Abstract
Peptidoglycan is an essential component of cell wall in Gram-positive bacteria with unknown architecture. In this review, we summarize solid-state NMR approaches to address some of the unknowns in the Gram-positive bacteria peptidoglycan architecture: 1) peptidoglycan backbone conformation, 2) PG-lattice structure, 3) variations in the peptidoglycan architecture and composition, 4) the effects of peptidoglycan bridge-length on the peptidoglycan architecture in Fem mutants, 5) the orientation of glycan strands respect to the membrane, and 6) the relationship between the peptidoglycan structure and the glycopeptide antibiotic mode of action. Solid-state NMR analyses of S. aureus cell wall show that peptidoglycan chains are surprisingly ordered and densely packed. The peptidoglycan disaccharide backbone adopts 4-fold screw helical symmetry with the disaccharide unit periodicity of 40 Å. Peptidoglycan lattice in S. aureus cell wall is formed by cross-linked PG stems that have parallel orientations. The structural characterization of Fem-mutants of S. aureus with varying lengths of bridge structures suggests that the PG-bridge length is an important determining factor for the PG architecture.
Keywords: Peptidoglycan, cell wall, REDOR, Staphylococcus aureus, solid-state NMR, oritavancin, architecture, glycopeptide antibiotic
1. Introduction
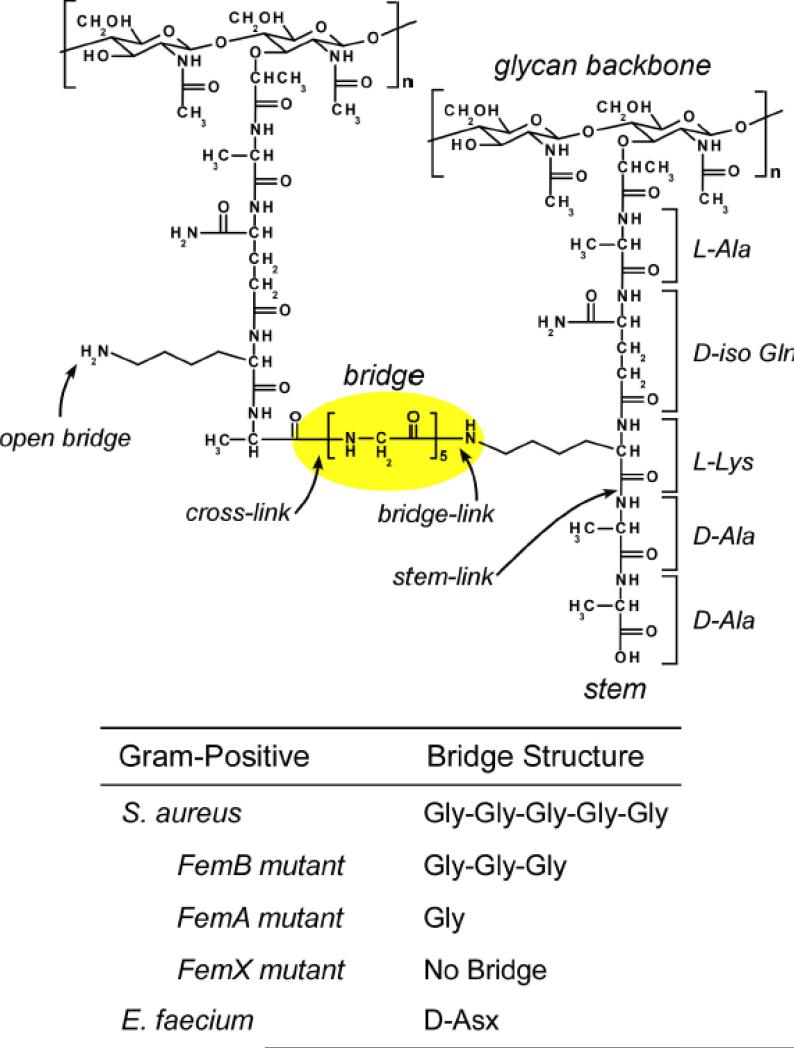
Cell wall (CW) is a common feature found in all bacteria with a few exceptions found in mycoplasmas, L-forms, and some archaebacteria [1]. In Gram-positive bacteria, the CW thickness varies from 20 to 40 nm. It functions as protective barrier against the external environment and allows bacteria to withstand physical extremes such as changes in osmotic pressure up to 25 atm [2]. The principal component of the CW is peptidoglycan (PG) that also serves as scaffold for the attaching proteins, polysaccharides, and ribitol-phosphate polymers. The chemical structure of a PG-repeat unit consists of disaccharide, pentapeptide-stem, and a bridge structure (Figure 1). The disaccharide, composed of N-acetyl-glucosamine (NAG) and N-acetyl-muramic acid (NAM), is conserved in all eubacteria; the pentapeptide-stem and bridge structure vary among species. The PG pentapeptide-stem chemical structure in Gram-positive pathogens Staphylococcus aureus and Enterococcus faecium are identical, L-Ala-D-iso-Gln-L-Lys-D-Ala-D-Ala, with the N-terminal L-Ala linked to the NAM of the disaccharide via a lactic moiety [3] (Figure 1). The only difference in the PG chemical structures of S. aureus and E. faecium is their bridge structure, where pentaglycine (Gly)5 is found in S. aureus and a single D-Asp in E. faecium. The C-terminus of the (Gly)5 bridge in S. aureus forms an amide bond to the side-chain nitrogen of the L-Lys (of pentapeptide stem), which is referred to as “bridge-link” [4]. In E. faecium the bridge-link is formed between the carboxyl side chain of the D-Asp (not the C-terminus) and the ε-nitrogen of L-Lys by D-Aspartate ligase [5].
Figure 1.
Chemical structure of S. aureus peptidoglycan. Peptidoglycan (PG) repeat unit (dotted box) consists of disaccharide, pentapeptide-stem, and bridge structure. The disaccharide consists of N-acetyl-glucosamine (NAG) and N-acetyl-muramic acid (NAM). The pentapeptides stem structure is L-Ala-D-Glu-L-Lys-D-Ala-D-Ala, with L-Ala attached to NAM of the disaccharide. The bridge structure in S. aureus is a pentaglycine which is attached to the ε nitrogen of L-Lys of the third position of the stem. Cross-link is formed between the N-terminus of glycine bridge to the D-Ala (4th position) carbonyl carbon of the adjacent stem. Fem-deletion mutants of S. aureus have different bridge lengths varying from triglycyl to monoglycyl-bridge structures. E. faecium have a D-Asp as a bridge structure.
The final steps of PG assembly is carried out by two enzymatic processes, transglycosylation and transpeptidation [6]. Transglycosylation is the polymerization of the PG disaccharide units (DU) by the formation of a β(1,4) glycosidic bond between the NAG and NAM (Figure 1). The length of polymerized disaccharides varies from one species to another. In S. aureus, 85% to 90% of the polymerized disaccharides are between 3 to 10 DU, with an average PG-chain length of only 6 DU. Glycan chains of 25 DU or longer are estimated to be less than 15% of the all PG units [7]. By comparison, other bacteria have much longer average chain lengths that range from 30-60 DU in E. coli [8] to over 500 DU in B. subtilis [9].
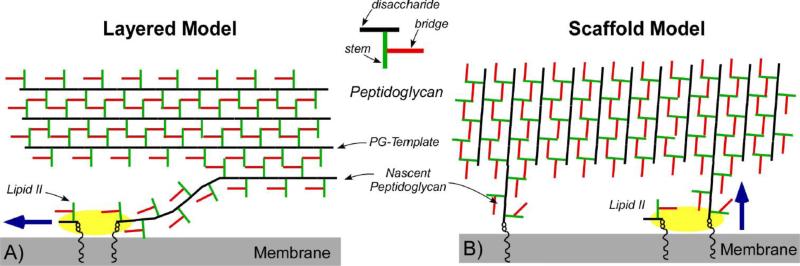
The second step in PG assembly is carried out by transpeptidases that incorporate the nascent PG to the mature CW by cross-linking the bridge N-terminus of one glycan chain to the D-Ala (4th amino acid of the pentapeptide stem) of the neighboring chain with a peptide bond. Transpeptidase activity is crucial for the formation of 3D PG architecture. The most extensive cross-linking is found in isolated cell walls of S. aureus with estimated 80% [10] to 90% [11] of PG units cross-linked, as based on the LC/MS analysis of mutanolysin-digested PG fragments. Mutanolysin is an N-acetyl-muramidase that cleaves the β(1,4) glycosidic bond between repeating disaccharide units but leaves cross-links intact. Extensive cross-linking is thought to physically compensate for the relatively short PG-chain lengths in S. aureus. In E. faecium cross-linking density is significantly less, as the mutanolysin-digested PG fragments are dominated by dimers (40 to 60%) and monomers (30 to 40%), with only few trimers (5 to 15%) and no oligomers of longer length [12]. The differences in the PG-chain lengths and cross-linking from S. aureus to E. faecium suggest that the PG architecture is likely to be very different between these two species despite the similarities in other parts of PG. Although the chemical structure of PG-repeat unit (Figure 1) is well known, the physical structure of assembled PG in CW remains unknown. This is because PG is an insoluble, complex, and heterogeneous supra-macromolecule incompatible with the conventional structural methods. Hence direct imaging methods such as cryo-electron tomography (CET) [13-15] and atomic force microscopy (AFM) [16-18] have recently played a key role in providing morphological insights into the PG architecture. There are two proposed PG models, layered and scaffold, that differ on the PG orientation in respect to the bacterial membrane (Figure 2). In the layered model, PG chains elongate in a plane parallel to the membrane; whereas in scaffold model the PG chains are aligned perpendicularly to the membrane. For Gram-negative bacteria, vertical alignment of PG chains (scaffold model) with average glycan chain lengths of 25 DU [19] would result in a sacculus with thickness that exceed 25 nm. This is not observed, as the average sacculus thickness of Gram-negative bacteria range from 3 nm (by AFM) [20] to 7 nm (by CET) [21]. The CET of Gram-negative bacteria C. crescentus by Jensen et al., [14] confirm the sheet-like sacculus structure formed by loosely packed and relatively disordered PG glycan chains oriented in parallel to the bacterial membrane [14]. The large pore structures in Gram-negative sacculus with 1-10 nm in diameter visible by AFM are also consistent with loosely connected PG chains organized as layers [16].
Figure 2.
Left) In the layered model PG chains elongate in a plane parallel to the membrane. The model shows the PG transglycosylation (highlighted in yellow) occur along the direction of the membrane as shown by the arrow. Right) In scaffold model the PG chains are oriented perpendicularly to the membrane. Layered model (left) shows the PG chains are oriented parallel to the bacterial cytoplasmic membrane. The arrows show the direction of PG chain elongation.
For the Gram-positive bacteria, the direct imaging methods cannot be used to differentiate from layered to scaffold models due to the substantially thickened CW that complicates the CET analysis [13] and limits the AFM to probing surface morphology. For S. aureus with 20 nm thick CW [12], the layered model predicts 20 layers of PG encapsulating the bacteria [22]. Equally plausible is the vertical arrangement of the PG chains (scaffold model), since the average PG length in S. aureus is only 6 DU. The combination of high degree of PG cross-linking and short PG chain lengths in S. aureus is thought to favor the scaffold model because the vertical alignment of highly cross-linked PG would be structurally more resistant to mechanical stress than the layer model with short PG strands aligned along the membrane [21].
In this review, we summarize solid-state NMR approaches to address some of the current unknowns in the Gram-positive bacteria PG architecture [11]. Solid-state NMR approach is unique in its ability to provide angstrom-resolution distance constraints necessary for structural characterization while also allowing the chemical compositional analysis of PG in intact whole-cells of bacteria [23]. Solid-state NMR addresses the following unknowns in the PG structure: 1) PG backbone conformation, 2) PG-lattice structure, 3) variations in the PG architecture and composition, 4) the effects of PG bridge-length on the PG architecture in Fem mutants, 5) the orientation of glycan strands respect to the membrane, and 6) the relationship between the PG structure and the glycopeptide antibiotic mode of action.
2 PG Backbone Conformation
2.1 Helical 2-fold and 4-fold axial symmetry
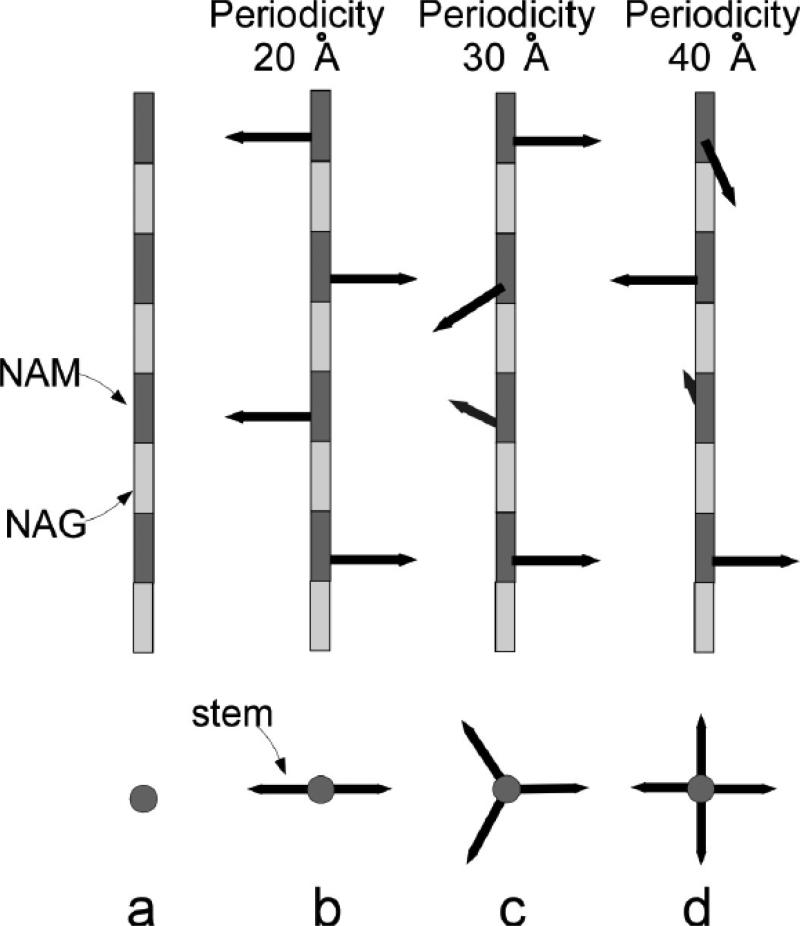
The PG disaccharide backbone conformation was initially thought to adopt chitin-like conformation with the disaccharide periodicity of 20 Å [24, 25] (Figure 3b). In this model, successive PG-stems are rotated 180° relative to previous stem orientation with a 2-fold screw symmetry. Hence the PG cross-links would result in a 2D-planar sheet-like structure. The chitin-like model was subsequently abandoned for 4-fold screw helical symmetry with 40 Å periodicity, based on the X-ray diffraction analysis of isolated PG [26, 27] and of whole-cells S. aureus [27]. The 4-fold screw helical symmetry allows the successive PG-stems on a glycan chain to rotate 90° relative to previous stem, allowing cross-links to form in all four directions (Figure 3d).
Figure 3.
Schematic representation of glycan backbone conformations. a) The repeating disaccharide, NAM (light box)-NAM (dark box), forms the PG backbone. b) The PG stems, represented as arrows, are rotated 180o with respect to the adjacent stem. The repeat disaccharide unit has a periodicity of the 20 Å. c) The PG stems are rotated 120o with respect to the adjacent stem with the disaccharide unit periodicity of 30 Å. d) The PG stems are rotated 90o with respect to the adjacent stem with the disaccharide unit periodicity of 40 Å. The figures are modified from reference [35].
2.2 Helical 3-fold axial symmetry by solution-state NMR
The structural characterization of PG by solution-state NMR has been impractical due to the difficulty in purifying soluble and homogenous PG fragments from CW. This limitation was circumvented by Meroueh et al. [28] through total synthesis of a solubilized PG-mimic consisting of two-repeat disaccharide units (NAG-NAM-NAG-NAM) with a stem, but without a bridge structure. The disaccharide backbone conformation was determined by measuring the glycosidic dihedral angles. The structure of PG mimic had a helical periodicity of 30 Å (3-fold screw symmetry) which implied that each successive PG-stem is rotated 120° in respect to the previous stem. The resulting PG-lattice is a hexagonal honeycomb-like structure with a 70-Å diameter pore in the middle of each hexagonal unit. This model assumes that the cross-links are formed between two neighboring antiparallel stems. Since the PG in CW disaccharide conformation is based on solubilized uncross-linked PG mimic, it is unlikely that the highly cross-linked and stress-bearing CW would adopt a PG-mimic-like conformation.
2.3 Helical 4-fold axial symmetry by solid-state NMR
In situ PG glycan conformation was investigated using a Centerband-Only Detection of Exchange (CODEX) carbon-13 spin diffusion experiment on [1-13C]Gly-labeled isolated CW from S. aureus strain BB255 [29]. The isolated CW was prepared from S. aureus grown in a defined medium containing [1-13C]Gly. For the mixing time of 840 ms, spin diffusion from the 13C in the [1-13C]Gly to surrounding 13C natural-abundance carbons within 5 Å are visible in the CODEX difference spectrum. As expected, the L-Lys and D-iso-Gln carbons in PG stem with [1-13C]Gly attached are visible in the CODEX difference spectrum. Unexpected was the visibility of 102-ppm anomeric carbon from the PG disaccharide in the CODEX difference spectrum. The intra-molecular distance between the anomeric carbons of disaccharide to the 13C in [1-13C]Gly of the same PG unit exceeds 10 Å. Hence the 102-ppm peak in the CODEX difference spectrum corresponds to inter-molecular distance, positioning the anomeric carbon in one PG unit within 5 Å of the 13C in [1-13C]Gly located on a different glycan chain. This 13C-13C spin-diffusion constrain implies the glycan chains in the CW of S. aureus are tightly packed in contrast to the proposed loosely packed PG-lattice model based on the solution-state NMR [28].
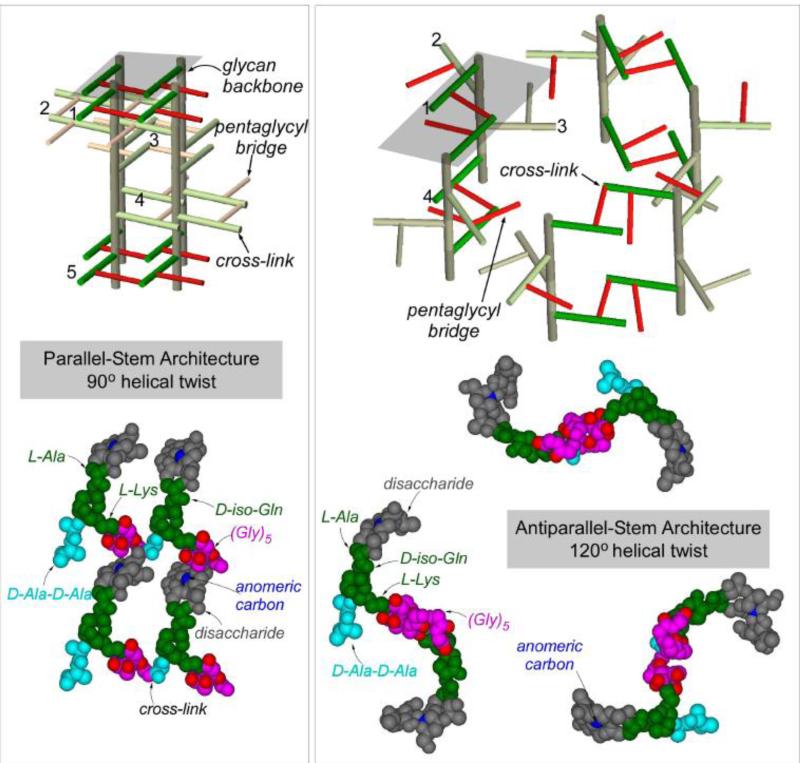
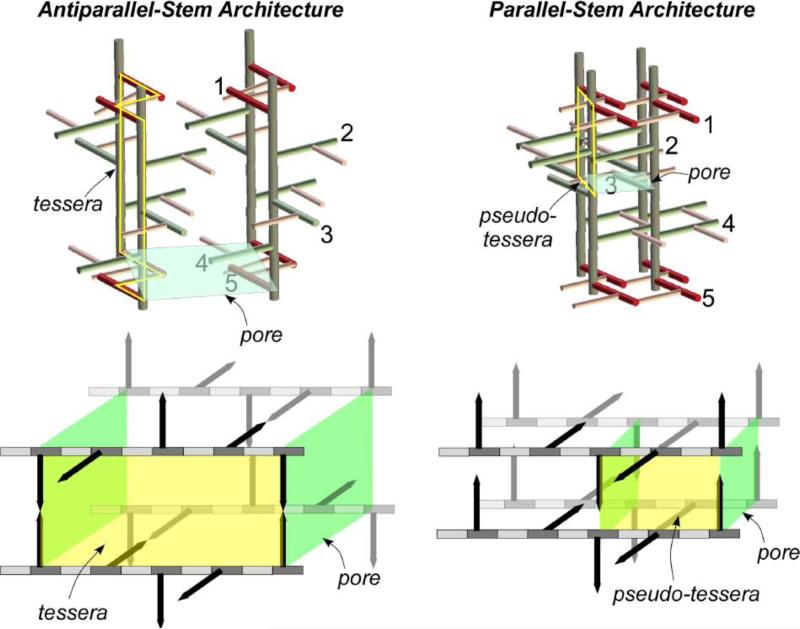
A 3D PG-lattice model of S. aureus based on 13C-13C spin-diffusion constrain is shown in Figure 4 (left), and a model for helical 3-fold axial symmetry based on solution-state NMR constrain in Figure 4 (right) [29]. The disaccharides are shown in gray, pentaglycyl bridge in red, and pentapeptide stem in green. The gray planes in Figure 4 (top) represent the cross section, with four glycan chains for the parallel-stem architecture (left) and eight glycan chains for the antiparallel-stem architecture (right). In parallel-stem architecture (left), the cross-links are formed between two adjacent parallel stems; whereas in antiparallel-stem architecture (right), only the antiparallel stems are cross-linked. The major differences between two models are as following: 1) The 13C labels in pentaglycyl bridge in the 4-fold axial symmetry model (Figure 4, left) are in close proximity (within 5 Å) to the anomeric sugar carbon of disaccharide in nearest glycan chain, while in 3-fold axial symmetry model the distances exceed 10 Å. 2) The glycan chains are densely packed in 4-fold axial symmetry model. 3) The 4-fold axial symmetry model allows for maximum 100% cross-linking, while 3-fold axial symmetry model allows only 50%. 4) The pore structure is absent from 4-fold axial symmetry model.
Figure 4.
Top) Peptidoglycan architecture of helical 4-fold axial symmetry with parallel PG stems (left), and helical 3-fold axial symmetry with antiparallel PG stems (right). The bridges are represented in pink, glycan backbones in brown, and stems in green. The maximum cross-linking possible for the 4-fold axial symmetry model (left) is 100% and for the 3-fold axial symmetry model (right) is 50%. Bottom) Space-filling models of PG architecture. In 4-fold axial symmetry model (left), the glycyl-carbonyl carbons of (Gly)5 (shown in red) are within 5 Å from anomeric carbon of disaccharide (shown in blue), consistent with the CODEX-measured 13C-13C diffusion constraint. In 3-fold axial symmetry model (right), the distances between glycyl-carbonyl carbons to the anomeric carbon exceed 10 Å. The dotted lines connecting the anomeric carbons in 3-fold axial symmetry model shows a large pore structure (right), which is absent in 4-fold axial symmetry model (left). The figure is adapted from reference [29].
The only model consistent with the CODEX-detected 13C-13C spin-diffusion constraint is helical 4-fold axial symmetry with parallel-stem PG architecture [29]. Although 3-fold axial screw symmetry parallel-stem architecture was considered, but the model showed crowding of glycan chains with implausible steric constraints for the formation of PG lattice structure. In addition, the PG cross-linking in isolated CW of S. aureus strain BB255 as measured by solid-state NMR was 75% [30], consistent with the 4-fold axial symmetry antiparallel stem architecture.
3. S. aureus PG-lattice structure
3.1 PG-stem architecture by REDOR NMR
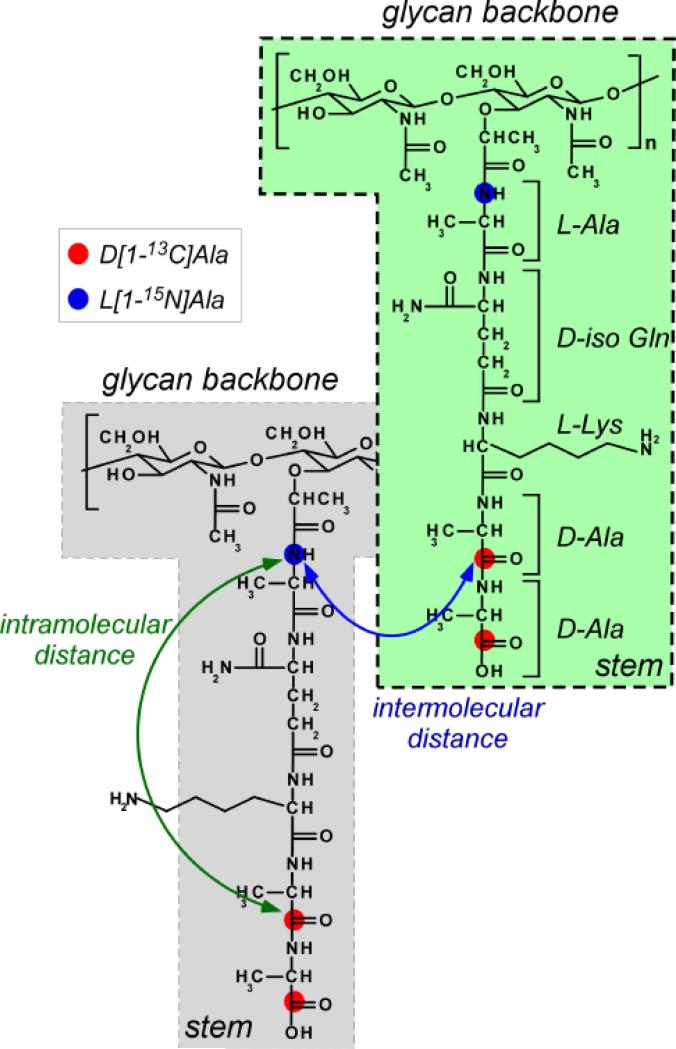
PG-lattice structure of intact whole-cell S. aureus was characterized using rotational-echo double resonance (REDOR) NMR [31, 32]. Samples were prepared by growing S. aureus in defined medium with D-[1-13C]alanine and L-[15N]alanine in the presence of alanine racemase inhibitor alaphosphin [33]. The 13C and 15N labeled positions are shown in Figure 5 (left). Inter-nuclear distance between 13C and 15N as measured by 13C{15N} and 15N{13C} REDOR NMR was 4.4 Å. Since the intra-molecular 13C-15N distance between the D-[1-13C]Ala and L-[15N]Ala on the same PG-stem must be over 10 Å, the measured distance can only be the inter-molecular distance between D-[1-13C]Ala and L-[15N]Ala of two different PG stems. These two stems cannot be from the same glycan chain, as each disaccharide unit on a single chain is separated from one another by approximately 10 Å (Figure 5, left). Thus the measured 13C-15N distance of 4.4 Å is the distance between D-[1-13C]Ala from a stem in one glycan chain to L-[15N]alanine in a stem of neighboring glycan chain.
Figure 5.
Isotope labeling strategy for REDOR characterization of S. aureus PG-lattice structure. Whole-cells of S. aureus were grown in defined media containing L-[15N]Ala and D-[1-13C]Ala in the presence of alanine racemase inhibitor alaphosphin (5 to 15 μg/ml). The incorporation of 15N (blue dots) and 13C (red dots) isotopic labels are shown to indicate that the intramolecular 13C-15N distances exceeds 10 Å, which beyond current 13C{15N} REDOR detection limit.
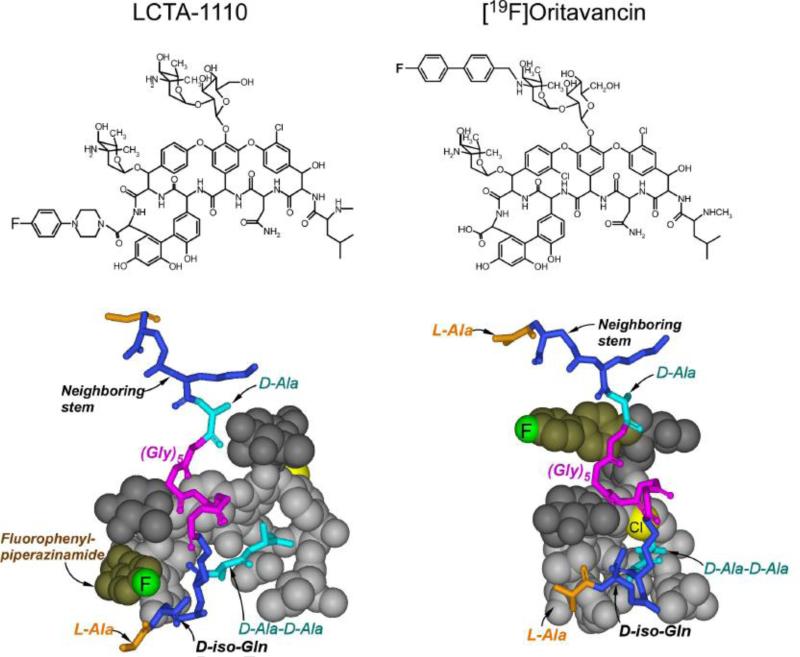
The PG-lattice structure was further characterized using 19F-labeled glycopeptide antibiotic LCTA-1110 (Figure 6, left top), which binds to the D-Ala-D-Ala terminus of uncross-linked PG stem. Using 19F on the arylpiperazine sidechain of LCTA-1110, 13C-19F and 15N-19F internuclear distances to nearby L-[15N]Ala and D-[1-13C]Ala in PG were measured. REDOR-measured 13C-19F distances were 5 and 9 Å, and the 15N-19F distance was 6 Å. The position of 19F in PG-bound LCTA-1110 complex (Figure 6, left middle) is at least 12 Å away from the 13C of the glycopeptide bound D-[1-13C]Ala-D-[1-13C]Ala [34]. Thus the 13C{19F} REDOR measured distances of 5 and 9 Å corresponds to the 13C-19F distances from the 19F of LCTA-1110 to the D-[1-13C]Ala-D-[1-13C]Ala of the nearest unbound PG-stems. On the other hand, measured 19F-15N distance of 6 Å is an intramolecular distance from the 19F of LCTA-1110 of the L-[15N]Ala on the same glycopeptide-bound PG stem (Figure 6, left bottom) [34].
Figure 6.
Top) Chemical structures of LCTA-1110 and [19F]oritavancin. LCTA-1110 is a C-terminus modified eremomycin and [19F]oritavancin is a disaccharide modified chloroeremomycin. Both glycopeptides contain fluorine but at different positions. Bottom) Space filling model of the LCTA-1110 and [19F]oritavancin complexed with the PG of S. aureus (shown as stick model). The 19F position satisfies all the REDOR-measured restraints. The 19F of LCTA-1110 is close to the L-Ala of the bound PG stem (red arrow), but far from the bound D-Ala-D-Ala, distance exceeding 10 Å, as shown by the black arrow.
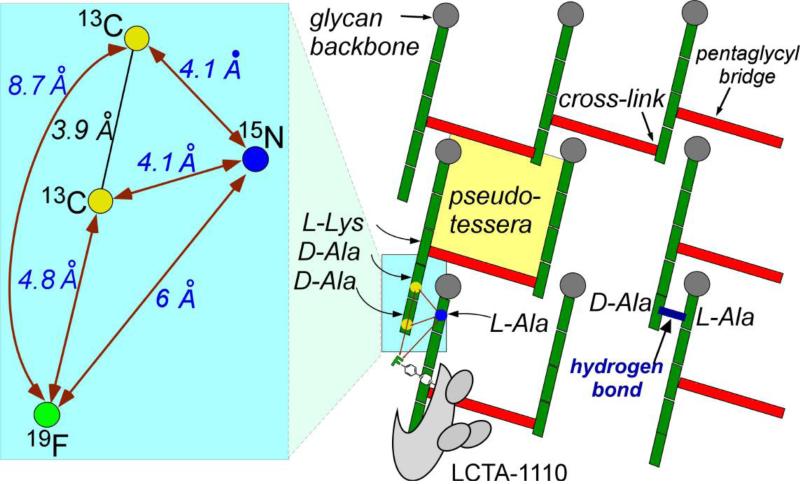
The REDOR-distance constraints for the PG-lattice are summarized in Figure 7 (left) with the measured distances in blue and the calculated geometric constraints in black. The measured distances place 19F of LCTA-1110 in a near-planar geometry to 15N of the L-[15N]Ala bound stem, and to the 13Cs in D-[1-13C]Ala-D-[1-13C]Ala of the nearest-neighboring stem. The short inter-glycan 13C-15N distance requires all coplanar PG-stems be parallel in respect to each other.
Figure 7.
Right) Cross-section of proposed peptidoglycan tertiary structure for S. aureus. Glycan backbones (gray circles) are propagating into the plane of the paper. The stems and bridges are represented by green and red rectangles, respectively. Bound drug is shown at the bottom left of the cross-section. The yellow square shows a minimum effective pore size, or pseudo-tessera. (Left) Details of the blue highlighted region of the right panel. Experimental REDOR distance measurements (dark-red) connect labels. Black lines and angles are calculated. The green circle represents the drug 19F. The Figure is adapted from the reference [35]
3.2 PG-lattice cross-section
Cross-sectional model of S. aureus PG-lattice, based on REDOR-distance constraints, is shown in Figure 7 (right) [35]. The glycan chains (gray circles) in a three-by-three matrix propagate perpendicularly to the plane of paper, with the PG stems (green) and bridges (red) forming cross-links in the plane. Some of the PG stems are missing bridges and D-Ala, which is consistent with the measured 85% bridge-linking degree [23] and 46% D-Ala-D-Ala stem termination in whole-cells of S. aureus [36]. The measured 13C-15N distance of 3.4 Å (Figure 7, blue inset) suggests a possible hydrogen bond (blue) between the carboxyl terminus of uncross-linked PG-stem (5th stem amino acid, D-[1-13C]Ala) and amide proton from L-Ala of adjacent stem (Figure 7, right). Thus uncross-linked PG stems that terminate in D-Ala-D-Ala can still contribute to overall CW stability with hydrogen bond formation. This provides a possible explanation for S. aureus CW, even with its short disaccharide chains with average length of 6 DU [7] and relatively low cross-linking density of 54% in intact whole cells [36], to exhibit remarkable mechanical and tensile strength.
3.3 Tessera and pseudo-tessera structures
In antiparallel-stem architecture, tessera describes an unit structure in the lattice of Gram-negative bacteria PG [37] with its quadrilateral boundary set by two glycan chains connected to a pair of cross-links (Figure 8, right yellow outline). The PG-lattice volume for the antiparallel-stem architecture is defined by the tessera structure and pore size, which is the cross-sectional area of the four cross-linked PG glycan chains (Figure 8, green square). The estimated diameter of tessera in antiparallel-stem architecture is approximately 45 Å, which is in agreement with the hypothetical tessera diameter of 52 Å [16] for the Gram-negative bacteria.
Figure 8.
Top) PG architecture for a 4-fold glycan symmetry axis with antiparallel (left) and parallel (right) stems. In parallel-stem architecture (right), the glycan chains are tightly packed with 100% cross-linking. In antiparallel-stem architecture, with loosely packed glycan chains the maximum cross-linking is 50%. Bottom) Tessera structures, a hypothetical PG cell-unit structure defined by two glycan chains connected by two pairs of cross-links, is shown in yellow for the antiparallel-stem architecture (left). Similar tesserae structure is not found in parallel-stem architecture of S. aureus. Pseudo-tessera, defined by two successive anti-parallel stems between two adjacent glycan chains, is significantly smaller than the tesserae structure shown on the left. The Figure is modified from the reference [35].
In parallel-stem architecture of S. aureus, tessera structure cannot be defined as the boundary defined by cross-linked antiparallel stems is absent. Instead a “pseudo-tessera” structure can be defined as an unit cell with the boundaries outlined by a pair of two successive antiparallel stems of two adjacent glycan chains (Figure 8, bottom right, yellow outline). The approximate diameter of a pseudo-tessera is 23 Å, significantly smaller than the tessera diameter of 52 Å. The estimated overall PG-lattice volume for the parallel-stem architecture is only an eighth of the PG-lattice volume in antiparallel-stem architecture.
4. Variation in the CW compositions and PG architectures in Gram-positive bacteria
4.1 S. aureus and E. faecium PG composition
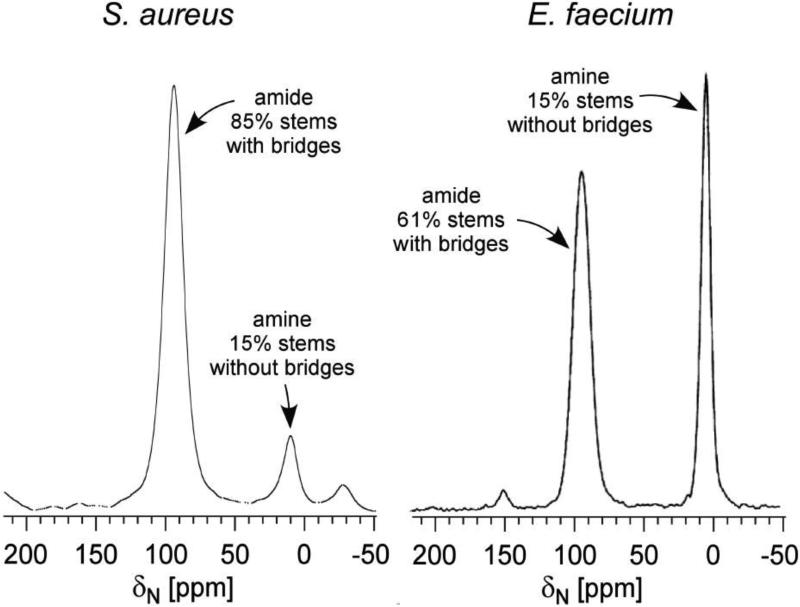
To investigate the effects of different PG chemical structure on the PG's tertiary structure, the composition of S. aureus and E. faecium CW were characterized using solid-state NMR [38, 39]. The PG chemical structures of S. aureus and E. faecium differ only in their bridge structure. In S. aureus the bridge is pentaglycine, whereas in E. faecium a single D-Asp is the bridge. This difference in the PG bridge length has profound effect on bridge-link and cross-link degrees in the CW of S. aureus and E. faecium. The degree of bridge-link was measured from isolated CWs of E. faecium and S. aureus labeled with L-[ε-15N]Lys. In the 15N-CPMAS spectra (Figure 9), PG stems with bridge attached are visible as a lysyl-amide (95-ppm peak), while PG stems without bridge appear as a lysyl-amine peak (5 ppm). The measured bridge-link density in E. faecium was 61% and S. aureus was 85% (Figure 9). Since the bridge structure is required for cross-linking, the measured bridge-link densities are also the limits of cross-linking frequencies.
Figure 9.
15N CPMAS echo-spectrum of isolated cell wall labeled with L-[ε-15N]Lys from S. aureus (left) and E. faecium (right). The 85% of the PG stems in S. arueus have have bridges [23], but only 61% in E. faecium [38].
The actual PG cross-linking densities were measured from isolated CWs of S. aureus and E. faecium with 13C-15N pair labeled cross-links. 13C{15N} and 15N{13C}REDOR after 1.2 ms dipolar evolution was used to select covalently bonded 13C-15N spin pair that can only be found at the cross-links. The measured cross-linking densities were 51% for E. faecium [38] and 67% for S. aureus [23]. Overall, PG cross-linking density in E. faecium was approximately 24% less than that of S. aureus.
Another key difference in S. aureus and E. faecium CW composition was the amount of PG stems that terminated in D-Ala-D-Ala. In E. faecium 64% of the PG stems terminated in either D-Ala-D-Ala or D-Ala [38]. Given that 51% of all the PG units are cross-linked (PG stems terminating in D-Ala), the remaining 13% (64% - 51%) of the PG stems terminated in D-Ala-D-Ala. In comparison, all PG stems in S. aureus terminated in either D-Ala-D-Ala or D-Ala [23], with 67% of them cross-linked. Thus in S. aureus 33% (100% - 67%) of the PG stems terminate in D-Ala-D-Ala. The PG stems terminating in D-Ala-D-Ala are glycopeptide-binding sites, and therefore glycopeptide-binding sites occur approximately 2.5 times more frequently in S. aureus CW than that of E. faecium.
5. Effects of PG bridge-length on PG architecture of Fem mutants
5.1 CW composition in Fem mutants
Classification of PG according Schleifer and Kandler [40] is shown in Table 1. Two major groups are Group A and Group B. Group A refers to PG with a bridge attached to the 3rd residue on the stem, and Group B with a bridge attached to the 2nd or 4th residue of the stem. The numbers are used to further specify the types of bridges. All Gram-positive bacteria in Group A have identical PG chemical structure save PG bridge structure. The PG-bridge lengths vary from 1 to 5 amino acids depending on the bacterial species. For example, the bridge in S. aureus is (Gly)5, E. faecalis is (L-Ala)3, S. pneumoniae is L-Ala-L-Ala (or L-Ser-L-Ala), and E. faecium is (D-Asp)1. To determine the effect of PG-bridge length on PG-lattice structures, CW composition of S. aureus and its isogeneic Fem mutants have been investigated using solid-state NMR [29, 30, 41]. The key advantage of using isogenic Fem-deletion mutants of S. aureus is that they retain identical PG biosynthesis machineries except for the fem deletions, which result in shortened bridge lengths (Table 2). The wild type S. aureus (BB255) has (Gly)5, FemB (UT34-2) has (Gly)3, and FemA (UK17) has (Gly)1 as its bridge structure. Summary of solid-state NMR measured bridge-link and cross-link densities in the isolated CWs of Fem-mutant S. aureus [29, 30, 41] are shown in Table 2.
Table 1.
Classification of peptidoglycan according to Schleifer and Kandler [40]. If the bridge is linked to the 3rd residue on the stem, it is classified as Group A, but if linked to the carboxyl group of the 2nd or 4th residue of the stem, it is classified as Group B. The numbers are used to specify the types of bridges.
| Organisms | PG-bridge structure | Group Type | Bridge Type (description) | Amino acid at bridge-link | Classification |
|---|---|---|---|---|---|
| E. coli | none | A | 1 (No bridge) | Meso-diaminopimelic acid | A1 |
| M. Luteus | L-Ala-D-Glu-Gly | A | 2 (polymerized subunits) | L-lysine | A2 |
| S. aureus | (Gly)5 | A | 3 (Mono carboxylic L-amino acids) | L-lysine | A3 |
| E. faecalis | (L-Ala)2,3 | A | 3 | L-lysine | A3 |
| E. faecium | D-Asn | A | 4 (Dicarboxylic amino acids) | L-lysine | A4 |
| M. lacticum | L-Lys-Gly | B | 1 (L-amino acid) | L-lysine | B1 |
| C. poinsettiae | D-Orn | B | 2 (diamino acid) | L-homoserine | B2 |
Table 2.
Solid-state NMR measured bridge-link and cross-link densities from isolated
| CWs of Gram-positive bacteria. The S. aureus (ATCC 6538P) measurements are from reference [23], BB255 and its isogenic Fem mutants are from references [29, 30, 41], and E. faecium are from [56]. | |||
|---|---|---|---|
| Organisms | PG-bridge structure | Bridge-links [%] | Cross-links [%] |
| S. aureus (ATCC 6538P) | Gly-Gly-Gly-Gly-Gly | 85 | 67 |
| Wild type S. aureus (BB255) | Gly-Gly-Gly-Gly-Gly | 94 | 75 |
| FemB (UT34-2) | Gly-Gly-Gly ^ | 92 | 70 |
| FemA (UK 17) | Gly | 91 | 50 |
| E. faecium (ATCC 49624) | D-Asp | 61 | 51 |
The bridge-link densities for all Fem mutants are constant, with all in the range of 91 to 94%. However, reducing the glycine-bridge lengths from 5 to 3 to 1 Gly has a dramatic effect on cross-linking densities, reducing it from 75% to 70% to 51% respectively. The decrease from 75% to 70% cross-linking density FemB compared to wildtype indicates that reduction from pentaglycyl to triglycyl is relatively well-tolerated in S. aureus. But when the bridge is further reduced to a single Gly in FemA, cross-linking density abruptly drops to 50%. Interestingly, E. faecium with mono-D-Asp bridge structure that is comparable in bridge length to FemA has nearly identical cross-linking density of 50% [38]. Such dramatic difference in CW composition suggests that the PG tertiary structure in FemA is fundamental different from wild type as the monoglycyl PG-bridge in FemA is too short to form parallel PG-stem architecture observed in wild type S. aureus [35]. Similarities in bridge lengths of FemA and E. faecium, along with cross-link and bridge-link densities, suggest that FemA and E. faecium may share similar PG-tertiary structure. Recent studies by Sharif et al. [41] have found that the PG of FemA does not exclusively consist of monoglycyl bridge PG, but is heterogeneous with monoand triglycyl bridged PG. This was confirmed by Transferred-Echo Double Resonance (TEDOR)-CODEX NMR experiment, where CW was labeled with [1-13C]Gly and [ε-15N]Lys. The deconvolution of 15N→13C TEDOR spectrum confirmed the presence of uncross-linked stems of both mono- and triglycyl-bridge structures in FemA. The TEDOR-CODEX difference spectrum indicated that the stems are found in a tight lattice structure.
5.2 PG architectures based on PG-bridge length
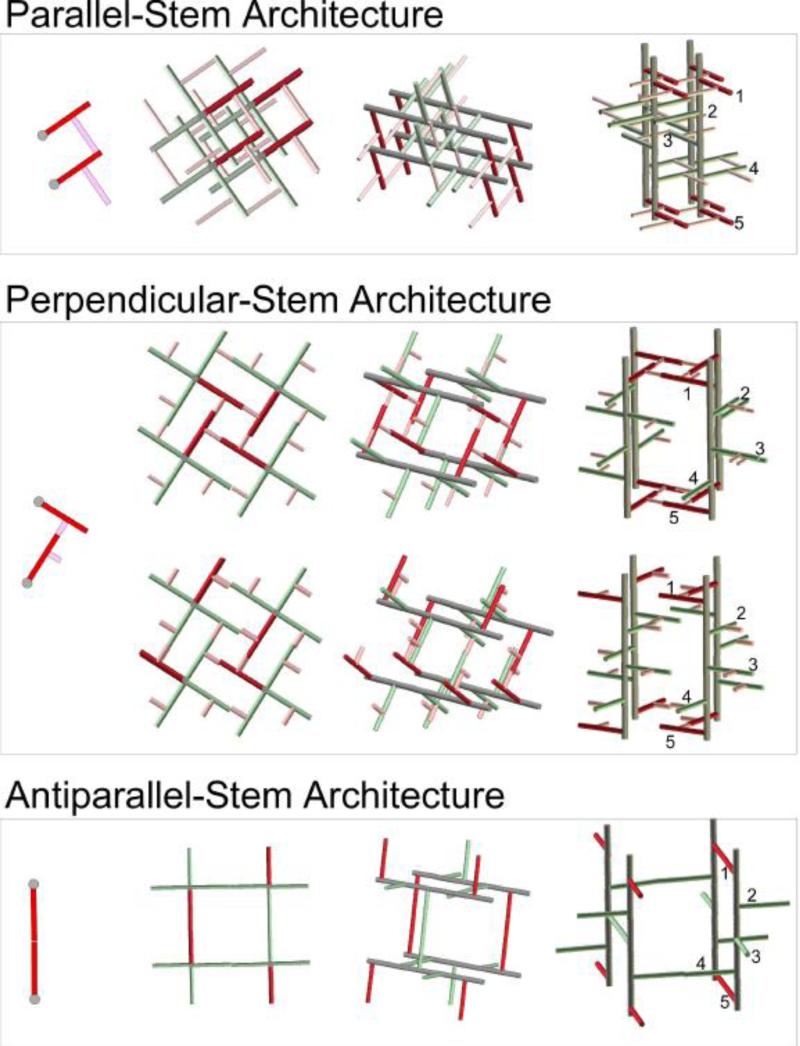
While long PG-bridge length in S. aureus is critical for the antiparallel-stem architecture, such architectures are not possible in bacteria with short bridges due to steric crowding. Thus we believe that the PG-bridge length is the key factor in determining the types of different PG architecture. Figure 10 illustrates proposed three principal PG architectures: parallel-, perpendicular-, and antiparallel-stem models. All models were built using right-handed glycan chains with 4-fold screw symmetry that align in parallel respect to each other, and the models assume all PG-repeat units have identical conformation [33]. The parallel-stem architecture (Figure 10, top) is suited for bacteria with long bridge length such as S. aureus. The maximum cross-linking density possible for this model is 100%, which can compensate for the short glycan chain lengths found in S. aureus. The parallel-stem architecture lacks tessera structure and it has the smallest pore size out of all three models. Perpendicular-stem architecture (Figure 10, middle) is proposed for bacteria with intermediate-bridge lengths too short for parallel-stem architecture, such as E. faecium and S. pneumonia. The perpendicular-stem architecture (Figure 10, bottom) has intermediate-pore size with intermediate glycan chain length. The antiparallel-stem architecture is for PG without bridge structure, such as E. coli and B. subtilis. This model has the largest pore size with tessera structure. Since the maximum cross-linking density is only 50%, long glycan chains are required for the bacteria with this type of architecture in order to compensate for lack of structural support from cross-linking. A summary of proposed properties for Gram-positive bacteria with varying chain lengths is shown in Table 3. Other possible architectures include hybrid models that combine multiple features from all three principal PG architectures. One example is a hybrid model proposed for FemA mutant of S. aureus, where antiparallel- and perpendicular-stem architectures are combined to form a repeating structural motif [42].
Figure 10.
PG-bridge length dependent architectures. The glycan backbones are shown in gray, stems in red, and bridges in pink. Top) Parallel-stem architecture where two stems that are cross-linked share same parallel orientation. Tessera structure is absent in this model. Middle) Antiparallel-stem architecture has cross-linked stems that are oriented perpendicular with respect to each other. Bottom) Antiparallel-stem architecture where cross-linked stems are oriented in opposite directions. Other possible architectures include hybrid models that combine multiple features are omitted for clarification.
Table 3.
Proposed properties of PG-bridge length dependent architectures.
| Architecture | Bacteria | Pore size and unit- cell volume | Cross- linking (theoretical maximum) | Glycan chain lengths | Muramidase digested maximum PG fragments |
|---|---|---|---|---|---|
| Parallel-Stem | S. aureus | Smallest | 100% | Shortest | oligomers |
| Perpendicular- Stem | E. faecium S. pneumonia | Intermediate | ~70% (model dependent) | Intermediate | tetramers |
| Antiparallel-Stem | E. coli B. subtilis | Largest | 50% | Longest | dimers |
6. Orientation of PG chains: Scaffold versus Layered Model
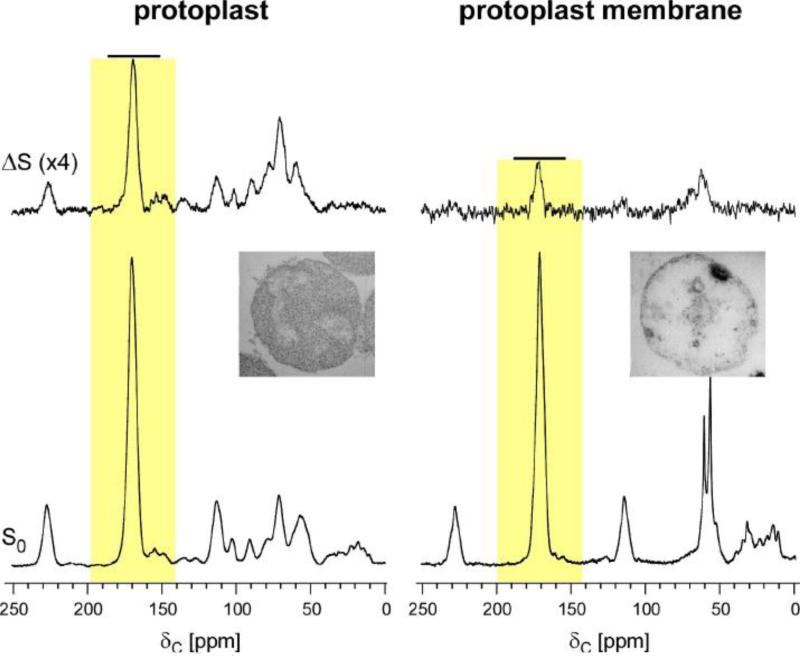
Solid-state NMR was used to investigate the orientation of PG chains in S. aureus to determine whether the PG chains are organized as scaffold or layered model in Gram-positive bacteria. The orientation of nascent PG in S. aureus protoplast was determined to align along the membrane, which is consistent with the layered model [33, 43]. This was determined by using 13C-labeled protoplasts prepared by lysostaphin digestion of whole-cells S. aureus grown in defined media with [1-13C]Gly. Lysostaphin is a glycylglycine endopeptidase which selectively cleaves the cross-linked pentaglycine bridge of the PG [44]. S. aureus in 1M sucrose was converted to protoplast within 15 min of adding lysostaphin (75 μg/ml) as monitored by turbidity assay (Figure 11, left inset). Lysostaphin rapidly cleaved the PG in mature CW, leaving the membrane-bound nascent PG. The isolated proptoplast membrane was prepared through osmotic rupture. 13C{31P} REDOR NMR was then used to examine the association of nascent PG to the protoplast membrane by measuring 13C-31P dipolar contact between the 13C of [1-13C]Gly in pentaglycyl bridge from nascent PG to the 31P in phospholipid bilayer of the membrane. Figure 11 shows the 13C{31P} REDOR spectra after 8.96 ms dipolar evolution from intact protoplast (left) and isolated protoplast membrane (right). The full-echo (S0) spectra (bottom) were normalized to equal 171-ppm peak intensity of the glycyl-carbonyl carbon from the labeled [1-13C]Gly. The 13C{31P} dephasing of the 171-ppm peak is 12% for protoplasts and only 4% for isolated protoplast membranes (Figure 11, yellow boxes). This reduction of 171-ppm peak intensity in the difference spectrum (ΔS) of the isolated protoplast membrane indicates that during the osmotic rupturing nascent-peptidoglycan loses its association with the bacterial cytoplasmic membrane. In other words, the nascent PG in protoplasts is organized in close association with the bacterial cytoplasmic membrane.
Figure 11.
Top) 13C{31P} REDOR difference (ΔS) spectra after a dipolar evolution time of 8.96 ms of intact protoplasts (left), and isolated protoplast membranes (right). The S. aureus cell wall was labeled with [1-13C]glycine. Bottom) The S0 13C{31P} REDOR spectra are shown. The S0 spectra were normalized for equal intensity of the 171-ppm peak. The 13C{31P} dephasing of the 171-ppm peak is 12% for protoplasts and only 4% for isolated protoplast membranes (yellow boxes). This indicates that the nascent PG in protoplast is organized close to the bacterial membrane, while such an organization is lost in isolated protoplast membrane. Inset) Transmission electron micrographs of cross sections of S. aureus protoplast prepared by treatment with 75 μg/mL lysostaphin (inset left), and isolated protoplast membrane (inset right). The Figure is modified from the reference [33, 43].
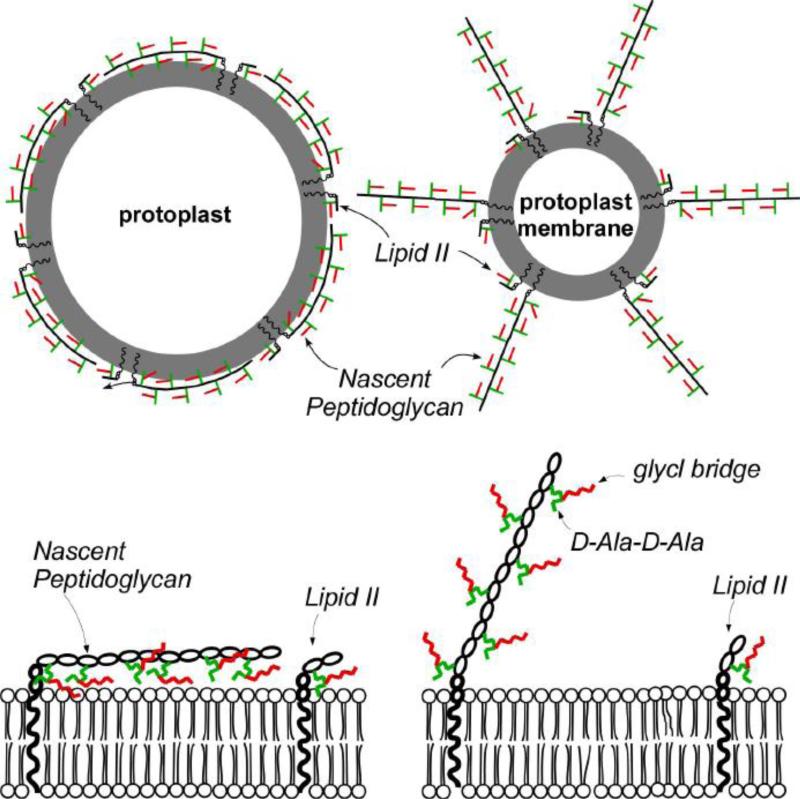
Possible nascent-peptidoglycan organization in protoplasts (left) and isolated protoplast membranes (right) consistent with the 13C{31P} REDOR results are illustrated in Figure 12 (top). In the figure, lipid bilayer is represented as gray circle, disaccharide in black line, PG stem in green line, and [1-13C]Gly in pentaglycine bridge in red line. Equal numbers of PG units (and lipid II) are depicted in both protoplasts and isolated protoplast membranes. In this arrangement all the [1-13C]Gly in the bridges of protoplasts contribute to the 13C{31P} REDOR dephasing. On the other hand, for isolated membranes of protoplast only the ends of the nascent peptidoglycan strand close to the membrane are 13C-31P dipolar coupled and contribute to the 171-ppm peak intensity in the ΔS spectrum.
Figure 12.
(Top) The gray circle represents the lipid bilayer of the cytoplasmic membrane. The black line represents the disaccharide of a peptidoglycan repeat unit, the green line represents the stem structure, and the red line represents the bridge structure. Equal quantities of lipid II and nascent peptidoglycan are shown in both intact protoplasts and isolated protoplast membranes. The nascent peptidoglycan strands are parallel with the membrane surface in intact protoplasts and perpendicular in isolated protoplast membranes. (Bottom) Close-up views of the nascent peptidoglycan organization are shown in the bottom panels of the figure. Color coding of the peptidoglycan units is the same in all panels. The arrow indicates the reduced of 13C-31P dipolar coupling through loss of PG-membrane contact. The Figure is modified from the reference [33, 43].
The 13C{31P} REDOR NMR result is consistent with the nascent PG organized in layer model. The 13C{31P} dephasing of the 171-ppm peak of 12% for protoplasts at 8.96 ms indicates that the majority of 13C are in close proximity to the phosphate 31P of the membrane, contrary to the extension of nascent PG perpendicular to the membrane as described in scaffold model. Furthermore, if the nascent PG were to adopt a scaffold model, then the 13C{31P} REDOR dephasing of 171-ppm peak will be constant independent of the membrane rupture.
7. The effects of PG structure on glycopeptide antibiotics mode of action
7.1 Glycopeptide antibiotic binding to PG
The pore size of pseudo-tessera in S. aureus provides insight into the glycopeptide antibiotic's mode of action. The disaccharide-modified glycopeptides exhibit strong dimerization and are found as dimers in solution [45, 46]. However, when six dimer-forming disaccharide-modified glycopeptides were added to the whole cells of S. aureus, all glycopeptides were found complexed to PG as monomers [23, 34, 47, 48] (Figure 6, right bottom). The result clearly indicated that in situ glycopeptide binding to PG prevents the dimer formation. The only possible explanation is that proposed PG-lattice structure in S. aureus is too small to accommodate the dimerized disaccharide-modified glycopeptides with bulky side chains. This size exclusion provides an insight into thus far unexplained reduced antimicrobial activity of covalently linked glycopeptide dimers against S. aureus [49], which may result from the bulky covalently linked glycopeptide dimer's inability to penetrate through the cell wall to reach the bacterial membrane.
Highly organized and tightly packed PG structure in S. aureus is also consistent with the proposed mechanism of vancomycin resistance in Mu50. Mu50 is a clinical isolate of methicillin-resistant S. aureus exhibiting vancomycin-intermediate resistance through increased concentration of uncross-linked PG stems. Increase in excess vancomycin-binding site at the outer layer of the CW of Mu50 is thought to facilitate the clogging of the PG pores when vancomycin is bound. This clogging of PG pores prevents further drug penetration from reaching the lipid II at the bacterial membrane [50]. This mechanism is possible only if the size of vancomycin is comparable to the diameter of PG pores. The diameter of vancomycin is approximately 20 Å, which is close to the diameter (23 Å) of the proposed pseudo-tessera structure in S. aureus.
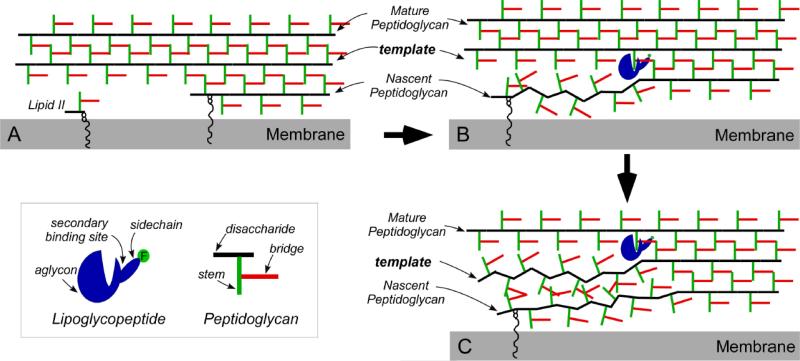
7.2 Dual mode of action of oritavancin against S. aureus
The mode of action of oritavancin at sub-inhibitory concentrations against intact whole cells of S. aureus has been investigated by measuring the change in bridge-link and cross-link densities using solid-state NMR. Oritavancin exhibits dual mode of action by inhibiting both transglycosylase and transpeptidase activities [34, 51] whereas vancomycin only inhibits transglycosylation [52]. Oritavancin's dual mode of action is attributed to its unique ability to target both lipid II (which results in transglycosylase inhibition), and nascent PG (transpeptidase inhibition). Oritavancin binding to nascent PG interferes with the PG template recognition by decoupling the transglycosylase and transpeptidase activities and thereby destroying the template (Figure 13). The subsequently formed PG without template has defects with the reduction in cross-liking, making bacteria susceptible to lyse. In order for oritavancin binding to have an effect on the template copying mechanism, the dimension of PG lattice separation must be close to that of oritavancin, which is consistent with the proposed parallel-stem architecture.
Figure 13.
A) Schematic representation of the effect on cell-wall biosynthesis. B) Lipoglycopeptide binding to the mature PG template. C) PG-template binding results in reductions in reduced cross-linking and the destruction of PG template. The figure is from reference [47].
8. Future Direction
Recently, there have been several exciting developments where solid-state NMR methods have been used to investigate cell wall and extracellular matrices compositions and dynamics. Various ribitol phosphates found in Gram-positive bacteria cell wall has been characterized by 31P NMR on hydrated cell wall [53]. The study provided a novel method for characterizing various types of wall teichoic acids found in Gram-positive bacteria. Wall teichoic acid is an anionic-polysacchairde polymer found on the cell surface of Gram-positive bacteria and it's one of the major components of cell wall. The use of dynamic nuclear polarization (DNP) solid-state NMR on intact whole-cells of B. subtilis has been reported by Takahashi et al. [54]. DNP selectively enhanced the PG-signal by a factor of 24, compared to conventional solid-state NMR, with a time saving factor of 600 [54]. Another significant approach is combining high-resolution solid-state NMR methods with isotope-labeling scheme to provide in situ structural information on an integral membrane protein in Gram-negative bacterial cell envelope [55]. These approaches highlight recent advancements in solid-state NMR as a tool for investigating the structures and compositions of bacterial cell wall which remains a formidable challenge by any other methods.
9. Conclusion
Solid-state NMR analyses of S. aureus cell wall provided unique structural and compositional insights into the PG architecture. The only PG disaccharide backbone conformation possible in S. aureus is 4-fold screw helical symmetry with the disaccharide unit periodicity of 40 Å, where each PG stems are rotated 90° respect to the previous stem. The cross-linked PG stems have parallel orientation, as the antiparallel-stem model is not consistent with constraints dictated by measured PG cross-linking density, CODEX-detected 13C-13C spin-diffusion, and REDOR. PG glycan chains in S. aureus are surprisingly ordered and densely packed without tessera structure. The characterization of Fem-mutants of S. aureus with varying lengths of bridge structures suggests that the PG-bridge length is an important determining factor for the PG architecture.
Highlights.
S. aureus peptidoglycan disaccharide adopts 4-fold screw helical symmetry.
The disaccharide unit has a periodicity of 40 Å.
The cross-linked peptidoglycan stems have parallel orientation.
Peptidoglycan bridge length is a determining factor for the architecture.
Acknowledgement
This paper is based on work supported by the National Institutes of Health under grant number EB002058. The authors thank Dr. Jacob Schaefer at Washington University in St. Louis for helpful discussions.
Abbreviations
- CW
cell wall
- 13C{15N}
observe carbon channel and dephase using nitrogen channel
- 15N{13C}
observe nitrogen channel and dephase using carbon channel
- 13C{19F}
observe carbon channel and dephase using fluorine channel
- 13C{31P}
observe carbon channel and dephase using phosphorus channel
- 15N{19F}
observe nitrogen channel and dephase using fluorine channel
- PG
peptidoglycan
- DNP
dynamic nuclear polarization
- REDOR
rotational-echo double resonance
- lipid II
N-acetylglucosamine-N-acetyl-muramyl-pentapeptide-pyrophosphoryl-undecaprenol
- LCTA-1110
N-4-(4-chlorophenylbenzyl)chloroeremomycin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rogers HJ, Ward JB, Perkins HR. Microbial cell walls and membranes. Chapman and Hall; London ; New York: 1980. [Google Scholar]
- 2.Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol. Mol. Biol. Rev. 1998;62:1371–1414. doi: 10.1128/mmbr.62.4.1371-1414.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong G, Pan Y, Dong H, Pryor R, Wilson GE, Schaefer J. Structure and dynamics of pentaglycyl bridges in the cell walls of Staphylococcus aureus by 13C-15N REDOR NMR. Biochemistry. 1997;36:9859–9866. doi: 10.1021/bi970495d. [DOI] [PubMed] [Google Scholar]
- 5.Bellais S, Arthur M, Dubost L, Hugonnet JE, Gutmann L, van Heijenoort J, Legrand R, Brouard JP, Rice L, Mainardi JL. Aslfm, the D-aspartate ligase responsible for the addition of D-aspartic acid onto the peptidoglycan precursor of Enterococcus faecium. J. Biol. Chem. 2006;281:11586–11594. doi: 10.1074/jbc.M600114200. [DOI] [PubMed] [Google Scholar]
- 6.Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annu. Rev. Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 7.Boneca IG, Huang ZH, Gage DA, Tomasz A. Characterization of Staphylococcus aureus cell wall glycan strands, evidence for a new beta-N-acetylglucosaminidase activity. J. Biol. Chem. 2000;275:9910–9918. doi: 10.1074/jbc.275.14.9910. [DOI] [PubMed] [Google Scholar]
- 8.Glauner B, Holtje JV. Growth pattern of the murein sacculus of Escherichia coli. J. Biol. Chem. 1990;265:18988–18996. [PubMed] [Google Scholar]
- 9.Hayhurst EJ, Kailas L, Hobbs JK, Foster SJ. Cell wall peptidoglycan architecture in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2008;105:14603–14608. doi: 10.1073/pnas.0804138105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gally D, Archibald AR. Cell wall assembly in Staphylococcus aureus: proposed absence of secondary crosslinking reactions. J. Gen. Microbiol. 1993;139:1907–1913. doi: 10.1099/00221287-139-8-1907. [DOI] [PubMed] [Google Scholar]
- 11.Vollmer W, Seligman SJ. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 2010;18:59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore MS. The enterococci: pathogenesis, molecular biology, and antibiotic resistance. ASM Press; Washington, D.C.: 2002. [Google Scholar]
- 13.Beeby M, Gumbart JC, Roux B, Jensen GJ. Architecture and assembly of the Gram-positive cell wall. Mol. Microbiol. 2013;88:664–672. doi: 10.1111/mmi.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc. Natl. Acad. Sci. USA. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tocheva EI, Lopez-Garrido J, Hughes HV, Fredlund J, Kuru E, Vannieuwenhze MS, Brun YV, Pogliano K, Jensen GJ. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol. Microbiol. 2013 doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turner RD, Hurd AF, Cadby A, Hobbs JK, Foster SJ. Cell wall elongation mode in Gram-negative bacteria is determined by peptidoglycan architecture. Nat. Commun. 2013;4:1496. doi: 10.1038/ncomms2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner RD, Ratcliffe EC, Wheeler R, Golestanian R, Hobbs JK, Foster SJ. Peptidoglycan architecture can specify division planes in Staphylococcus aureus. Nat. Commun. 2010;1:26. doi: 10.1038/ncomms1025. [DOI] [PubMed] [Google Scholar]
- 18.Turner RD, Vollmer W, Foster SJ. Different walls for rods and balls: the diversity of peptidoglycan. Mol. Microbiol. 2014 doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vollmer W, Holtje JV. The architecture of the murein (peptidoglycan) in gram-negative bacteria: vertical scaffold or horizontal layer(s)? J. Bacteriol. 2004;186:5978–5987. doi: 10.1128/JB.186.18.5978-5987.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao X, Jericho M, Pink D, Beveridge T. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol. 1999;181:6865–6875. doi: 10.1128/jb.181.22.6865-6875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dmitriev BA, Toukach FV, Schaper KJ, Holst O, Rietschel ET, Ehlers S. Tertiary structure of bacterial murein: the scaffold model. J. Bacteriol. 2003;185:3458–3468. doi: 10.1128/JB.185.11.3458-3468.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beveridge TJ. Ultrastructure, chemistry, and function of the bacterial wall. Int. Rev. Cytol. 1981;72:229–317. doi: 10.1016/s0074-7696(08)61198-5. [DOI] [PubMed] [Google Scholar]
- 23.Kim SJ, Cegelski L, Studelska DR, O'Connor RD, Mehta AK, Schaefer J. Rotational-echo double resonance characterization of vancomycin binding sites in Staphylococcus aureus. Biochemistry. 2002;41:6967–6977. doi: 10.1021/bi0121407. [DOI] [PubMed] [Google Scholar]
- 24.Kelemen MV, Rogers HJ. Three-dimensional molecular models of bacterial cell wall mucopeptides (peptidoglycans) Proc. Natl. Acad. Sci. USA. 1971;68:992–996. doi: 10.1073/pnas.68.5.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun V, Gnirke H, Henning U, Rehn K. Model for the structure of the shape-maintaining layer of the Escherichia coli cell envelope. J. Bacteriol. 1973;114:1264–1270. doi: 10.1128/jb.114.3.1264-1270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burge RE, Fowler AG, Reaveley DA. Structure of the peptidogylcan of bacterial cell walls. I. J. Mol. Biol. 1977;117:927–953. doi: 10.1016/s0022-2836(77)80006-5. [DOI] [PubMed] [Google Scholar]
- 27.Labischinski H, Barnickel G, Bradaczek H, Giesbrecht P. On the secondary and tertiary structure of murein. Low and medium-angle X-ray evidence against chitin-based conformations of bacterial peptidoglycan. Eur. J. Biochem. 1979;95:147–155. doi: 10.1111/j.1432-1033.1979.tb12949.x. [DOI] [PubMed] [Google Scholar]
- 28.Meroueh SO, Bencze KZ, Hesek D, Lee M, Fisher JF, Stemmler TL, Mobashery S. Three-dimensional structure of the bacterial cell wall peptidoglycan. Proc. Natl. Acad. Sci. USA. 2006;103:4404–4409. doi: 10.1073/pnas.0510182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharif S, Singh M, Kim SJ, Schaefer J. Staphylococcus aureus peptidoglycan tertiary structure from carbon-13 spin diffusion. J. Am. Chem. Soc. 2009;131:7023–7030. doi: 10.1021/ja808971c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharif S, Kim SJ, Labischinski H, Schaefer J. Characterization of peptidoglycan in fem-deletion mutants of methicillin-resistant Staphylococcus aureus by solid-state NMR. Biochemistry. 2009;48:3100–3108. doi: 10.1021/bi801750u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gullion T, Schaefer J. Detection of weak heteronuclear dipolar coupling by rotational-echo double-resonance nuclear magnetic resonance. Adv. Magn. Reson. 1989;13:57–83. [Google Scholar]
- 32.Gullion T, Schaefer J. Rotational-echo double-resonance NMR. J. Magn. Reson. 1989;81:196–200. doi: 10.1016/j.jmr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Kim SJ, Preobrazhenskaya M, Schaefer J. Characterization of Staphylococcus aureus peptidoglycan tertiary structure using glycopeptides as probe by solid-state NMR. Interscience Conference on Antimicrobial Agents and Chemotherapy Washington DC. 2008:F2–2073. [Google Scholar]
- 34.Kim SJ, Cegelski L, Preobrazhenskaya M, Schaefer J. Structures of Staphylococcus aureus cell-wall complexes with vancomycin, eremomycin, and chloroeremomycin derivatives by 13C{19F} and 15N{19F} rotational-echo double resonance. Biochemistry. 2006;45:5235–5250. doi: 10.1021/bi052660s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim SJ, Singh M, Preobrazhenskaya M, Schaefer J. Staphylococcus aureus peptidoglycan stem packing by rotational-echo double resonance NMR spectroscopy. Biochemistry. 2013;52:3651–3659. doi: 10.1021/bi4005039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cegelski L, Steuber D, Mehta AK, Kulp DW, Axelsen PH, Schaefer J. Conformational and quantitative characterization of oritavancin-peptidoglycan complexes in whole cells of Staphylococcus aureus by in vivo 13C and 15N labeling. J. Mol. Biol. 2006;357:1253–1262. doi: 10.1016/j.jmb.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 37.Koch AL. The three-for-one model for gram-negative wall growth: a problem and a possible solution. FEMS Microbiol. Lett. 1998;162:127–134. doi: 10.1111/j.1574-6968.1998.tb12989.x. [DOI] [PubMed] [Google Scholar]
- 38.Patti GJ, Kim SJ, Schaefer J. Characterization of the peptidoglycan of vancomycin-susceptible Enterococcus faecium. Biochemistry. 2008;47:8378–8385. doi: 10.1021/bi8008032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patti GJ, Kim SJ, Yu TY, Dietrich E, Tanaka KS, Parr TR, Jr., Far AR, Schaefer J. Vancomycin and oritavancin have different modes of action in Enterococcus faecium. J. Mol. Biol. 2009;392:1178–1191. doi: 10.1016/j.jmb.2009.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharif S, Kim SJ, Labischinski H, Chen J, Schaefer J. Uniformity of glycyl bridge lengths in the mature cell walls of Fem mutants of methicillin-resistant Staphylococcus aureus. J. Bacteriol. 2013;195:1421–1427. doi: 10.1128/JB.01471-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim SJ, Singh M, Sharif S, Schaefer J. Cross-link formation and peptidoglycan lattice assembly in the FemA mutant of Staphylococcus aureus. Biochemistry. 2014;53:1420–1427. doi: 10.1021/bi4016742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim SJ, Singh M, Schaefer J. Oritavancin binds to isolated protoplast membranes but not intact protoplasts of Staphylococcus aureus. J. Mol. Biol. 2009;391:414–425. doi: 10.1016/j.jmb.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grundling A, Schneewind O. Cross-linked peptidoglycan mediates lysostaphin binding to the cell wall envelope of Staphylococcus aureus. J. Bacteriol. 2006;188:2463–2472. doi: 10.1128/JB.188.7.2463-2472.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen NE, LeTourneau DL, Hobbs JN., Jr. The role of hydrophobic side chains as determinants of antibacterial activity of semisynthetic glycopeptide antibiotics. J. Antibiot. 1997;50:677–684. doi: 10.7164/antibiotics.50.677. [DOI] [PubMed] [Google Scholar]
- 46.LeTourneau DL, Allen NE. Use of capillary electrophoresis to measure dimerization of glycopeptide antibiotics. Analytical Biochemistry. 1997;246:62–66. doi: 10.1006/abio.1997.2004. [DOI] [PubMed] [Google Scholar]
- 47.Kim SJ, Schaefer J. Hydrophobic side-chain length determines activity and conformational heterogeneity of a vancomycin derivative bound to the cell wall of Staphylococcus aureus. Biochemistry. 2008;47:10155–10161. doi: 10.1021/bi800838c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SJ, Tanaka KS, Dietrich E, Rafai Far A, Schaefer J. Locations of the hydrophobic side chains of lipoglycopeptides bound to the peptidoglycan of Staphylococcus aureus. Biochemistry. 2013;52:3405–3414. doi: 10.1021/bi400054p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicolaou KC, Hughes R, Cho SY, Winssinger N, Labischinski H, Endermann R. Synthesis and biological evaluation of vancomycin dimers with potent activity against vancomycin-resistant bacteria: target-accelerated combinatorial synthesis. Chemistry. 2001;7:3824–3843. doi: 10.1002/1521-3765(20010903)7:17<3824::aid-chem3824>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 50.Cui L, Murakami H, Kuwahara-Arai K, Hanaki H, Hiramatsu K. Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. Antimicrob. Agents Chemother. 2000;44:2276–2285. doi: 10.1128/aac.44.9.2276-2285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim SJ, Matsuoka S, Patti GJ, Schaefer J. Vancomycin derivative with damaged D-Ala-D-Ala binding cleft binds to cross-linked peptidoglycan in the cell wall of Staphylococcus aureus. Biochemistry. 2008;47:3822–3831. doi: 10.1021/bi702232a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cegelski L, Kim SJ, Hing AW, Studelska DR, O'Connor RD, Mehta AK, Schaefer J. Rotational-echo double resonance characterization of the effects of vancomycin on cell wall synthesis in Staphylococcus aureus. Biochemistry. 2002;41:13053–13058. doi: 10.1021/bi0202326. [DOI] [PubMed] [Google Scholar]
- 53.Kern T, Giffard M, Hediger S, Amoroso A, Giustini C, Bui NK, Joris B, Bougault C, Vollmer W, Simorre JP. Dynamics characterization of fully hydrated bacterial cell walls by solid-state NMR: evidence for cooperative binding of metal ions. J. Am. Chem. Soc. 2010;132:10911–10919. doi: 10.1021/ja104533w. [DOI] [PubMed] [Google Scholar]
- 54.Takahashi H, Ayala I, Bardet M, De Paepe G, Simorre JP, Hediger S. Solid-State NMR on bacterial cells: Selective cell wall signal enhancement and resolution improvement using dynamic nuclear polarization. J. Am. Chem. Soc. 2013;135:5105–5110. doi: 10.1021/ja312501d. [DOI] [PubMed] [Google Scholar]
- 55.Renault M, Tommassen-van Boxtel R, Bos MP, Post JA, Tommassen J, Baldus M. Cellular solid-state nuclear magnetic resonance spectroscopy. Proc. Natl. Acad. Sci. USA. 2014;109:4863–4868. doi: 10.1073/pnas.1116478109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patti GJ, Chen J, Schaefer J, Gross ML. Characterization of structural variations in the peptidoglycan of vancomycin-susceptible Enterococcus faecium: understanding glycopeptide-antibiotic binding sites using mass spectrometry. J. Am. Soc. Mass Spectrom. 2008;19:1467–1475. doi: 10.1016/j.jasms.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]