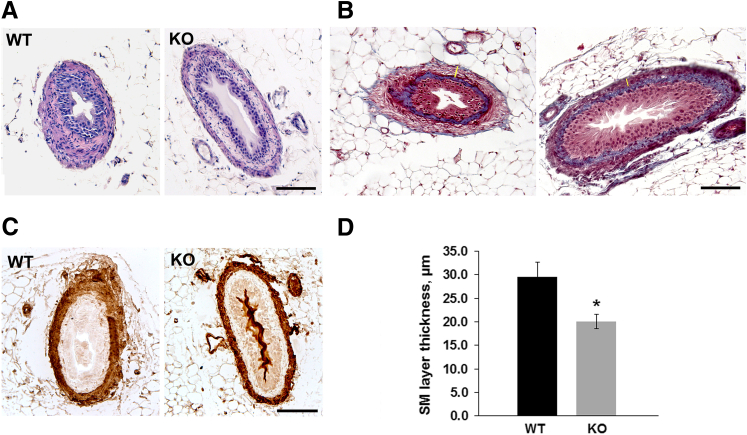
Next, we sought to examine the morphology of ureters from miR-143/145 KO mice. Histologic analysis of ureter sections by hematoxylin and eosin and Masson's trichrome staining as well as immunostaining for α-SMA confirmed that all cell layers were present in mutant mice (Figure 3, A–C). However, frequent distension of the proximal ureter (Figures 3, A and B, and 4) and a significant thinning of the smooth muscle layer was evident in miR-143/145 KO mice (Figure 3). Interestingly, reduced thickness of the smooth muscle layer also has been observed in the aorta of miR-143/145 KO mice.16 By direct anatomic examination, we did not find evidence of an abnormal connection between the kidney and the ureter or a physical obstruction of the ureter (not shown), suggesting that hydronephrosis may be caused by a functional obstruction of the urinary system.
Figure 3.
Histologic analysis of ureters from miR-143/145–deficient mice. Hematoxylin and eosin (A), Masson's trichrome (B), and α-SMA (C) staining of ureter sections from WT and miR-143/145 KO mice. Histologic analysis shows frequent dilatation of the proximal ureter and thinning of the smooth muscle layer in KO mice. All cell layers are present in the KO ureters. D: Quantitation of ureter smooth muscle layer thickness. Quantitation was performed in three Masson's trichrome–stained sections from each mouse. Examples of where measurements were performed are indicated by yellow bars (B). ∗P < 0.05. WT, n = 4; KO, n = 3. Scale bars: 100 μm.
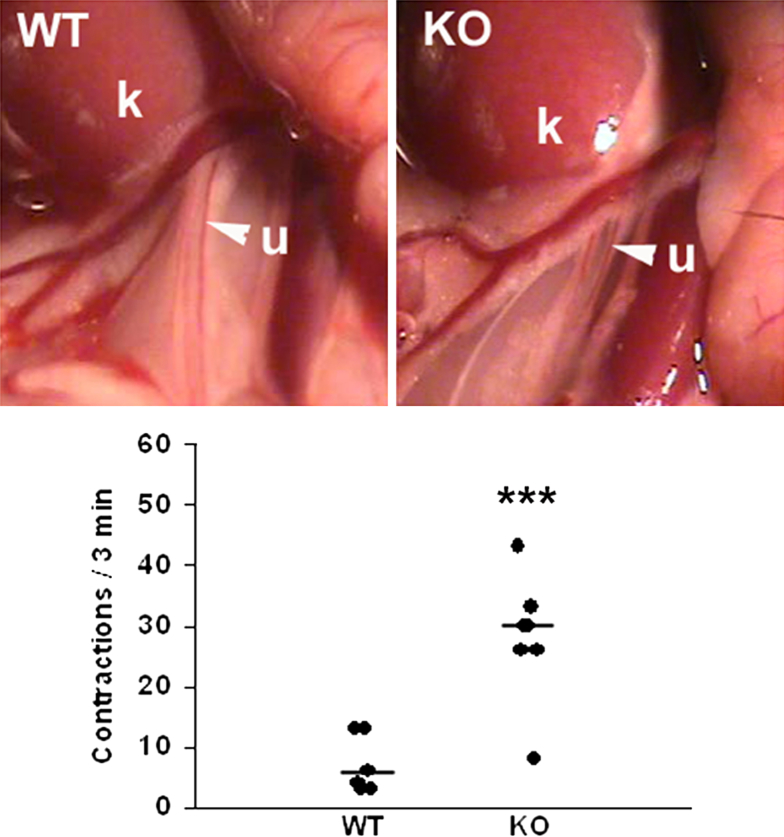
Figure 4.
Ureters of miR-143/145–deficient mice show abnormal contractility. Exposed ureters (arrowheads) of anesthetized 3-month-old WT and miR-143/145 KO female mice. Quantification of peristaltic activity. The number of ureteral contractions in a 3-minute period was significantly higher in miR-143/145 KO compared with WT mice. Peristalsis in WT mice occurred in normal, wave-like fashion from the proximal (renal) ureter toward the bladder, whereas in KO mice it was characterized by frequent, incomplete contractions that failed to propagate in a proximal-distal direction. ∗∗∗P < 0.001. k, kidney; u, ureter.
We found that hydronephrosis occurred in both male and female mutant mice as early as 2 months of age (Table 2). In addition, hydronephrosis developed gradually during postnatal life in miR-143/145–deficient mice. We observed that kidneys from younger animals (ie, 2 weeks of age) had no major abnormalities (not shown), suggesting a progressive effect of miR-143/145 deficiency on the kidney.
Table 2.
Occurrence of Hydronephrosis in WT and miR-143/145 KO Mice
| Sex | WT | miR-143/145 KO |
|---|---|---|
| Female | 0/8 | 6/6 |
| Male | 0/9 | 10/10 |
| Total | 0/17 | 16/16 |
Age of mice ranged from 2 to 8 months.
To determine whether miR-143/145 deficiency leads to impaired renal function, we measured blood urea nitrogen and plasma creatinine levels of mutant mice.22 Both urea nitrogen and creatinine levels were slightly, but not significantly, increased in mutant mice compared with WT littermates up to 8 months of age (Supplemental Figure S2). These results are consistent with the overall normal appearance of the kidney cortex and the absence of any sign of early health decline or reduced life span of miR-143/145–deficient mice.
Ureters of miR-143/145–Deficient Mice Show Abnormal Contractility
Given the apparent lack of physical obstruction in mice deficient for miR-143/145 we sought to determine whether abnormal ureteric peristalsis contributes to the hydronephrosis phenotype. The number of peristaltic waves recorded in exposed ureters of anesthetized animals was significantly higher in miR-143/145 KO mice compared with WT controls (Figure 4 and Supplemental Videos S1 and S2). Moreover, peristaltic waves were short, more frequent, and failed to completely propagate in a proximal-distal direction (Supplemental Videos S1 and S2). We next sought to analyze peristaltic activity in isolated ureters using an in vitro culture system of embryonic ureters.23 Embryonic day 16.5 ureter explants from WT mice grown for 5 days on membranes in the air-medium interface acquired the ability to generate peristaltic waves that propagated in a proximal-distal direction. Ureters from miR-143/145–deficient embryos also developed peristalsis in culture and showed a slight, but not significant, increase in the number of contractions measured over a period of 2 minutes (Supplemental Figure S3). These results indicate that abnormal peristalsis progresses gradually and becomes evident later in postnatal life.
Taken together, these data indicate that hydronephrosis in miR-143/145–deficient mice likely is the result of the abnormal peristaltic activity of the ureter and that this pathologic condition progresses gradually with age.
Ureters from miR-143/145–Deficient Mice Have an Altered Pattern of Gene Expression
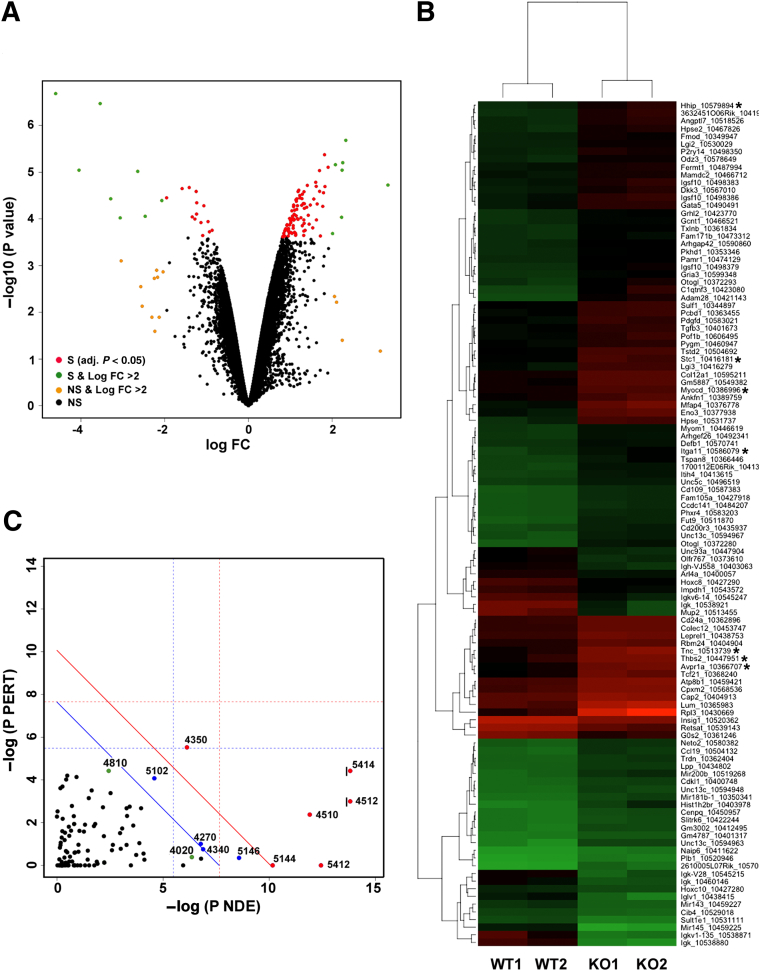
To gain insight into the mechanism of impaired peristalsis leading to hydronephrosis we sought to identify genes with altered expression in miR-143/145–deficient ureters. According to miRWalk and other databases (http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk, last accessed May 5, 2014), miR-143 and miR-145 have the potential to targeting more than 1000 genes by binding to putative binding sites in the 3′ untranslated region of these genes. We conducted a gene expression profiling analysis on total RNA isolated from ureters of WT and miR-143/145 KO mice. Figure 5 provides an overview of the gene expression pattern of WT and mutant ureters. We found that 108 genes were expressed differentially (using an adjusted PFDR < 0.05 as the threshold for significance). Ninety genes were up-regulated and 18 genes were down-regulated in the miR-143/145 KO ureters. The fact that most genes (83%) were up-regulated is consistent with the well-established negative effect of miRNAs on gene expression. Figure 5A shows the results as a volcano plot, which illustrates the fold-difference in the expression, and the significance of the difference, for all genes in the arrays. We conducted a hierarchical cluster analysis of genes which showed significant differential expression (PFDR < 0.05) with the purpose of grouping genes with similar expression patterns (Figure 5B). This analysis showed a few significantly dysregulated genes involved in smooth muscle development and/or function (eg, myocardin,24,25 stanniocalcin-1,26 hedgehog-interacting protein,27 tenascin C,28 arginine vasopressin receptor 1A,29 thrombospondin 2,30 and α11 integrin31).
Figure 5.
Gene expression profiling in ureters from WT and miR-143/145 KO mice. Total RNA samples from WT and mutant ureters were analyzed using Affymetrix GeneChip Mouse Gene 1.0 ST arrays. A: Volcano plot shows the fold-difference in the expression and the significance of the difference for all genes in the arrays. B: Heatmap shows differentially expressed genes in ureters from miR-143/145 KO mice. Of the 108 differentially expressed genes, 90 were up-regulated and 18 were down-regulated. Each column represents results from an independent animal (indicated as WT1, WT2, KO1, and KO2). Each row corresponds to a single gene probe. The color indicates the level of mRNA expression (red, high expression; green, low expression). We used hierarchical cluster analysis of gene expression to group genes with similar expression patterns as indicated at the left side of the map. C: SPIA of genes showing differential expression in miR-143/145 KO ureters. A two-dimensional plot shows the relationship between the gene over-representation (x axis) and the perturbation of the pathways (y axis) analyzed by SPIA. We considered the 500 most differentially expressed genes for the analysis. Each point represents a pathway. The x axis shows the P value on the over-representation of differentially expressed genes in the given pathway: P(NDE). The y axis shows the significance of the overall perturbation of the pathway: P(PERT). Pathways above the red line are significant at 5% after Bonferroni correction, and pathways above the blue line are significant at 5% after FDR correction. The vertical and horizontal thresholds represent the same corrections for the two types of evidence considered individually. Indicated in green are relevant pathways that did not meet both the over-representation and perturbation criteria but were above a 10% cut-off of the FDR-corrected overall P value: P(G)FDR. For pathways 4512 and 5414, the actual −log P(NDE) values were 22 and 14.5, respectively. These values were cut off from the plot as indicated by the black vertical line on the left side of the point. Numbers are the Kyoto Encyclopedia of Genes and Genomes (Kegg) database identification number for the pathway: 4512, ECM-receptor interaction; 5414, dilated cardiomyopathy; 4510, focal adhesion; 5412, arrhythmogenic right ventricular cardiomyopathy; 4350, transforming growth factor-β signaling pathway; 4270, vascular smooth muscle contraction; 4340, hedgehog signaling pathway; 4810, regulation of actin cytoskeleton; 4020, calcium signaling pathway; 5146, amebiasis; 5144, malaria; and 5202, transcriptional dysregulation in cancer. Genes related to smooth muscle biology are marked with asterisks. FC, fold-change; NS, not significant (P > 0.05); S, significant (P < 0.05).
SPIA Shows Several Significantly Perturbed Pathways in the Ureters of miR-143/145 KO Mice
Considering that miRNAs often regulate biological processes by their modest effects on numerous targets,32,33 we sought to identify biological pathways that may be altered in the miR-143/145 KO ureters by performing SPIA21 on the annotated Kyoto Encyclopedia of Genes and Genomes pathway database. To identify perturbed networks of genes and pathways, SPIA takes into consideration the over-representation and the function of dysregulated genes in a given pathway as well as the extent of gene expression changes. By taking into account both the directionality and the topology of the gene networks, SPIA can detect perturbed pathways even in situations in which the number of genes or the change in expression of the genes in a given pathway is relatively low. The SPIA analysis output of our gene array data is shown in Figure 5C. We found 12 significantly perturbed and enriched pathways (Table 3). Interestingly, nine of these pathways corresponded to biological processes that involved genes that were associated directly or indirectly to smooth muscle biology (Table 3). Table 4 shows the genes represented in each perturbed pathway. We found that almost half of the 108 dysregulated genes (46 genes: 40 up-regulated and 6 down-regulated) accounted for these perturbed pathways.
Table 3.
Dysregulated Pathways in miR-143/145–Deficient Ureters
| Pathway name | pGFdr | Status | Kegg pathway ID |
|---|---|---|---|
| Pathways related to smooth muscle biology | |||
| ECM-receptor interaction | 3.99 × 10−08 | Activated | 4512 |
| Dilated cardiomyopathy | 6.34 × 10−06 | Inhibited | 5414 |
| Focal adhesion | 0.0003 | Activated | 4510 |
| Arrhythmogenic right ventricular cardiomyopathy | 0.0014 | Inhibited | 5412 |
| TGF-β signaling pathway | 0.0023 | Activated | 4350 |
| Vascular smooth muscle contraction | 0.0432 | Activated | 4270 |
| Hedgehog signaling pathway | 0.0440 | Inhibited | 4340 |
| Regulation of actin cytoskeleton | 0.0728 | Activated | 4810 |
| Calcium signaling pathway | 0.0746 | Inhibited | 4020 |
| Other pathways | |||
| Malaria | 0.0076 | Inhibited | 5144 |
| Amebiasis | 0.020 | Activated | 5146 |
| Transcriptional dysregulation in cancer | 0.022 | Inhibited | 5202 |
Microarray data were subjected to SPIA using the 500 most differentially expressed genes. Data above a 10% cut-off of the FDR-corrected overall P-value pGFdr are shown.
ECM, extracellular matrix; Kegg, Kyoto Encyclopedia of Genes and Genomes database; TGF, transforming growth factor.
Table 4.
Dysregulated Genes in Altered Pathways
| Gene symbol | Gene name | FC | ECM-receptor interaction | Dilated cardiomyopathy | Focal adhesion | ARVC | TGF-β signaling pathway | Vascular smooth muscle contraction | Hedgehog signaling pathway | Regulation of actin cytoskeleton | Calcium signaling pathway |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cacna1d | Calcium channel, L-type α1D subunit | 1.6 | • | • | • | • | |||||
| Prkch | Protein kinase C, η | 1.9 | • | ||||||||
| Adra1d | Adrenergic receptor α1d | 0.6 | • | • | |||||||
| Ppp1r14a | Protein phosphatase 1, regulatory subunit 14A | 1.7 | • | ||||||||
| Kcnma1 | Potassium large-conductance calcium-activated, M, α member 1 | 1.9 | • | ||||||||
| Pla2g4a | Phospholipase A2, group IVA | 2.0 | • | ||||||||
| Adora2b | Adenosine A2b receptor | 1.8 | • | • | |||||||
| Avpr1a | Arginine vasopressin receptor 1A | 3.5 | • | • | |||||||
| Ramp1 | Receptor G-protein–coupled, activity-modifying protein 1 | 1.8 | • | ||||||||
| Itgα3 | Integrin α3 | 1.7 | • | • | • | • | • | ||||
| Itgα4 | Integrin α4 | 1.6 | • | • | • | • | • | ||||
| Itgα8 | Integrin α8 | 1.8 | • | • | • | • | • | ||||
| Itgα11 | Integrin α11 | 2.5 | • | • | • | • | • | ||||
| Itgβ6 | Integrin β6 | 1.9 | • | • | • | • | • | ||||
| Lama2 | Laminin, α2 | 1.8 | • | • | • | • | |||||
| Lama5 | Laminin, α5 | 1.7 | • | • | |||||||
| Lamb3 | Laminin, β3 | 1.8 | • | • | |||||||
| Col6a3 | Collagen, type VI, α3 | 1.5 | • | • | |||||||
| Col1a1 | Collagen, type I, α1 | 1.9 | • | • | |||||||
| Col1a2 | Collagen, type I, α2 | 1.7 | • | • | |||||||
| Tnc | Tenascin C | 3.5 | • | • | |||||||
| Thbs1 | Thrombospondin 1 | 1.9 | • | • | • | ||||||
| Thbs2 | Thrombospondin 2 | 2.9 | • | • | • | ||||||
| Npnt | Nephronectin | 1.9 | • | ||||||||
| BclII | B-cell leukemia/lymphoma 2 | 1.6 | • | ||||||||
| Pdgfd | Platelet-derived growth factor D | 2.2 | • | • | |||||||
| Cdh2 | Cadherin 2 | 0.5 | • | ||||||||
| Sgcg | Sarcoglycan, γ | 0.4 | • | • | |||||||
| Dmd | Dystrophin | 2.0 | • | • | |||||||
| Tgfb2 | Transforming growth factor, β2 | 1.7 | • | • | |||||||
| Tgfb3 | Transforming growth factor, β3 | 2.0 | • | • | |||||||
| Bmp2 | Bone morphogenetic protein 2 | 2.0 | • | • | |||||||
| Bmp4 | Bone morphogenetic protein 4 | 2.2 | • | • | |||||||
| Bmp5 | Bone morphogenetic protein 5 | 3.5 | • | • | |||||||
| Ptch1 | Patched homolog 1 | 1.6 | • | ||||||||
| Wnt5a | Wingless-related MMTV integration site family, member 5A | 1.7 | • | ||||||||
| Hhip | Hedgehog interacting protein | 3.1 | • | ||||||||
| Fgf7 | Fibroblast growth factor 7 | 1.8 | • | ||||||||
| Myh10 | Myosin, heavy polypeptide 10, nonmuscle | 1.7 | • | ||||||||
| Itpka | Inositol-trisphosphate 3-kinase A | 0.5 | • | ||||||||
| Slc8a3 | Solute carrier family 8 (sodium/calcium exchanger), member 3 | 1.9 | • | ||||||||
| Ppp3ca | Protein phosphatase 3, catalytic subunit, α isozyme | 1.6 | • | ||||||||
| Igh/Igh-VJ558 | Immunoglobulin heavy chain (J558 family) | 0.3 | • | • | |||||||
| Gna15 | Guanine nucleotide binding protein (G protein), α15 (Gq class) | 1.9 | • | ||||||||
| Ptger1 | Prostaglandin E receptor 1 (subtype EP1) | 1.6 | • | ||||||||
| Ptger3 | Prostaglandin E receptor 3 (subtype EP3) | 0.5 | • |
ARVC, arrhythmogenic right ventricular cardiomyopathy; ECM, extracellular matrix; FC, fold-change; Kegg, Kyoto Encyclopedia of Genes and Genomes database; TGF, transforming growth factor.
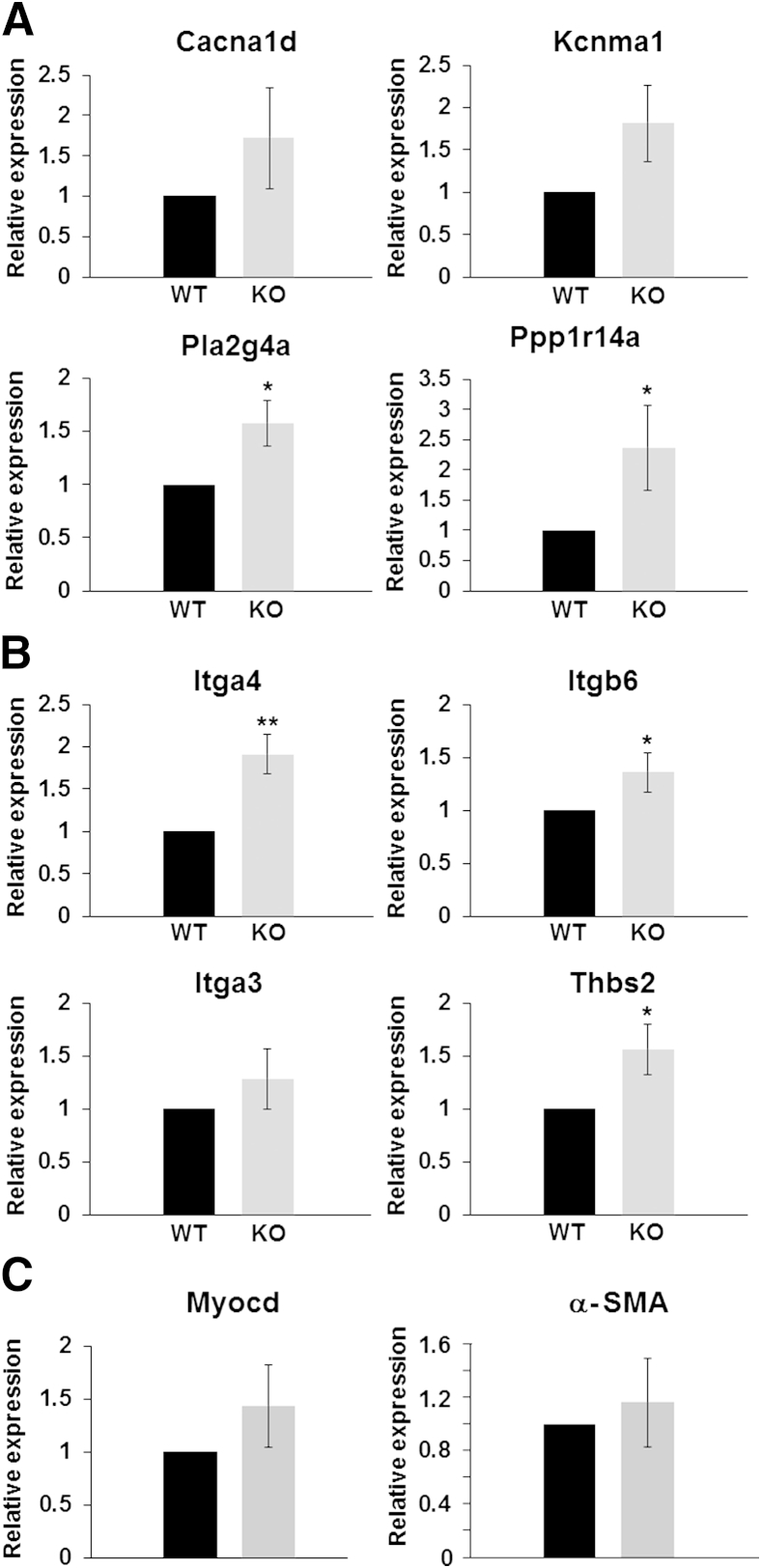
Altered Expression of Selected Genes in Ureters from miR143/145 KO Mice as Assessed by qPCR
To corroborate the microarray and SPIA results, we measured the expression of selected mRNAs by qPCR. We focused on genes involved directly in smooth muscle contraction because of the abnormal ureter peristaltic activity of miR-143/145 KO mice, and on genes involved in ECM-receptor interaction because this pathway is important for smooth muscle function34 and is the most dysregulated pathway in our analysis with a FDR-corrected overall P value (pGFdr) of 3.99 × 10−08. We found that the expression levels of all selected genes were higher in ureters from miR-143/145 KO mice compared with their WT counterparts (Figure 6). In the cases of Cacna1d, Kcnma1, and Itga3, the differences in expression were not significant, which was expected as a result of the modest effect of these miRNAs shown by our microarray analysis. We also measured the levels of Myocd and its target α-SMA (Acta2) in ureters of WT and miR-143/145 KO mice by qPCR. The results showed a slight but nonsignificant increase in myocardin in miR-143/145 KO mice. The levels of α-SMA were not different between the two groups. In agreement with these findings, normal levels of myocardin and α-SMA also were reported in SMCs of aorta from miR-143/145 KO mice.16
Figure 6.
mRNA levels of genes related to smooth muscle biology in ureters from miR-143/145 mice. Expression levels of selected genes were measured by qPCR. The selection of genes was based on SPIA results. Expression levels of genes involved in vascular smooth muscle contraction (A), ECM-receptor interaction (B), and smooth muscle transcriptional regulator myocardin and its target α-SMA (C). Values are means ± SEM of four animals. Statistical analysis was performed using the Student's t-test. In most cases the differences between WT and miR-143/145 KO mice were significant. For Cacna1d, Kcnma1, Itga3, and Myocd the differences were not significant but levels were slightly higher in the mutant ureters, in agreement with the microarray results. Levels of α-SMA were not changed. Primer sequences and qPCR conditions are listed in Table 1. The expression of glyceraldehyde-3-phosphate dehydrogenase was used as an internal control. ∗P < 0.05, ∗∗P < 0.01 versus WT.
Overall, these results corroborate the microarray and the in silico data and support the concept that miR-143/145 deficiency results in a pattern of gene dysregulation in pathways that are important for normal ureteric peristalsis.
Discussion
Here, we show that miR-143/145, a miRNA cluster that regulates the phenotype and fate of smooth muscle cells in vascular tissue,15,16 is crucial for the normal function of the urinary tract. We found that mice lacking whole-body expression of miR-143/145 show compromised ureter peristaltic activity, which results in hydronephrosis as early as 2 months postnatally.
The phenotype of miR-143/145–deficient mice is characterized by severe papillary atrophy and marked dilatation of the pelvicalyceal system and ureter. These major phenotypic abnormalities indicate that these two miRNAs are indispensable for the maintenance of the structure and function of the urinary tract. Despite the high expression of miR-145-5p in the JG cells and vasculature of the kidney,17 we did not find any evident alteration in the structure of either renal vessels or the JG area by light microscopy examination. The structure of the tubules and glomeruli also appeared normal in miR143/145 KO mice. Most importantly, we did not find changes in the pattern of α-SMA staining in the blood vessels. Immunostaining for renin showed that JG cells in KO mice are plump compared with controls, suggesting increased renin production. It is likely that this increase is in response to the low blood pressure that results from the reduced vascular tone shown in miR143/145 KO mice.16 Alternatively, the existence of a putative miR-145-5p binding site in the 3′ untranslated region of renin, as predicted by miRWalk and other databases, suggests the possibility of a direct effect of miR-145-5p on renin expression. Ultrastructural studies16,35 on large arteries of miR-143/145–deficient mice (eg, the aorta and femoral artery) have shown thinning of the smooth muscle layer of the vessels, a reduced number of actin-based stress fibers, and a switch to a synthetic smooth muscle phenotype. Therefore, additional studies at the electron microscope level are necessary to determine whether the kidney vasculature is affected similarly.
Given that we did not find any indication of a physical obstruction of the urinary tract associated with the hydronephrosis phenotype, we hypothesized that the lack of miR-143/145 was the result of a functional obstruction caused by abnormal ureter smooth muscle contraction and resulting in impaired peristalsis. We confirmed this hypothesis by observing that miR-143/145 KO mice showed an abnormal pattern of peristaltic waves characterized by frequent and incomplete ureteral contractions. The fact that peristalsis was not altered in embryonic ureter explants (ie, mutant ureters showed peristaltic waves similar to those of WT embryos) indicates that the effect of miR-143/145 deficiency progresses gradually with age. Therefore, it is possible that in the absence of miR-143/145 the smooth muscle of the ureter does not contract efficiently, resulting in a gradual accumulation of fluid in the kidney with age. Our finding of several dysregulated genes and pathways related to smooth muscle supports this idea. On the other hand, it can be argued that the effect may not be entirely intrinsic to the ureter and therefore is not evident in the detached organ (ie, embryo explants). Although myogenic mechanisms are key regulators of ureteral peristalsis, neurogenic factors also play an important role in this process.36 Therefore, it is possible that neurogenic factors contribute to the miR-143/145 KO phenotype. For example, SPIA analysis showed inhibition of the cholinergic synapse pathway [FDR-corrected overall P value (pGFdr) = 0.105] in the miR-143/145 KO ureters. Further studies involving pharmacologic manipulations of isolated ureters are necessary to elucidate the mechanism of altered peristalsis in miR-143/145–deficient mice.
Although some of the genes and pathways that control normal ureteric function are known,36 the involvement of specific miRNAs has not been reported previously. It is well accepted that normal smooth muscle contraction requires a fine control of expression of many genes. Given that miRNAs in general, and miR-143/145 in particular, have the potential to influence the activity of thousands of genes, it is likely that the impaired peristalsis shown by miR-143/145–deficient mice could be the result of the dysregulation of several genes that control and maintain ureteric function. Although we found a significant number of up-regulated genes, which is consistent with the well-known inhibitory effect of miRNAs in gene expression, only a few dysregulated genes were related to smooth muscle structure and function. We reasoned that because individual miRNAs can act on entire gene networks instead of on single genes alone, even moderate or minimal changes in expression of these genes could have a great effect on a given pathway or biological process. In agreement with our prediction, the results of the SPIA analysis determined several pathways that were altered in the miR-143/145 KO ureters. More importantly, some of these pathways and biological processes are associated with smooth muscle biology. Therefore, we were able to identify affected genes that otherwise would not have been chosen for further investigation owing to modest changes in their expression. These results stress the importance of using pathway analysis when performing miRNA KO studies.
There likely are multiple mechanisms underlying the changes we found in the kidneys and ureters of mutant mice. It is important to recognize that it is unlikely that the dysregulation of a single gene can account solely for the observed phenotype. To our knowledge, there are no studies of overexpression of the genes that we found up-regulated in our arrays that resulted in a similar phenotype as the one described in this study. miRNAs generally act as negative post-transcriptional regulators of mRNAs, therefore suppressing the expression of miRNAs should produce changes that are opposite to those of their predicted target mRNAs. We found that up-regulated genes are related not only to smooth muscle contraction, but also to ECM-receptor interaction. Surface adhesion receptors and their ligands are known regulators of SMC functions, including contraction.34 Interestingly, altered expression of integrins and other ECM proteins has been reported in patients with ureteropelvic junction obstruction.37 Further studies aimed at establishing the role of the ECM-receptor pathway in the smooth muscle cells of the ureter in miR-143/145 KO mice may help to elucidate the pathogenic mechanism underlying the phenotype described here.
The findings reported here are consistent with a model in which miR-143/145 fine-tune the expression of multiple genes related to smooth muscle contraction and that deficiency of these miRNAs results in a chronic impairment in peristalsis and a gradual development of hydronephrosis. This phenotype closely resembles findings encountered in patients with nonobstructive (functional) hydronephrosis. Currently, there are no predictive indicators of nonobstructive hydronephrosis in humans. Down-regulation of miR-145 recently was documented in patients affected by chronic kidney disease38 and renal cell carcinoma.39,40 At present, there are no reports on miR-143/145 expression in patients affected by hydronephrosis. Our findings suggest that the miR-143/145 cluster should be considered a potential underlying factor in the development of nonobstructive hydronephrosis in humans. Future work needs to be performed to determine whether miR-143/145 offer an opportunity for the diagnosis and management of functional hydronephrosis.
Acknowledgments
We thank Dr. Eric Olson for providing the miR143/145 KO mice; Dr. Stephen Turner for the microarray data analysis; Minghong Li, Kimberly Hilsen, Takele Yazew, and Danielle Stumbo for technical assistance with the mouse work; and Theodore Mehalic for assistance with video imaging.
Footnotes
Supported by NIH grants HL096735 (R.A.G.) and DK075481 (M.L.S.S.-L.), and a University of Virginia Children's Hospital Grant-In-Aid (S.M.).
Disclosures: None declared.
Supplemental Data
Immunostaining for renin and α-SMA in the kidney cortex of 3-month-old male WT and miR-143/145 KO mice. Kidneys of mutant mice showed renin expression in JG cells (arrowheads) and α-SMA in the arterioles (arrows). Renin cells are present in similar numbers and locations, but are plump in KO compared with WT kidneys. The pattern of α-SMA labeling of blood vessels and the interstitium is not altered in KO kidneys. Tubules and glomeruli (asterisks) structure also appear normal. Renin and α-SMA staining is shown in brown. Scale bars: 100 μm.
Blood urea nitrogen and plasma creatinine levels. Urea nitrogen and creatinine levels are slightly, but not significantly, increased in miR-143/145–deficient mice compared with WT littermates.
Peristalsis in embryonic ureters in culture. Quantitation of in vitro peristalsis over a 2-minute period showed a slight, but not significant, increase in the number of contractions in ureters from miR-143/145–deficient mice.
WT mouse. Ureter contractions are visible at 7, 24, 43, 64, and 84 seconds.
miR-143/145 mouse. Ureter contractions are visible at 2, 9, 16, 20, 27, 32, 37, 43, 49, 54, 58, 64, 70, 75, and 79 seconds.
References
- 1.Ek S., Lidefeldt K.J., Varricio L. Fetal hydronephrosis; prevalence, natural history and postnatal consequences in an unselected population. Acta Obstet Gynecol Scand. 2007;86:1463–1466. doi: 10.1080/00016340701714802. [DOI] [PubMed] [Google Scholar]
- 2.Johnson C.E., Elder J.S., Judge N.E., Adeeb F.N., Grisoni E.R., Fattlar D.C. The accuracy of antenatal ultrasonography in identifying renal abnormalities. Am J Dis Child. 1992;146:1181–1184. doi: 10.1001/archpedi.1992.02160220067024. [DOI] [PubMed] [Google Scholar]
- 3.Dudley J.A., Haworth J.M., McGraw M.E., Frank J.D., Tizard E.J. Clinical relevance and implications of antenatal hydronephrosis. Arch Dis Child Fetal Neonatal Ed. 1997;76:F31–F34. doi: 10.1136/fn.76.1.f31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 5.Friedman R.C., Farh K.K., Burge C.B., Bartel D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cano A., Nieto M.A. Non-coding RNAs take centre stage in epithelial-to-mesenchymal transition. Trends Cell Biol. 2008;18:357–359. doi: 10.1016/j.tcb.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Heuston E.F., Lemon K.T., Arceci R.J. The beginning of the road for non-coding RNAs in normal hematopoiesis and hematologic malignancies. Front Genet. 2011;2:94. doi: 10.3389/fgene.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Rourke J.R., Georges S.A., Seay H.R., Tapscott S.J., McManus M.T., Goldhamer D.J., Swanson M.S., Harfe B.D. Essential role for Dicer during skeletal muscle development. Dev Biol. 2007;311:359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perruisseau-Carrier C., Jurga M., Forraz N., McGuckin C.P. miRNAs stem cell reprogramming for neuronal induction and differentiation. Mol Neurobiol. 2011;43:215–227. doi: 10.1007/s12035-011-8179-z. [DOI] [PubMed] [Google Scholar]
- 11.Qi J., Yu J.Y., Shcherbata H.R., Mathieu J., Wang A.J., Seal S., Zhou W., Stadler B.M., Bourgin D., Wang L., Nelson A., Ware C., Raymond C., Lim L.P., Magnus J., Ivanovska I., Diaz R., Ball A., Cleary M.A., Ruohola-Baker H. microRNAs regulate human embryonic stem cell division. Cell Cycle. 2009;8:3729–3741. doi: 10.4161/cc.8.22.10033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh R., Saini N. Downregulation of BCL2 by miRNAs augments drug induced apoptosis: combined computational and experimental approach. J Cell Sci. 2012;125:1568–1578. doi: 10.1242/jcs.095976. [DOI] [PubMed] [Google Scholar]
- 13.Wang S., Olson E.N. AngiomiRs–key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19:205–211. doi: 10.1016/j.gde.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 15.Cordes K.R., Sheehy N.T., White M.P., Berry E.C., Morton S.U., Muth A.N., Lee T.H., Miano J.M., Ivey K.N., Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin M., Small E.M., Sutherland L.B., Qi X., McAnally J., Plato C.F., Richardson J.A., Bassel-Duby R., Olson E.N. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sequeira-Lopez M.L., Weatherford E.T., Borges G.R., Monteagudo M.C., Pentz E.S., Harfe B.D., Carretero O., Sigmund C.D., Gomez R.A. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol. 2010;21:460–467. doi: 10.1681/ASN.2009090964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellanos Rivera R.M., Monteagudo M.C., Pentz E.S., Glenn S.T., Gross K.W., Carretero O., Sequeira-Lopez M.L., Gomez R.A. Transcriptional regulator RBP-J regulates the number and plasticity of renin cells. Physiol Genomics. 2011;43:1021–1028. doi: 10.1152/physiolgenomics.00061.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reiner A., Yekutieli D., Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 21.Tarca A.L., Draghici S., Khatri P., Hassan S.S., Mittal P., Kim J.S., Kim C.J., Kusanovic J.P., Romero R. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogura A., Asano T., Suzuki O., Yamamoto Y., Noguchi Y., Kawaguchi H., Yamaguchi Y. Hereditary nephrotic syndrome with progression to renal failure in a mouse model (ICGN strain): clinical study. Nephron. 1994;68:239–244. doi: 10.1159/000188264. [DOI] [PubMed] [Google Scholar]
- 23.Cain J.E., Islam E., Haxho F., Blake J., Rosenblum N.D. GLI3 repressor controls functional development of the mouse ureter. J Clin Invest. 2011;121:1199–1206. doi: 10.1172/JCI45523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du K.L., Ip H.S., Li J., Chen M., Dandre F., Yu W., Lu M.M., Owens G.K., Parmacek M.S. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z., Wang D.Z., Pipes G.C., Olson E.N. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheikh-Hamad D., Bick R., Wu G.Y., Christensen B.M., Razeghi P., Poindexter B., Taegtmeyer H., Wamsley A., Padda R., Entman M., Nielsen S., Youker K. Stanniocalcin-1 is a naturally occurring L-channel inhibitor in cardiomyocytes: relevance to human heart failure. Am J Physiol Heart Circ Physiol. 2003;285:H442–H448. doi: 10.1152/ajpheart.01071.2002. [DOI] [PubMed] [Google Scholar]
- 27.Ren Y., Cowan R.G., Migone F.F., Quirk S.M. Overactivation of hedgehog signaling alters development of the ovarian vasculature in mice. Biol Reprod. 2012;86:174. doi: 10.1095/biolreprod.112.099176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimoto M., Suzuki H., Shiba M., Shimojo N., Imanaka-Yoshida K., Yoshida T., Kanamaru K., Matsushima S., Taki W. Tenascin-C induces prolonged constriction of cerebral arteries in rats. Neurobiol Dis. 2013;55:104–109. doi: 10.1016/j.nbd.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 29.Ostrowski N.L., Young W.S., III, Knepper M.A., Lolait S.J. Expression of vasopressin V1a and V2 receptor messenger ribonucleic acid in the liver and kidney of embryonic, developing, and adult rats. Endocrinology. 1993;133:1849–1859. doi: 10.1210/endo.133.4.8404628. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell E.K., Taylor D.F., Woods K., Davis M.J., Nelson A.L., Teasdale R.D., Grimmond S.M., Little M.H., Bertram J.F., Caruana G. Differential gene expression in the developing mouse ureter. Gene Expr Patterns. 2006;6:519–538. doi: 10.1016/j.modgep.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 31.Velling T., Kusche-Gullberg M., Sejersen T., Gullberg D. cDNA cloning and chromosomal localization of human alpha(11) integrin. A collagen-binding, I domain-containing, beta(1)-associated integrin alpha-chain present in muscle tissues. J Biol Chem. 1999;274:25735–25742. doi: 10.1074/jbc.274.36.25735. [DOI] [PubMed] [Google Scholar]
- 32.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 34.Moiseeva E.P. Adhesion receptors of vascular smooth muscle cells and their functions. Cardiovasc Res. 2001;52:372–386. doi: 10.1016/s0008-6363(01)00399-6. [DOI] [PubMed] [Google Scholar]
- 35.Boettger T., Beetz N., Kostin S., Schneider J., Kruger M., Hein L., Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santicioli P., Maggi C.A. Myogenic and neurogenic factors in the control of pyeloureteral motility and ureteral peristalsis. Pharmacol Rev. 1998;50:683–722. [PubMed] [Google Scholar]
- 37.Cutroneo G., Arena S., Anastasi G., Cervellione R.M., Grimaldi S., Di M.D., Speciale F., Arena F., Di B.V., Favaloro A., Magno C. Altered cytoskeletal structure of smooth muscle cells in ureteropelvic junction obstruction. J Urol. 2011;185:2314–2319. doi: 10.1016/j.juro.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 38.Chen N.X., Kiattisunthorn K., O'Neill K.D., Chen X., Moorthi R.N., Gattone V.H., Allen M.R., Moe S.M. Decreased microRNA is involved in the vascular remodeling abnormalities in chronic kidney disease (CKD) PLoS One. 2013;8:e64558. doi: 10.1371/journal.pone.0064558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu R., Ji Z., Li X., Zhai Q., Zhao C., Jiang Z., Zhang S., Nie L., Yu Z. miR-145 functions as tumor suppressor and targets two oncogenes, ANGPT2 and NEDD9, in renal cell carcinoma. J Cancer Res Clin Oncol. 2014;140:387–397. doi: 10.1007/s00432-013-1577-z. [DOI] [PubMed] [Google Scholar]
- 40.Yoshino H., Enokida H., Itesako T., Kojima S., Kinoshita T., Tatarano S., Chiyomaru T., Nakagawa M., Seki N. Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci. 2013;104:1567–1574. doi: 10.1111/cas.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunostaining for renin and α-SMA in the kidney cortex of 3-month-old male WT and miR-143/145 KO mice. Kidneys of mutant mice showed renin expression in JG cells (arrowheads) and α-SMA in the arterioles (arrows). Renin cells are present in similar numbers and locations, but are plump in KO compared with WT kidneys. The pattern of α-SMA labeling of blood vessels and the interstitium is not altered in KO kidneys. Tubules and glomeruli (asterisks) structure also appear normal. Renin and α-SMA staining is shown in brown. Scale bars: 100 μm.
Blood urea nitrogen and plasma creatinine levels. Urea nitrogen and creatinine levels are slightly, but not significantly, increased in miR-143/145–deficient mice compared with WT littermates.
Peristalsis in embryonic ureters in culture. Quantitation of in vitro peristalsis over a 2-minute period showed a slight, but not significant, increase in the number of contractions in ureters from miR-143/145–deficient mice.
WT mouse. Ureter contractions are visible at 7, 24, 43, 64, and 84 seconds.
miR-143/145 mouse. Ureter contractions are visible at 2, 9, 16, 20, 27, 32, 37, 43, 49, 54, 58, 64, 70, 75, and 79 seconds.