Abstract
Class-switch DNA recombination (CSR) and somatic hypermutation (SHM), which require AID, and plasma cell differentiation, which requires Blimp-1, are critical for the generation of class-switched and hypermutated (mature) antibody and autoantibody responses. We showed here that the histone deacetylase (HDAC) inhibitors (HDI) valproic acid (VPA) and butyrate upregulated miR-155, miR-181b and miR-361, which silenced AICDA/Aicda (AID) mRNA, and miR-23b, miR-30a and miR-125b, which silenced PRDM1/Prdm1 (Blimp-1) mRNA, in human and mouse B cells. This led to downregulation of AID, Blimp-1 and Xbp-1 expression, thereby dampening CSR, SHM and plasma cell differentiation without altering B cell viability or proliferation. The selectivity of HDI-mediated silencing of AICDA/Aicda and PRDM1/Prdm1 was emphasized by unchanged expression of HoxC4 and Irf4 (important inducers/modulators of AICDA/Aicda), Rev1 and Ung (central elements for CSR/SHM), and Bcl6, Bach2 or Pax5 (repressors of PRDM1/Prdm1 expression), as well as unchanged expression of miR-19a/b, miR-20a and miR-25, which are not known to regulate AICDA/Aicda or PRDM1/Prdm1. Through these B cell intrinsic epigenetic mechanisms, VPA blunted class-switched and hypermutated T-dependent and T-independent antibody responses in C57BL/6 mice. In addition, it decreased class-switched and hypermutated autoantibodies, ameliorated disease and extended survival in lupus MRL/Faslpr/lpr mice. Our findings outline epigenetic mechanisms that modulate expression of an enzyme (AID) and transcription factors (Blimp-1 and Xbp-1) that critical to the B cell differentiation processes that underpin antibody and autoantibody responses. They also provide therapeutics proof-of-principle in autoantibody-mediated autoimmunity.
Introduction
Ig class switch DNA recombination (CSR) and somatic hypermutation (SHM) are critical for the production of protective antibodies against microbial pathogens, IgE in allergic responses as well as pathogenic autoantibodies in autoimmune diseases. CSR recombines S region DNA located upstream of exons of constant heavy-chain (CH) regions, thereby encoding new Ig CH regions that endow antibodies with new biological effector functions (1). SHM introduces mainly point-mutations in Ig variable regions, thereby providing the structural substrate for antigen-mediated selection of B cell mutants with higher affinity BCRs (2, 3). CSR and SHM require activation-induced cytidine deaminase (AID, encoded by Aicda in mice and AICDA in humans), which is expressed in a B cell differentiation stage-specific fashion (1, 2). Class-switched and hypermutated B cells further differentiate into antibody-secreting plasma cells in a fashion critically dependent on B lymphocyte-induced maturation protein 1 (Blimp-1, encoded by Prdm1 in mice and PRDM1 in humans) (4).
As a potent DNA mutator, AID must be tightly regulated in order to prevent off-target effects, which may result in mutations in non-Ig genes, genomic instability and interchromosomal translocations (5, 6). Accordingly, AID is expressed in B cells and in a B cell differentiation stage-specific fashion. This is made possible by stringent transcriptional, post-transcriptional and post-translational control (2). In response to T-dependent and T-independent “primary” stimuli (e.g., CD154, LPS, BAFF and APRIL) (7-10), AID expression is induced by multiple transcription factors including HoxC4, NF-κB, Pax5, Irf4, Irf8, Oct1/Oct2, Sp1/Sp3 and E47 (2, 11-14). AID expression is further upregulated by secondary stimuli (e.g., IL-4, TGF-β, IFN-γ), which induce selected IH-S-CH germline transcription, thereby directing CSR to specific isotypes (1, 15). It is then downregulated in memory B cells and plasma cells to preserve the specificity, affinity and isotype of the expressed BCR and antibody (1, 2). Terminal plasma cell differentiation is critically dependent on the transcriptional repressor Blimp-1. This extinguishes the proliferative mature B cell gene expression program and drives the expression of the X-box binding protein 1 (Xbp-1), which induces secretory pathway genes for Ig secretion (4, 16, 17).
As we have contended, an additional and critical level of regulation of AID expression would occur through epigenetic modifications and factors (2, 3, 18). As we have also contended, epigenetic modifications and factors, including histone posttranslational modifications, DNA methylation and microRNAs (miRNAs), interact with genetic programs to regulate B cell CSR, SHM and plasma cell differentiation, thereby informing the antibody response (3). Accordingly, we have suggested that, in addition to DNA methylation of the Aicda promoter (19) and histone acetylation of the Aicda locus (20), selected miRNAs provide a more important mechanism of modulation of AID expression (2, 3, 18). miRNAs likely play important roles in B cell development and peripheral differentiation, as well as T cell stage-specific differentiation and autoimmunity (18, 21-26). Some miRNAs, including miR-155, miR-181b and miR-361, would negatively regulate AID expression (2, 27-30), whereas miR-30a (31) and miR-125b (32, 33) would negatively regulate Blimp-1 expression. These miRNAs bind to evolutionarily conserved miRNA target sites in the 3’UTR of Aicda and Prdm1 mRNAs and cause degradation of the mRNA transcripts and/or inhibit their translation.
Histone deacetylase (HDAC) inhibitors (HDI) have been shown to alter gene expression by altering chromatin accessibility (34-37). In immune cells, these epigenetic modifiers exert modulatory effects even at moderate concentrations. By using two well-characterized short-chain fatty acid (SCFA) HDI, valproic acid or sodium valproate (VPA, 2-propyl-pentanoic acid sodium) (38) and sodium butyrate (butyrate) (39), we tested the hypothesis that HDI regulate intrinsic B cell functions that are critical in shaping effective antibody and autoantibody responses. VPA is an FDA-approved drug, which, as marketed under different brand names, is widely used as an anticonvulsant and a mood-stabilizer. It selectively inhibits class I HDACs, particularly, HDAC1 and HDAC2, and, less effectively, class IIa HDACs, of the four HDAC classes identified in mammals (38, 40). Butyrate is a major metabolite in the digestive tract, arising from bacterial fermentation of dietary fibers (41, 42), and it is widely available as a dietary supplement. Butyrate modulates gene expression by selectively inhibiting HDAC1, and, less effectively, other members of class I and class IIa HDACs (39).
We addressed the ability of VPA and butyrate to modulate AID and Blimp-1 expression, CSR, SHM and plasma cell differentiation, in human and mouse B cells in vivo and in vitro. In addition, we analyzed the role of HDI as epigenetic modifiers of selected B cell miRNAs that silence Aicda and Prdm1, in antibody and autoantibody responses. Finally, we evaluated the impact of VPA on NP-CGG and NP-LPS class-switched and hypermutated antibody responses in normal mice, and on the autoantibody response in lupus-prone MRL/Faslpr/lpr mice. Our findings outline important modalities of epigenetic regulation of AID and Blimp-1 expression, unveil new approaches to modulation of T-dependent and T-independent antibody responses, and provide a proof-of-principle therapeutics study in autoantibody-mediated autoimmunity.
Materials and Methods
HDI
For in vivo studies, valproic acid sodium salt (Sigma-Aldrich) was dissolved in drinking water at 0.8% w/v. This yields a stable VPA serum level (400 – 600 μM) in mice, comparable to the serum concentration in humans under long-term VPA treatment (300 – 900 μM) (43, 44). Drinking water containing VPA at the above concentration (HDI-water) was always well accepted by C57BL/6 and MRL/Faslpr/lpr mice. VPA and butyrate (obtained as sodium butyrate from Sigma-Aldrich) were directly diluted in culture media for in vitro experiments, at molarities ranging from 125 μM to 1,000 μM. Trichostatin A (TSA) (Cayman Chemical) was dissolved in DMSO and diluted in culture media at 10 or 20 nM (~3 or 6 ng/ml) for in vitro experiments.
Modulation of T-dependent and T-independent antibody responses by HDI
C57BL/6 mice (Jackson Laboratory) were housed in a pathogen-free vivarium and provided with autoclaved food and deionized water. To study the T-dependent antibody response, 16 eight-week-old C57BL/6 mice received an i.p. injection of 100 μg NP16-CGG (Biosearch Technologies) in alum (Imject® alum, Pierce). Eight of the 16 mice were on HDI-water ad libitum, starting the day before and continuing throughout the time length of the experiment. The remaining 8 mice were on untreated drinking water. Five mice drinking HDI-water and 5 drinking untreated water were sacrificed at day 10, the remaining 6 mice were given a ‘booster’ i.p. injection of 100 µg NP16-CGG (Biosearch Technologies) in alum at day 21, and sacrificed at day 28. To study the T-independent antibody response, 8 eight-week-old C57BL/6 mice received an i.p. injection of 25 μg NP–LPS (Biosearch Technologies) in PBS. Four of these mice were on HDI-water and 4 untreated water throughout the time length of the experiment. All 8 mice were given a ‘booster’ injection of 25 μg of NP-LPS at day 21, and then sacrificed at day 28. The Institutional Animal Care and Use Committees of University of California, Irvine and The University of Texas Health Science Center, San Antonio approved all animal protocols.
Mouse B cells, CSR and plasma cell differentiation
Naïve IgD+ B cells were isolated from 8-week-old C57BL/6 as described (9). B cells were resuspended in RPMI 1640 medium with 10% FBS, 50 mM β-mercaptoethanol and 1x antibiotic-antimycotic mixture (15240-062; Invitrogen) (FBS-RPMI) at 37°C in 48-well plates and stimulated with: LPS (5 μg/ml) from Escherichia coli (055:B5; Sigma-Aldrich) for CSR to IgG3; LPS (3 μg/ml) or CD154 (1 U/ml; obtained from membrane fragments of baculovirus-infected Sf21 insect cells (45)) plus IL-4 (5 ng/ml; R&D Systems) for CSR to IgG1/IgE and plasma cell differentiation; LPS (3 μg/ml) or CD154 (1 U/ml) plus TGF-β (2 ng/ml; R&D Systems), IL-4 (5 ng/ml), IL-5 (3 ng/ml; R&D Systems) and anti-IgD dextran-Ab (Fina Biosolutions) for CSR to IgA. Nil, VPA (125-1,000 μM; doses that were similar to or below serum concentrations of VPA-treated mice (46)) or butyrate (125-1,000 μM) was then added to cultures and cells or supernatants were collected at various times.
Human B cells, CSR and plasma cell differentiation
Human naïve IgD+ B cells (over 95% pure) were purified by negative selection using the EasySepTM human naive B cell enrichment kit (19254; StemCell Technologies) from PBMCs, following manufacturer’s instructions. Naïve B cells were then cultured in FBS-RPMI and stimulate with CD154 (10 U/ml), IL-4 (20 ng/ml; R&D Systems) and IL-21 (50 ng/ml; R&D Systems) or CD154 (10 U/ml), IL-21 (50 ng/ml) and TGF-β (0.5 ng/ml) in the presence of nil, VPA or butyrate for 120 h. B cells were then stained with 7-AAD, FITC-anti-IgM mAb (F5384; Sigma-Aldrich), PE-anti-CD19 mAb (HIB19; BioLegend) and allophycocyanin (APC)-anti-IgG mAb (G18-145; BD Biosciences) or 7-AAD, FITC-anti-IgA mAb (F5259; Sigma-Aldrich), PE-anti-CD19 mAb (HIB19; BioLegend) and biotin-F(ab’)2 anti-IgM (2022-08; Southern Biotech), followed by APC-streptavidin and analyzed by flow cytometry. For plasma cell differentiation, cells were stained with 7-aminoactinomycin D (7-AAD; BD Biosciences), APC-anti-CD27 (LG.3A10; BioLegend) and FITC-anti-CD38 (31014X; BD Biosciences) and analyzed by flow cytometry.
Flow cytometry
For surface staining, B cells were reacted with PE-anti-B220 mAb (CD45R; RA3-6B2; eBioscience), Alexa Fluor® 647-peanut agglutinin (PNA; Invitrogen), PE-anti-IgM mAb (AF6-78; BD Biosciences), FITC-anti-IgG1 mAb (A85-1; BD Biosciences), FITC-anti-IgG3 mAb (R40-82; BD Biosciences), FITC-anti-IgA mAb (C10-3; BD Biosciences), 7-AAD, biotin-anti-CD138 mAb (281-2; BD Biosciences) followed by FITC-streptavidin (11-4317-87; eBioscience) or PE-streptavidin (12-4317-87; eBioscience), FITC-anti-CD3 mAb (17A2; BioLegend), FITC-anti-CD4 (GK1.5; BioLegend) and/or APC-anti-CD8 mAb (53-6.7; BD Biosciences). For intracellular staining, B cells were fixed in 150 μl of formaldehyde (3.6%) for 10 min at 25°C. In the case of IgE intracellular staining, cells were trypsinized and then fixed (47). Cells were then permeabilized in cold methanol (90%) for 30 min on ice before staining with biotin-anti-CD138 mAb followed by PE-streptavidin, PE-Cy7-anti-B220 mAb (RA3-6B2; eBioscience), FITC-anti-IgM mAb (II/41; BD Biosciences), APC-anti-IgG1 mAb (A85-1; BD Biosciences) and/or PE-anti-IgE mAb (23G3; eBioscience). FACS was performed on single spleen cell suspensions. Cells were surface stained with PE-anti-CD19 (1D3; BD Biosciences), 7-AAD and biotin-anti-CD138 followed by FITC-streptavidin. CD19+CD138− B cells (comprised a large proportion of GC B cells, referred to as “total” B cells) and CD19loCD138+ plasma cells were then sorted using a Cytomation MoFlo® cell sorter (Beckman Coulter) and frozen at −80°C until used for gene expression analysis. Annexin V analysis for apoptotic cells was performed using the Annexin V:FITC Apoptosis Detection Kit II (556570; BD Biosciences), according to the manufacturer’s protocol. In all flow cytometry experiments, cells were appropriately gated on forward scatter and side scatter to exclude dead cells and debris. Cell analyses were performed using a FACSCalibur™ flow cytometer (BD Biosciences); data were analyzed using FlowJo software (Tree Star). All experiments were performed in triplicate.
B cell proliferation, cell cycle and cell division
To analyze in vivo C57BL/6 B cell proliferation, mice previously i.p. injected with NP16-CGG, were i.p. injected 10 d later (twice within 16 h) with BrdU (1 mg), and sacrificed 4 h after the last injection. For in vivo MRL/Faslpr/lpr B cell proliferation, mice were provided with drinking water containing BrdU (0.8 mg/ml) for 10 d before sacrificing them. Spleen cells were stained with PE-anti-B220 mAb. Incorporated (intracellular) BrdU was stained with APC-anti-BrdU mAb using the APC BrdU Flow Kit (BD Biosciences) and analyzed by flow cytometry. B cell cycle was analyzed using the APC BrdU Flow Kit (BD Biosciences) according to manufacturer’s instructions. Briefly, spleen B cells were stimulated and cultured as previously described, except for that during the last 30 min of culture, the cells were pulse-labeled with 10 μM BrdU. The cells were then stained with PE-anti-B220 mAb before being fixed/permeabilized and stained for total DNA content with 7-AAD and BrdU incorporation with APC-anti-BrdU mAb. B cell division was analyzed by carboxyfluorescein succinimidyl ester (CFSE) dilution using the CellTrace™ CFSE Cell Proliferation Kit (Invitrogen). Briefly, B cells were incubated for 3 min at 37°C in 1 ml PBS with 2.5 μM CFSE at a density of 1 × 106 cells/ml and then washed in FBS-RPMI. Cells were then cultured in the presence LPS alone or LPS plus IL-4 for 4 d, before being stained with PE-anti-B220 mAb, 7-AAD and APC-anti-IgG1 mAb (X56; BD Biosciences) or biotin-anti-IgG3 mAb (R40-82; BD Biosciences) followed by APC-streptavidin (554067; BD Biosciences), and then analyzed by flow cytometry.
Secreted Ig and ELISPOT
Titers of IgG1, IgG3, IgA and IgE in cell culture supernatants of in vitro-stimulated B cells or in vivo titers of circulating total and/or NP-binding IgM, IgG1, IgG2b and IgG3 were measured using specific ELISAs, as we described (12, 14, 48). Titers of NP30- or NP3-binding IgM, IgG1 and IgG3 were also determined as previously described (12, 14, 48) and expressed as relative units (RU), defined as the dilution factor needed to reach 50% saturation binding, as calculated using GraphPad Prism® software (GraphPad Software). ELISAs for human Ig were performed as previously described (12, 14, 48), except that plates were coated with F(ab’)2 anti-Ig (H+L) Ab (2012-01; Southern Biotech) and Ig were detected by biotin-rabbit F(ab’)2 anti-IgG (2042-08; Southern Biotech) or biotin-rabbit F(ab’)2 anti-IgA (2052-08; Southern Biotech).
MultiScreen® ELISPOT plates (MAIPS4510; Millipore) were activated with ethanol (35%), washed four times with PBS and coated with 100 μl of NP3-BSA (5 μg/ml; N-5050L; Biosearch Technologies) in PBS overnight at 4°C. The plates were then washed six times with PBS, blocked with BSA (0.5%) in RPMI/HEPES+L-glutamine for 1 h at room temperature. Single cell suspensions from spleens of NP-LPS-injected mice were cultured in the plates at 37°C for 16 h in FBS-RPMI at concentrations of 250,000, 125,000 and 75,000 cells/ml. The cultures were then removed, the plates were washed six times, incubated with biotin-anti-IgG3 (R40-82; BD Biosciences) for 2 h on a shaker at room temperature, washed, incubated with horseradish peroxidase (HRP)-streptavidin (Santa Cruz Biotech) for 1 h on a shaker at room temperature, washed again and developed using the Vectastain AEC peroxidase substrate kit following manufacturer’s protocol (SK-4200; Vector Laboratories). Plates were imaged and quantified using a CTL-ImmunoSpot® Analyzer and software.
Fluorescence and confocal fluorescence microscopy
To analyze IgG1-producing cells, B cells stimulated with LPS plus IL-4 or LPS alone were immobilized on acid-washed cover slips coated with poly-D-lysine. Cells were then fixed with formaldehyde (3.6%) for 10 min at 25°C, washed three times with TBS, permeabilized with Triton X-100 (0.5%) in TBS for 10 min at 25°C, washed three times with Triton X-100 (0.1%) in TBS, and then stained with FITC-anti-IgG1 mAb and biotin-anti-CD138 mAb followed by APC-streptavidin or anti-Blimp-1 mAb (6D3; eBioscience), followed by Alexa Fluor 647®-anti-IgG mAb (4418; Cell Signaling Technology) for 1 h at 25°C. After washing three times with Triton X-100 (0.1%) in TBS, cover slips were mounted with ProLong® Gold Antifade Reagent using DAPI (Invitrogen). Fluorescent images were captured using a 40x objective lens with an Olympus FluoView FV1000 Laser Scanning Confocal microscope. To analyze GC structure, 8 μm spleen sections were prepared by cryostat and loaded onto positively charged slides, fixed in cold acetone and stained with FITC-PNA and PE-anti-B220 mAb for 1 h at 25°C in a moist chamber. Cover slips were then mounted using ProLong® Gold Antifade Reagent with DAPI, before examination with an Olympus CKX41 fluorescence microscope.
Immunoblotting
B cells from C57BL/6 mice were stimulated with LPS (5 μg/ml), LPS (3 μg/ml) plus IL-4 (4 ng/ml) or 1 U/ml CD154 plus IL-4 (5 ng/ml) in the presence of nil, VPA (1,000 μM) or butyrate (1,000 μM) for 60 h. Cells were harvested and lysed in Laemmli buffer. Cell extracts containing equal amounts of protein (20 μg) were fractionated through SDS-PAGE (10%). The fractionated proteins were transferred onto polyvinylidene difluoride membranes (Bio-Rad Laboratories) overnight (30 V) at 4°C. After blocking and overnight incubation at 4°C with anti-AID (ZA001; Invitrogen), anti-Blimp-1 mAb (6D3, eBioscience) or anti-β-Actin mAb (AC-15; Sigma-Aldrich), the membranes were incubated with HRP-conjugated secondary Abs. After washing with PBS-Tween 20 (0.05%), bound HRP-conjugated Abs were detected using Western Lightning® Plus-Enhanced Chemiluminescence reagents (PerkinElmer Life and Analytical Sciences).
IgH locus SHM and deletions
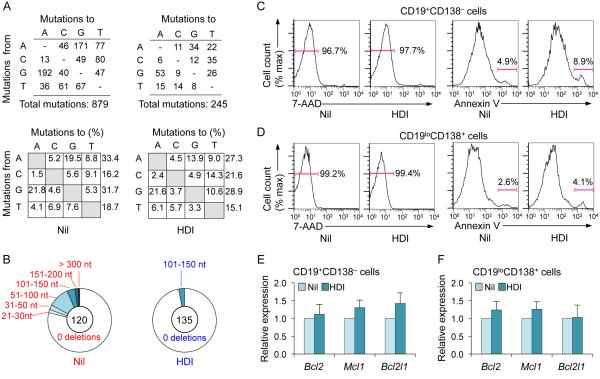
To analyze Ig SHM induced in response to NP-immunization, spleen B cells were isolated from C57BL/6 mice that were immunized with NP16-CGG and given untreated water or HDI-water. Rearranged V186.2DJH-Cγ1 cDNA encoding the anti-NP IgH chain was amplified using a V186.2 leader-specific forward primer together with a reverse Cγ1-specific primer (13) and Phusion™ high fidelity DNA polymerase (New England BioLabs). PCR conditions were 98°C for 10 s, 60°C for 45 s and 72°C for 1 min for 30 cycles. To analyze spontaneous SHM and deletion in MRL/Faslpr/lpr mice, CD19+PNAhi GC B cells were isolated from Peyer's patches and used for extraction of genomic DNA. The intronic JH4-iEμ DNA region (downstream of rearranged VJ558DJH4), which is targeted by SHM but not subjected to any positive or negative selection pressure, was amplified by nested PCR using Phusion™ DNA polymerase and two VJ558 framework region 3–specific forward primers and two reverse primers specific for sequences downstream of JH4, which yielded DNA of approximately 960 bp if a JH4 rearrangement occurred (49). Amplification condition for both the first and second round PCRs were 30 cycles of 98°C for 10 s, 60°C for 45 s and 72°C for 1 min. PCR products were cloned into the pCR™-Blunt II-TOPO® vector (Invitrogen) and sequenced. Sequences were compared to the germline V186.2 or intronic JH4-iEμ DNA region sequences using MacVector software (MacVector, Inc.) for analysis of point-mutations or deletions. For intronic JH4-iEμ DNA region, only a 700 bp region from rearrangements involving JH4-iEμ were analyzed.
Quantitative RT-PCR (qRT-PCR) of mRNAs and miRNAs
For quantification of mRNA, pri-miRNA, germline IH-CH, post-recombination Iμ-CH and mature VHDJH-CH transcripts, RNA was extracted from 0.2-5 × 106 cells using either Trizol® Reagent (Invitrogen) or RNeasy Plus Mini Kit (Qiagen). Residual DNA was removed from the extracted RNA with gDNA eliminator columns (Qiagen). cDNA was synthesized from total RNA with the SuperScript™ III First-Strand Synthesis System (Invitrogen) using oligo-dT primer. Transcript expression was measured by qRT-PCR with the appropriate primers (Supplemental Table 1) using a Bio-Rad MyiQ™ Real-Time PCR Detection System (Bio-Rad Laboratories) to measure SYBR Green (IQ™ SYBR® Green Supermix, Bio-Rad Laboratories) incorporation with the following protocol: 95°C for 15 sec, 40 cycles of 94°C for 10 sec, 60°C for 30 sec, 72°C for 30 sec. Data acquisition was performed during 72°C extension step. Melting curve analysis was performed from 72°C-95°C. For quantification of mature miRNA transcripts, RNA was extracted from 0.2-5 × 106 cells using miRNeasy® Mini Kit (Qiagen) and then reverse-transcribed with miScript II RT Kit (Qiagen) using the miScript HiSpec buffer. A Bio-Rad MyiQ™ Real-Time PCR Detection System was used to measure SYBR Green (miScript SYBR Green PCR Kit; Qiagen) incorporation according to manufacturer’s instructions. Mature miRNA forward primers (Supplemental Table 1) were used at 250 nM in conjunction with the Qiagen miScript Universal Primer and normalized to expression of small nuclear/nucleolar RNAs Rnu6/RNU61/2, Snord61/SNORD61, Snord68/SNORD68, and Snord70/SNORD70. The ΔΔCt method was used for data analysis of qRT-PCR experiments.
Histone acetylation of miRNA host genes and Aicda promoter by ChIP and qPCR
ChIP assays were performed as previously described (15, 48, 50). B cells (1 × 107) were treated with formaldehyde (1% v/v) for 10 min at 25°C to crosslink chromatin, washed once in cold PBS with protease inhibitors (Roche) and resuspended in lysis buffer (20 mM Tris-HCl, 200 mM NaCl, 2 mM EDTA, 0.1% w/v SDS and protease inhibitors, pH 8.0). Chromatin was sonicated to yield DNA fragments (about 200 to 1,000 bp in length), pre-cleared with protein A agarose beads (Pierce) and incubated with anti-acetyl-histone H3 mAb (H3K9ac/K14ac; 17-615; Millipore) at 4°C overnight. Immune complexes were precipitated by Protein A agarose beads, washed and eluted (50 mM Tris-HCl, 0.5% SDS, 200 mM NaCl, 100 μg/ml proteinase K, pH 8.0), followed by incubation at 65°C for 4 h. DNA was purified using a QIAquick PCR purification kit (Qiagen). The miRNA host gene promoter region DNA was amplified from immunoprecipitated chromatin by qPCR using appropriate primers (Supplemental Table 1). Data were normalized to input chromatin DNA and depicted as relative abundance of each amplicon.
Luciferase 3’UTR reporter assays
The partial 3’UTRs of Aicda mRNA (nucleotides 691 - 1168 of NM_009645.2, NCBI) and Prdm1 mRNA (nucleotides 2652 - 5101 of NM_007548.3, NCBI) were PCR amplified from spleen B cell cDNA and cloned into the pMIR-REPORT™ miRNA Expression Reporter Vector System (Invitrogen), which allows for analysis of 3’UTR-mediated regulation of firefly luciferase activity. The mut Aicda 3’UTR containing point mutations (as diagramed in Supplemental Fig. 2E) was generated by PCR-based mutagenesis of the Aicda 3’UTR pMIR-REPORT™ vector using Phusion™ DNA polymerase (New England BioLabs.). Point-mutations were introduced into the Prdm1 3’UTR, as described above, together with an additional deletion (nucleotides 3680 to 5101 were deleted by PCR) to generate mut Prdm1 3’UTR. The sequence of constructs was confirmed by two independent sequencing reactions. Reporter constructs were co-transfected with the pRL-TK vector (Promega), which drives constitutive expression of Renilla reniformis luciferase, into mouse CH12F3 B cells by electroporation (250V and 900 Ω) with a Gene Pulser II™ (BioRad Laboratories). Transfected CH12F3 B cells were then cultured in FBS-RPMI for 1.5 h to allow for reporter gene expression. The ability of VPA to repress reporter activity was determined by firefly luciferase activity and normalized to Renilla luciferase activity according to manufacturer’s instructions using the Dual-Luciferase Reporter Assay System® (Promega).
Methylation analysis of Aicda promoter DNA
Genomic DNA was treated with sodium bisulfite using the EpiTect Bisulfite Kit® (Qiagen) according to the manufacturer’s instructions. Bisulfite-treated DNA was amplified by PCR using GoTaq® Hot Start Polymerase (Promega). The primers for bisulfite sequencing PCR (Supplemental Table 1) were designed using MethPrimer (http://www.urogene.org/methprimer/index1.html). PCR products were purified with QIAquick® PCR purification kit (Qiagen) and sequenced before or after cloned into the pCR™-Blunt II-TOPO® vector.
Lupus mice: autoantibodies, pathology and disease
MRL/Faslpr/lpr mice (Jackson Laboratory) were housed in the UC Irvine and UT Health Science Center vivaria and provided with autoclaved food and deionized water. MRL/Faslpr/lpr mice were started on HDI-water ad libitum at 6- or 17-weeks of age, or were on untreated water throughout their life and scarified when moribund. Anti-nuclear antibody (ANA) and anti-dsDNA antibody titers were determined in sera. For ANA assays, sera were serially diluted in PBS (from 1:40 to 1:160), incubated on antinuclear Ab substrate slides (HEp-2 cell-coated slides, MBL-BION) and detected with a 1:1 mixture of FITC-anti-IgG1 and FITC-anti-IgG2a mAbs (R19-15; BD Biosciences). Images were acquired with a 40x objective on an Olympus CKX41 fluorescence microscope. Anti-dsDNA IgG and IgG2a antibody titers were measured in sera of MRL/Faslpr/lpr mice by ELISA as previously described (14). Titers were expressed in RU, defined as the dilution factor needed to reach 50% of binding saturation, calculated using GraphPad Prism® software (GraphPad Software, Inc.). Skin lesions were scored on a scale of 0 to 3, with 0 = none, 1 = mild (snout and ears), 2 = moderate (< 2 cm snout, ears and intrascapular), and 3 = severe (> 2 cm snout, ears and intrascapular). To assess kidney pathology, kidneys from MRL/Faslpr/lpr mice were either frozen in Tissue-Tek® O.C.T. compound (Sakura Finetek USA) on dry ice for immunofluorescence or fixed in PFA (4%) and paraffin embedded for hematoxylin and eosin (H & E) staining. For immunofluorescence, 4 μm sections were prepared by cryostat, loaded onto positively charged slides, fixed in cold acetone and stained with a mixture of FITC-labeled rat mAb to mouse IgG1 and FITC-labeled rat mAb to mouse IgG2a. Cover slips were then mounted using ProLong® Gold Antifade Reagent with DAPI, before examination with an Olympus CKX41 fluorescence microscope. For H & E staining, kidneys were fixed overnight in PFA (4%), serially passed into ethanol (30%) for 1 h, ethanol (50%) for 1 h, ethanol (70%) overnight, paraffin embedded and then sections cut (4 μm) for H & E staining.
Statistical analyses
All statistical analyses were performed using Excel (Microsoft) or GraphPad Prism® software. Differences in Ig titers, CSR and RNA transcript expression were analyzed with Student’s paired (in vitro) and unpaired (in vivo) t-test assuming two-tailed distributions. Differences in the frequency and spectrum of somatic point-mutations were analyzed with χ2 tests. Differences in lifespan between mice that were administered untreated water or HDI-water were compared by Kaplan-Meier curves and calculated using the Mantel-Cox log-rank test.
Results
CSR and SHM in antibody responses are inhibited by HDI
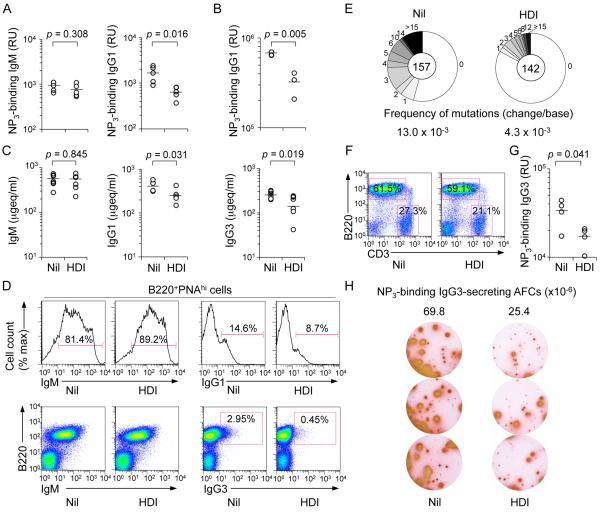
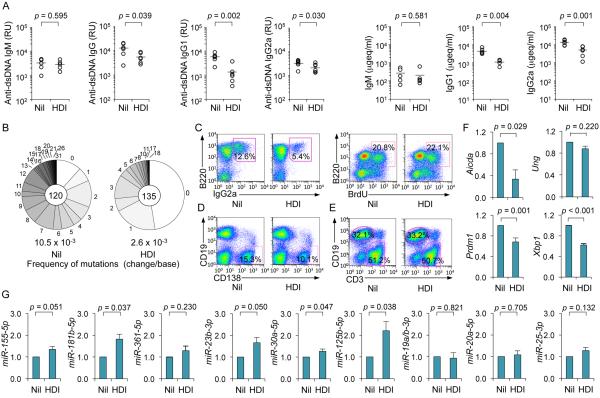
To address the effect of HDI on a T-dependent response, we injected C57BL/6 mice with NP16-CGG, which preferentially induces NP-binding IgG1, one day after starting them on the HDI VPA in drinking water (HDI-water) – these mice drank HDI-water at a comparable or higher rate than that of mice drinking untreated water. Mice drinking HDI-water showed a reduced IgG1 (including high affinity NP3-binding IgG1), but not IgM response to NP, even after a second NP16-CGG injection (Fig. 1A, 1B); reduced class-switched NP-binding IgG1 titers occurred in the context of reduced total IgG1, IgG3 and IgG2b (IgG2b data not shown), but not IgM (Fig. 1C). They were associated with reduced proportions of germinal center (GC) IgG1+, but not IgM+ B cells and reduced frequency of somatic point-mutations (by more than 65%) in V186.2DJH-Cγ1 transcripts, with no significant alteration in spectrum of the residual mutations (Fig. 1D and 1E, Supplemental Fig. 1A), suggesting that this HDI reduced AID expression to impair the class-switched and hypermutated antibody response. In these mice, HDI had no significant effect on T (CD3+) or B (B220+) cell number (Fig. 1F). A “direct” HDI effect on B cells was indicated by the reduced T-independent IgG3 response in HDI-treated mice injected with T-independent NP-LPS. These mice showed reduced titers of (high-affinity) NP3-binding IgG3, fewer total IgG3+ B cells, and NP3-binding IgG3 antibody-forming cells (Fig. 1G, 1H). Thus, HDI can dampen class-switched specific T-dependent and T-independent antibody responses.
FIGURE 1.
HDI reduce CSR and SHM in antibody responses to NP-CGG and NP-LPS. 8-week-old C57BL/6 mice were started on HDI-water one day before injection with NP16-CGG or NP-LPS. (A) Titers of high-affinity NP3-binding IgM or IgG1 (relative units, RU) in serum 10 d after NP16-CGG injection. Each symbol represents an individual mouse; n = 5; p values, unpaired t-test. (B) NP3-binding IgG1 titers (RU) in serum 28 d after initial NP16-CGG injection (7 d after ‘booster’ NP16-CGG injection). Each symbol represents an individual mouse; n = 3; p values, unpaired t-test. (C) Titers of total IgM, IgG1 or IgG3 in serum 10 d after NP16-CGG injection, depicted as μgeq/ml (n is at least 5 mice). p values, unpaired t-test. (D) Surface IgM and IgG1 expression in spleen B220+PNAhi GC B cells 10 d after NP16-CGG injection. Data are representative of three independent experiments. (E) Somatic point-mutations in the V186.2 region of V186.2DJH-Cγ1 transcripts amplified from spleen B cells 10 d after NP16-CGG injection. Sequence data were pooled from three mice in each group. Pie charts depict the proportions of sequences that carry 1, 2, 3, etc. point-mutations over the 294 bp V186.2 region of V186.2DJH-Cγ1 transcripts. In the center are the numbers of independent sequences analyzed; listed below the pie charts is the overall mutation frequency (changes/base). (F) Surface B220 and CD3 expression in spleen cells 10 d after NP16-CGG injection. Data are representative of three independent experiments. (G) NP3-binding IgG3 titers (RU) in serum 28 d after initial NP-LPS injection (7 d after ‘booster’ NP-LPS injection). Each symbol represents an individual mouse; n = 4; p values, unpaired t-test. (H) ELISPOT analysis of NP3-binding IgG3 antibody-forming cells (AFCs) in spleens 28 d after initial NP-LPS injection (7 d after ‘booster’ NP-LPS injection). Data are from three independent experiments. Numbers indicate NP3-binding IgG3 AFCs per 1 × 106 spleen cells (n = 3 mice).
HDI inhibit CSR without altering B cell viability or proliferation
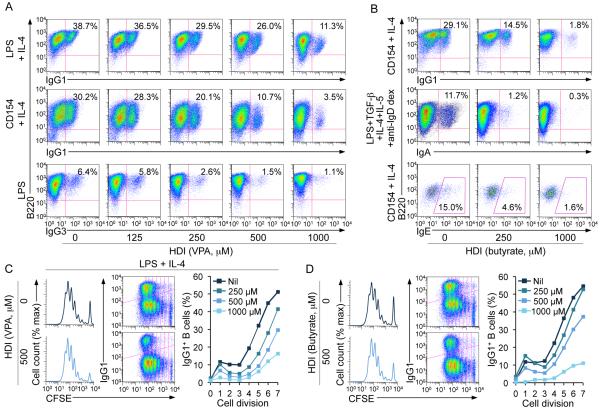
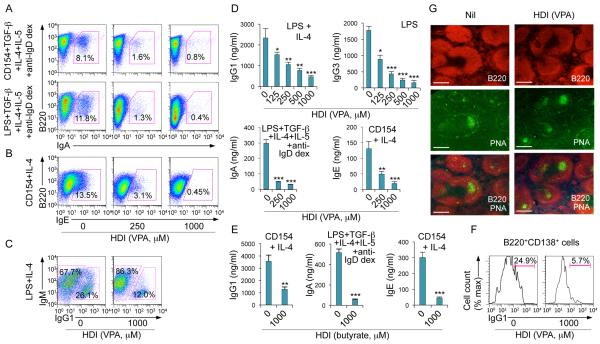
To further define the impact of HDI on CSR, we used appropriate stimuli to induce B cells to switch to IgG1, IgG3, IgA or IgE, in the presence of VPA, butyrate (0 - 1,000 μM) or TSA (0 - 20 nM). These HDI reduced in a dose-dependent fashion CSR to IgG1, IgG3, IgA and IgE (Fig. 2A, 2B, 3A-3C, Supplemental Fig. 1B), without affecting B cell division (Fig. 2C, 2D), viability or apoptosis in vivo or in vitro (Fig. 4A; Supplemental Fig. 1C, 1D). Expression of the anti-apoptotic genes Bcl2, Mcl1 and Bcl2l1 (Bcl2l1 encodes Bcl-xL), which enhance B cell and plasma cell survival, was unaltered or increased by HDI in vivo and in vitro (Supplemental Fig. 1F, 1H). The reduction in IgG1+ and IgG3+ B cells reflected a lower proportion of class-switched cells per round of cell division (Fig. 2C, 2D; Supplemental Fig. 2A) and was associated with decreased titers of IgG1, IgG3, IgA and IgE in culture fluids (Fig. 3D, 3E). HDI inhibition of CSR was further confirmed by decreased mature VHDJH-Cγ1, VHDJH-Cγ3, VHDJH-Cα and VHDJH-Cε transcripts and post-recombination Iμ-Cγ1, Iμ-Cγ1, Iμ-Cγ3, Iμ-Cα and Iμ-Cε transcripts in the presence of normal levels of the respective germline IH-CH transcripts, which are necessary for initiation of CSR (Supplemental Fig. 2B-2D). Thus, HDI significantly reduce CSR, without altering B cell viability or proliferation.
FIGURE 2.
Dose-dependent inhibition of CSR by HDI in B cells. (A) Surface expression of B220 and IgG1 or IgG3 on B cells stimulated with LPS plus IL-4 or CD154 plus IL-4 (for CSR to IgG1) or LPS (for CSR to IgG3) in the presence of nil or increasing doses of VPA (concentrations comparable to or lower than serum concentrations of VPA-treated mice) for 4 d. (B) Surface expression of B220 and IgG1 or IgA, or intracellular expression of B220 and IgE in B cells stimulated with CD154 plus IL-4 (for CSR to IgG1 and IgE) or LPS plus TGF-β, IL-4, IL-5 and anti-IgD dextran (for CSR to IgA) in the presence of nil or increasing doses of butyrate for 4 d. (C and D) Proliferation of lymph node B220+ B cells labeled with CFSE and stimulated for 4 d with LPS plus IL-4 in the presence of nil, (C) VPA (500 μM) or (D) butyrate (500 μM). Progressive left shift of fluorescence intensity indicates B220+ B cell division (left panels). Cell divisions are plotted by vertical lines versus IgG1 surface density, among B220+ cells (middle panels). The percentages of IgG1+B220+ B cells among total B220+ B cells that had completed the same number of divisions when cultured 4 d in the presence of nil, VPA or butyrate are depicted by scatter plots (right panels). Data are representative of three independent experiments.
FIGURE 3.
HDI inhibit CSR, and IgG, IgA and IgE production in dose-dependent fashion, but not GC formation. (A) Surface expression of B220 and IgA in B cells stimulated with CD154 or LPS plus TGF-β, IL-4, IL-5 and anti-IgD dextran in the presence of nil or increasing doses of VPA for 4 d. (B) Intracellular expression of IgE in B cells stimulated with CD154 plus IL-4 in the presence of nil or increasing doses of VPA for 4 d. (C) Intracellular expression of IgM and IgG1 in B cells stimulated with LPS plus IL-4 for 4 d in the presence of nil or VPA. (D) IgG1, IgG3, IgA or IgE in culture fluids of B cells stimulated with LPS plus IL-4, LPS alone, LPS plus TGF-β, IL-4, IL-5 and anti-IgD dextran or CD154 plus IL-4, respectively, for 7 d in the presence of nil or increasing doses of VPA. (E) IgG1, IgA and IgE in culture fluids of B cells stimulated with CD154 plus IL-4 or LPS plus TGF-β, IL-4, IL-5 and anti-IgD dextran for 7 d in the presence of nil or butyrate. Data are from three independent experiments (mean and SEM). *p < 0.05, **p < 0.01, ***p < 0.001, unpaired t-test. (F) Proportions of IgG1+ plasmablasts (B220+CD138+), as measured by surface expression of B220, CD138 and IgG1 in B cells stimulated with LPS plus IL-4 for 7 d in the presence of nil or VPA. (G) GC structure, as visualized by fluorescent microscopy (B220, red; PNA-binding lectin, green) in the spleens of mice that were on HDI-water or untreated water and injected with NP16-CGG for 10 d (as in Figure 1). Data are representative of three independent experiments; scale bars, 50 μm.
FIGURE 4.
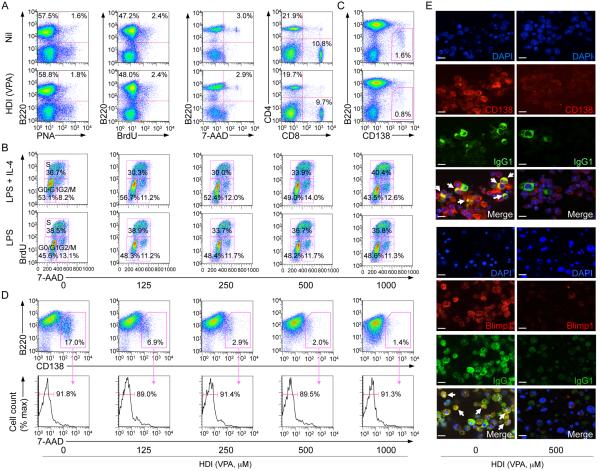
HDI inhibit plasma cell differentiation. (A) Proportions of: B220+PNAhi GC B cells, proliferating B cells (BrdU-stained B220+ B cells), viable (7-AAD−) B220+ B cells and CD4+ and CD8+ T cells in spleen cells from mice that were on HDI-water or untreated water and injected with NP16-CGG 10 d before analysis. (B) HDI does not alter B cell cycle. Mouse IgD+ B cells were stimulated for 48 h with LPS or LPS plus IL-4 in the presence of nil or VPA, during the last 30 min of culture, the cells were pulse-labeled with 10 μM BrdU. The cells were then surface stained for B220 before intracellular staining with anti-BrdU mAb and 7-AAD. B220+ cells are displayed with gates indicating the percentage of cells in G0/G1, S and G2/M phase. (C) Proportions of B220loCD138+ (plasma) cells in spleen cells from C57BL/6 mice that were on HDI-water or untreated water were analyzed 10 d after NP16-CGG injection. (D) Dose-dependent inhibition by VPA of plasma cell (B220loCD138+) differentiation (upper row) in B cells stimulated for 4 d with LPS plus IL-4, without alteration of plasma cell viability, as analyzed by 7-AAD staining (lower row, proportions of 7-AAD− viable cell among B220loCD138+ cells are indicated). (E) IgG1-producing plasma cells (IgG1+CD138+ or IgG1+Blimp-1+) are reduced in cultures of IgD+ B cells stimulated for 7 d with LPS plus IL-4 in the presence of VPA (500 μM), as shown by confocal fluorescence microscopy. Cells were permeabilized and stained with DAPI (blue) to visualize nuclei and fluorescent mAbs to visualize IgG1 (green) and CD138 (red, top set of panels) or Blimp-1 (red, bottom set of panels). Arrows indicate IgG1-producing cells (yellow; CD138+/Blimp-1+IgG1+). Data are representative of three independent experiments; scale bars, 10 μm.
HDI inhibit plasma cell differentiation but not survival
HDI-mediated impairment of antibody responses was not due to alteration of GC development, as GC structure, GC B cell proportion, B cell viability and proliferation were unaltered in NP-CGG-injected mice treated with VPA (Fig. 3G; Fig. 4A), nor did it stem from alteration of cell cycle, as shown by the normal proportion of B cells in G0/G1, S or G2/M, upon stimulation in vitro (Fig. 4B). Rather, it reflected an HDI-mediated inhibition of B cell differentiation into plasma cells in vivo and in vitro (Fig. 4C-4E). This was not associated with decreased plasma cell survival, as shown by normal plasma cell viability and apoptosis as well as elevated Bcl2, Mcl1 and Bcl2l1 expression (Fig. 4D; Supplemental Fig. 1E, 1G, 1I) and normal transcripts of Il6 (not shown), which enhances Blimp-1 expression (51). Thus, HDI inhibit plasma cell differentiation without altering plasma cell survival.
Aicda, Prdm1 and Xbp-1 are silenced by HDI
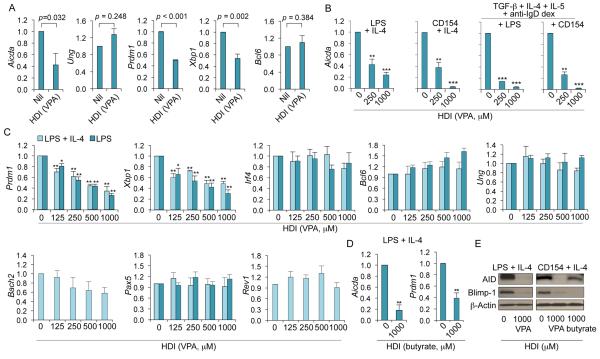
The HDI-impaired antibody response was associated with reduced CSR and SHM, which are initiated by AID, and reduced plasma cell differentiation, which is orchestrated by Blimp-1. Expression of Aicda, Prdm1 and Xbp1 (Xbp1 is under control of Blimp-1 and its gene product promotes Ig secretion in plasma cells (16)) was significantly reduced by HDI in vivo, while expression of Ung, which encodes for Ung that plays a role downstream of AID in CSR and SHM (1), and Bcl6, which encode Bcl-6, a master regulator of the GC reaction and a Prdm1 repressor (52, 53), were unchanged (Fig. 5A). Consistent with these in vivo findings, the expression of Aicda, Prdm1 and Xbp1 was silenced in a dose-dependent fashion by HDI in stimulated B cells in vitro (Fig. 5B-5D). This contrasted with the unchanged expression of Irf4 (encoding Irf4, a transcription factor that regulates CSR, SHM and plasma cell differentiation) (54), Bcl6, Ung, HoxC4 (encoding the AID-inducing HoxC4 transcription factor) (12-14), Rev1 (encoding the CSR scaffold protein Rev1) (48) and Bach2 and Pax5 (encoding Bach2 and Pax5, both repressors of Prdm1) (55, 56) (Fig. 5C and not shown). The downregulation of Aicda and Prdm1 transcripts greatly affected the expression of AID and Blimp-1 proteins (Fig. 5E). Thus, the inhibition of CSR, SHM and plasma cell differentiation by HDI reflects the HDI-mediated downregulation of AID and Blimp-1.
FIGURE 5.
HDI reduce expression of AID (Aicda), Blimp-1 (Prdm1) and Xbp-1 (Xbp1) in a dose-dependent fashion in B cells. (A) Aicda, Ung, Prdm1 Xbp1 and Bcl6 transcripts in spleen B cells from the C57BL/6 mice that were on HDI-water or untreated water and injected with NP16-CGG 10 d before analysis, as in Figure 1, measured by qRT-PCR and normalized to Gapdh expression. Values in the B cells from HDI (VPA)-treated mice are depicted as relative to the expression of each transcript in the B cells from untreated (nil) mice, set as 1. Data are presented as mean and SEM from at least three pairs of mice. p values, unpaired t-test. (B) Aicda transcripts in purified B cells treated with nil (0 μM of VPA) or VPA (250 or 1,000 μM) and stimulated for 60 h with LPS or CD154 plus IL-4, or LPS or CD154 plus TGF-β, IL-4, IL-5 and anti-IgD dextran, measured by qRT-PCR and normalized to Cd79b expression. (C) Prdm1, Xbp1, Irf4, Bcl-6, Ung, Rev1, Bach2 and Pax5 transcripts in purified B cells treated with nil or increasing doses of VPA and stimulated for 60 h with LPS or LPS plus IL-4, measured by qRT-PCR and normalized to Cd79b expression. (D) Aicda and Prdm1 transcripts in B cells treated with nil or butyrate (1,000 μM) and stimulated for 60 h with LPS plus IL-4, measured by qRT-PCR and normalized to Cd79b expression. Values in B cells cultured in the presence of HDI are depicted as relative to the expression of each transcript in B cells cultured in the absence of HDI, set as 1. Data are presented as mean and SEM from three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, unpaired t-test. (E) AID, Blimp-1 and β-Actin proteins in cell lysates from B cells cultured for 60 h with LPS plus IL-4 or CD154 plus IL-4 in the presence of nil or HDI (1,000 μM of VPA or butyrate) were detected by immunoblotting. Data are representative of three independent experiments.
HDI upregulate miRNAs that target Aicda and Prdm1 transcripts in B cells
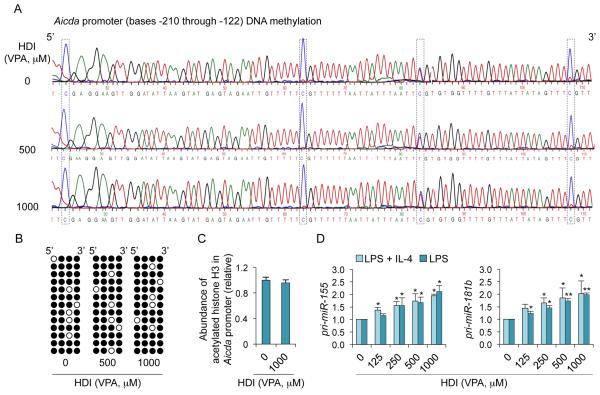
Enhancement of histone acetylation, a function of HDI, which generally upregulate gene expression, is at odds with the decreased expression of AID/Aicda and Blimp-1/Prdm1 by HDI in vivo and in vitro (Fig. 5). In fact, in B cells induced to undergo CSR, HDI did not alter histone H3 acetylation in the Aicda promoter, nor did they alter DNA methylation (HDACs have been suggested to interact with DNA methyltransferases (57)) (Fig. 6A-6C), raising the possibility that HDI upregulated the expression of gene(s), which, in turn, negatively regulated Aicda and Prdm1. HDI can modulate expression of miRNAs (36), which silence target mRNAs by inducing their degradation and/or reducing their translation. In this context, Aicda can be silenced by miR-155, miR-181b and miR-361 (2, 3, 18, 27-30); Prdm1 can be targeted by miR-23b (our prediction, using TargetScan.org, miRNA.org and miRbase.org) and has been suggested to be targeted by miR-30a and miR-125b (3, 18, 31-33) (Supplemental Fig. 2E). We found that miR-155, miR-181b, miR-361, miR-23b, miR-30a and miR-125b were upregulated by VPA in B cells in vivo, and in purified B cells induced to undergo CSR and plasma cell differentiation in vitro – irrelevant miRNAs miR-19a/b, miR-20a and miR-25, which are not known to regulate Aicda, Prdm1 or Xbp1, were unchanged in B cells in vivo and in vitro (Fig. 7A, 7B). Primary miRNA transcripts pri-miR-155 and pri-miR-181b, which are processed by Drosha and Dicer to give rise to mature miRNAs, were also upregulated, suggesting that HDI upregulate miRNA host gene transcription (Fig. 6D). Accordingly, miRNA upregulation in stimulated B cells was accompanied by increased overall histone H3 acetylation of the host genes encoding these miRNAs (Fig. 7C). Thus, HDI upregulate miRNAs that target Aicda and Prdm1 mRNA 3’UTRs and, therefore, can silence these mRNAs.
FIGURE 6.
HDI do not alter DNA methylation and histone acetylation in the Aicda promoter. (A and B) CpG DNA methylation of the Aicda promoter was analyzed by bisulfite sequencing of genomic DNA from B cells stimulated for 4 d with LPS plus IL-4 in the presence of increasing doses of VPA. (A) DNA sequencing of PCR products of bisulfite-treated genomic DNA. The sequence signal from dCs in CpG motifs is framed. As unmethylated dC nucleotides can be converted to dU (read as dT in DNA sequence), while methylated dC cannot, the ratio of dC (blue) to dT (red) signal indicates the level of methylated dC at any given positions. (B) Methylation pattern at each of the four dCs within CpG motifs from individually cloned sequences (each row is a unique sequence and each dC is represented by a column of circles) is shown as an array of circles (closed circles represent methylated dCs; open circles represent unmethylated dCs). (C) Abundance of acetylated histone H3 (H3K9ac/K14ac) in the Aicda promoter in B cells stimulated with LPS plus IL-4 for 60 h in the presence of nil or VPA (1,000 μM) was measured by ChIP and qPCR. (D) Primary (pri-) miRNA transcripts of miR-155 and miR-181b in B cells cultured for 60 h with LPS or LPS plus IL-4 in the presence of nil or increasing doses of VPA, were measured by qRT-PCR and normalized to Cd79b expression. Values in B cells cultured in medium containing VPA are depicted as relative to the values in B cells cultured in the absence of HDI, set as 1. Data are presented as mean and SEM from three independent experiments. *p < 0.05, **p < 0.01, unpaired t-test.
FIGURE 7.
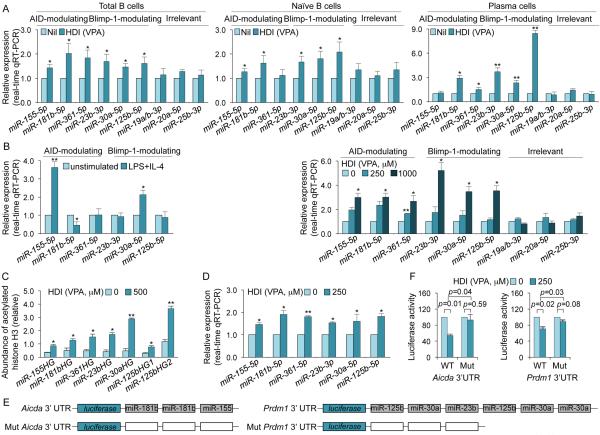
HDI hyperacetylate miRNA host genes to upregulate miRNAs that silence Aicda and Prdm1 mRNAs. (A) Upregulation of miRNAs that modulate AID and Blimp-1 in “total” (spleen CD19+CD138−) B cells, naïve B cells and plasma cells from mice that were on HDI-water or untreated water and injected with NP16-CGG (as in Figure 1), measured by qRT-PCR. (B) Left panel, expression of miRNAs that modulate AID and Blimp-1 in freshly isolated naïve B cells and in B cells stimulated with LPS plus IL-4 for 60 h. Right panel, dose-dependent upregulation of miRNAs that modulate AID and Blimp-1 in B cells cultured with LPS plus IL-4 for 60 h in the presence of nil or VPA (250 or 1,000 μM). miRNA expression was normalized to expression of small nuclear/nucleolar RNAs Rnu6, Snord61, Snord68, and Snord70. miRNA expression in B cells isolated from mice that were on HDI-water, or B cells cultured with HDI are depicted as relative to the expression of each miRNA in B cells isolated from mice that were on untreated water, or B cells cultured in the absence of HDI, respectively, set as 1. Data are mean and SEM from at least three mice or three independent experiments. (C) Relative abundance of acetylated histone H3 (H3K9ac/K14ac) in miRNA host genes (HGs) in purified B cells stimulated with LPS plus IL-4 in the presence of nil or VPA (500 μM), as analyzed by ChIP and qPCR. (D) Mature miRNA expression in CH12F3 B cells treated with nil or VPA (250 μM) for 24 h, as measured by qRT-PCR. Values in B cells treated with VPA are depicted as relative to the expression of each miRNA in B cells treated with nil, set as 1. *p < 0.05, **p < 0.001, unpaired t-test. (E) Schematic diagram of the 3’UTRs of Aicda and Prdm1 mRNAs and their mutant (mut) counterparts that were cloned into the pMIR-REPORT luciferase reporter vector. Open boxes indicate point-mutation sequences that are putative miRNA target sites, as detailed in Supplementary Figure 2E. mut Prdm1 3’UTR consists of a DNA deletion. (F) Luciferase activity in CH12F3 cells transfected with wildtype or mutated Aicda or Prdm1 3’UTRs (cloned into pMIR-REPORT luciferase reporter vector) after 24 h treatment with nil or VPA (250 μM). Luciferase activity was measured 1.5 h after transfection. Transfection efficiency was controlled for by normalizing to signal from co-transfected Renilla reniformis luciferase vector. Luciferase activity in B cells treated with VPA is depicted as relative to values in B cells cultured in the absence of VPA, set as 1. Data are presented as mean and SEM from three independent experiments. p values, unpaired t-test.
Selected B cell miRNAs upregulated by HDI silence Aicda and Prdm1 transcripts
To prove that HDI inhibition of AID and Blimp-1 expression was mediated by upregulation of miRNAs that directly target Aicda and Prdm1 mRNA 3’UTRs, we cloned either wildtype or mutant (mut) 3’UTRs of Aicda and Prdm1 mRNAs (mut 3’UTRs were constructed by mutation or deletion of the target sites of miR-155 and miR-181b in Aicda mRNA 3’UTR or miR-23b, miR-30a and miR-125b in Prdm1 mRNA 3’UTR) into pMIR-REPORT™ luciferase reporter vectors. These were used to transfect mouse CH12F3 B cells that can be induced to undergo CSR at high rate. Like in primary B cells, VPA upregulated miR-155, miR-181b, miR-361, miR-23b, miR-30a and miR-125b expression in CH12F3 B cells induced to undergo CSR (Fig. 7D). We then measured the ability of VPA to repress luciferase activity using reporter constructs containing wildtype or mut 3’UTRs of Aicda and Prdm1 mRNAs that were transfected into CH12F3 B cells (Fig. 7E; Supplemental Fig. 2E). The luciferase reporter activity was reduced by VPA in B cells transfected with reporter constructs containing wildtype Aicda or Prdm1 3’UTRs but not those transfected with reporter constructs containing Aicda or Prdm1 3’UTR mutants (Fig. 7F). In B cells transfected with reporter constructs containing wildtype Aicda or Prdm1 3’UTRs, the degree of inhibition of luciferase activity was significant, in spite of the relatively low dose of VPA used (250 μM) and the omission of some additional miRNA target sites in the 3’UTR of Aicda mRNA, thereby emphasizing the potency of the HDI-mediated upregulation of miRNAs on B cell gene expression. Thus, VPA silences AID and Blimp-1 expression in B cells through upregulation of selected miRNAs that directly target Aicda and Prdm1 mRNA 3’UTRs.
HDI silence AICDA and PRDM1 in human B cells to inhibit CSR and plasma cell differentiation
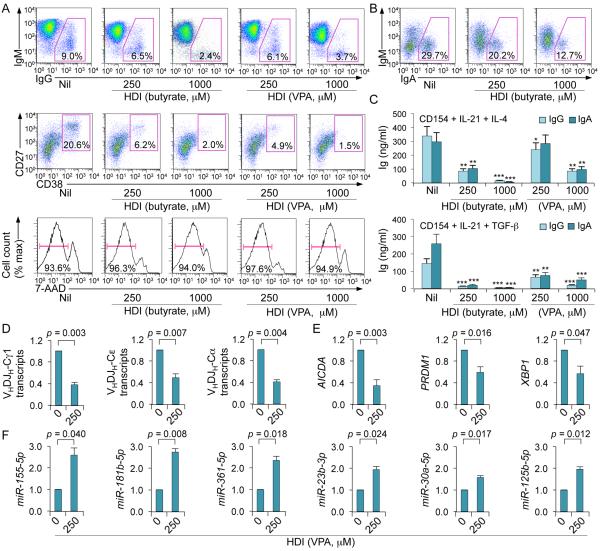
Next, we determined whether HDI also inhibit CSR and plasma cell differentiation in human B cells. We stimulated purified human IgD+ B cells with CD154 plus IL-21 and IL-4 or TGF-β. Like in mouse B cells, VPA and butyrate effectively inhibited CSR to IgG, IgA and IgE in human B lymphocytes, as well as plasma cell differentiation in a dose-dependent fashion without altering B cell viability (Fig. 8A-8D). VPA increased expression of miR-155, miR-181b, miR-361, miR-23b, miR-30a and miR-125b, which (as reported or as predicted by us (2, 3, 18, 27-33)) target miRNA binding sites in the 3’UTR of human and mouse AICDA/Aicda or PRDM1/Prdm1 3’UTRs, and decreased expression of AICDA, PRDM1 and XBP1 transcripts (Fig. 8E, 8F). Thus, HDI (VPA and butyrate) modulate human B cell class-switching and plasma cell differentiation, as they do in mouse B cells.
FIGURE 8.
HDI inhibit CSR and plasma cell differentiation increases selected miRNAs, and decreases target AICDA and PRDM1 mRNAs as well as XBP1 mRNA, in human B cells. Human peripheral blood IgD+ B cells were stimulated with CD154, hIL-4 and hIL-21 (for CSR to IgG1 and IgE and plasma cell differentiation) or CD154, hIL-21 and TGF-β (for CSR to IgA) in the presence of nil, VPA or butyrate for 60 h (for transcript or miRNA analysis) or 120 h (for flow cytometry or analysis of Ig titers in supernatants). (A) Proportions of IgG+ B cells, plasma cells (CD27+CD38+) or viable (7-AAD−) CD19+ cells. (B) Proportions of IgA+ B cells. (C) IgG and IgA titers in supernatants of B cells stimulated with CD154, hIL-4 and hIL-21 (top) or CD154, hIL-21 and TGF-β (bottom) and cultured in the presence of VPA or butyrate. Data are from three independent experiments (mean and SEM). *p < 0.05, **p < 0.01, ***p < 0.001, unpaired t-test. (D) Mature VHDJH-Cα (in cells stimulated by CD154, hIL-21 and TGF-β), VHDJH-Cγ1 and VHDJH-Cε (in cells stimulated with CD154, hIL-4 and hIL-21) transcripts were analyzed by qRT-PCR and normalized to HPRT1 transcripts. (E) AICDA, PRDM1 and XBP1 transcripts (in cells stimulated with CD154, hIL-4 and hIL-21) were analyzed by qRT-PCR and normalized to HPRT1 transcripts. (F) miRNA expression was analyzed by qRT-PCR and normalized to expression of small nuclear/nucleolar RNAs RNU6-1/2, SNORD61, SNORD68, and SNORD70. Values in B cells cultured in the presence of HDI are depicted as relative to the expression of each transcript or miRNA in B cells cultured in the absence of HDI, set as 1. Data are presented as mean and SEM from three independent experiments. p values, unpaired t-test.
HDI dampen the autoantibody response, ameliorates disease and increase survival in lupus mice
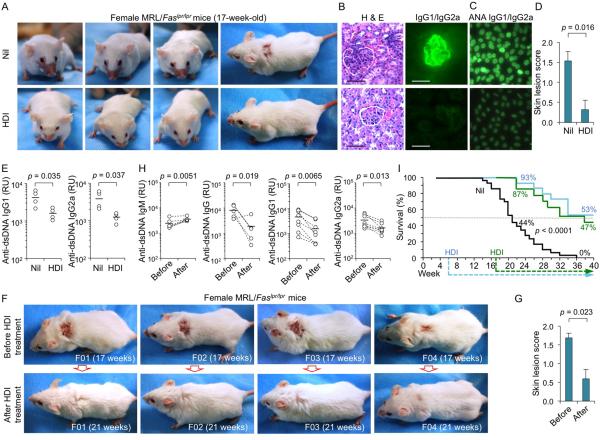
Lupus-prone MRL/Faslpr/lpr mice spontaneously upregulate AID, Blimp-1, CSR and SHM, and generate great numbers of plasma cells, which produce large amounts of anti-dsDNA IgG and other autoantibodies, and develop age-dependent disease, which includes skin lesions and kidney pathology (2, 14, 49). In our female MRL/Faslpr/lpr mice, ANA and anti-dsDNA IgG autoantibodies appeared at 6-weeks of age and reached high levels at 12 wk, in association with significant loads of point-mutations and DNA deletions in the IgH locus. At 17 wk, all these mice show severe kidney immunopathology and 75% of them display various skin lesions, which in 55% of the cases include the characteristic “butterfly” rash. To determine whether HDI could inhibit the lupus class-switched and hypermutated autoantibody response, we assigned 80 three-week-old female MRL/Faslpr/lpr mice to three groups: (i) a “non-treatment” group consisting of 50 mice that were given untreated water throughout their lives; (ii) an “early treatment” group of 15 mice that were given untreated water for the first 6 wk of life, at which age they were started on HDI-water; and, (iii) a “late treatment” group of 15 mice that were given untreated water for the first 17 wk of life, at which age they were started on HDI-water.
At 12 wk of age, “early” HDI-treated MRL/Faslpr/lpr mice displayed reduced levels of anti-dsDNA IgG, IgG1, IgG2a and ANA (not shown), but not IgM autoantibodies, reduced overall IgG1/IgG2a but not IgM, significantly reduced (by over 75%) somatic point-mutations, with no significant alteration in spectrum, and a reduced load of DNA deletions in the IgH locus (Fig. 9A, 9B, 10A, 10B; Supplemental Fig. 3). They also showed reduced numbers of IgG2a+ B cells as well as reduced plasma cell differentiation, in the presence of normal B220+ cell proliferation and normal B and T cell numbers (Fig. 9C-9E). Early treated MRL/Faslpr/lpr mice downregulated Aicda, Prdm1 and Xbp1, but not Ung expression in B cells or plasma cells (Fig. 9F), and increased miR-155, miR-181b, miR-361, miR-23b, miR-30a and miR-125 in B cells (irrelevant miR-19a/b, miR-20a and miR-25 expression was unchanged) (Fig. 9G). This did not result from altered cell viability/apoptosis (Fig. 10C, 10D) or decreased expression of anti-apoptotic genes (Fig. 10E, 10F). At 17 wk of age, “early” HDI-treated MRL/Faslpr/lpr mice showed reduced skin lesions, including absence of “butterfly” rash, reduced kidney pathology (Fig. 11A, 11B, 11D) and size of spleen and lymph nodes (not shown). They also continued to show reduced ANA and anti-dsDNA IgG1/IgG2a autoantibodies (Fig. 11C, 11E). Decreased anti-dsDNA IgG, IgG1 and IgG2a (but not IgM) autoantibodies and amelioration of pathology (not shown), in association with complete healing of extensive skin lesions, were also observed in the late treatment group mice, started on HDI-water at 17 wk, an age at which our MRL/Faslpr/lpr mice already showed significant anti-dsDNA IgG autoantibodies and disease (Fig. 11F-11H). The reduced IgG autoantibody levels and disease activity in early and late HDI-treated mice resulted in significantly extended lifespan (p < 0.0001), with 8 out of the 15 early treated mice and 7 of the 15 mice late treated mice still alive and apparently healthy at 40 wk of age. Only one of the 50 “non-treated” mice made it to 32 wk; this mouse died at 36 wk (Fig. 11I). Thus, in lupus-prone mice, HDI upregulate selected miRNAs, which can silence B cell AID or Blimp-1 expression, thereby reducing CSR, SHM and plasma cell differentiation, and dampening the class-switched and hypermutated autoantibody response, immunopathology and disease, and significantly prolonging life.
FIGURE 9.
HDI impair class-switched and hypermutated autoantibody response, increase selected miRNAs and decrease target Aicda and Prdm1 mRNAs, as well as Xbp1 mRNA in MRL/Faslpr/lpr mice. Female MRL/Faslpr/lpr mice were started on HDI-water or untreated water at the age of 6 wk, and analyzed at the age of 12 wk. (A) Titers of circulating anti-dsDNA IgM, IgG, IgG1 and IgG2a, and total IgM, IgG1 and IgG2a (n is at least 5 mice per group). p values, unpaired t-test. (B) Frequency of somatic point-mutations was reduced by 75% in MRL/Faslpr/lpr mice given HDI-water. A 700 bp intronic JH4-iEμ DNA in CD19+PNAhi Peyer’s patch GC B cells was amplified and sequenced (sequence data were pooled from three mice in each group). Pie charts depict the proportions of sequences that carried 1, 2, 3, etc. point-mutations (center of pie: number of independent sequences analyzed). Listed below the pie charts are overall mutation frequencies (changes/base). (C) Proportions of IgG2a+ B cells, and proliferation of spleen B220+ cells (analyzed by administration of BrdU in drinking water at 10 wk of age). (D) CD19loCD138+ plasma cells in spleens. Data are representative of three independent experiments. (E) Surface CD19 and CD3 expression in spleen cells. Data are from one of three independent experiments yielding comparable results. (F) Aicda and Ung transcripts (in spleen CD19+CD138− B cells, normalized to Cd79b transcript) and Prdm1 and Xbp1 transcripts (in spleen CD19loCD138+ plasma cells, normalized to Gapdh transcript) were measured by qRT-PCR. Values in mice that were on HDI-water are depicted as relative to the expression of each transcript in mice that were on untreated water, set as 1. Data are mean and SEM from four independent experiments. (G) miRNA expression in CD19+CD138− spleen B cells was measured by qRT-PCR and normalized to expression of small nuclear/nucleolar RNAs RNU6-1/2, SNORD61, SNORD68, and SNORD70. miRNA expression in the B cells from mice that were on HDI-water are depicted as relative to the expression of each miRNA in the B cells from mice that were on untreated water, set as 1. Data are mean and SEM from three independent experiments. p values, unpaired t-test.
FIGURE 10.
Somatic point-mutations and DNA deletions are significantly reduced in frequency, while the spectrum of point-mutations is unaltered; viability and apoptosis of B cells and plasma cells are not altered in MRL/Faslpr/lpr mice treated with HDI. Female MRL/Faslpr/lpr mice that were on HDI-water or untreated water (starting from 6 wk of age until 12 wk, at which age mice were sacrificed, as in Figure 9 and Supplemental Figure 3). (A and B) Point-mutations and DNA deletions in intronic JH4-iEμ DNA sequences of Peyer’s patches CD19+PNAhi (GC) B cells. (A) The spectrum of the residual mutations is unchanged by HDI treatment. Values are expressed as actual numbers of different nucleotide substitutions (top panels) or as percentage of total mutations (bottom panels). (B) The number of IgH locus DNA deletions is greatly reduced in mice that were on HDI-water. Pie charts depict the proportions of sequences that contain differently sized deletions in the IgH locus. In the center are the numbers of independent sequences analyzed. (C-F) Spleen cells from MRL/Faslpr/lpr mice that were on HDI-water or untreated water were gated to analyze 7-AAD− viable cells or Annexin V+ apoptotic cells among (C) CD19+C138− B cells and (D) CD19loCD138+ plasma cells. Expression of Bcl2, Mcl1 and Bcl2l1 transcripts in (E) CD19+CD138− B cells and (F) CD19loCD138+ plasma cells were measured by qRT-PCR and normalized to Gapdh expression. Values in the cells from mice that were on HDI-water are depicted as relative to the expression of each transcript in the cells from mice that were on untreated water, set as 1. Data are presented as mean and SEM from three independent experiments.
FIGURE 11.
HDI reduce autoantibodies, immunopathology, disease symptoms, and extend lifespan of MRL/Faslpr/lpr mice. (A-E) Female MRL/Faslpr/lpr mice were started on HDI-water or untreated water at the age of 6 wk, and analyzed at the age of 17 wk. (A) Rostral and dorsal images show characteristic lupus of skin lesion, including a “butterfly” rash. (B) Kidney pathology. The representative photomicrographs of kidney sections stained with hematoxylin and eosin (H & E, left) or FITC-labeled rat mAb to mouse IgG1 and IgG2a (right); scale bars, 100 μm. Data are from one of three independent experiments yielding similar results. (C) ANA visualized by indirect immunofluorescence on HEp-2 cells that were incubated with sera (1:40 dilution) from the MRL/Faslpr/lpr mice using FITC-labeled rat mAb to mouse IgG1 and IgG2a; scale bars, 20 μm. Data are from one of three independent experiments yielding similar results. (D) Skin lesion score in the MRL/Faslpr/lpr mice (n = 15; 11 of 15 untreated mice, but only 4 of 15 HDI-treated mice, developed skin lesions). p values, unpaired t-test. (E) Titers of circulating anti-dsDNA IgG1 and IgG2a. p values, unpaired t-test. (F-I) Fifty female MRL/Faslpr/lpr mice were on untreated water throughout their lives; 15 female MRL/Faslpr/lpr mice were on untreated water for the first 6 wk of life, at which age they were started on HDI-water; 15 female MRL/Faslpr/lpr mice were on untreated water for the first 17 wk of life, at which age they were showing active disease (skin lesions, proteinuria, anti-dsDNA autoantibodies) and were started on HDI-water. (F) Dorsal images showing severe skin lesions in four female MRL/Faslpr/lpr mice before (top, 17 wk of age) and after 4 wk of HDI administration (bottom, 21 wk of age). (G) Skin lesion scores in 15 female MRL/Faslpr/lpr mice before (17 wk of age) and after 4 wk of HDI administration (21 wk of age). p values, paired t-test. (H) anti-dsDNA IgM, IgG, IgG1 and IgG2a autoantibody titers in female MRL/Faslpr/lpr mice before (17 wk of age) and after 4 wk of HDI administration (21 wk of age). p values, paired t-test. (I) Survival of the 50 female MRL/Faslpr/lpr mice given untreated water throughout their lives (black line); the 15 female MRL/Faslpr/lpr mice started on HDI-water at 6 wk of age (blue line), and the 15 female MRL/Faslpr/lpr mice started on HDI-water at 17 wk of age when they showed already active disease (green line).
Discussion
As we have argued, epigenetic changes, such as histone posttranslation modifications and DNA methylation, and epigenetic factors, such as miRNAs, can interact with genetic programs to regulate B cell functions, including CSR, SHM and plasma cell differentiation, thereby informing antibody responses, which are critical for the defense against microbial pathogens and tumor cells, as well as autoantibody responses, which mediate autoimmunity and disease (3). We showed here that VPA and butyrate, two SCFA HDI, inhibited CSR, SHM and plasma cell differentiation by modulating intrinsic B cell mechanisms. They repressed AID and Blimp-1 expression in mouse and human B cells by upregulating selected miRNAs that silenced AICDA/Aicda and PRDM1/Prdm-1 mRNAs, as demonstrated by multiple qRT-PCRs (these data) and further confirmed by mRNA-Seq and microRNA-Seq (Zan and Casali, preliminary experiments). AID expression and CSR were also inhibited by TSA, a hydroxamic acid HDI. The doses at which VPA and butyrate inhibited B cell class-switching, hypermutation and plasma cell differentiation were within the range of those measured in humans for these HDI (41, 43). By inhibiting AID and Blimp-1 expression, VPA dampened class-switched and hypermutated antibodies in specific T-dependent and T-independent antibody responses in normal mice. This HDI also dampened class-switched and hypermutated autoantibody levels, reduced immunopathology and extended survival in autoimmune MRL/Faslpr/lpr mice, a well-studied model of human lupus.
SCFA HDI have been suggested to display significant selectivity for different HDACs (58). For example, VPA targets class I HDACs, particularly, HDAC1 and HDAC2, and, less effectively, class IIa HDACs, and butyrate targets class I, mainly HDAC1, and, less effectively, other members of class I and class IIa HDACs (38, 39). HDAC activity is mostly associated with multiprotein complexes, the role and composition of which are often cell type-specific. HDAC-associated proteins would specify the selectivity of HDI, which display different affinities for different HDAC/co-factor complexes. HDI with diverse chemical properties target different HDACs and HDAC/co-factor complexes, thereby regulating gene expression in a locus- and cell type-specific fashion (58). Our findings indicate that in B cells, HDI modulate miRNAs selectively, possibly as a result of HDACs existing in unique contexts of HDAC/co-factor complexes, as occurring in these lymphocytes, particularly when in an activated state.
HDI may also indirectly modulate antibody responses or mitigate autoimmunity by affecting elements other than B cells, such as innate immune cells (59) and T cells (Treg, TH1 and TH17 cells), or inhibit proinflammatory cytokines (37, 46, 60, 61). As shown here, however, HDI directly regulate B cell genes that are central to peripheral differentiation of these lymphocytes and maturation of antibody and autoantibody responses. Silencing AICDA/Aicda by HDI was intrinsic to B lymphocytes and independent of other cellular elements, as shown by our in vitro experiments using purified human and mouse B cells, as well as our in vivo studies of the T-independent response to NP-LPS. In both in vivo and in vitro B cells, the HDI-mediated downregulation of AICDA/Aicda expression was associated with a concomitant increase of the respective targeting miR-155, miR-181b and miR-361 (2), in a tight dose-dependent fashion. Our findings extend those suggesting a role of miR-155 in downregulating AID expression (28, 30, 62), in agreement with the demonstration that repression of this miRNA provides a mechanism of Bcl6-promoted positive regulation of AID and increased GC gene expression (30). As we shown, silencing of PRDM1/Prdm1 (and XBP1/Xbp1) by HDI was also intrinsic to B cells and independent of other cells. Like for Aicda, HDI-mediated downregulation of PRDM1/Prdm1 was associated with a concomitant increase of the respective B cell targeting miRNAs (miR-23b, miR-30a and miR-125b) (2), in vivo and in vitro, and in a tight dose-dependent fashion. HDI-induced downregulation of XBP1/Xbp1 could be secondary to decreased Blimp-1 expression and/or upregulation of selected miRNAs that we have tentatively identified as silencers of XBP1/Xbp1 (Zan and Casali, unpublished). That HDI downregulate Blimp-1 expression by upregulating miR-23b, miR-30a and miR-125b that silence Prdm1 was further supported by our demonstration that HDI slightly reduced or did not essentially altered the Prdm1 repressor genes Bach2, Bcl6 or Pax5.
The selectivity of HDI-mediated silencing of AICDA/Aicda and PRDM1/Prdm1 in B cells was further emphasized by the unchanged expression of HoxC4, Irf4, Rev1 or Ung, which play important roles in AICDA/Aicda regulation and/or CSR, as well as of miR-19a/b, miR-20a and miR-25, which are not known to regulate AICDA/Aicda or PRDM1/Prdm1. We could not rule out the possibility that HDI regulated other B cell factors (e.g. NF-κB or Id2/3), which contributed to the reduction of AID or Blimp-1. The relief of HDI-mediated repression of luciferase activity under the control of Aicda and Prdm1 mRNA 3’UTRs bearing mutated miR-155, miR-181b, miR-23b, miR-30a and miR-125b target sites demonstrated that miRNAs are indeed direct effectors of the HDI-mediated repression of such selected genes in B cells. The role of B cell miRNAs in mediating HDI suppression of AID and Blimp-1 expression, and in dampening antibody and autoantibody responses could be further addressed by using an integrated three-prong approach, involving generation of in vivo Argonaute-miRNA-Aicda or Prdm1 mRNA ternary complexes, knockin mice lacking specific miRNA targeting sites in Aicda or Prdm1 3’UTR or mice with B cells specifically expressing “sponge” inhibitors of miR-155, miR-181b and miR-361, or miR-23b, miR-30a and miR-125b. HDI had no direct effect on the epigenetic status of the Aicda locus, as our acetylated histone ChIP and bisulfite sequencing experiments showed no alteration of histone acetylation or methylation of the Aicda promoter by VPA. While it is possible that HDI could modify protein functions by also increasing acetylation of non-chromatin proteins, our findings allow us to conclude that modulation of miRNAs leading to silencing of selected mRNAs is the mechanism by which HDI mediate inhibition of the B cell differentiation processes that underpin the maturation of antibody responses.
Our findings in human and mouse B cells, in vitro and in vivo, greatly extend and provide a mechanistic underpinning for the limited data by Kienzler and coll. (63), suggesting that VPA reduces human naïve B cell differentiation to (CD27hiCD38hi) plasmablasts and reduces IgG and IgA expression. At odd with our findings, however, these authors seemed to show no decrease of CSR induction in human B cells in vitro by VPA. This could be explained by the different designs of their experiments and ours. Kienzler and coll. stimulated human B cells with CD154 plus IL-21; we stimulated human naïve B cells with CD154 plus IL-21 plus IL-4 or CD154 plus IL-21 plus TGF-β. IL-4 or TGF-β, both critical CSR-inducing stimuli, was missing in Kienzler’s experiments, possibly resulting in relatively lower AICDA and PRDM1 expression. Analysis of the molecular events underpinning CSR was also missing, making it virtually impossible to provide a thorough explanation for the putative discrepancy between the data by Kienzler and coll. and ours.
In vivo HDI-inhibition of CSR and plasma cell differentiation would “freeze” B cells at an IgM+ stage, as indicated by the higher proportion of IgM+ lymphocytes in human or mouse B cell cultures exposed to HDI. Whether these IgM+ B cells remained “naïve” B cells or underwent some degree of memory B cell differentiation is unclear. The much-reduced IgH locus mutational load in HDI-treated normal and lupus-prone mice supports the contention that those IgM+ B cells expressed mainly unmutated IgM natural antibodies. These would have been still available for the response to microbial pathogens and might have played a protective role in systemic autoimmunity (64). In addition, in both normal and autoimmune mice, HDI treatment allowed for some residual AID expression, which resulted in a significant reduction but not ablation of secondary antibody isotypes. These can, even at low titers, mediate a protective anti-microbial immunity, as suggested by the apparently normal risk of infections in Aicda+/– mice with reduced AID levels (65). Thus, in normal and autoimmune mice, HDI dampen the antibody or autoantibody response by efficiently inhibiting CSR, SHM and plasma cell differentiation, while leaving an intact or even increased IgM pool as well as residual IgG and IgA levels that may be sufficient for immune protection.
At higher doses than those used in our study, HDI inhibit proliferation and induce apoptosis in cancer cells (66). They do so partially through induction of DNA damage, which healthy cells can repair, but cancer cells cannot (67). In our in vitro and in vivo experiments, even the highest HDI doses did not reduce B cell and plasma cell viability or increase B cell or plasma cell apoptosis. Consistent with a recent finding that butyrate did not affect survival or proliferation of T cells (42), this SCFA HDI did not affect viability or proliferation of B lymphocytes at the concentrations used here. Similarly, VPA did not affect B cell viability, proliferation, cell cycle or apoptosis, nor did it affect viability or survival of plasma cells. The reduction in proportion of class-switched B cells per round of cell division (which is required for CSR) in the context of an unchanged overall cell number at each division, further indicated that HDI inhibited CSR without interfering with cell viability. This is consistent with the normal levels of lymphocytes in patients treated with VPA (46), in whom lymphopoenia would have been expected to occur if this HDI impaired B and/or T cell viability.
Epigenetic dysregulation can compound genetic susceptibility to mediate autoantibody responses and autoimmunity (3). Epigenetic changes associated with autoimmune responses have been investigated in T cells, but only marginally in B cells (68). We and others have previously shown that highly upregulated AID and Blimp-1 expression is an important feature of lupus patients and lupus-prone mice, including MRL/Faslpr/lpr mice (14, 69). In these mice, dysregulation of AID and Blimp-1 causes aberrant rates of CSR and SHM, leading to increased loads of somatic point-mutations and deletions/insertions in the IgH locus, as well as heightened Ig secretion rates that result in abundant production of pathogenic autoantibodies. Accordingly, increased AID and Blimp-1 expression in lupus patients is associated with high levels of mutated IgG autoantibodies, which heighten disease activity (14, 69). Conversely, AID deficiency in MRL/Faslpr/lpr Aicda–/– protected mice against disease (64, 70); and, decreased AID expression in MRL/Faslpr/lpr HoxC4+/– and MRL/Faslpr/lpr Aicda+/– mice, which display 30-60% of the AID level of MRL/Faslpr/lpr Aicda+/+ mice, reduced autoantibody titers and delayed disease (14, 71). A role in promoting GC formation and generation of class-switched autoantibodies has been suggested for miR-155 (62, 72, 73), and reduced autoantibody production and autoimmunity have been reported in miR-155-deficient B6/Faslpr/lpr mice (74). This, however, likely resulted from dysregulation of a variety of genes in multiple immune cells, including derepressed expression of SHIP-1 in B cells, which led to mitigation of B cell activation, proliferation and autoantibody production. In our hands, HDI did not yield obvious alteration in GC formation or B cell SHIP-1 transcripts (not shown).
PRDM1 has been identified as one of the risk loci for lupus in human genetic association studies (75, 76) and increased Blimp-1 expression has been shown to parallel a surge in circulating plasma cells during disease flares (77). Blimp-1 is required for generation of short-lived and long-lived plasma cells. Unlike short-lived plasma cells, long-lived autoreactive, malignant or allergen-specific (IgE+) plasma cells are refractory to immunosuppression and irradiation (78). In our MRL/Faslpr/lpr mice, HDI-mediated upregulation of miR-30a and miR-125b, which are highly expressed in GC B cells and downregulated in plasma cells (32, 79), along with HDI-mediated upregulation of miR-23b, which is reduced in lupus (80), silenced Blimp-1 expression, thereby impairing plasma cell differentiation and compounding the negative effect of decreased CSR and SHM on the production of high-affinity class-switched autoantibodies. As Blimp-1 is required not only for formation but also for maintenance of plasma cells (4), HDI-mediated downregulation of Blimp-1 can lead to significantly long-lasting lower levels of autoantibodies by decreasing autoantibody-producing plasma cells. Accordingly, our preliminary data (not shown) suggest that the HDI beneficial effect is long lasting after withdrawal of the drug from treated autoimmune mice.
Butyrate is one of the SCFAs produced by gut commensal bacteria through dietary fiber fermentation (41) and is the most potent HDI among those SCFAs. It modulates the function of intestinal macrophages (81) and acts on naïve T cells to promote epigenetic changes that regulate the expression of genes responsible for differentiation into Treg cells and IL-10-producing T cells (42, 82). Our demonstration that butyrate (at a dose as low as 250 μM) modulates AID expression, CSR to IgG, IgA and IgE, as well as plasma cell differentiation through direct activity on B cells indicates that this HDI can play an important role in modulating antibody responses of gut lymphoid organs (in which butyrate occurs at 1-20 mM). This would be particularly true of the caecal patch, a major intestinal lymphoid organ in the proximal colon, as well as in Peyer’s patches, which are highly represented in the ileum, the portion of intestine that is immediately afferent to the proximal colon. Both the caecal patch and Peyer’s patches contain vast numbers of B cells committed to the production of IgA, and to a lesser extent, IgE (83, 84). Butyrate may also play an important role in limiting AID expression in the inflamed colonic mucosa, in which AID is induced by proinflammatory cytokines (85). By suppressing AID, butyrate could suppress inflammation-mediated neoplastic transformation leading to colorectal cancer (86), a process in which AID-mediated oncogenic mutagenesis play a significant role.
Metabolites from intestinal microbiota are key determinants of host-microbe mutualism and, consequently, the health or disease of not only the intestinal tract but also other organs and tissues (87-89). It has been suggested that SCFAs produced by gut commensal bacteria can distribute systemically and shape the immunological environment in the lung, thereby influencing the severity of allergic inflammation. Mice fed a high-fiber diet had increased circulating levels of SCFAs and were protected against allergic inflammation (mediated by IgE) of the lung, whereas a low-fiber diet decreased levels of SCFAs and increased allergic airway disease (87). A diverse microbial population, which would produce appropriate amount of SCFA HDI, particularly, butyrate, is required to maintain a baseline immune regulatory state, including IgG, IgA and IgE levels. Elevated serum IgE and CSR to IgE in B cells at mucosal sites in the absence of microbial colonization in germ-free mice and in mice with low-diversity gut microbiota would further emphasize an important role for gut commensal bacteria-produced butyrate in modulating IgE levels (90, 91). Altered composition and decreased bacterial diversity of gut microbiota would lead to changes in absolute and relative levels of SCFA HDI (89) and, therefore, changes in systemic IgG, IgA and IgE specificities and levels, and contribute to altered immunity and increased susceptibility to immune-mediated diseases.
Despite involving no human patient, our study provides a strong rationale and a mechanistic basis for use of HDI as epigenetic modulators of antibody responses, and as therapeutics for systemic autoimmunity and, possibly, IgE-mediated allergic responses. In our experiments, HDI was administered in drinking water, rather than parenterally as in previous studies (46), resulting in steady and well-tolerated therapeutic HDI levels. These were comparable to those of patients taking HDI per os (43) and without the concentration spikes associated with HDI injections (46). HDI administration in drinking water likely contributed to the effectiveness of modulation of T-dependent and T-independent antibody responses in healthy mice as well as to the dampening of the systemic autoantibody response, reduction in immunopathology and extended survival in lupus mice. Overall, our studies suggest a new and important therapeutic indication for VPA and butyrate, and, likely, other HDI, such as TSA and SAHA (suberoylanilide hydroxamic acid, approved by the FDA for the treatment of cutaneous T cell lymphoma). They also provide novel and significant mechanistic insights into epigenetic mechanisms of immunoregulation, as mediated by direct modulation of B cell-intrinsic functions, thereby offering new cues for further therapeutic approaches, as specifically targeted to B cells.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Christie Mortales, Julia Taylor, Isabella Stenmark, Sonia Aghera and Eun Jee Kim for their assistance in performing some experiments, and Dr. Irene Pederson for the pMIR-REPORT luciferase vector.
This work was supported by US National Institutes of Health grants AI079705, AI105813 and AI060573 (to P.C.), the Alliance for Lupus Research Target Identification in Lupus Grant ALR 295955 (to P.C.) and the Arthritis National Research Foundation grant (to H.Z.).
Abbreviations
- AID
activation-induced cytidine deaminase
- ANA
anti-nuclear antibody
- Blimp-1
B lymphocyte-induced maturation protein-1
- CFSE
carboxyfluorescein succinimidyl ester CSR, class switch DNA recombination
- HDAC
histone deacetylase
- HDI
histone deacetylase inhibitor
- miRNA
microRNA
- SCFA
short-chain fatty acid
- SHM
somatic hypermutation
- Ung
uracil DNA glycosylase
- VPA
valproic acid
- Xbp-1
X-box binding protein 1
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures and one table and can be found with this article online.
References
- 1.Xu Z, Zan H, Pone EJ, Mai T, Casali P. Immunoglobulin class-switch DNA recombination: induction, targeting and beyond. Nat. Rev. Immunol. 2012;12:517–531. doi: 10.1038/nri3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013;46:83–101. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G, Zan H, Xu Z, Casali P. Epigenetics of the antibody response. Trends Immunol. 2013;34:460–470. doi: 10.1016/j.it.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 5.Liu M, Duke JL, Richter DJ, Vinuesa CG, Goodnow CC, Kleinstein SH, Schatz DG. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–845. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 6.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–181. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 7.He B, Santamaria R, Xu W, Cols M, Chen K, Puga I, Shan M, Xiong H, Bussel JB, Chiu A, Puel A, Reichenbach J, Marodi L, Doffinger R, Vasconcelos J, Issekutz A, Krause J, Davies G, Li X, Grimbacher B, Plebani A, Meffre E, Picard C, Cunningham-Rundles C, Casanova JL, Cerutti A. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat. Immunol. 2010;11:836–845. doi: 10.1038/ni.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pone EJ, Xu Z, White CA, Zan H, Casali P. B cell TLRs and induction of immunoglobulin class-switch DNA recombination. Front. Biol. 2012;17:2594–2615. doi: 10.2741/4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura JK, Patel PJ, Al-Qahtani A, Zan H, Xu Z, Casali P. BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-kappaB pathway. Nat. Commun. 2012;3:767. doi: 10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerutti A, Cols M, Puga I. Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat. Rev. Immunol. 2013;13:118–132. doi: 10.1038/nri3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sayegh CE, Quong MW, Agata Y, Murre C. E-proteins directly regulate expression of activation-induced deaminase in mature B cells. Nat. Immunol. 2003;4:586–593. doi: 10.1038/ni923. [DOI] [PubMed] [Google Scholar]
- 12.Park SR, Zan H, Pal Z, Zhang J, Al-Qahtani A, Pone EJ, Xu Z, Mai T, Casali P. HoxC4 binds to the promoter of the cytidine deaminase AID gene to induce AID expression, class-switch DNA recombination and somatic hypermutation. Nat. Immunol. 2009;10:540–550. doi: 10.1038/ni.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mai T, Zan H, Zhang J, Hawkins JS, Xu Z, Casali P. Estrogen receptors bind to and activate the HOXC4/HoxC4 promoter to potentiate HoxC4-mediated activation-induced cytosine deaminase induction, immunoglobulin class switch DNA recombination, and somatic hypermutation. J. Biol. Chem. 2010;285:37797–37810. doi: 10.1074/jbc.M110.169086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White CA, Seth Hawkins J, Pone EJ, Yu ES, Al-Qahtani A, Mai T, Zan H, Casali P. AID dysregulation in lupus-prone MRL/Faslpr/lpr mice increases class switch DNA recombination and promotes interchromosomal c-Myc/IgH loci translocations: modulation by HoxC4. Autoimmunity. 2011;44:585–598. doi: 10.3109/08916934.2011.577128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, White CA, Lam T, Pone EJ, Tran DC, Hayama KL, Zan H, Xu Z, Casali P. Combinatorial H3K9acS10ph histone modification in IgH locus S regions targets 14-3-3 adaptors and AID to specify antibody class-switch DNA recombination. Cell Rep. 2013;5:702–714. doi: 10.1016/j.celrep.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, Hurt EM, Petroulakis E, Sonenberg N, Yewdell JW, Calame K, Glimcher LH, Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zan H, Tat C, Casali P. MicroRNA in lupus. Autoimmunity. 2014;47:272–285. doi: 10.3109/08916934.2014.915955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujimura S, Matsui T, Kuwahara K, Maeda K, Sakaguchi N. Germinal center B-cell-associated DNA hypomethylation at transcriptional regions of the AID gene. Mol. Immunol. 2008;45:1712–1719. doi: 10.1016/j.molimm.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Crouch EE, Li Z, Takizawa M, Fichtner-Feigl S, Gourzi P, Montaño C, Feigenbaum L, Wilson P, Janz S, Papavasiliou FN, Casellas R. Regulation of AID expression in the immune response. J. Exp. Med. 2007;204:1145–1156. doi: 10.1084/jem.20061952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taganov KD, Boldin MP, Baltimore D. MicroRNAs and immunity: tiny players in a big field. Immunity. 2007;26:133–137. doi: 10.1016/j.immuni.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 23.Belver L, de Yebenes VG, Ramiro AR. MicroRNAs prevent the generation of autoreactive antibodies. Immunity. 2010;33:713–722. doi: 10.1016/j.immuni.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, Hu-Li J, Lu K, Dubois W, Kitamura Y, Charles N, Sun HW, Muljo S, Schwartzberg PL, Paul WE, O'Shea J, Rajewsky K, Casellas R. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Yebenes VG, Bartolome-Izquierdo N, Ramiro AR. Regulation of B-cell development and function by microRNAs. Immunol. Rev. 2013;253:25–39. doi: 10.1111/imr.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat. Immunol. 2014;15:484–491. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 27.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Yebenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basso K, Schneider C, Shen Q, Holmes AB, Setty M, Leslie C, Dalla-Favera R. BCL6 positively regulates AID and germinal center gene expression via repression of miR-155. J. Exp. Med. 2012;209:2455–2465. doi: 10.1084/jem.20121387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Wang K, Han L, Zhang A, Shi Z, Zhang K, Zhang H, Yang S, Pu P, Shen C, Yu C, Kang C. PRDM1 is directly targeted by miR-30a-5p and modulates the Wnt/beta-catenin pathway in a Dkk1-dependent manner during glioma growth. Cancer Lett. 2013;331:211–219. doi: 10.1016/j.canlet.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Gururajan M, Haga CL, Das S, Leu CM, Hodson D, Josson S, Turner M, Cooper MD. MicroRNA 125b inhibition of B cell differentiation in germinal centers. Int. Immunol. 2010;22:583–592. doi: 10.1093/intimm/dxq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJ, Birolo RS, Moro M, Crosti MC, Gruarin P, Maglie S, Marabita F, Mascheroni D, Parente V, Comelli M, Trabucchi E, De Francesco R, Geginat J, Abrignani S, Pagani M. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat. Immunol. 2011;12:796–803. doi: 10.1038/ni.2057. [DOI] [PubMed] [Google Scholar]
- 34.Nambu Y, Sugai M, Gonda H, Lee CG, Katakai T, Agata Y, Yokota Y, Shimizu A. Transcription-coupled events associating with immunoglobulin switch region chromatin. Science. 2003;302:2137–2140. doi: 10.1126/science.1092481. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, Wuerffel R, Feldman S, Khamlichi AA, Kenter AL. S region sequence, RNA polymerase II, and histone modifications create chromatin accessibility during class switch recombination. J. Exp. Med. 2009;206:1817–1830. doi: 10.1084/jem.20081678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delcuve GP, Khan DH, Davie JR. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin. Epigenetics. 2012;4:5. doi: 10.1186/1868-7083-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reilly CM, Regna N, Mishra N. HDAC inhibition in lupus models. Mol. Med. 2011;17:417–425. doi: 10.2119/molmed.2011.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gottlicher M, Minucci S, Zhu P, Kramer OH, Schimpf A, Giavara S, Sleeman JP, Lo Coco F, Nervi C, Pelicci PG, Heinzel T. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20:6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davie JR. Inhibition of histone deacetylase activity by butyrate. J. Nutr. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 40.Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 41.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- 42.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 43.Atmaca A, Al-Batran SE, Maurer A, Neumann A, Heinzel T, Hentsch B, Schwarz SE, Hovelmann S, Gottlicher M, Knuth A, Jager E. Valproic acid (VPA) in patients with refractory advanced cancer: a dose escalating phase I clinical trial. Br. J. Cancer. 2007;97:177–182. doi: 10.1038/sj.bjc.6603851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Munster P, Marchion D, Bicaku E, Schmitt M, Lee JH, DeConti R, Simon G, Fishman M, Minton S, Garrett C, Chiappori A, Lush R, Sullivan D, Daud A. Phase I trial of histone deacetylase inhibition by valproic acid followed by the topoisomerase II inhibitor epirubicin in advanced solid tumors: a clinical and translational study. J. Clin. Oncol. 2007;25:1979–1985. doi: 10.1200/JCO.2006.08.6165. [DOI] [PubMed] [Google Scholar]
- 45.Pone EJ, Zan H, Zhang J, Al-Qahtani A, Xu Z, Casali P. Toll-like receptors and B-cell receptors synergize to induce immunoglobulin class-switch DNA recombination: relevance to microbial antibody responses. Crit. Rev. Immunol. 2010;30:1–29. doi: 10.1615/critrevimmunol.v30.i1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dowdell KC, Pesnicak L, Hoffmann V, Steadman K, Remaley AT, Cohen JI, Straus SE, Rao VK. Valproic acid (VPA), a histone deacetylase (HDAC) inhibitor, diminishes lymphoproliferation in the Fas -deficient MRL/lpr−/− murine model of autoimmune lymphoproliferative syndrome (ALPS) Exp. Hematol. 2009;37:487–494. doi: 10.1016/j.exphem.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wesemann DR, Magee JM, Boboila C, Calado DP, Gallagher MP, Portuguese AJ, Manis JP, Zhou X, Recher M, Rajewsky K, Notarangelo LD, Alt FW. Immature B cells preferentially switch to IgE with increased direct Smu to Sepsilon recombination. J. Exp. Med. 2011;208:2733–2746. doi: 10.1084/jem.20111155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zan H, White CA, Thomas LM, Mai T, Li G, Xu Z, Zhang J, Casali P. Rev1 recruits Ung to switch regions and enhances dU glycosylation for immunoglobulin class switch DNA recombination. Cell Rep. 2012;2:1220–1232. doi: 10.1016/j.celrep.2012.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zan H, Zhang J, Ardeshna S, Xu Z, Park SR, Casali P. Lupus-prone MRL/Faslpr/lpr mice display increased AID expression and extensive DNA lesions, comprising deletions and insertions, in the immunoglobulin locus: concurrent upregulation of somatic hypermutation and class switch DNA recombination. Autoimmunity. 2009;42:89–103. doi: 10.1080/08916930802629554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu Z, Fulop Z, Wu G, Pone EJ, Zhang J, Mai T, Thomas LM, Al-Qahtani A, White CA, Park SR, Steinacker P, Li Z, Yates J. r., Herron B, Otto M, Zan H, Fu H, Casali P. 14-3-3 adaptor proteins recruit AID to 5'-AGCT-3'-rich switch regions for class switch recombination. Nat. Struct. Mol. Biol. 2010;17:1124–1135. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalez-Garcia I, Ocana E, Jimenez-Gomez G, Campos-Caro A, Brieva JA. Immunization-induced perturbation of human blood plasma cell pool: progressive maturation, IL-6 responsiveness, and high PRDI-BF1/BLIMP1 expression are critical distinctions between antigen-specific and nonspecific plasma cells. J. Immunol. 2006;176:4042–4050. doi: 10.4049/jimmunol.176.7.4042. [DOI] [PubMed] [Google Scholar]
- 52.Tunyaplin C, Shaffer AL, Angelin-Duclos CD, Yu X, Staudt LM, Calame KL. Direct repression of prdm1 by Bcl-6 inhibits plasmacytic differentiation. J. Immunol. 2004;173:1158–1165. doi: 10.4049/jimmunol.173.2.1158. [DOI] [PubMed] [Google Scholar]
- 53.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Adv. Immunol. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 54.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, Chong AS, Klein U, Dinner AR, Singh H, Sciammas R. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38:918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ochiai K, Katoh Y, Ikura T, Hoshikawa Y, Noda T, Karasuyama H, Tashiro S, Muto A, Igarashi K. Plasmacytic transcription factor Blimp-1 is repressed by Bach2 in B cells. J. Biol. Chem. 2006;281:38226–38234. doi: 10.1074/jbc.M607592200. [DOI] [PubMed] [Google Scholar]
- 56.Yasuda T, Hayakawa F, Kurahashi S, Sugimoto K, Minami Y, Tomita A, Naoe T. B cell receptor-ERK1/2 signal cancels PAX5-dependent repression of BLIMP1 through PAX5 phosphorylation: a mechanism of antigen-triggering plasma cell differentiation. J. Immunol. 2012;188:6127–6134. doi: 10.4049/jimmunol.1103039. [DOI] [PubMed] [Google Scholar]
- 57.Arzenani MK, Zade AE, Ming Y, Vijverberg SJ, Zhang Z, Khan Z, Sadique S, Kallenbach L, Hu L, Vukojevic V, Ekstrom TJ. Genomic DNA hypomethylation by histone deacetylase inhibition implicates DNMT1 nuclear dynamics. Mol. Cell Biol. 2011;31:4119–4128. doi: 10.1128/MCB.01304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, Boesche M, Delling M, Dumpelfeld B, Eberhard D, Huthmacher C, Mathieson T, Poeckel D, Reader V, Strunk K, Sweetman G, Kruse U, Neubauer G, Ramsden NG, Drewes G. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 2011;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 59.Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, Koessler T, Ding XC, Chanson AL, Reymond MK, Miconnet I, Schrenzel J, François P, Calandra T. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood. 2011;117:1205–1217. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
- 60.Villagra A, Sotomayor EM, Seto E. Histone deacetylases and the immunological network: implications in cancer and inflammation. Oncogene. 2010;29:157–173. doi: 10.1038/onc.2009.334. [DOI] [PubMed] [Google Scholar]
- 61.Akimova T, Beier UH, Liu Y, Wang L, Hancock WW. Histone/protein deacetylases and T-cell immune responses. Blood. 2012;119:2443–2451. doi: 10.1182/blood-2011-10-292003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kienzler AK, Rizzi M, Reith M, Nutt SL, Eibel H. Inhibition of human B-cell development into plasmablasts by histone deacetylase inhibitor valproic acid. J. Allergy Clin. Immunol. 2013;131:1695–1699. doi: 10.1016/j.jaci.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 64.Jiang C, L. Zhao M, Scearce RM, Diaz M. Activation-induced deaminase-deficient MRL/lpr mice secrete high levels of protective antibodies against lupus nephritis. Arthritis Rheum. 2011;63:1086–1096. doi: 10.1002/art.30230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harada Y, Muramatsu M, Shibata T, Honjo T, Kuroda K. Unmutated immunoglobulin M can protect mice from death by influenza virus infection. J. Exp. Med. 2003;197:1779–1185. doi: 10.1084/jem.20021457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang J, Yan H, Zhuang S. Histone deacetylases as targets for treatment of multiple diseases. Clin. Sci. 2013;124:651–662. doi: 10.1042/CS20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee JH, Choy ML, Ngo L, S. Foster S, Marks PA. Histone deacetylase inhibitor induces DNA damage, which normal but not transformed cells can repair. Proc. Natl. Acad. Sci. USA. 2010;107:14639–14644. doi: 10.1073/pnas.1008522107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol. Med. 2011;17:714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luo J, Niu X, Liu H, Zhang M, Chen M, Deng S. Up-regulation of transcription factor Blimp1 in systemic lupus erythematosus. Mol. Immunol. 2013;56:574–582. doi: 10.1016/j.molimm.2013.05.241. [DOI] [PubMed] [Google Scholar]
- 70.Jiang C, Foley J, Clayton N, Kissling G, Jokinen M, Herbert R, Diaz M. Abrogation of lupus nephritis in activation-induced deaminase-deficient MRL/lpr mice. J. Immunol. 2007;178:7422–7431. doi: 10.4049/jimmunol.178.11.7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jiang C, Zhao ML, Diaz M. Activation-induced deaminase heterozygous MRL/lpr mice are delayed in the production of high-affinity pathogenic antibodies and in the development of lupus nephritis. Immunology. 2009;126:102–113. doi: 10.1111/j.1365-2567.2008.02882.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 74.Thai TH, Patterson HC, Pham DH, Kis-Toth K, Kaminski DA, Tsokos GC. Deletion of microRNA-155 reduces autoantibody responses and alleviates lupus-like disease in the Fas(lpr) mouse. Proc. Natl. Acad. Sci. USA. 2013;110:20194–20199. doi: 10.1073/pnas.1317632110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Odendahl M, Jacobi A, Hansen A, Feist E, Hiepe F, Burmester GR, Lipsky PE, Radbruch A, Dorner T. Disturbed peripheral B lymphocyte homeostasis in systemic lupus erythematosus. J. Immunol. 2000;165:5970–5979. doi: 10.4049/jimmunol.165.10.5970. [DOI] [PubMed] [Google Scholar]
- 76.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Rantapaa-Dahlqvist S, Baechler EC, Brown EE, Alarcon GS, Edberg JC, Ramsey-Goldman R, McGwin G, Jr., Reveille JD, Vila LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Ronnblom L, Criswell LA, Syvanen AC, Behrens TW, Graham RR. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat. Genet. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guerra SG, Vyse TJ, Cunninghame Graham DS. The genetics of lupus: a functional perspective. Arthritis Res. Ther. 2012;14:211. doi: 10.1186/ar3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hiepe F, Dorner T, Hauser AE, Hoyer BF, Mei H, Radbruch A. Long-lived autoreactive plasma cells drive persistent autoimmune inflammation. Nat. Rev. Rheumatol. 2011;7:170–178. doi: 10.1038/nrrheum.2011.1. [DOI] [PubMed] [Google Scholar]
- 79.Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri DA, Friedman DR, Weinberg JB, Lipsky PE, Dave SS. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113:4586–4594. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu S, Pan W, Song X, Liu Y, Shao X, Tang Y, Liang D, He D, Wang H, Liu W, Shi Y, Harley JB, Shen N, Qian Y. The microRNA miR-23b suppresses IL-17-associated autoimmune inflammation by targeting TAB2, TAB3 and IKK-alpha. Nat. Med. 2012;18:1077–1086. doi: 10.1038/nm.2815. [DOI] [PubMed] [Google Scholar]
- 81.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA. 2014;111:2247–2252. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, Ganapathy V. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cerutti A. The regulation of IgA class switching. Nat. Rev. Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cerutti A, Chen K, Chorny A. Immunoglobulin responses at the mucosal interface. Annu. Rev. Immunol. 2011;29:273–293. doi: 10.1146/annurev-immunol-031210-101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Takai A, Marusawa H, Minaki Y, Watanabe T, Nakase H, Kinoshita K, Tsujimoto G, Chiba T. Targeting activation-induced cytidine deaminase prevents colon cancer development despite persistent colonic inflammation. Oncogene. 2012;31:1733–1742. doi: 10.1038/onc.2011.352. [DOI] [PubMed] [Google Scholar]
- 86.Goncalves P, Martel F. Butyrate and colorectal cancer: the role of butyrate transport. Curr. Drug Metab. 2013;14:994–1008. doi: 10.2174/1389200211314090006. [DOI] [PubMed] [Google Scholar]
- 87.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, Marsland BJ. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 88.Dorrestein PC, Mazmanian SK, Knight R. Finding the missing links among metabolites, microbes, and the host. Immunity. 2014;40:824–832. doi: 10.1016/j.immuni.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thorburn AN, Macia L, Mackay CR. Diet, Metabolites, and “Western-Lifestyle” Inflammatory Diseases. Immunity. 2014;40:833–842. doi: 10.1016/j.immuni.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 90.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, Bushman FD, Artis D. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013;14:559–570. doi: 10.1016/j.chom.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.