Abstract
Cells can adapt to their environment and develop distinct identities by rewiring their transcriptional networks to regulate the output of key biological pathways without concomitant mutations to the underlying genes. These alterations, called epigenetic changes, persist stably through mitotic or, in some instances, meiotic cell divisions. In eukaryotes, heritable changes to chromatin structure are a prominent, but not exclusive, mechanism by which epigenetic changes are mediated. These changes are initiated by sequence-specific events, which trigger a cascade of molecular interactions resulting in feedback mechanisms, alterations in chromatin structure, histone posttranslational modifications (PTMs), and ultimately establishment of distinct transcriptional states. In recent years, advances in next generation sequencing have led to the discovery of several novel classes of noncoding RNAs (ncRNAs). In addition to their well-established cytoplasmic roles in posttranscriptional regulation of gene expression, ncRNAs have emerged as key regulators of epigenetic changes via chromatin-dependent mechanisms in organisms ranging from yeast to man. They function by affecting chromatin structure, histone PTMs, and the recruitment of transcriptional activating or repressing complexes. Among histone PTMs, lysine methylation serves as the binding substrate for the recruitment of key protein complexes involved in regulation of genome architecture, stability, and gene expression. In this review, we will outline the known mechanisms by which ncRNAs of different origins regulate histone methylation, and in doing so contribute to a variety of genome regulatory functions in eukaryotes.
Keywords: noncoding RNA, histone methylation, epigenetics, lncRNA, siRNA, piRNA
Introduction
Cells exhibit a variety of phenotypes by altering the expression patterns of their genes. The accurate propagation of these phenotypes to progeny cells is critical for survival and adaptability. In all organisms the stable inheritance of phenotypic states is achieved by a combination of genetic and epigenetic means [1]–[3]. Precise DNA replication and chromosome segregation mechanisms transmit a complete blueprint of genes from parental to progeny cells through mitotic and meiotic cell divisions [4]–[6]. The accuracy of this process is of paramount importance for survival. Environmental mutagens, DNA replication and chromosome segregation errors, and activation of mobile genetic elements threaten the fidelity of this process, and endanger the stability of genomes [7]–[11]. In face of these, organisms have evolved rigorous mechanisms to repair their damaged DNA, correct replication and chromosome segregation errors, safeguard their genomes against mobilization of transposable elements, and prevent spurious recombination among repetitive sequences, the loss of which can lead to the erosion of genetic information [12]–[14]. Together these mechanisms accurately duplicate and transfer the genetic information to progeny cells upon which transcriptional regulatory mechanisms act.
Epigenetic mechanisms also contribute to the stable transmission of cellular phenotypes through mitotic, and in some instances, meiotic cell divisions [15]–[17]. Unlike genetic alterations, epigenetic changes occur in the absence of mutations to the underlying genes. Once triggered, these alterations persist through numerous cell divisions independently of the original inducing signal. The establishment, maintenance and transmission of stable epigenetic properties enable organisms to develop distinct cellular identities during development, resist stress by optimizing gene expression patterns, and, broadly, display properties favoring survival. All epigenetic changes are inherently reversible, thus equipping cells with dynamic adaptive responses, whose potential is limited by the combination of the available genetic information and epigenetic systems regulating gene expression [16], [18], [19]. Disruptions to epigenetic pathways can have serious pathological consequences including malignancies [20] infertility [21] neurological disorders [22] and metabolic diseases [23]–[26]. Epigenetic changes occur by a variety of mechanisms, frequently involving changes to gene expression patterns, but, examples in which stable epigenetic alterations occur with no apparent change to gene expression patterns also exist [27]–[29]. For this review, we will focus our discussion on RNA-mediated epigenetic changes that involve alterations to gene expression patterns via histone methylation.
Three features are common to all epigenetic pathways which operate by altering gene expression programs [19]. (1) Sequence-dependent binding events (e.g. proteins binding to a specific DNA or RNA sequence) lead to the recruitment of regulatory factors to a target genomic site; (2) cooperative interactions of various strengths among regulatory and sequence-dependent factors enhance and reinforce specificity; (3) these interactions trigger positive and negative feedback mechanisms which stabilize regulatory factor-mediated transcriptional changes at the target locus, and ensure its propagation through cell division.
Eukaryotic chromatin
In eukaryotes, long tracks of DNA are packaged and organized into a nucleoprotein structure called chromatin. The functional unit of chromatin is a nucleosome: an octameric DNA-protein complex composed of two copies of each histone (H2A, H2B, H3 and H4) around which roughly 147 base-pairs (bp) of DNA wraps. Nucleosomes are the basic repetitive unit of chromatin, which are assembled into long linear arrays separated by a 10–70 bp linker DNA region. Nucleosome density, compaction, and nuclear localization profoundly impact chromatin structure and the transcriptional state of the underlying genes. In addition to transcription, chromatin structure also regulates other key DNA metabolic activities including repair, recombination and replication [30]. Accordingly, eukaryotic cells have evolved highly conserved mechanisms to alter chromatin structure of specific parts of their genomes as an amenable and programmable means for regulating their genomes. But how is this achieved?
The crystal structure of a nucleosome reveals that the N- (and in some cases C-) termini of individual histones protrude out of the nucleosomal globular core domain. These ‘tails’ are accessible for interaction with other proteins and are marked a variety of posttranslational modifications (PTMs), the best characterized of which are acetylation, methylation, ubiquitination, sumoylation, and phosphorylation [31]. These modifications regulate chromatin structure, function, and nuclear localization by helping recruit or restrict chromatin regulatory complexes to specific regions of the chromosome.
Several conserved protein families cooperate to regulate chromatin. Because of their biochemical activities, they are classified into three broad categories: histone-modifying, histone-binding and chromatin remodeling protein families. Histone-modifying proteins catalyze the addition or removal of specific PTMs (e.g. methyl groups) on different histone residues. Often these proteins are targeted to different parts of the genome by a variety of mechanisms, and their activity profoundly impacts the transcriptional outcome of the affected chromosomal region [19]. Histone-binding proteins recognize and bind to specific histone PTMs [32], [33], and their binding often recruits other regulatory complexes through their protein-protein interactions [31]. Chromatin remodelers, as their name suggests, alter the overall structure and packaging of chromatin, by using the energy of ATP hydrolysis to reposition, evict or change the histone composition of nucleosomes [34]. These activities together impact chromatin compaction, organization and positioning within the eukaryotic nucleus, and regulate the DNA accessibility and transcriptional state of the underlying genes. Targeting and temporal coordination of these activities allows cells to establish, maintain and duplicate chromatin and transcriptional properties of specific parts of their genome.
Chromatin-based epigenetic mechanisms
In eukaryotes, many, but not all (for example [35]), epigenetic mechanisms involve heritable alterations of chromatin states, which regulate the transcription of the underlying genes. Several cis- and/or trans-acting factors cooperate to establish these chromatin states whose duplication and propagation after DNA replication is critical for epigenetic inheritance. DNA replication poses a special challenge to inheritance of chromatin states because histones must dissociate from chromosomes during the passage of replication forks, and newly synthesized nucleosomes are incorporated to restore proper nucleosome density on the newly replicated daughter strands. Pioneering work in 70’s and 80’s [36]–[40] revealed that parental histones are retained and distributed randomly to newly replicated DNA molecules. However, newly incorporated nucleosomes carry different PTMs than the parental histones [41]; therefore, a mechanism for restoring the parental histone PTMs must exist for propagation of chromatin states following replication. These observations were confirmed by recent work [42] suggesting that at least some nucleosomes, carrying parental histone PTMs, are transferred onto the newly replicated chromosomes. Below we will discuss several models for inheritance of chromatin states, a few of which suggest that the parentally inherited nucleosomes may contribute to the efficiency of reestablishment of chromatin states after DNA replication.
Models of inheritance of chromatin states
Several models for inheritance of chromatin states have been proposed, but the exact mechanism by which chromatin states are reestablished after DNA replication remains poorly understood. One model proposes that chromatin states are duplicated by histone-modifying complexes that bind to the same PTM which they catalyze on a neighboring nucleosome [23]–[25]. According to this model, the PTMs inherited from the parental nucleosomes deposited on newly replicated DNA provide sufficient specificity to reestablish chromatin states independently of sequence-specific events. A prediction of this would be that histone PTMs alone are epigenetic (self-perpetuating) marks. Other models propose that either sequence-specific events alone [43] or (at least partially) assisted by chromatin-based mechanisms [44] restore parental histone PTMs by targeting and recruiting chromatin-modifying proteins after each round of DNA replication. According to these models, histone PTMs alone do not provide sufficient specificity to mediate their own duplication; instead specificity is encoded by sequence-dependent interactions which recruit histone-modifying activities to specific parts of the genome. Experiments in yeast and flies which directly tested these two models fail to provide support for the first model [45]–[50], and demonstrate that histone PTMs alone are incapable of self-perpetuation. Rather, these results show that sequence-specific events are upstream of histone PTMs, and are critical for the duplication and reestablishment of chromatin states. The requirement for sequence-specificity can be considered a ‘check’, which may serve as a corrective mechanism to ensure that aberrant histone modifications on unintended parts of the genome are not propagated indefinitely. Without sequence-specific events to reinforce PTMs, these marks would disappear quickly after a few rounds of DNA replication. Thus only the chromatin state of the targeted parts of the genome, dictated by sequence-dependent interactions, are duplicated.
Regardless of the initiating event, histone PTMs act as binding substrates for the recruitment of other chromatin-regulatory complexes, whose activities regulate gene expression of the target region. Iterative cycles of trans recruitment of regulatory complexes create a positive feedback mechanism, establishing chromatin modifications locally, which can spread in cis to the neighboring chromosomal regions forming large tracks of transcriptionally co-regulated chromosomal domains. Similar initiation and spreading mechanisms appear to govern activating (e.g. Drosophila dosage compensation [51]) or repressing transcriptional events (e.g. mammalian dosage compensation [52]).
Histone methylation
Among the different histone tail PTMs, lysine methylation has emerged as a key histone mark involved in several important nuclear functions including transcription, heterochromatin formation, replication and DNA repair. Highly conserved lysine methyltransferases (KMTs) and demethylases (KDMs) catalyze the addition and removal of methyl marks from specific lysine residues, respectively [53]. Lysines can be mono-, di- or tri-methylated and the degree and specificity of methylation is exquisitely regulated by a combination of enzymatic specificity of KMTs/KDMs and their expression and recruitment mechanism to chromatin. Several chromatin-binding proteins can bind to these methylated lysine residues through their methyl lysine-binding motifs [54] and target the recruitment of regulatory proteins through their protein-protein interactions [31].
In this review we will focus on noncoding (nc)RNA-regulated lysine methylation events. These include histone H3 lysine 9 methylation (H3K9me), the hallmark of eukaryotic heterochromatin, histone H3 lysine 4 methylation (H3K4me), the ubiquitous mark of active transcription, and histone H3 lysine 27 methylation (H3K27me), the polycomb group protein modification which demarcates transcriptional repression of developmentally regulated genes [53]. Together these histone modifications impact critical biological processes as diverse as maintenance of genomic stability to regulation of gene expression during development.
ncRNAs and histone methylation
In recent years, the multitude of cellular functions attributed to ncRNAs has been growing rapidly. This is mainly due to the discovery of the RNA interference (RNAi) pathway, which revealed that (antisense) noncoding transcripts have critical gene regulatory functions. This discovery brought renewed research interest and urgency to the ncRNA field, which at the time was mostly focused on highly abundant and easily detectable ncRNAs (e.g rRNA, snoRNA, etc) with roles in RNA processing and protein translation. Breakthrough advances in next-generation sequencing technologies uncovered several new classes of ncRNAs and the surprisingly pervasive (albeit low-level) transcriptional landscape of the noncoding regions of the eukaryotic genomes. Recent studies have revealed that many of the newly identified classes of ncRNAs are regulators of several chromatin-dependent nuclear functions in eukaryotes ranging from yeast to man [55]–[57]. These functions include regulation of gene expression, transposon silencing, dosage compensation, genomic stability and heterochromatin formation. Work from several organisms has shown that ncRNAs mediate some of these functions by regulating histone methylation at various genomic loci. Mechanistically, ncRNAs directly regulate histone methylation by acting as (1) specificity factors for the trans recruitment of KMTs, (2) molecular scaffolds which act in cis upon which KMTs and other chromatin regulatory complexes assemble, or (3) structural components of KMT complexes. Indirectly, (4) inducible transcription of ncRNAs can regulate histone methylation of promoters of overlapping protein-coding genes. Even though in these instances ncRNA molecules themselves do not contribute to histone methylation directly, their transcription recruits KMTs, which methylate histones of overlapping promoters, regulating their activity in cis (Figure 2). We will discuss at least one example for each mechanism in detail in the sections below.
Figure 2.
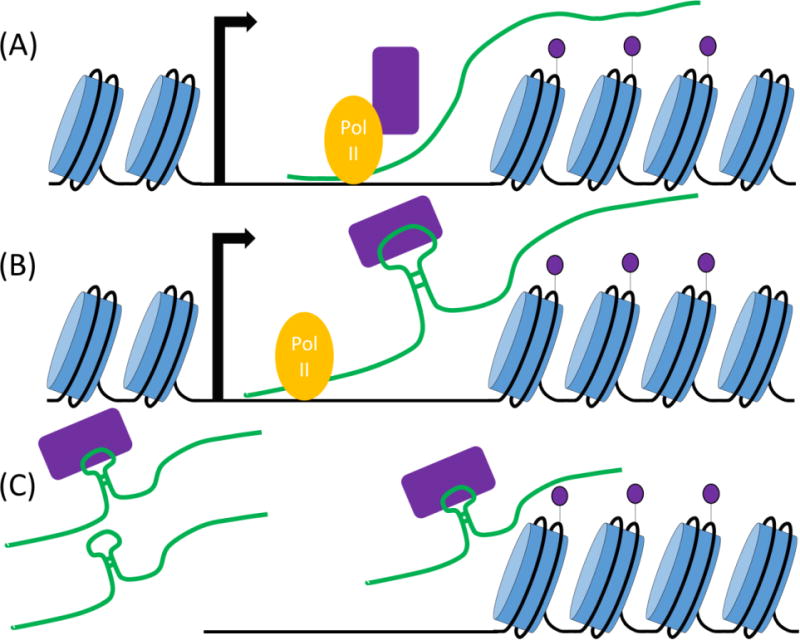
Models of lncRNA-mediated histone methylation. (A) RNA Pol II-mediated transcription of lncRNAs leads to the recruitment of KMTs, and alterations in histone methylation within the transcriptional unit of the lncRNA itself. In these instances, lncRNAs are not directly involved in KMT recruitment; instead their transcription results in recruitment of chromatin-regulatory complexes that alter local chromatin structure and regulate the transcription of neighboring genes in cis. (B) Chromatin-associated lncRNAs work as specificity factors and scaffolds for the cis recruitment of multiple histone-modifying complexes, including histone methyltransferases. RNA-protein (or lncRNA:smallRNA/Ago) interactions recruit KMT complexes and lead to coordinated modification of histone tails locally. In some instances, these modifications can spread to the neighboring nontranscribed regions. Some ncRNAs are known to interact with multiple proteins by distinct binding domains. (C) lncRNAs, bound to histone-modifying complexes, target different genomic loci in trans. In some instances, lncRNAs may act both as specificity factors, and as structural components of the KMT complexes.
Classes of ncRNAs
ncRNAs fall under two categories: small RNAs and long noncoding RNAs (lncRNAs). Small ncRNAs are derived from the degradation of larger endogenous or exogenous transcripts, which range in size from 21–30 nucleotides. lncRNAs are defined as endogenous transcripts greater than 200 base pairs (bp) long, with no known or predicted protein products. Both types of RNAs have been shown to regulate histone methylation in the nucleus of a variety of eukaryotic cells.
1. Small RNA-mediated histone methylation
Three different classes of small ncRNAs have been described: microRNAs (miRNAs), small interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs). miRNAs are Argonaute (Ago)-associated small RNAs, derived from processing of RNA hairpins, which once loaded onto the Ago family of proteins can mediate PTGS by degrading or inhibiting the translation of cytoplasmic RNAs. Even though implicated in epigenetic phenomena, no direct role for this class of small RNAs have been described for histone methylation in the nucleus, thus we refer the readers interested in this topic to other excellent reviews covering miRNA biology [58]–[60].
siRNAs are 20–25nt long degradation products of long exogenous or endogenous double-stranded (ds) RNAs. Long dsRNAs are substrates for the highly conserved RNase III family of proteins called Dicer (DCR), which degrade these transcripts into short dsRNA species. These are loaded onto effector Ago-containing complexes [60] whose slicer activity cleaves the passenger strand of the ds siRNAs, liberating the guide strand for basepairing interactions with complementary RNA molecules. siRNAs require perfect complementarity for target recognition, and can act as guides for the recruitment of a variety of chromatin-modifying complexes, including KMTs, to complementary sequences. In organisms such as S. pombe, N. crassa, C. elegans and plants, another class of proteins, RNA-dependent RNA polymerases (RdRP) are used to convert endogenous single-stranded (ss) RNAs into dsRNAs, thus availing these molecules as substrates for Dicer degradation. These proteins are used to target [61] and/or amplify the RNAi signal [62]–[64], generating positive feedback loops, a central feature of epigenetic regulation.
The last class of small ncRNAs discussed in this review is the PIWI-interacting small ncRNAs (piRNAs). piRNAs are 22–30bp long and are found associated with the PIWI clade of the Argonaute family of proteins [65]. Originally described in Drosophila, piRNAs and Piwi proteins protect the genome against germline activation of transposable elements by repressing transposon activity.
a. siRNAs in Schizosaccharoymces pombe
The discovery that small ncRNAs can direct the methylation of histone lysine residues in the fission yeast, Schizosaccharomyces pombe, was made in 2002 [62]. In this organism like most other eukaryotes, H3K9me demarcates heterochromatin, which is found at centromeres, telomeres, rDNA, and in the fission yeast mating-type region. Interestingly, RNAi proteins Dicer (Dcr1), Argonaute (Ago1) and RNA-dependent RNA polymerase (Rdp1) homologs are required for H3K9 methylation at centromeres by the sole KMT1 histone methyltransferase protein, Clr4 [62] – a member of the Suv39h family of proteins. This discovery raised the intriguing possibility that RNAi proteins and possibly siRNAs (which mostly were known for their cytoplasmic PTGS role at the time) target histone modifications to heterochromatin and directly or indirectly participate in transcriptional gene silencing (TGS) mechanisms in the nucleus. The first clue about how siRNAs function in TGS came with the discovery of the RNA-induced Transcriptional Gene silencing complex (RITS) [66]. RITS physically connects the RNAi pathway to heterochromatin. It is composed of Ago1, the sole Argonaute homolog which associates with heterochromatic siRNAs, Chp1, a chromodomain protein which specifically binds to H3K9me [67] and Tas3, a GW motif protein [68] which links Chp1 and Ago1 [69] and whose self-polymerization is required for spreading of RITS-dependent silencing to non-transcribed regions of centromeres [70]. Efficient RITS recruitment to heterochromatin requires siRNAs and H3K9me [66] suggesting that RITS is a bivalent complex whose binding to H3K9me (via Chp1) and siRNA basepairing interactions with centromeric (cen) sequences target RITS to heterochromatin. RITS physically interacts with the Rdp1-bearing complex, RDRC [63] which converts RNA Pol II-transcribed single-stranded cen RNAs into dsRNAs. RDRC, in turn, physically associates with Dcr1, thus targeting Dcr1 activity to heterochromatic dsRNAs [71] creating a positive feedback loop for siRNA production. Furthermore, RITS, RDRC and Dcr1 physically interact with cen RNAs in an H3K9me-dependent manner. Together these observations led to the proposal of the Nascent Transcript Model in which long centromeric ncRNAs act as specificity platforms for the recruitment of histone-modifying and -binding proteins in cis [63] (Figure 1A). Many key features of this model have emerged as conserved components of all small RNA or long RNA-mediated histone methylation studied in other systems including humans (see below).
Figure 1.
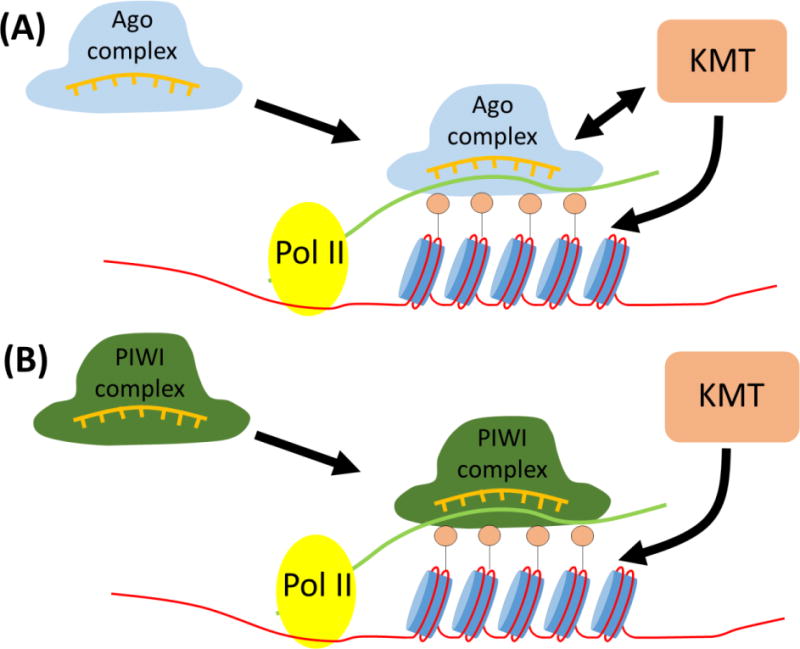
Small RNA-directed histone methylation. (A) siRNAs derived from processing of endogenous dsRNAs transcripts are loaded onto Argonaute proteins. Ago:siRNA complexes recognize complementary chromatin-associated transcripts by base-pairing interactions and binding to H3K9me via Chp1 component of RITS and target the recruitment of lysine methyltransferase (KMT) to the genome via direct protein-protein interactions. Iterative cycles of dsRNA generation in organisms which possess RNA-dependent RNA polymerases (RdRPs) leads to a positive feedback loop which amplifies the dsRNA/siRNA signal and reinforces H3K9me in a sequence-specific manner (B) Similar to above, piRNAs recognize transposable element (TE)-derived RNAs to promote methylation of TE loci in gonadal (and maybe somatic) tissues. Instead of RdRPs, ‘Ping-Pong’ cycles of cleavage activity by different Piwi clade members of the Argonaute family amplify the piRNA signal. Primary antisense piRNA clusters generate the complementary RNAs used in feed forward loop. piRNA:Piwi complexes target H3K9me to TEs in a sequence-specific manner, but whether this is through direct protein-protein interactions, by a linker protein, or other means remains undetermined. Small circles above nucleosomes represent methylated histone tails.
How do siRNAs guide CLRC recruitment?
Clr4 exists in the Clr4-Rik1-Cul4 (CLRC) complex [72]–[76] and is solely responsible for the methylation of H3K9 in S. pombe. Four observations support the siRNA-mediated mechanism for CLRC targeting: (1) siRNAs guide CLRC to complementary centromere-like sequences throughout the genome including the ones found at the fission yeast mating-type region [77] and telomeres [78]; (2) CLRC physically interacts with the RNAi complexes RITS, RDRC [49], [50], [79] and Dcr1 [80] through a complicated series of cooperative interactions which are assisted in part by the LIM domain protein, Stc1, siRNAs, centromeric RNAs and H3K9me; (3) RITS programmed with ectopic siRNAs generated from hairpins can establish H3K9me and silencing of a reporter gene with complementary sequences [81], [82]; and (4) artificial tethering of the Tas3 subunit of RITS to a euchromatic transcript can induce H3K9me-dependent silencing at the locus [83]. Together these data demonstrate the CLRC interaction with RNAi complexes targets its activity to regions of siRNA complementarity in the genome (Figure 1A).
A novel class of small ncRNAs and their role in H3K9me spreading
A recent report has identified a novel class of siRNA-sized ncRNAs, which are not Ago1-associated, but appear to be the Dcr1 degradation product of a lncRNA called BORDERLINE. This lncRNA is expressed at the heterochromatin-euchromatin border of the fission yeast pericentromeric regions, and its siRNA-sized degradation products appears to prevent the spreading of H3K9me marks outside of these borders [84]. Even though BORDERLINE is essential for boundary function, it operates in a sequence-independent manner such that expression of an ectopic RNA also can provide the barrier function at this locus. These small RNAs are thought to disrupt the cis spreading of heterochromatin by sequestering chromatin-binding or -modifying proteins away from chromatin. Even though the exact mechanism remains to be determined, these data potentially reveal a new class of ncRNAs which are products of Dcr1, but are not associated with Ago1 complexes [84]. It will be interesting to determine whether the two inverted repeats (IRs) which circumscribe the fission yeast mating-type region also mediate their boundary function by a similar mechanism. In this instance though, the IRs prevent the spreading of euchromatin into heterochromatic domains [85].
How do centromeric (cen) lncRNAs guide CLRC recruitment?
Current models propose that cen nascent lncRNAs are tethered to chromatin and act as scaffolds [63] or epigenetic sensors [44] for the assembly of chromatin- and RNA-regulatory complexes. In support of this, RNA Pol II-mediated transcription of cen lncRNAs is required for siRNA production, H3K9 methylation and full transcriptional repression at centromeres [86], [87], which operates by two parallel mechanisms [88], [89]. Together these data suggest that siRNAs carry the sequence-dependent transcriptional memory of their complementary lncRNA sequences, and which, in concert with H3K9me inherited from parental nucleosomes, stabilize RITS interaction with heterochromatic domains [90] promote CLRC recruitment, and rapidly reestablish chromatin states after DNA replication [91], [92]. Overall these observations suggest that cis-acting lncRNAs provide the scaffold and sequence specificity required for the recruitment and reestablishment of chromatin states after DNA replication (Figure 2B,D). This seems to be a general mechanism by which all cis-acting lncRNA elements in eukaryotes contribute to epigenetic propagation of chromatin states (See section on cis lncRNAs).
b. siRNAs in C. elegans
In C. elegans, a similar RNA-dependent mechanism is required for epigenetic inheritance of H3K9 methylation. In this organism, cytoplasmic and nuclear RNAi pathways cooperate to mediate epigenetic changes to chromatin modifications. Trigger dsRNAs introduced into animals via soaking, feeding or microinjection induce multigenerational gene silencing, which in some instances is concomitant with heritable changes to H3K9 methylation at the complementary gene [93], [94]. In this system, exogenous dsRNAs are sliced into primary siRNAs by the activities of DICER homologs DCR-1 [95] and RDE-4 [96] in the cytoplasm. Primary siRNAs are bound by the Argonaute homolog, RDE-1, guiding this protein to its target mRNA [97] by base-pairing interactions. RdRP homologs EGO-1 [98] and RRF-1 [99] are then recruited to the target transcript amplifying the dsRNA signal, and leading to the formation of secondary siRNAs after processing by DICER. Secondary siRNAs are incorporated into another group of Argonaute proteins [100] whose accumulation results in drastic increase of H3K9me marks [101]. Recent reports have shown that the nucleocytoplasmic Argonaute NRDE-3 directly binds cytoplasmic secondary siRNAs and transports them into the nucleus. This leads to the recruitment of other nuclear RNAi factors NRDE-1, NRDE-2 and NRDE-4 [93], [94], [102] which together are thought to associate with the nascent complementary sequences in the nucleus. NRDE interaction with the complementary nuclear transcripts is thought to lead to the H3K9me-dependent transcriptional repression of target sequences. These results suggest that siRNA-NRDE complexes can target nascent transcripts or DNA, and recruit histone-modifying enzymes directly. However, mechanistic details of this pathway remain largely unknown. It is important to note that siRNA generation precedes H3K9me, suggesting that siRNAs reinforce the fidelity of H3K9me state in each generation, and carry the memory of the RNAi-dependent silencing event from the previous generation.
Interestingly, a recent genetic screen identified another germline-specific nuclear Argonaute, HRDE-1, which associates with exogenous siRNAs and is required for multigeneration RNAi inheritance. In hrde-1 mutant animals, RNAi-dependent silencing operates in the parental worms, however their F1 progeny exhibit no memory of their exposure to trigger dsRNAs [103]. This is unlike wild-type animals in which silencing of the target transcript is maintained for up to five generations after the removal of the dsRNA-inducing signal. Kennedy and colleague show that HRDE-1 operates in the NRDE pathway, is loaded with exogenous siRNA, and is required for NRDE-2 interaction with nascent transcripts. Interestingly, deep sequencing of HRDE-1-assoicated siRNAs revealed that HRDE-1 also interacts with a set of endogenous siRNAs, complementary to transcripts whose expression is repressed by the H3K9me-dependent NRDE pathway. Loss of HRDE-1 similar to other NRDE proteins results in the up-regulation of these endogenous transcripts, loss of H3K9me and infertility. Collectively these data reveal that the nuclear HRDE-1 is a key mediator of a mutligenerational epigenetic inheritance mechanism whose memory is transmitted via siRNAs to the next generation. Whether the NRDE proteins along with HRDE-1 directly recruit KMTs to these genes remains to be determined, but the current model posits such a mechanism, similar to the co-transcriptional silencing mechanism described in the fission yeast [103].
c. piRNAs in Drosophila
As mentioned above, piRNAs are another class of Argonaute-associated small ncRNAs which may regulate histone methylation by guiding KMTs to their target sites around the genome via basepairing interactions. Uniquely, piRNAs are found mostly in germ cells, and their primary role seems to be protection of the genome against activation of transposable elements (TEs) [104]. piRNAs specifically associate with the PIWI clade of Argonaute proteins. PIWI proteins are critical for germ cell development and fertility in organisms ranging from flies to mammals, and their loss or reduction in activity results in genomic instability, loss of transposon silencing, infertility and DNA damage [105]. In addition to their role in germline development, recent work has revealed that piRNAs and PIWI proteins also contribute to important biological functions, including epigenetic programming and stem cell functions in somatic tissues of different organisms [106].
piRNAs were originally discovered in Drosophila, in which three germline-specific PIWI members, Aubergine (Aub), Piwi, and Argonaute3 (Ago3) associate with piRNAs. In this organism, piRNAs are 23–29nt long and have a strong preference for a 5′ terminal uridine. Unlike miRNAs and siRNAs, primary piRNA biogenesis is thought to occur independently of Dicer [107] and the subsequent generation of secondary piRNAs go through the so called ‘ping-pong’ mechanism in the cytoplasm [108], [109]. Numerous excellent reviews have described piRNA biogenesis pathways in detail [55], [104], [105] and we encourage the readers to refer to these papers for additional details. However we will briefly describe the ‘ping-pong’ model below.
According to the ‘ping-pong’ model transcription of piRNA clusters, whose transcripts are antisense to active transposons, is a key initiating step in piRNA biogenesis. Both piRNA cluster transcripts and the complementary TE RNAs are exported to the cytoplasm. piRNA cluster transcripts are then processed into primary antisense piRNAs by an unknown mechanism. These primary piRNAs associate with Piwi or Aub proteins, and target the sense strand of complementary TE RNAs. This results in the cleavage of the TE transcript, thus generating the 5′ end of a new ‘sense’ piRNA. Ago3 then binds to sense piRNAs and targets the cleavage of antisense piRNA cluster transcripts, producing more antisense piRNAs [108], [109]. Iterative cycles of antisense-sense binding and cleavage of respective transcripts creates a positive feed forward loop, which amplifies the piRNA signal and represses TE expression in germ cells. Whether the ping-pong model describes all instances of piRNA biogenesis remains untested. Several new reports suggest that piRNA formation can occur independently of the ping-pong pathway, however the details of this alternative mechanism for piRNA biogenesis remains largely unknown [106], [110]–[112].
Germline TGS
Even though piRNAs can repress the expression of TE RNAs via cytoplasmic PTGS mechanisms, recent studies suggest that piRNAs also contribute to silencing of TE genes by a chromatin-based TGS pathway. Several observations support such a model. (1) Piwi physically interacts with the Drosophila HP1a protein which itself associates with heterochromatin via binding to H3K9me [111], [113]. (2) Piwi depletion results in loss of silencing and H3K9me at transposons in ovarian somatic follicle cell lines [114]. (3) Piwi proteins programmed with piRNAs complementary to a reporter construct cause a decrease in Pol II occupancy, increase in H3K9me3 and HP1 localization, and induce transcriptional silencing of the reporter gene [115]. (4) Insertion of transposable elements at euchromatic sites leads to the formation of Piwi-mediated H3K9me3 islands in these regions [116]. Together these data suggest that Piwi-bound piRNAs interact with nascent transposon transcripts and directly or indirectly recruit TGS complexes, including KMTs (Figure 1B) leading to H3K9me at the TE genes. A similar mechanism of chromatin-based piRNA-dependent TGS seems to operate in C. elegans [117], [118].
Somatic nuclear RNAi in flies
In addition to its role in gametes, Drosophila nuclear RNAi may promote transposon repression and heterochromatin maintenance in somatic cells via an H3K9me-dependent pathway. Transposable element (TE)-derived siRNAs mapping to complementary regions of overlapping transcripts have been detected in flies [119]. The size of these TE-derived small RNAs shifts from 25nts in germ cells to 21nts in somatic cells during development, suggesting that endogenous TE-derived siRNAs may exist in the adult animal [120]. Also, absence of Dcr2, and Ago2 leads to marked reduction of centromeric H3K9me and TE silencing in this organism [121]. These data suggest that RNAi may play a role in maintaining H3K9me mark at constitutive heterochromatin in flies, but the mechanism of this process remains largely unknown.
Small ncRNA-mediated mechanisms of silencing are not limited to the organisms or the examples described above. We refer the readers to several recent reviews covering siRNA- and piRNA-mediated pathways in plants [56] and DNA elimination in tetrahymena [122], [123]. These are additional examples of how small RNA-dependent mechanisms target different parts of eukaryotic genomes by mechanisms similar to the ones described above.
2. lncRNA-dependent histone methylation
The majority of the eukaryotic genome is transcribed, and untranslated. Major advances in RNA-Seq technologies have led to discovery of a rapidly increasing number of lncRNAs in eukaryotes. lncRNAs are defined as RNA species longer than 200 nucleotides for which no protein products are predicted [57], [124] or have been detected [125], [126] based on current protein prediction and detection technologies, respectively. Even though lncRNAs are similar to protein-coding genes in length, histone modification profile of their open reading frames, and mechanism of transcription and splicing [127] they differ from protein-coding genes in several important aspects. (A) Majority of lncRNAs show little sequence conservation, but they are found in syntenic regions among species. Interestingly, these transcripts regulate the expression of similar, often neighboring, sets of genes [128], [129]. (B) Their inter- [130] or intra-species [124] mutational patterns differ from canonical protein coding genes, supporting their noncoding nature. (C) Most lncRNAs are nuclear, chromatin-associated, and interact with chromatin proteins [131]. Together these observations suggest that one of the main functions of the noncoding lncRNAs is to provide an additional layer of transcriptional regulation to the protein-coding regions of the genome. A spate of new papers has revealed that lncRNAs mediate this effect by different mechanisms such as targeting the recruitment of chromatin modifiers, DNA methylation machinery or transcriptional regulators by direct or indirect means [55], [57].
Below, we will focus our discussion on lncRNAs whose activities alter histone methylation of their target genes both directly and indirectly (Figure 2). Three broad categories are discussed. First, we will discuss instances in which inducible transcription of ncRNAs regulates histone methylation of promoters of overlapping protein-coding genes. (Figure 2A). Second we will discuss how lncRNAs can act as specificity factors for recruitment of KMTs in cis or trans to various regions around the genome (Figure 2B). Lastly, we will consider ribonucleoprotein complexes in which lncRNAs act as scaffolds to for association of different chromatin-modifying complexes, coordinating their recruitment and activities of their target loci. Considering the growing number of lncRNAs, we discuss a few examples per mechanism to illustrate the details of each pathway.
a. Pol II-mediated transcription of lncRNAs alters methylation state of neighboring genes
Proper positioning and distribution of nucleosomes over eukaryotic genomes is important for regulation of gene expression. This is especially important in gene promoters where nucleosome positioning and their histone modifications have a large impact on promoter activity. Upstream of all transcription start sites is the AT-rich nucleosome-devoid region (NDR), to which transcription factors often bind. Some lncRNAs overlap promoters and their transcription disrupts NDRs and histone modifications of nucleosomes in promoters [132]–[134].
During transcription in the budding yeast, histone H3 lysine 4 (H3K4) and lysine 36 (H3K36) are methylated by Set1 and Set2 KMTs, respectively. These marks are critical for maintaining the coding region in a hypoacetylated and repressive state [135] which prevents spurious transcription by cryptic promoters within these sequences. An example of a lncRNA whose transcription regulates the expression of a neighboring gene is IRT1. IRT1 resides within the promoter region of the master regulator of meiosis, Ime1. Ime1 is only active during meiotic events and its expression is inhibited in cis by IRT1. Ime1 silencing requires not only transcription of IRT1, but also the SET2 methyltransferase (H3K36me) and SET3 histone deacetylase complexes [134]. During IRT1 transcription, SET1 and SET2 methylate H3K4 and H3K36 cotranscriptionally [136], [137], which leads to the recruitment of the repressive histone deacetylase complex RPD3C [137]. Together these complexes disrupt nucleosome positioning and create a repressive chromatin state within the Ime1 promoter which prevents its expression. There are several other examples of this type of cis regulation of protein-coding genes by the expression of neighboring lncRNAs, but overall, transcription of lncRNAs residing within promoters of protein-coding genes leads to deposition of methylation marks and recruitment of repressive complexes, which prevent proper promoter activity.
b. lncRNAs recruit histone-modifying enzymes in cis
As described above transcription of lncRNAs can impact the methylation and therefore transcriptional state of their neighboring genes. In addition to this, there are a growing number of examples where lncRNAs directly associate with histone-modifying enzymes, and similar to lncRNAs in the fission yeast, recruit KMTs to chromatin. This way, lncRNAs can serve as the key specificity factors which regulate histone PMTs in a locus-specific manner [57], [138].
lncRNAs regulate vernalization in plants
Vernalization is an adaptive switch that regulates the timing of flowering in plants. Consistent exposure to cold temperatures in winter triggers a chromatin-based epigenetic silencing mechanism which targets the repressors of flowering genes in plants [139]. This mechanism ensures proper timing and efficient flowering during spring. In Arabidopsis, two lncRNAs found at a potent repressor of flowering called Flowering Locus C (FLC) mediate its repression. One of the two lncRNAs is an antisense transcript found at the 3′ end of FLC locus, called COOLAIR, which is strongly induced upon exposure to cold and silences the sense FLC transcription by promoter interference similar to what was described in 2a (see above) [140]. The other lncRNA, COLD ASSISTED INTRONIC NONCODING RNA (COLDAIR), is transcribed in the sense orientation to FLC transcript and represses FLC by directly interacting with the Polycomb Repressive Complex 2 (PRC2). COLDAIR binds PRC2 via its Enhancer of Zest (E(z)) homolog CURLY LEAF (CLF), and knockdown of COLDAIR greatly reduces the repressive H3K27me3 marks in the neighboring region [141]. This suggests that COLDAIR represses FLC expression in cis via recruiting PRC2 to the FLC locus specifically (Figure 2B).
X chromosome inactivation in mammalian cells
Many mammalian lncRNAs also associate directly with histone-modifying complexes. An example of such a mechanism is dosage compensation or X-chromosome inactivation (XCI) in mammals. XCI maintains comparable levels of X chromosome gene expression between males (XY) and females (XX). The details for this mechanism have been studied extensively and we refer the readers to other recent reviews for details [142], [143]. Here we will use mammalian X-chromosome inactivation as an example of how cis-acting lncRNAs interact with KMTs and affect the transcriptional fate of, in this case, an entire chromosome.
One copy of X chromosomes in females is inactivated during development. The inactive X (Xi) is ‘decorated’ with a lncRNA called X-inactive-specific transcript (Xist), which is only expressed on Xi and mediates its silencing in cis [144]–[146]. Xist resides in the cis-acting silencing region called X-Inactivation Center (Xic), which is necessary and sufficient to mediate chromosome-wide transcriptional repression [143]. Several other lncRNAs are expressed from Xic in addition to Xist [147] but we will limit our discussion to those which physically interact with KMTs . Xist, which coats Xi [148] interacts with the PRC2 complex and targets its recruitment and H3K27me to Xi [149]. RepA, a 1.6kb lncRNA within Xist, binds to EZH2, the KMT member of the PRC2 complex and promotes EZH2-mediated H3K27me3 of the Xi [149]. PRC2-Xist interaction and binding to Xi is regulated by several factors, one of which is a bivalent transcription factor called Yin Yang 1 (YY1). YY1 binds to both DNA (three YY1 binding sites at Xic) and RNA (Xist) sequences through its sequence-specific binding domains [150]. These interactions are thought to tether the PRC2-Xist to Xic cotranscriptionally [150]. Recent studies have demonstrated that JARID2, a cofactor of PRC2, binds to lncRNAs including Xist and is required for efficient PRC2 recruitment to Xi [151], [152]. Together these data suggest that YY1 anchors Xist-PRC2 to Xic and promotes its interaction with JARID2, which leads to H3K27me3 at Xi. However, how silencing spread to 100 megabases scale remains a topic of intense research. Overall the model presented for cis-acting lncRNAs (Figure 2B) shares many conserved mechanisms to the one proposed in the fission yeast (Figure 1A), in which lncRNAs tethered to chromatin coordinate histone methylation by recruiting KMTs to their site of synthesis.
c. lncRNA-mediated methylation in trans
Hox genes regulate the development of body patterning in metazoans and their expression is tightly regulated during development and in adults. Hox genes are found in clusters on different chromosomes in humans and contain many lncRNAs that display differential expression during development. Overexpression of Hox genes correlates with cancer [153]. A well-studied trans-acting lncRNA is the 2.2kb long Hox Antisense Intergenic RNA, called HOTAIR. HOTAIR is expressed from within the HoxC gene cluster and represses transcription of the HoxD genes in trans by regulating PRC2 recruitment. The 5′ end of HOTAIR binds to EZH2 component of the PRC2 complex, and mediates its recruitment to the 40kb region of HoxD cluster on another chromosome. Disruptions to HOTAIR expression correlate with breast and colorectal cancers [154]–[156]. These data suggest that lncRNAs can recruit KMTs in trans to other genomic sites.
H19 lncRNA has been shown to regulate H3K9 methylation of nine genes in the Imprinted Gene Network (IGN), which are important for the control of embryonic growth [157]. RNA immunoprecipitation has shown that H19 lncRNA interacts with MBD1, a protein involved in the recruitment of H3K9 KMTs SETDB1 and SUV39H1 [158], [159]. MBD1 is necessary for the repression of five genes in the IGN repressed by H19 lncRNA and has been shown to directly interact with differentially methylated regions (DMR) of three IGN genes. These data suggest that H19-MBD1 interaction recruits KMTs to maintain the repressive chromatin mark on these genes.
Recent work also has implicated lncRNAs in transcriptional activation of several genes, however the mechanistic details of their involvements remains to be determined [129], [160], [161].
d. Scaffold lncRNAs recruit multiple histone-modifying complexes
Activation and repression of target genes often requires coordinated modification of histone PTMs such as methylation/deacetylation and acetylation/demethylation. These are primarily mediated by interactions between chromatin complexes. Interestingly, recent studies have shown that lncRNAs sometimes have multiple binding sites for distinct histone-modifying complexes, and similar to a scaffold bring together two or more protein complexes [162]. As previously discussed, 5′ end of the HOTAIR transcript binds to the EZH2 component of the PRC2 complex [156]. Recently it was shown that the 3′ end of HOTAIR transcripts binds to the LSD1 component of the REST/CoREST complex, which has H3K4 demethylation activity [163]. Knockdown of HOTAIR leads to reduction in both H3K27me and increased H3K4me at target regions. These data suggest that HOTAIR through its interaction with both PRC2 and REST/CoREST complexes simultaneously coordinates H3K27 methylation and H3K4 demethylation at target loci around the genome.
Antisense noncoding RNA in the INK4 locus (ANRIL) is another example of a cis-acting scaffold lncRNA, whose expression is initiated from the INK4A-ARF-INK4B locus. This locus encodes three tumor suppressor proteins, p16(INK4A), p14(ARF) and p15(INK4B) whose expression are repressed by polycomb during normal cell growth and activated in cancers by oncogenes such as Ras [164]. In leukemia and prostate cancer cells, ANRIL overexpression has been shown to silence p15(INK4B) tumor suppressor gene [165]. In addition to its interaction with the SUZ12 component of PRC2 complex, ANRIL binds to CBX7, a component of the PRC1 complex. PRC1 contains an ubiquitin ligase RING1B protein which ubiquitinates lysine 119 of histone H2A (H2AK119ub). This suggests that ANRIL coordinates H3K27 methylation, H3K27me binding and H2AK119 ubiquitination by bringing together PRC1 and PRC2 complexes [164], [166]. Overall, these examples demonstrate that lncRNAs can be coopted to serve as multivalent scaffolds for the assembly and coordination of multiple chromatin-modifying complexes activities. These illustrate the versatility of RNA, which through its sequence-specific binding interactions can assemble, coordinate and potentially target histone-modifying activities to a variety of specific genomic regions in eukaryotes.
In Summary
The general picture that emerges from our current mechanistic understanding of how ncRNAs contribute to chromatin-based epigenetic pathways demonstrate that they can act as specificity factors and/or molecular scaffolds for the targeted recruitment and assembly of chromatin-regulatory complexes in eukaryotic genomes. lncRNAs can function as multivalent bridges, coordinating (and potentially targeting) several chromatin activities together in one larger ribonucleoprotein complex. These observations highlight the polytropic nature of RNA as a biological macromolecule, and illustrate its usefulness in propagating chromatin-based epigenetic pathways.
ncRNAs provide a wide-range of nuclear functions from maintaining genomic stability to coordinating DNA replication. Recently, work in Neurospora, plants and animals has revealed that a new class of small Argonaute-associated ncRNAs arise in response to double-strand breaks (DSBs) [167]–[169] which contributes to DSB repair directly or indirectly. It may be that damage-induced siRNAs directly assist in recruiting repair and chromatin complexes to sites of DSB, or indirectly activate or repress the expression of other repair factors. Regardless of the details, these findings illustrate the surprising capacities by which ncRNAs can mediate nuclear functions and make it difficult to predict what other roles they may perform in the cell.
The regulatory and functional roles of lncRNAs are only now beginning to be understood. Their diversity, low expression level and sequence divergence pose a challenge to studying their functions. Advances in computational technologies, combined with improvements in our predictive abilities for finding their protein or nucleic acid binding partners would greatly enhance the speed by which we unravel their functions. Recent studies have shown that RNAs can be posttrascriptionally methylated like histones [170], [171], which may add an extra layer of complexity in RNA-mediated gene regulation. How these RNA modifications affect their regulatory functions is largely unknown, and may hold a few additional surprises in the coming years. Also, lncRNAs have been associated with many human pathologies and have emerged as potential diagnostic targets for a variety of diseases including cancers [154], [172]–[174]. These may yield some novel insights about how ncRNAs contribute to pathogenesis. How the story of ncRNAs will unfold in the future is as unpredictable as their diverse functions in the cell.
Highlights.
Noncoding RNAs (ncRNAs) regulate histone methylation in a variety organisms
Both small and large noncoding RNAs are implicated in histone methylation
ncRNAs act as platforms or specificity factors for histone methyltransferase recruitment
Acknowledgments
We thank the members of the lab for discussion and the critical reading of the manuscript. Research in the lab is supported by NCI Proton Beam Grant and a V Scholar Award to MM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006 Sep;70(3):830–56. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol. 2007 Jan;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007 Feb;128(4):747–62. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Murray AW, Szostak JW. Chromosome segregation in mitosis and meiosis. Annu Rev Cell Biol. 1985 Jan;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- 5.Marians KJ. Prokaryotic DNA replication. Annu Rev Biochem. 1992 Jan;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 6.Draper GC, Gober JW. Bacterial chromosome segregation. Annu Rev Microbiol. 2002 Jan;56:567–97. doi: 10.1146/annurev.micro.56.012302.160729. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, Geldmacher DS, Herrup K. DNA replication precedes neuronal cell death in Alzheimer’s disease. J Neurosci. 2001 Apr;21(8):2661–2668. doi: 10.1523/JNEUROSCI.21-08-02661.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassold T, Sherman S. Down syndrome: genetic recombination and the origin of the extra chromosome 21. Clin Genet. 2001 Dec;57(2):95–100. doi: 10.1034/j.1399-0004.2000.570201.x. [DOI] [PubMed] [Google Scholar]
- 9.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nar Rev Cancer. 2001 Nov;1(2):109–17. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 10.Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004 Mar;303(5664):1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 11.Wu X, Gu J, Spitz MR. Mutagen sensitivity: a genetic predisposition factor for cancer. Cancer Res. 2007 Apr;67(8):3493–5. doi: 10.1158/0008-5472.CAN-06-4137. [DOI] [PubMed] [Google Scholar]
- 12.Griffin CS, Simpson PJ, Wilson CR, Thacker J. Mammalian recombination-repair genes XRCC2 and XRCC3 promote correct chromosome segregation. Nat Cell Biol. 2000 Oct;2(10):757–61. doi: 10.1038/35036399. [DOI] [PubMed] [Google Scholar]
- 13.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004 Jan;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 14.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007 Nov;318(5851):761–4. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 15.Grewal SI, Klar AJ. Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell. 1996 Jul;86(1):95–101. doi: 10.1016/s0092-8674(00)80080-x. [DOI] [PubMed] [Google Scholar]
- 16.Cavalli G, Paro R. The drosophila Fab-7 chromosomal element conveys epigenetic inheritance during mitosis and meiosis. Cell. 1998 May;93(4):505–518. doi: 10.1016/s0092-8674(00)81181-2. [DOI] [PubMed] [Google Scholar]
- 17.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999 Nov;23(3):314–8. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 18.Kaeppler SM, Kaeppler HF, Rhee Y. Epigenetic aspects of somaclonal variation in plants. Plant Mol Biol. 2000 Jun;43(3):179–188. doi: 10.1023/a:1006423110134. [DOI] [PubMed] [Google Scholar]
- 19.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 20.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012 Jul;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Rajender S, Avery K, Agarwal A. Epigenetics, spermatogenesis and male infertility. Mutat Res. 2011;727(3):62–71. doi: 10.1016/j.mrrev.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 22.Kwok JBJ. Role of epigenetics in Alzheimer’s and Parkinson’s disease. Epigenomics. 2010 Oct;2(5):671–82. doi: 10.2217/epi.10.43. [DOI] [PubMed] [Google Scholar]
- 23.Kouzarides T. Chromatin modifications and their function. Cell. 2007 Feb;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008 Nov;10(11):1291–300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- 25.Kaufman PD, Rando OJ. Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol. 2010 Jun;22(3):284–90. doi: 10.1016/j.ceb.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sugii S, Evans RM. Epigenetic codes of PPARγ in metabolic disease. FEBS Lett. 2011 Jul;585(13):2121–8. doi: 10.1016/j.febslet.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saijo E, Kang HE, Bian J, Bowling KG, Browning S, Kim S, Hunter N, Telling GC. Epigenetic dominance of prion conformers. PLoS Pathog. 2013 Oct;9(10):e1003692. doi: 10.1371/journal.ppat.1003692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen T. Multigenerational cortical inheritance of the Rax2 protein in orienting polarity and division in yeast. Science. 2000 Dec;290(5498):1975–1978. doi: 10.1126/science.290.5498.1975. [DOI] [PubMed] [Google Scholar]
- 29.Beisson J, Sonneborn TM. Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. Proc Natl Acad Sci USA. 1965 Feb;53:275–82. doi: 10.1073/pnas.53.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007 Feb;128(4):721–33. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 31.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011 Mar;21(3):381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yap KL, Zhou MM. Keeping it in the family: diverse histone recognition by conserved structural folds. Crit Rev Biochem Mol Biol. 2010 Dec;45(6):488–505. doi: 10.3109/10409238.2010.512001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel DJ, Wang Z. Readout of epigenetic modifications. Annu Rev Biochem. 2013 Jan;82:81–118. doi: 10.1146/annurev-biochem-072711-165700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009 Jan;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 35.Zacharioudakis I, Gligoris T, Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr Biol. 2007 Dec;17(23):2041–6. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 36.Jackson V, Granner DK, Chalkley R. Deposition of histones onto replicating chromosomes. Proc Natl Acad Sci USA. 1975 Nov;72(11):4440–4. doi: 10.1073/pnas.72.11.4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jackson V, Chalkley R. Histone segregation on replicating chromatin. Biochemistry. 1985 Nov;24(24):6930–8. doi: 10.1021/bi00345a027. [DOI] [PubMed] [Google Scholar]
- 38.Sogo JM, Stahl H, Koller T, Knippers R. Structure of replicating simian virus 40 minichromosomes. The replication fork, core histone segregation and terminal structures. J Mol Biol. 1986 May;189(1):189–204. doi: 10.1016/0022-2836(86)90390-6. [DOI] [PubMed] [Google Scholar]
- 39.Jackson V. Deposition of newly synthesized histones: new histones H2A and H2B do not deposit in the same nucleosome with new histones H3 and H4. Biochemistry. 1987 Apr;26(8):2315–25. doi: 10.1021/bi00382a037. [DOI] [PubMed] [Google Scholar]
- 40.Jackson V. Deposition of newly synthesized histones: hybrid nucleosomes are not tandemly arranged on daughter DNA strands. Biochemistry. 1988 Mar;27(6):2109–20. doi: 10.1021/bi00406a044. [DOI] [PubMed] [Google Scholar]
- 41.Russev G, Hancock R. Assembly of new histones into nucleosomes and their distribution in replicating chromatin. Proc Natl Acad Sci USA. 1982 May;79(10):3143–3147. doi: 10.1073/pnas.79.10.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Radman-Livaja M, Verzijlbergen KF, Weiner A, van Welsem T, Friedman N, Rando OJ, van Leeuwen F. Patterns and mechanisms of ancestral histone protein inheritance in budding yeast. PLoS Biol. 2011 Jun;9(6):e1001075. doi: 10.1371/journal.pbio.1001075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ptashne M. Binding reactions: epigenetic switches, signal transduction and cancer. Curr Biol. 2009 Mar;19(6):R234–41. doi: 10.1016/j.cub.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 44.Moazed D. Mechanisms for the inheritance of chromatin states. Cell. 2011 Aug;146(4):510–8. doi: 10.1016/j.cell.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holmes SG, Broach JR. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 1996 Apr;10(8):1021–32. doi: 10.1101/gad.10.8.1021. [DOI] [PubMed] [Google Scholar]
- 46.Busturia A, Wightman CD, Sakonju S. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development. 1997 Nov;124(21):4343–50. doi: 10.1242/dev.124.21.4343. [DOI] [PubMed] [Google Scholar]
- 47.Cheng TH, Gartenberg MR. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 2000 Feb;14(4):452–63. [PMC free article] [PubMed] [Google Scholar]
- 48.Sengupta AK, Kuhrs A, Müller J. General transcriptional silencing by a Polycomb response element in Drosophila. Development. 2004 May;131(9):1959–65. doi: 10.1242/dev.01084. [DOI] [PubMed] [Google Scholar]
- 49.Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, Allshire RC. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010 Mar;140(5):666–77. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerace EL, Halic M, Moazed D. The methyltransferase activity of Clr4Suv39h triggers RNAi independently of histone H3K9 methylation. Mol Cell. 2010 Aug;39(3):360–72. doi: 10.1016/j.molcel.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet. 2011 Feb;13(2):123–34. doi: 10.1038/nrg3124. [DOI] [PubMed] [Google Scholar]
- 52.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008 Jan;42:733–72. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 53.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012 Nov;48(4):491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkinson AW, Gozani O. Histone-binding domains: Strategies for discovery and characterization. BBA Gene Reg Mech. 2014 Feb; doi: 10.1016/j.bbagrm.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabin LR, Delás MJ, Hannon GJ. Dogma derailed: the many influences of RNA on the genome. Mol Cell. 2013 Mar;49(5):783–94. doi: 10.1016/j.molcel.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013 Feb;14(2):100–12. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012 Jan;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinforma. 2009 Dec;7(4):147–54. doi: 10.1016/S1672-0229(08)60044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004 Jul;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 60.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009 Feb;136(4):642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wirén M, Silverstein RA, Sinha I, Walfridsson J, Lee HM, Laurenson P, Pillus L, Robyr D, Grunstein M, Ekwall K. Genomewide analysis of nucleosome density histone acetylation and HDAC function in fission yeast. EMBO J. 2005 Aug;24(16):2906–18. doi: 10.1038/sj.emboj.7600758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002 Sep;297(5588):1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- 63.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004 Dec;119(6):789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 64.Sugiyama T, Cam H, Verdel A, Moazed D, Grewal SIS. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc Natl Acad Sci USA. 2005 Jan;102(1):152–7. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008 Jan;135(1):3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- 66.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004 Jan;303(5658):672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Partridge JF, Scott KSC, Bannister AJ, Kouzarides T, Allshire RC. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002 Oct;12(19):1652–60. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- 68.Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007 Oct;14(10):897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 69.Debeauchamp JL, Moses A, Noffsinger VJP, Ulrich DL, Job G, Kosinski AM, Partridge JF. Chp1-Tas3 interaction is required to recruit RITS to fission yeast centromeres and for maintenance of centromeric heterochromatin. Mol Cell Biol. 2008 Apr;28(7):2154–66. doi: 10.1128/MCB.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, Motamedi MR, Yip CK, Wang Z, Walz T, Patel DJ, Moazed D. An alpha motif at Tas3 C terminus mediates RITS cis spreading and promotes heterochromatic gene silencing. Mol Cell. 2009 Apr;34(2):155–67. doi: 10.1016/j.molcel.2009.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colmenares SU, Buker SM, Buhler M, Dlakid M, Moazed D. Coupling of double-stranded RNA synthesis and siRNA generation in fission yeast RNAi. Mol Cell. 2007 Aug;27(3):449–61. doi: 10.1016/j.molcel.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 72.Hong EJE, Villén J, Gerace EL, Gygi SP, Moazed D. A cullin E3 ubiquitin ligase complex associates with Rik1 and the Clr4 histone H3-K9 methyltransferase and is required for RNAi-mediated heterochromatin formation. RNA Biol. 2005;2(3):106–11. doi: 10.4161/rna.2.3.2131. [DOI] [PubMed] [Google Scholar]
- 73.Horn PJ, Bastie JN, Peterson CL. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev. 2005 Jul;19(14):1705–14. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia S, Kobayashi R, Grewal SIS. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol. 2005 Oct;7(10):1007–13. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 75.Li F, Goto DB, Zaratiegui M, Tang X, Martienssen R, Cande WZ. Two novel proteins, dos1 and dos2, interact with rik1 to regulate heterochromatic RNA interference and histone modification. Curr Biol. 2005 Aug;15(16):1448–57. doi: 10.1016/j.cub.2005.07.021. [DOI] [PubMed] [Google Scholar]
- 76.Thon G, Hansen KR, Altes SP, Sidhu D, Singh G, Verhein-Hansen J, Bonaduce MJ, Klar AJS. The Clr7 and Clr8 directionality factors and the Pcu4 cullin mediate heterochromatin formation in the fission yeast Schizosaccharomyces pombe. Genetics. 2005 Dec;171(4):1583–95. doi: 10.1534/genetics.105.048298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jia S, Noma K, Grewal SIS. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004 Jun;304(5679):1971–6. doi: 10.1126/science.1099035. [DOI] [PubMed] [Google Scholar]
- 78.Kanoh J, Sadaie M, Urano T, Ishikawa F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr Biol. 2005 Oct;15(20):1808–19. doi: 10.1016/j.cub.2005.09.041. [DOI] [PubMed] [Google Scholar]
- 79.Zhang K, Mosch K, Fischle W, Grewal SIS. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008 Apr;15(4):381–8. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 80.Yu R, Jih G, Iglesias N, Moazed D. Determinants of heterochromatic siRNA biogenesis and function. Mol Cell. 2014 Dec;53(2):262–76. doi: 10.1016/j.molcel.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iida T, Nakayama J, Moazed D. siRNA-mediated heterochromatin establishment requires HP1 and is associated with antisense transcription. Mol Cell. 2008 Jul;31(2):178–89. doi: 10.1016/j.molcel.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Simmer F, Buscaino A, Kos-Braun IC, Kagansky A, Boukaba A, Urano T, Kerr ARW, Allshire RC. Hairpin RNA induces secondary small interfering RNA synthesis and silencing in trans in fission yeast. EMBO Rep. 2010 Feb;11(2):112–8. doi: 10.1038/embor.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bühler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006 Jun;125(5):873–86. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 84.Keller C, Kulasegaran-Shylini R, Shimada Y, Hotz HR, Bühler M. Noncoding RNAs prevent spreading of a repressive histone mark. Nat Struct Mol Biol. 2013 Aug;20(8):994–1000. doi: 10.1038/nsmb.2619. [DOI] [PubMed] [Google Scholar]
- 85.Thon G, Bjerling P, Bünner CM, Verhein-Hansen J. Expression-state boundaries in the mating-type region of fission yeast. Genetics. 2002 Jun;161(2):611–22. doi: 10.1093/genetics/161.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005 Jul;309(5733):467–9. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- 87.Djupedal I, Portoso M, Spåhr H, Bonilla C, Gustafsson CM, Allshire RC, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005 Oct;19(19):2301–6. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Motamedi MR, Hong EJE, Li X, Gerber S, Denison C, Gygi S, Moazed D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008 Dec;32(6):778–90. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Buscaino A, Lejeune E, Audergon P, Hamilton G, Pidoux A, Allshire RC. Distinct roles for Sir2 and RNAi in centromeric heterochromatin nucleation, spreading and maintenance. EMBO J. 2013 May;32(9):1250–64. doi: 10.1038/emboj.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Noma K, Sugiyama T, Cam H, Verdel A, Zofall M, Jia S, Moazed D, Grewal SIS. RITS acts in cis to promote RNA interference-mediated transcriptional and post-transcriptional silencing. Nat Genet. 2004 Nov;36(11):1174–80. doi: 10.1038/ng1452. [DOI] [PubMed] [Google Scholar]
- 91.Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SIS. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008 Feb;451(7179):734–7. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- 92.Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008 Apr;18(7):490–5. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burkhart KB, Guang S, Buckley BA, Wong L, Bochner AF, Kennedy S. A pre-mRNA-associating factor links endogenous siRNAs to chromatin regulation. PLoS Genet. 2011 Aug;7(8):e1002249. doi: 10.1371/journal.pgen.1002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010 Jun;465(7301):1097–101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001 Jan;409(6818):363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 96.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-Box helicase to direct RNAi in C. elegans. Cell. 2002 Jun;109(7):861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 97.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999 Oct;99(2):123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 98.Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000 Feb;10(4):169–78. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 99.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001 Nov;107(4):465–76. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 100.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CCG, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006 Nov;127(4):747–57. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 101.Gu SG, Pak J, Guang S, Maniar JM, Kennedy S, Fire A. Amplification of siRNA in Caenorhabditis elegans generates a transgenerational sequence-targeted histone H3 lysine 9 methylation footprint. Nat Genet. 2012 Feb;44(2):157–64. doi: 10.1038/ng.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Burton NO, Burkhart KB, Kennedy S. Nuclear RNAi maintains heritable gene silencing in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011 Dec;108(49):19683–8. doi: 10.1073/pnas.1113310108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012 Sep;489(7416):447–51. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011 Apr;12(4):246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 105.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009 Feb;136(4):656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014 Jan;505(7483):353–9. doi: 10.1038/nature12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006 Jul;313(5785):320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- 108.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell. 2007;128(6):1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 109.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science. 2007 Mar;315(5818):1587–90. doi: 10.1126/science.1140494. [DOI] [PubMed] [Google Scholar]
- 110.Yin H, Lin H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nature. 2007 Nov;450(7167):304–8. doi: 10.1038/nature06263. [DOI] [PubMed] [Google Scholar]
- 111.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SCR, Lin H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007 Sep;21(18):2300–11. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beyret E, Liu N, Lin H. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 2012 Oct;22(10):1429–39. doi: 10.1038/cr.2012.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mendez DL, Kim D, Chruszcz M, Stephens GE, Minor W, Khorasanizadeh S, Elgin SCR. The HP1a disordered C terminus and chromo shadow domain cooperate to select target peptide partners. Chembiochem. 2011 May;12(7):1084–96. doi: 10.1002/cbic.201000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Moshkovich N, Lei EP. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010 Mar;6(3):e1000880. doi: 10.1371/journal.pgen.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013 Feb;27(4):390–9. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sienski G, Dönertas D, Brennecke J. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151(5):964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shirayama M, Seth M, Lee HC, Gu W, Ishidate T, Conte D, Mello CC. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell. 2012 Jul;150(1):65–77. doi: 10.1016/j.cell.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ashe A, Sapetschnig A, Weick EM, Mitchell J, Bagijn MP, Cording AC, Doebley AL, Goldstein LD, Lehrbach NJ, Le Pen J, Pintacuda G, Sakaguchi A, Sarkies P, Ahmed S, Miska EA. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012 Jul;150(1):88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008 May;320(5879):1077–81. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fagegaltier D, Bougé AL, Berry B, Poisot E, Sismeiro O, Coppée JY, Théodore L, Voinnet O, Antoniewski C. The endogenous siRNA pathway is involved in heterochromatin formation in Drosophila. Proc Natl Acad Sci USA. 2009 Dec;106(50):21258–63. doi: 10.1073/pnas.0809208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007 Jan;9(1):25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coyne RS, Lhuillier-Akakpo M, Duharcourt S. RNA-guided DNA rearrangements in ciliates: is the best genome defence a good offence? Biol Cell. 2012 Jun;104(6):309–25. doi: 10.1111/boc.201100057. [DOI] [PubMed] [Google Scholar]
- 123.Mochizuki K. DNA rearrangements directed by non-coding RNAs in ciliates. Wiley Interdiscip Rev RNA. 1(3):376–87. doi: 10.1002/wrna.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013 Jul;154(1):240–51. doi: 10.1016/j.cell.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Kundaje A, Gunawardena HP, Yu Y, Xie L, Krajewski K, Strahl BD, Chen X, Bickel P, Giddings MC, Brown JB, Lipovich L. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012 Sep;22(9):1646–57. doi: 10.1101/gr.134767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Slavoff SA, Mitchell AJ, Schwaid AG, Cabili MN, Ma J, Levin JZ, Karger AD, Budnik BA, Rinn JL, Saghatelian A. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat Chem Biol. 2013 Jan;9(1):59–64. doi: 10.1038/nchembio.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigó R. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012 Sep;22(9):1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011 Dec;147(7):1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Carpenter S, Aiello D, Atianand MK, Ricci EP, Gandhi P, Hall LL, Byron M, Monks B, Henry-Bezy M, Lawrence JB, O’Neill LAJ, Moore MJ, Caffrey DR, Fitzgerald KA. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013 Aug;341(6147):789–92. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009 Mar;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011 Sep;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008 Dec;32(5):685–95. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009 Jun;28(12):1697–707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Werven FJ, Neuert G, Hendrick N, Lardenois A, Buratowski S, van Oudenaarden A, Primig M, Amon A. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012 Sep;150(6):1170–81. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Venkatesh S, Workman JL, Smolle M. UpSETing chromatin during non-coding RNA production. Epigenet Chromatin. 2013 Jan;6(1):16. doi: 10.1186/1756-8935-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Boa S, Coert C, Patterton HG. Saccharomyces cerevisiae Set1p is a methyltransferase specific for lysine 4 of histone H3 and is required for efficient gene expression. Yeast. 2003 Jul;20(9):827–35. doi: 10.1002/yea.995. [DOI] [PubMed] [Google Scholar]
- 137.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005 Nov;123(4):581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 138.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013 Mar;152(6):1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kim DH, Doyle MR, Sung S, Amasino RM. Vernalization: winter and the timing of flowering in plants. Annu Rev Cell Dev Biol. 2009 Jan;25:277–99. doi: 10.1146/annurev.cellbio.042308.113411. [DOI] [PubMed] [Google Scholar]
- 140.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009 Dec;462(7274):799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 141.Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011 Jan;331(6013):76–9. doi: 10.1126/science.1197349. [DOI] [PubMed] [Google Scholar]
- 142.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011 Jun;12(6):429–42. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- 143.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013 Mar;152(6):1308–23. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 144.Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996 Feb;6(2):149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 145.Brockdorff N. X-chromosome inactivation: closing in on proteins that bind Xist RNA. Trends Genet. 2002 Jul;18(7):352–358. doi: 10.1016/s0168-9525(02)02717-8. [DOI] [PubMed] [Google Scholar]
- 146.Lessing D, Anguera MC, Lee JT. X chromosome inactivation and epigenetic responses to cellular reprogramming. Anu Rev Genom Hum Genet. 2013 Jan;14:85–110. doi: 10.1146/annurev-genom-091212-153530. [DOI] [PubMed] [Google Scholar]
- 147.Romito A, Rougeulle C. Origin and evolution of the long non-coding genes in the X-inactivation center. Biochimie. 2011 Nov;93(11):1935–42. doi: 10.1016/j.biochi.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 148.Ng K, Pullirsch D, Leeb M, Wutz A. Xist and the order of silencing. EMBO Rep. 2007 Jan;8(1):34–9. doi: 10.1038/sj.embor.7400871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008 Oct;322(5902):750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011 Jul;146(1):119–33. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, Kaneko S, Helin K, Reinberg D, Stewart AF, Wutz A, Margueron R, Heard E. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell. 2014 Jan;53(2):301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 152.Kaneko S, Bonasio R, Saldaña-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and Noncoding RNAs Regulate PRC2 Recruitment to Chromatin. Mol Cell. 2013 Dec;53(2):290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Starkova J, Zamostna B, Mejstrikova E, Krejci R, Drabkin HA, Trka J. HOX gene expression in phenotypic and genotypic subgroups and low HOXA gene expression as an adverse prognostic factor in pediatric ALL. Ped Blood Cancer. 2010 Dec;55(6):1072–82. doi: 10.1002/pbc.22749. [DOI] [PubMed] [Google Scholar]
- 154.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010 Apr;464(7291):1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, Miyano S, Mori M. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011 Oct;71(20):6320–6. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 156.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007 Jun;129(7):1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, Pavlidis P, Journot L. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006 Nov;11(5):711–22. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 158.Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004 Aug;15(4):595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 159.Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem. 2003 Jun;278(26):24132–8. doi: 10.1074/jbc.M302283200. [DOI] [PubMed] [Google Scholar]
- 160.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, Guigo R, Shiekhattar R. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010 Oct;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011 Apr;472(7341):120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Spitale RC, Tsai MC, Chang HY. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics. 2011 May;6(5):539–543. doi: 10.4161/epi.6.5.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010 Aug;329(5992):689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kotake Y, Nakagawa T, Kitagawa K, Suzuki S, Liu N, Kitagawa M, Xiong Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011 Apr;30(16):1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008 Jan;451(7175):202–6. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010 Jun;38(5):662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang Y-G, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012 Mar;149(1):101–12. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 168.Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d’Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012 Aug;488(7410):231–5. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lee HC, Chang SS, Choudhary S, Aalto AP, Maiti M, Bamford DH, Liu Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009 May;459(7244):274–7. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010 Aug;24(15):1590–5. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zheng G, Dahl JA, Niu Y, Fu Y, Klungland A, Yang YG, He C. Sprouts of RNA epigenetics: the discovery of mammalian RNA demethylases. RNA Biol. 2013 Jun;10(6):915–8. doi: 10.4161/rna.24711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009 Feb;136(4):629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 173.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009 Mar;10(3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 174.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso CS, Kominsky HD, Cao X, Jing X, Wang X, Siddiqui J, Wei JT, Robinson D, Iyer HK, Palanisamy N, Maher CA, Chinnaiyan AM. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotech. 2011 Aug;29(8):742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]


