Abstract
Amyotrophic Lateral Sclerosis has been linked to the gain of aberrant function of superoxide dismutase, Cu,Zn-SOD1 upon protein misfolding. The mechanism of SOD1 misfolding is thought to involve mutations leading to the loss of Zn, followed by protein unfolding, and aggregation. We show that the removal of Zn from SOD1 may not lead to an immediate unfolding, but immediately deactivates the enzyme through a combination of subtle structural and electronic effects. Using Quantum Mechanics/Discrete Molecular Dynamics, we showed that Zn-less wild type SOD1 and its D124N mutant that does not bind Zn both have at least metastable folded states. In those states, the reduction potential of Cu increases, leading to the presence of detectable amounts of Cu(I) instead of Cu(II) in the active site, as confirmed experimentally. The Cu(I) protein cannot participate in the catalytic Cu(I) – Cu(II) cycle. However, even without the full reduction to Cu(I), the Cu site in the Zn-less variants of SOD1 is shown to be catalytically incompetent: unable to bind superoxide in a way comparable to the wild type SOD1. The changes are more radical and different in the D124N Zn-less mutant than in the Zn-less wild type SOD1, suggesting D124N being perhaps not the most adequate model for Zn-less SOD1. Overall, Zn in SOD1 appears to be influencing the Cu site directly by adjusting its reduction potential and geometry. Thus, the role of Zn in SOD1 is not just structural, as was previously thought; it is a vital part of the catalytic machinery.
I. Introduction
Superoxide dismutase, SOD1, is an enzyme responsible for the removal of superoxide, (O2)−, (Eq. 1), a toxic product linked to aging and other negative effects in the body.
| (1) |
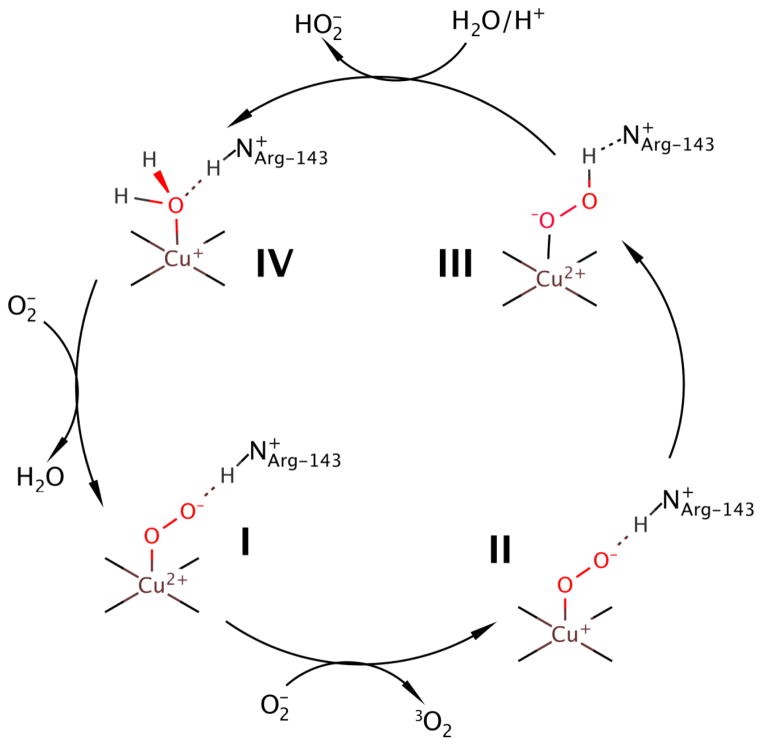
The dismutation/disproportionation mechanism is shown in Scheme 1. (1–3) In this mechanism, superoxide is guided to the Cu(II) site by the electrostatic loop (Figure 1) via the Arg-143 or R143 residue (I in Scheme 1).(4–7) Upon oxidation of superoxide to triplet oxygen dimer, the Cu(II) reduces to Cu(I) (II in Scheme 1), which is followed by proton transfer to the substrate and oxidation of Cu(I) to Cu(II) (III in Scheme 1). A proton is delivered to the active site and hydroperoxide anion leaves, while Cu coordinates a water molecule, and the protonation state of R143 gets restored (IV in Scheme 1). Lastly, substitution of the water molecule by another superoxide causes the reforming of the initial complex. The critical aspect of SOD1 functionality is the efficient cycling of the Cu center between Cu(I) and Cu(II) forms.
Scheme 1.
Schematic showing the dismutation mechanism of superoxide in WT-SOD1.(1)
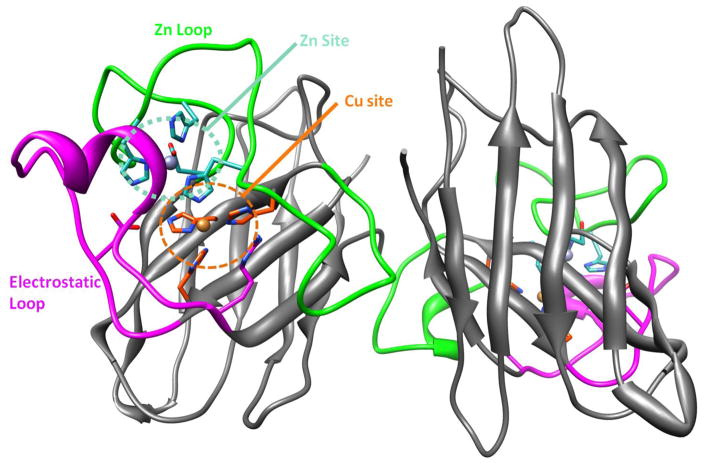
Figure 1.
WT-SOD1 dimer showing the Cu site (H46, H48, H120, Cu (large orange ball); orange region), Zn site (H63, H80, H71, D83, Zn (large gray ball); teal region), Zn loop (residues 50 to 83; bright green region), and electrostatic loop (residues 121 to 142; magenta region); R143 is shows at the end of the electrostatic loop.
Aggregation and loss of function of SOD1 is known to be associated with Amyotrophic Lateral Sclerosis, or ALS, a neurodegenerative disease that affects the motor neurons of afflicted patients.(8–12) Aggregation results primarily from mutations in SOD1, and over 100 mutant forms of SOD1 have been linked to ALS.(8,13–15)
SOD1 is a dimer (Figure 1) and aggregation is initiated by first the formation of monomers followed by the release of metals and, finally, aggregation due to loss of key structural support in the enzyme.(13) In human SOD1, catalysis occurs at the Cu site, whereas the Zn site is believed to be important for the support of the whole protein structure.(16) The Zn-less monomer structure is the precursor to SOD1 aggregates. The effect of Zn loss has been addressed experimentally.(16) Kinetic studies have shown that, under dialysis conditions, dimer dissociation, and the aggregation of the monomers are primarily coupled to metal loss.(13) The X-ray structures of Zn-less wild type (WT) SOD1 have distortions of the Zn loop and electrostatic loop primarily near to the Zn site, whereas Cu-deficient SOD1 largely retains its structure.(7) Previous computational studies on the SOD1 enzyme and its variants replicated the experiment in predicting an aggregation prone misfolded Zn-less state.(17,18) These studies also show that there is a strong positive correlation in the motion between residues on the active site loop, which may explain the ease of aggregation upon single mutations.(8) Thus, the structural role of Zn was established. However, the Zn site is located very near the Cu site and shares the bridging H63 residue. Thus, Zn is likely to be important also for the enforcement of a particular coordination of Cu, and influence the Cu site electrostatically. Hence, Zn might be a vital part of the catalytic machinery. In this work, we address this aspect.
We focus on the effect of Zn removal on the protein structure, electronic structure of the Cu site, and catalytic functionality of SOD1. Unlike the earlier studies that used classical force-field based methods, we use mixed quantum-classical methodology to more rigorously assess the behavior of the Cu site. In experimental studies of Zn-less SOD1, mutants unable to bind or retain Zn are used, since removing Zn from the wild type (WT) SOD1 appears to be difficult without also losing Cu.(7) Computationally, we have access to the WT Zn-less SOD1. We employ mixed quantum-classical simulations (QM/DMD(19)), and ab initio electronic structure calculations, described in Section Materials and Methods. The variants studied here are the WT-SOD1 (WT-SOD1), Zn-less WT-SOD1, and the Zn-less D124N variant of SOD1 (D124N-SOD1). D124N-SOD1 was chosen because it is one of the primary mutants used in experiments as a model of Zn-less SOD1; D124 is a residue coordinating Zn, and it is also a secondary bridging residue between the Cu and Zn sites. We test our theoretical predictions experimentally.
II. Results and Discussion
Zn-less WT- and D124N-SOD1 structures
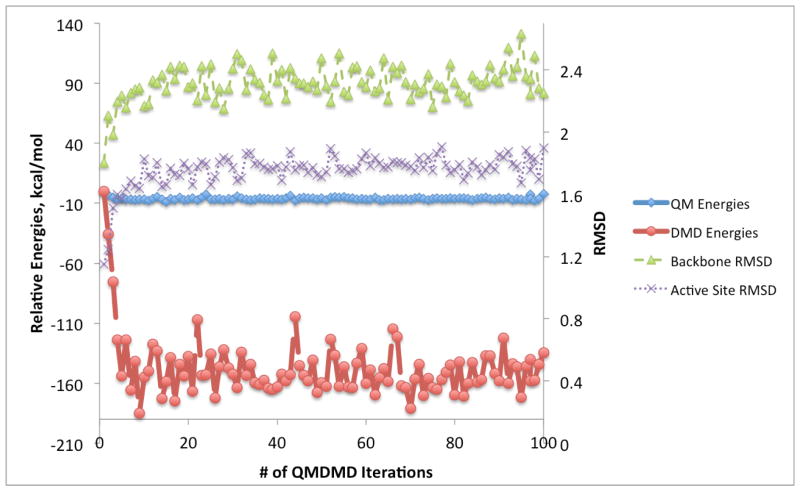
In Figure 2, the structures of the active sites and portions of the protein treated at the QM and DMD levels are shown, and detailed are described in the Section Materials and Methods. The results of QM/DMD simulations for the Zn-less WT-SOD1 (WT-SOD1), and Zn-less D124N-SOD1 are shown in Figures 3 and 4, respectively. These are the graphs showing changes in QM energies, DMD energies, backbone RMSD, and active site all-atom RMSD in the course of the simulations. The initial input structure taken from PDB and modified for our purposes is used as a reference in these plots. The convergence of the results is apparent from Figure 3 and 4. The fairly large fluctuations of the DMD energies are typical, in view of the abrupt square-well potentials used. In a number of previous studies we have shown that this is adequate and does not prevent QM/DMD from recapitulating native protein structures in agreement with the experiment.(19, 29, 31, 32) Importantly, the fluctuations of the QM energies and RMSDs of the QM region are small at convergence.
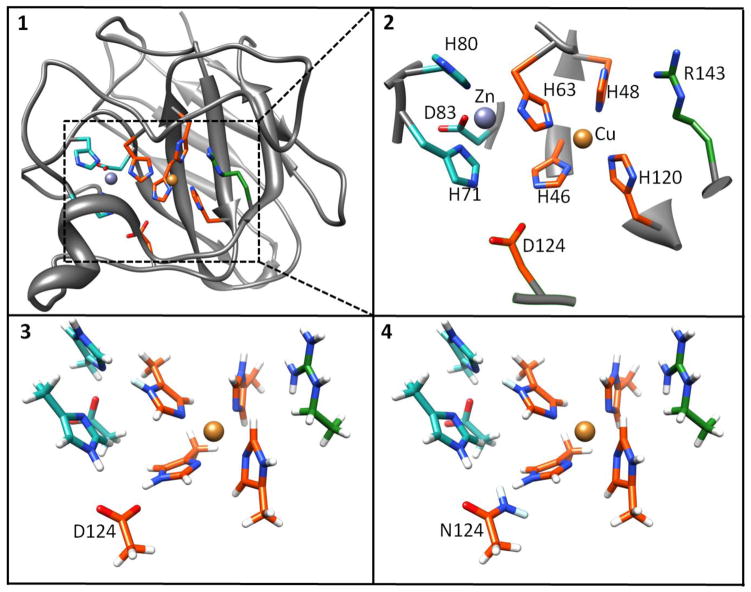
Figure 2.
1. SOD1 monomer with active site highlighted and boxed. 2. Expanded box showing the active site region; the orange zone represents the assigned QM region used for QM/DMD calculations, the included green regions were added for QM-only studies of properties. 3. Side-chain truncated and hydrogen capped expanded active site for Zn-less WT-SOD1 4. Side-chain truncated and hydrogen capped expanded active site for Zn-less D124N-SOD1 mutant. The coordinates of the capping hydrogen and its attached carbon are frozen during QM region optimizations.
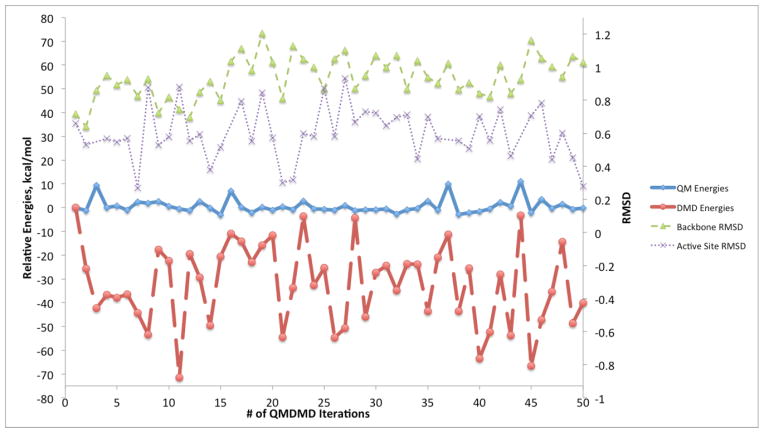
Figure 3.
Graph showing convergence of QM/DMD simulations in terms of the QM energies of the active site (bold blue solid line), DMD energies (bold red dashed line), backbone RMSD (thin green dashed line), and active site RMSD (thin purple dotted line) for Zn-less WT-SOD1. RMSD units are in Å.
Figure 4.
Graph showing convergence of QM/DMD simulations in terms of the QM energies of the active site (bold blue solid line), DMD energies (bold red dashed line), backbone RMSD (thin green dashed line), and active site RMSD (thin purple dotted line) for Zn-less D124N-SOD1 mutant. RMSD units are in Å.
From Figure 3, the QM energies for Zn-less WT-SOD1 mostly decrease by less than 10 kcal/mol. Most of the deviation in energy is seen in the DMD energies, which show energy decreases ranging from −5 to −70 kcal/mol, with an average decrease of approximately −35 kcal/mol over the course of 50 iterations. This indicates the rearrangements in the DMD region and developing deviations from the X-Ray structure. The backbone RMSD stabilizes after 20 iterations at ca. 1.0 Å. The active site all-atom RMSD stabilizes at ca. 0.6 Å. The overlaid structures for Iterations 20 to 50 are shown in the Supporting Information (Figure S1). QM/DMD simulations predict a clear shift in the structure of the Zn-less WT-SOD1 relative to the starting X-ray structure of WT-SOD1, and stabilization in a new basin. Based on what was previously known about the Zn-less SOD1, it is surprising that the changes in the structure of the protein are not more dramatic. No unfolding was observed in the time-scale of the simulation. The found folded state thus must have a finite life-time, though perhaps it is still metastable.
The QM/DMD simulation for the D124N mutant took longer to converge. In Figure 4, the QM energies for Zn-less D124N-SOD1 decreased by less than 15 kcal/mol. In fact, there are greater deviations in all energies and RMSDs as compared to the Zn-less WT-SOD1. The DMD energies decreased by ca. 160 kcal/mol relative to the initial structure, where these DMD energy changes range from −100 to −180 kcal/mol. For the backbone, the RMSD value stabilized after about 50 iterations at around 2.3 Å, which is a representation of a larger change in the backbone structure as compared to that for Zn-less WT-SOD1. The active site RMSD stabilizes at ca. 1.8 Å after iteration 50. The overlaid structures for Iterations 50 to 100 are shown in the Supporting Information. The Zn-less D124N-SOD1 shows a clear stabilization in the new well, different from both WT-SOD1, and Zn-less WT-SOD1. Still, the change in structure is rather modest, considering the anticipated unfolding. These results again suggest that there is at least a metastable minimum corresponding to the folded Zn-less D124N-SOD1. It is also curious that the changes in the two Zn-less variants are quite different, potentially alerting against the adequacy of the D147N mutant as a model for Zn-less SOD1.
From the converged QM/DMD iterations, initial structures for Zn-less WT-SOD1, and Zn-less D124N-SOD1 were selected for bond and charge analysis, as well as for the study of binding of superoxide to the Cu site.
Details of changes in the active sites SOD1 variants upon Zn removal
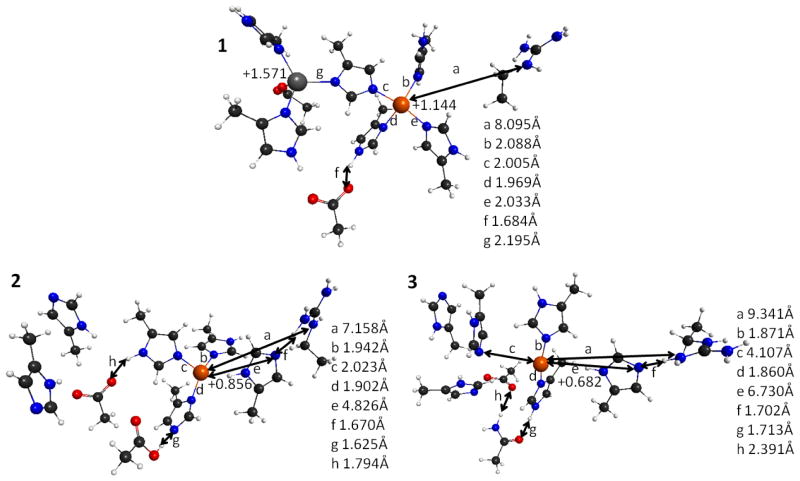
The effects of Zn-deficiency on bond lengths around the Cu site, as well as the Cu charge without the superoxide substrate are shown in Figure 5. The reference WT-SOD1 structure was obtained by cutting the larger active site containing both Zn and Cu with their coordination spheres out of the PDB structure, fixing the points of attachment to the rest of the protein, and optimizing at the QM level. This structure is in close agreement with the X-Ray data. There are four ligands coordinating Cu in a distorted square planar form (H46, H48, H63, and H120), with bond lengths shorter than 2.1 Å. However, Zn-less WT-SOD1 loses one of these ligands (H120), with the coordinating nitrogen on the imidazole being 4.826 Å away from Cu (label e in Figure 5.2). The Zn-less D124N-SOD1 variant differs from WT-SOD1 even more dramatically: the coordinating nitrogen on the H120 imidazole is now 6.730 Å away from Cu (label e in Figure 5.3), and the coordinating nitrogen on the H63 imidazole is 4.107 Å away (label c in Figure 5.3). Indeed, Zn-less WT-SOD1 and its D124N-SOD1 variant exhibit very different changes in the coordination of Cu.
Figure 5.
Representative structures of the Cu and Zn sites of the SOD1 systems resulted from QM/DMD equilibration (X-ray structure for WT-SOD1), showing specific bond lengths and NPA atomic charges. 1. WT-SOD1 2. Zn-less WT-SOD1. 3. Zn-less D124N-SOD1. Charges are labeled on the structures, whereas bond lengths are shown in the legends. Coordinates and all atomic charges are given in the Supporting Information.
Focusing on the R143 residue, in the WT-SOD1 this residue is far from the Cu site (ca. 8 Å) and adopts a sterically favored position (label a in Figure 5.1). This residue comes into play during the catalytic mechanism (Scheme 1). However, in both Zn-less WT-SOD1 and Zn-less D124N-SOD1, R143 develops a H-bonding contact with H120 departed from the Cu site (label f in Figures 5.2 and 5.3).
The Zn cation in the WT-SOD1 shows coordination with the four ligands (H63, H71, H80, and D83), H63 being a bridge to the Cu site. Once Zn is removed, this site restructures: residues that used to coordinate Zn start interacting with each other and also with the residue D124 (in the Zn-less WT-SOD1). D83 now forms a hydrogen bond of 1.794Å with the bridging H63 (label h in Figure 5.2). D124 forms a hydrogen bond of 1.624Å with H46 (label g in Figure 5.2), and engages in the shared proton relationship, with H now preferentially residing on D124 rather than on H46. H46 is coordinated to Cu, and the fact that it easily gives away its second proton indicates a change in the electronic structure of Cu (see discussion below).
The distortions are more dramatic for Zn-less D124N-SOD1. The bridging H63 no longer coordinates with Cu (label g in Figure 5.2). The H-bonding network is formed with mutant secondary bridging residue N124. There is a H-bond between N124 and H46 of 1.713Å (label g in Figure 5.3) but without H-transfer, and there is an H-bond of 2.391Å between N124 and D83 (label h in Figure 5.3).
The geometric changes in Zn-less WT-SOD1 and D124N-SOD1 are accompanied by changes in the electronic structure of the rest of the active site. Notice that in order to be catalytic, Cu needs to remain Cu(II) before binding superoxide (Scheme 1, I), and maintain the proper reduction potential for the dismutation mechanism to work. The NPA charge on the Cu site in WT-SOD1 as +1.144 (Figure 5.1), corresponding to the formal oxidation state of Cu(II). However, in both Zn-less SOD1 variants the charge on Cu drops significantly (Figure 5.2, 5.3), to +0.856 in Zn-less WT-SOD1, and +0.682 in Zn-less D124N. Notice again the appreciable difference between these two Zn-less proteins. Since the number of electrons remains unchanged in our calculations, and Cu remains formally Cu(II), then the change of charge indicates the change in the reduction potential, i.e. the Cu site in Zn-less proteins seems to be more readily reduced.
As an estimate for the reduction potentials for SOD1 variants, the energies of the LUMO and HOMO-LUMO gaps were calculated (Table 1). Also, the HOMO, and LUMO isosurfaces are plotted in Figure 6. The HF LUMO energies will be used to represent the reduction potential (E0) based on, E0 ~ −EA (Electron Affinity) = ELUMO, in accord with Koopmans theorem. (20–27) The WT-SOD1 has the lowest E0 (the energy of the LUMO is −9.995eV), whereas the Zn-less WT-SOD1 and Zn-less D124N-SOD1 have much higher E0 values (the energies of the LUMO are −1.076 eV and −1.629 eV, respectively). This trend indicates that the Zn-less forms of SOD1 are much more easily reduced, i.e. go from Cu(II) to Cu(I) in the presence of a suitable source of electrons. This correlates with the decrease of the positive charge on Cu in the Zn-less forms; the Cu centers pull electrons from the surroundings.
Table 1.
HOMO-LUMO gaps, LUMO energies, and NPA charges on Cu for WT-SOD1, Zn-less WT-SOD1, and Zn-less D124N-SOD1, without and with superoxide bound. Results obtained with TPSSh. For charges: TPSSh without solvent, with implicit solvent (square brackets), B3-LYP (italics) and B2-PLYP (bold italics) charges.
| SOD variants | HOMO-LUMO gap, eV | LUMO, eV | Q(Cu) |
|---|---|---|---|
|
| |||
| WT-SOD1 | 3.534 | −9.995 | 1.144 [1.233], 1.145, 1.268 |
| WT-SOD1 + Superoxide | 0.003 | −8.885 | 1.165 [1.257], 1.193, 1.482 |
| Zn-less WT-SOD1 | 6.229 | −1.076 | 0.856 [1.101], 0.841, 0.784 |
| Zn-less WT-SOD1 + Superoxide | 7.63 | 1.443 | 1.162 [1.225], 1.214, 1.498 |
| Zn-less D124N-SOD1 | 9.48 | −1.629 | 0.682 [0.688], 0.679, 0.726 |
| Zn-less D124N-SOD1 + Superoxide | 8.548 | −0.951 | 0.883 [0.972], 0.868, 0.796 |
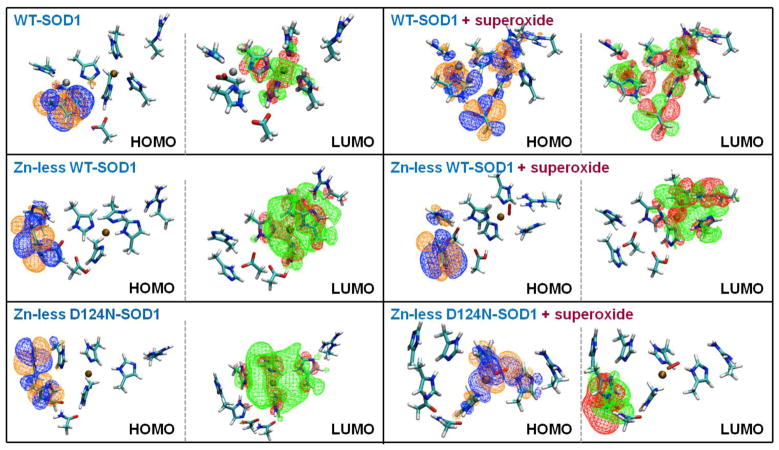
Figure 6.
The HOMOs and the LUMOs of WT-SOD1, Zn-less WT-SOD1, and Zn-less D124N-SOD1, without (left) and with (right) superoxide bound.
The changes in the electronic structure of the active sites upon Zn removal are further illustrated by the HOMOs and LUMOs shown in Figure 6 (left panel). Notice in particular the change of the shape of the LUMO upon Zn removal. The electron density is spread over a much larger region (including H46, H48, H63, H120, and R143), indicating the mixing of the atomic orbitals of the Cu site with the states of the ligands. This mixing is inaccessible in the WT-SOD1, where the LUMO concentrates on the Cu site. The HOMOs in all protein variants is localized on the H71 and D83 residues, (former) ligands of the Zn cation.
The change in the reduction potential of the Cu site must have direct consequences for the catalytic activity of the Zn-less SOD1 variants, since the enzyme depends on the efficient cycling between Cu(II) and Cu(I) (Scheme 1). If the Cu site preferentially exists as Cu(I), the catalytic efficiency will be harmed. To experimentally test for the possibility of Cu(I) forming in the active site of Zn-less SOD1, we used a colorimetric assay for Cu(I) detection.
Experimental confirmation of Cu(I) in D124N-SOD1
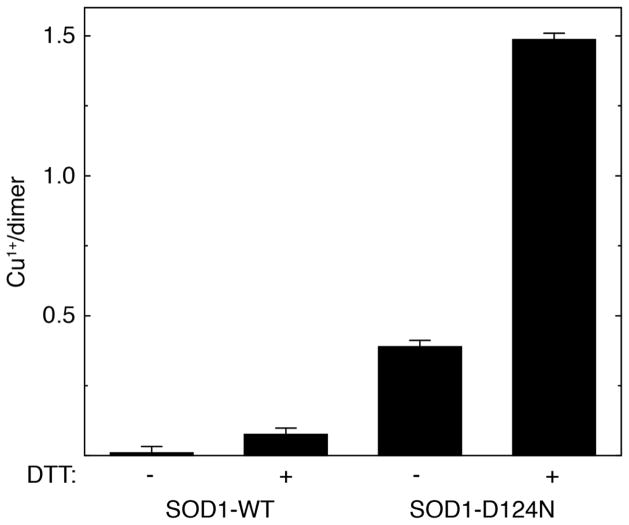
Experimental results are shown in Figure 7. The Zn-less D124N-SOD1 variant contains an estimated 0.4 Cu(I) per dimer (corresponding to a reduction of approximately 23% of the total bound copper) prior to the addition of reducing agent. Incubation of D124N-SOD1 with 1 mM DTT increases [Cu(I)] to ~1.5 per dimer, over 85% of the total copper. As previously observed,(28) very little Cu(I) can be detected in metal-replete WT-SOD1, even following incubation with 1 mM DTT. These results demonstrate the susceptibility of Cu(II) to be reduced in Zn-less SOD1, even in the absence of reducing agents in the solvent.
Figure 7.
Cu(I) content of Zn-replete WT-SOD1 (labeled SOD1-WT) and Zn-less D124N-SOD1 (labeled SOD1-D124N) in the absence (−) and presence (+) of a reducing agent (1 mM DTT). Bars show average values ± S. D. from triplicate experiments.
Thus, indeed, Zn-less SOD1 is prone to deactivation due to the reduction of Cu(II) to Cu(I), without the need for unfolding and aggregation of the protein. However, most importantly, this experiment confirms the dramatic change in the reduction potential of Cu, which in itself should impact the catalytic mechanism, even without the complete reduction of Cu in the resting state of the protein.
Effect of Zn removal on binding the superoxide substrate in the catalytic mechanism of SOD1
In order to determine how the large structural distortions and changes in the electronic structure of the Cu site would affect the catalytic mechanism of Zn-less SOD1, we tested the binding of superoxide to the Cu site in the considered protein variants. The full mechanistic investigation is formidable and beyond the scope of the present study. The coordination of O2− must be assisted by the interaction with the residue R143, as in the WT enzyme (Scheme 1), and therefore, R143 is included in these calculations. The structures of the complexes of the SOD1 variants with superoxide are shown in Figure 8. These were obtained by taking representative structures from the equilibrated QM/DMD ensembles (the x-ray structure in the case of the WT-SOD1), binding the substrate, and optimizing the complexes at the QM level, keeping the atoms attaching the complexes to the protein backbone fixed. The WT-SOD1 has the lowest binding energy (−10.991eV), compared to Zn-less WT-SOD1 (−7.867eV) and Zn-less D124N-SOD1 (−6.895eV), i.e. the WT-SOD1-superoxide complex is the most stable of the three.
Figure 8.
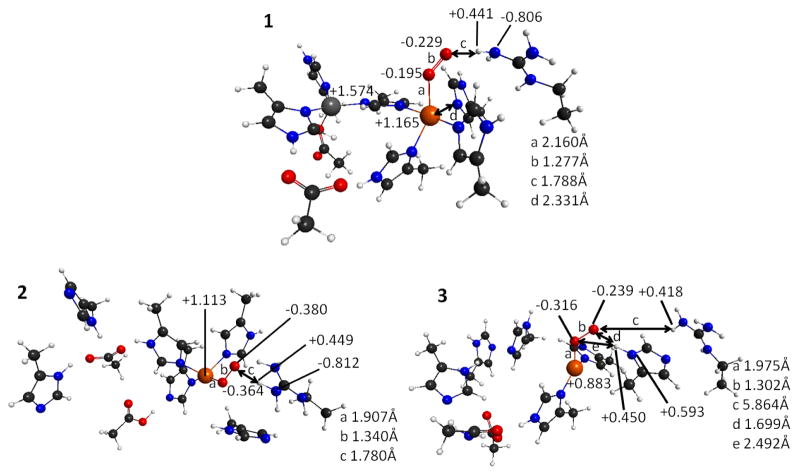
Structures of the active sites of the SOD1 systems with superoxide bound. 1. WT-SOD1. 2. Zn-less WT-SOD1. 3. Zn-less D124N-SOD1. Charges are labeled on the structure, whereas bond lengths are shown in the legend using labels a, b, c etc. More details on structures and charges are given in the Supporting Information.
There are significant differences between the WT-SOD1 and the Zn-less variants, and also between the two Zn-less variants, in how they bind the substrate. For the WT-SOD1 (Figure 8.1), the Cu site maintains the highly positive partial charge of +1.165, experiencing only a small change of +0.021 after adding superoxide. These positive charges are indicative of Cu(II), which is the Cu form required in the initial binding of superoxide mechanism. The superoxide substrate replaces the H120 ligand of Cu, which moves to 2.331Å (d in Figure 8.1) away from Cu. The Cu site thus keeps the original distorted square planar coordination with the remaining Cu ligands of H46, H48, and H63 and superoxide bound. It is noted that the calculated bond lengths for the unbound superoxide and neutral triplet oxygen dimer are 1.360Å and 1.215Å respectively. The bond length in superoxide bound to the WT-SOD1 is 1.277Å, i.e. between the calculated bond lengths for unbound superoxide and neutral triplet oxygen dimer. This distance indicates that the Cu(II) already engages in electron transfer and transitions the superoxide towards the neutral oxygen dimer. The charges on the superoxide are as follows: the oxygen attached to the Cu (OCu1) is charged −0.195, and the other oxygen (OCu2) is charged −0.229. OCu2 forms a 1.788Å H-bond to the NH-group of R143, and the H on that group holds the charge of +0.441. The dihedral angle Cu-OCu1-OCu2-H-N-(R143) is 4°, i.e. these atoms coordinate in a plane, as in I (Scheme 1). The structure is completely consistent with the proposed catalytic mechanism (Scheme 1).
Zn-less WT-SOD1 and Zn-less D124N-SOD1 produce very different coordination modes with superoxide. In Zn-less WT-SOD1 (Figure 8.2), Cu holds a partial charge of +1.113 (lower than in WT-SOD1), and exhibits a greater change in charge of +0.257 after adding superoxide (unlike the negligible change seen in WT-SOD1). The distorted square planar coordination of Cu changes in the same way as in the WT-SOD1. However, the superoxide bond length is 1.340 Å, i.e. nearly identical to that in the unbound superoxide (1.360 Å), consistent with the weaker binding to Cu in this protein variant. The charge separation between OCu1 and OCu2 is now very small, 0.016, i.e. about half that for WT-SOD1. The Cu-OCu1-OCu2-H-(N-R143) dihedral angle is 156°, which is reasonably close to 110° in structure III (Scheme 1)(1), and definitely very different from the almost planar dihedral angle found in WT-SOD1 with bound superoxide (Figure 8.1) and I (Scheme 1). The H from R143 does not spontaneously transfer to superoxide as in III (Scheme 1), however.
These observations confirm that the redox behavior of Zn-less WT-SOD1 is radically different from that of WT-SOD1, and in a way akin to the protein containing Cu(I) in the resting state. The similarity between the structure with the superoxide bound to Zn-less WT-SOD1 and structure III indicates that its catalytic functionality is depleted, since the steps in the oxidation of superoxide between I and II cannot occur.
Zn-less D124N-SOD1 (Figure 8.3) behaves completely differently from both WT-SOD1 and Zn-less TW-SOD1, and that indicates significant mechanistic differences, and again alerts against the adequacy of this system as a model for Zn-less SOD1. There is a complete loss of interaction between the superoxide and R143, and instead there is a new H-bond between the superoxide and H120. The partial charge on Cu is +0.883 (lower than that for WT-SOD1 and Zn-less WT-SOD1), indicating that Cu is prone to become Cu(I) in this Zn-less variant, as also seen in our experiment. The change of charge upon adding superoxide is +0.201 (quite large as compared to WT-SOD1). The coordination of Cu is no longer distorted square planar but almost trigonal planar, more characteristic of Cu(I) than Cu(II). The superoxide bond length upon coordination is 1.302 Å. The partial charges are swapped on the superoxide oxygens, where the charge on OCu1 is now more negative (−0.316) than that on OCu2 (−0.239). The Cu-OCu1-OCu2-H-(N-Arg-143) dihedral angle is 26°, i.e. not resembling III (Scheme 1) as in the case of Zn-less WT-SOD1. This behavior, though convoluted, where the redox properties of the Cu site and the structure are completely different from WT-SOD1 with or without Zn, again suggests that the Zn-less D124N-SOD1 would be catalytically inactive, even without protein unfolding and aggregation.
Electronic changes in the active sites with superoxide bound support the described observations, and provide further insight into the inactivity of the Zn-less SOD1 variants. Table 1 shows the related LUMO energies. From those, WT-SOD1 has the lowest E0, whereas the Zn-less WT-SOD1 and Zn-less D124N-SOD1 have relatively high E0 values (like in the case of SOD1 without superoxide). Still, in all cases, the reduction potentials are higher than those for the variants without superoxide. Furthermore, WT-SOD1 has an almost non-existent HOMO-LUMO gap (+0.003 eV), indicating the readiness of the system for the next step in the catalytic cycle. The HOMO-LUMO gap for Zn-less WT-SOD1 is +7.630 eV, whereas the HOMO-LUMO gap for Zn-less D124N-SOD1 is +8.548 eV, i.e. nearly an order of magnitude larger than that in WT-SOD1.
In Figure 6 (right panel), the HOMO and LUMO of these superoxide bound systems are shown, and one may see how inconsistent these frontier orbitals responsible for the reactivity and redox cycle look among the three studied variants. For WT-SOD1, both the HOMO, and the LUMO show extensive electron delocalization across all residues of the active site. The close proximity and overlap of the HOMO and LUMO orbitals correlate with the negligible HOMO-LUMO gap. For Zn-less WT-SOD1, the HOMO changed very little upon adding superoxide, and it still mostly covers H71, H80, and D83. The LUMO stays on the Cu site and additionally incorporates superoxide and R143. The loss of more extended delocalization and overlap between the HOMO and LUMO goes together with the larger HOMO-LUMO gap. For Zn-less D124N-SOD1, the HOMO and LUMO are now localized on different parts of the active site, and appear to switch as compared to the other two systems, suggesting that the ground state of Zn-less D124N-SOD1 roughly corresponds to the first excited electronic state for WT-SOD1 with or without Zn. These observations suggest a complete incompetence of the active site for its catalytic function.
Overall, our findings indicate that the Zn-less SOD1 variants do not immediately unfold and can be found in metastable folded states. However, they undergo significant changes that eliminate their catalytic activity at the electronic level. It is thus obvious that Zn not only plays a structural role, but also directly influences the catalysis; Zn is an important component of the active site enabling the required redox behavior of Cu. In view of our experimental finding that there is a good chance that Cu exist as Cu(I) in the resting state of the Zn-less proteins, it is important to notice that, for Cu(I)-containing Zn-less SOD1 would be unable to return to the Cu(II) state required for catalysis. Thus, with or without a complete reduction of Cu(II) to Cu(I), Zn-less forms of SOD1 are not catalytic.
To summarize, a joint theoretical and experimental approach was used to assess the structural and electronic consequences of Zn removal on SOD1. Two Zn-less SOD1 variants were considered, the one with the native sequence, and the D124N mutant typically used in experimental studies of SOD1 without Zn. Mixed quantum-classical dynamics simulations showed that these Zn-less proteins have at least metastable states with appreciable life-times in which the SOD1 fold is mostly preserved. This is in contrast to the idea that the loss of Zn leads to the immediate unfolding of the protein. The structural changes found in these folded Zn-less states are localized to the active sites, and the catalytic Cu sites in particular. Both the coordination sphere and the electronic structure of the Cu center change upon Zn removal. Through electronic structure calculations and molecular orbital analysis, we show that the redox behavior of Cu changes, and it is likely to exist as Cu(I) in the resting state of the Zn-less enzyme, instead of Cu(II) required for the catalytic cycle. This prediction is confirmed experimentally: in the Zn-less D124N-SOD1, there is a noticeable amount of Cu(I), which becomes nearly 85% Cu(I) in the presence of a reducing agent. For the WT-SOD1, the presence of a reducing agent does not change the oxidation state of Cu, which thus remains Cu(II). Once in the Cu(I) form, SOD1 becomes catalytically inactive, according to the known mechanism (Scheme 1) where Cu cycles between Cu(II) (in the resting state) and Cu(I).
We further show that, even without the complete transition to Cu(I), the catalytic activity of the Zn-less proteins is eliminated. The effect of the structural and electronic changes in the Cu site on its interaction with the superoxide substrate was assessed. The substrate binding, resultant charge distribution and charge separation in it, HOMO-LUMO gaps in the SOD1-superoxide complexes, and H-bonding networks are all disrupted in the Zn-less variants. Also, the changes are quite different between the Zn-less SOD1 with the native sequence and its D124N mutant; in the mutant they are more radical. The binding of superoxide to Zn-less WT-SOD1 is much weaker than to WT-SOD1, and it is even weaker in Zn-less D124N-SOD1. The residue R143, critical for H-transfer in the catalytic mechanism (Scheme 1), is oriented differently with respect to the bound substrate in Zn-less WT-SOD1, and is not in coordination with the substrate in the Zn-less D124N variant. The HOMO-LUMO gap in the complex of native WT-SOD1 and superoxide is nearly zero, indicating the readiness of the system to undergo the required election transfer, but in the Zn-less variants the HOMO-LUMO gaps are insurmountable, on the order of 8 eV. The D124N-variant further exhibits state-crossing as compared to WT-SOD1, with or without Zn. The electron density distributions in the frontier orbitals of the complexes support the differences in the HOMO-LUMO gaps. In both Zn-less proteins, the oxidation of superoxide to triplet oxygen dimer is, thus, predicted to be abolished.
Overall, we show that Zn not only plays a structural role in SOD1, but also directly influences the catalysis, enabling the proper coordination and reduction potential of the Cu site. Removal of Zn causes the elimination of the catalytic activity of SOD1 even without protein unfolding and aggregation. It also makes the Cu center prone to deactivation due to immediate reduction to Cu(I) in the resting state.
III. Materials and Methods
Computational Details
The SOD1 enzyme was characterized using our quantum mechanic/discrete molecular dynamics, QM/DMD, method.(19) In a number of works we demonstrated its successful performance for the assessment of structure, mechanisms, conformational changes, and effects of metal replacement in metalloenzymes.(19,29–32) In Figure 2, we show the QM/DMD partitioning of the protein. The Cu site with its immediate ligands and part of second coordination spheres are treated at the QM level: we used the density functional theory method, TPSS,(33–36) with the resolution of the identity(37–39) and Grimme’s D3 dispersion correction(40). The nonhybrid functionals are known to do well in structure prediction, and we used it for this purpose in order to save the computational cost. For further properties assessment, we relied on hybrid functionals (see below). The def2/SVP(41) basis set was used for all atoms other than Cu, and for Cu we used the def2-TZVPP(42) basis set with effective core potential. The TURBOMOLE program(43) was used for all QM calculations. QM/DMD is an iterative dynamical method, as described in details elsewhere. During the DMD(44,45) stage of the simulations, the QM-DMD boundary shrinks down to the tiny coordination area around Cu, so that there would be no need to treat the Cu site using the classical force field, and yet it would be possible to provide maximal sampling on all of the protein including most of the active site. At the QM stage, the QM-DMD boundary expands to the larger and chemically meaningful active site shown in Figure 2. The points where the structure is attached to the rest of the protein are capped with H and fixed, and the resultant structures are ranked and optimized at the QM level. They then get reinstalled in the protein, the QM-DMD boundary shrinks again, and simulations continue with the DMD phase.
The number of QM/DMD iterations ranged from 50 to 100 depending on the protein variant considered, which roughly corresponds to 25–50 ns of dynamics. (19) The timing is approximate because not the entire QM region is treated dynamically, and because of the light coarse-graining in the DMD machinery. QM/DMD is used here for sampling, and the assessment of the equilibrium protein structures. The simulations were running to convergence in terms of the total backbone RMSD, all-atoms RMSD of the active site, QM energies of the active site, and DMD energies of the DMD region.
The structure of the WT-SOD1 with Zn was obtained from pdb.org (PDB ID: 1SPD; see Figure 2, panels 1 and 2); the Zn-less WT-SOD1 was obtained by removing the Zn from 1SPD (QM region is in orange in Figure 2, panel 3 (Residues: Cu, H46, H48, H63, H120, D124)) and equilibrating the resultant protein; and Zn-less D124N-SOD1 was obtained by removing Zn and performing the D124N mutation to the monomer of 1SPD (QM region is in orange in Figure 2, panel 4 (Residues: Cu, H46, H48, H63, H120, N124)), followed by equilibration. The protonation states of amino acids were assumed to correspond to pH=7, e.g. Asp and Glu were deprotonated, His was singly-protonated, unless bound to both metals in which case it was doubly-deprotonated, etc. The effect of the pH was not considered any further in this work. All residues included in the QM region were truncated by cutting the Cα-Cβ bond and capping the Cβ atom with H pointing along the Cβ-Cα vector at a fixed Cβ-H distance.
For the property calculations that followed equilibration, we used exclusively QM. The more extended Cu-containing active site, the Zn site, and R143 were included in the expanded QM region (Figure 2, panels 2–4). R143 is important for the catalytic mechanism (see Scheme 1).(1,6) Additionally, R143 shares the electrostatic loop with the D124 residue, which is a secondary bridge between the Cu and Zn sites (Figure 1). The Zn loop and Cu site are involved in maintaining the dimer structure, and the Zn loop also interacts with the electrostatic loop (Figure 2). Partial atomic charges were obtained using the Natural Population Analysis (NPA)(46) at the TPSSh/def2-SVP,def2-TZVPP, B3-LYP(33,34,47–50)/def2-SVP,def2-TZVPP, and B2-PLYP(51)/def2-SVP,def2-TZVPP levels of theory. The density functionals of TPSSh, B3-LYP and B2-PLYP are acceptable choices for charge analysis.(52) In order to determine the solvent effects on the SOD1 variants, single point total energies at zero Kelvin were performed using TURBOMOLE’s version of the Conductor-like Screening Model (COSMO).(53) The dielectric constant for water used was 20.(54) Molecular orbitals were constructed at the Hartree-Fock level of theory, HF/def2-SVP,def2-TZVPP, as the HF orbitals are one-electron eigenstates whose energies have the meaning of Koopmans’ theorem.(55)
Experimental details
Experiment was used to detect Cu(I) in the active site of Zn-less SOD1, as per our theoretical prediction. Expression of human D124N-SOD1 in S. cerevisiae and purification, with the exclusion of the anion exchange chromatography step, were performed as described elsewhere.(11) Following hydrophobic interaction chromatography, fractions containing SOD1 at >95% purity (as confirmed by SDS-PAGE) were dialyzed against 20 mM Tris pH 7.4 to remove (NH4)2SO4. Removal of residual metal and Cu(II) loading were then performed by successive dialysis at 4°C against 50 mM acetic acid, pH 5.5 for 30 min.; 100 mM acetic acid pH 3.8, 10 mM EDTA for 120 min.; 100 mM acetic acid pH 5.5, 10 mM MgCl2 for 60 min.; and 100 mM acetic acid pH 5.5 for 60 min., after which the contents of the dialysis tubing were stirred overnight at 4°C with excess CuSO4. Remetallated SOD1-D124N was concentrated and buffer was changed to 20 mM Tris, pH 7.4, 150 mM NaCl using an Amicon Ultra 10,000 MWCO centrifugal filter. Metal occupancy in purified D124N-SOD1 was measured using an established colorimetric assay utilizing Zincon(56) and calculated to be 1.73 Cu(II) and 0.13 Zn(II) per SOD1 dimer. Metal-replete WT-SOD1 was generated by purification of SOD1 from human erythrocytes as described previously.(11) Cu(I) was detected by addition of bathocuproine disulfonate (BCDS), which binds to Cu(I) to form a complex with maximal absorbance at 482 nm.(28) To 25 μM Zn-less D124N-SOD1 or metal-replete WT-SOD1, BCDS was added to a final concentration of 0.1 mM in the presence and absence of 1 mM DTT. Following a 50 min. room temperature incubation, absorbance at 482 nm was measured using a ND-1000 Nanodrop spectrophotometer. Cu(I) content of SOD1 samples was quantified by comparison with a standard curve of A482 vs. [Cu(I)] from 0 – 70 μM, a range in which the relationship between A482 and [Cu(I)] is linear. For generation of the standard curve, Cu(I) was produced by incubating CuSO4 with 1 mM DTT for 50 min. at room temperature, after which absorbance was measured at 482 nm in the presence of 0.1 mM BCDS.
Supplementary Material
The role of Zn in the active site of Cu,Zn-superoxide dismutase (SOD1) is explicated computationally and confirmed experimentally
Contrary to the old hypothesis, Zn is not merely a structural cation, but an active modifier of the Cu site
Zn influences the structure and reduction potential on Cu, and without Zn those become noncatalytic
Experiment and theory show that Cu in Zn-less SOD1 is highly prone to becoming inactive Cu(I)
Acknowledgments
This work was supported by the DARPA Young Faculty Award N66001-11-1-4138 and Alfred P. Sloan Research Fellowship to ANA. The calculations were carried out on the computers of the Hoffman2 cluster at the Institute for Digital Research and Education (IDRE) located in the University of California, Los Angeles.
This work was also supported by the National Institutes of Health grant R01GM080742 to N.V.D. R.L.R. was supported by the National Institutes of Health Predoctoral Fellowship F31NS073435 from the National Institute of Neurological Disorders and Stroke. E.A.P. was supported by the National Institutes of Health Predoctoral Fellowship F31AG039266 from the National Institute on Aging.
We thank Prof. Joan S. Valentine for providing the EG118 strain of S. cerevisiae and the YEp351-hwtSOD1 expression construct.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could a3ect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Osman R, Basch H. On the Mechanism of Action of Superoxide-Dismutase - a Theoretical-Study. J Am Chem Soc. 1984;106:5710–5714. [Google Scholar]
- 2.Hart PJ, Balbirnie MM, Ogihara NL, Nersissian AM, Weiss MS, Valentine JS, Eisenberg D. A structure-based mechanism for copper-zinc seperoxide dismutase. Biochemistry-Us. 1999;38:2167–2178. doi: 10.1021/bi982284u. [DOI] [PubMed] [Google Scholar]
- 3.Abreua IA, Cabelli DE. Superoxide dismutases—a review of the metal-associated mechanistic variations. Biochimica et Biophysica Acta. 2010;1804:263–274. doi: 10.1016/j.bbapap.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nature Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 5.Cudd A, Fridovich I. Electrostatic Interactions in the Reaction-Mechanism of Bovine Erythrocyte Superoxide-Dismutase. J Biol Chem. 1982;257:1443–1447. [PubMed] [Google Scholar]
- 6.Fisher CL, Cabelli DE, Tainer JA, Hallewell RA, Getzoff ED. The Role of Arginine-143 in the Electrostatics and Mechanism of Cu,Zn Superoxide-Dismutase - Computational and Experimental Evaluation by Mutational Analysis. Proteins-Structure Function and Genetics. 1994;19:24–34. doi: 10.1002/prot.340190105. [DOI] [PubMed] [Google Scholar]
- 7.Strange RW, Antonyuk S, Hough MA, Doucette PA, Rodriguez JA, Hart PJ, Hayward LJ, Valentine JS, Hasnain SS. The structure of holo and metal-deficient wild-type human Cu, Zn superoxide dismutase and its relevance to familial amyotrophic lateral sclerosis. J Mol Biol. 2003;328:877–891. doi: 10.1016/s0022-2836(03)00355-3. [DOI] [PubMed] [Google Scholar]
- 8.Khare SD, Dokholyan NV. Common dynamical signatures of familial amyotrophic lateral sclerosis-associated structurally diverse Cu, Zn superoxide dismutase mutants. P Natl Acad Sci USA. 2006;103:3147–3152. doi: 10.1073/pnas.0511266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardo CA, Xu ZS, Borchelt DR, Price DL, Sisodia SS, Cleveland DW. Superoxide-Dismutase Is an Abundant Component in Cell-Bodies, Dendrites, and Axons of Motor-Neurons and in a Subset of Other Neurons. P Natl Acad Sci USA. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valentine JS, Hart PJ. Misfolded CuZnSOD and amyotrophic lateral sclerosis. P Natl Acad Sci USA. 2003;100:3617–3622. doi: 10.1073/pnas.0730423100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilcox KC, Zhou L, Jordon JK, Huang Y, Yu YB, Redler RL, Chen X, Caplow M, Dokholyan NV. Modifications of Superoxide Dismutase (SOD1) in Human Erythrocytes A POSSIBLE ROLE IN AMYOTROPHIC LATERAL SCLEROSIS. J Biol Chem. 2009;284:13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller AF, Teixeira M, Valentine JS. Superoxide Dismutases and Superoxide Reductases. Chem Rev. 2014;114:3854–3918. doi: 10.1021/cr4005296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khare SD, Caplow M, Dokholyan NV. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. P Natl Acad Sci USA. 2004;101:15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, Oregan JP, Deng HX, Rahmani Z, Krizus A, Mckennayasek D, Cayabyab A, Gaston SM, Berger R, Tanzi RE, Halperin JJ, Herzfeldt B, Vandenbergh R, Hung WY, Bird T, Deng G, Mulder DW, Smyth C, Laing NG, Soriano E, Pericakvance MA, Haines J, Rouleau GA, Gusella JS, Horvitz HR, Brown RH. Mutations in Cu/Zn Superoxide-Dismutase Gene Are Associated with Familial Amyotrophic-Lateral-Sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 15.Kim J, Lee H, Lee JH, Kwon D-Y, Genovesio A, Fenistein D, Ogier A, Brondani V, Grailhe R. Dimerization, oligomerization, and aggregation of human Amyotrophic lateral sclerosis Cu/Zn-superoxide dismutase 1 mutant forms in live cells. J Biol Chem. 2014 doi: 10.1074/jbc.M113.542613. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lyons TJ, Liu HB, Goto JJ, Nersissian A, Roe JA, Graden JA, Cafe C, Ellerby LM, Bredesen DE, Gralla EB, Valentine JS. Mutations in copper-zinc superoxide dismutase that cause amyotrophic lateral sclerosis alter the zinc binding site and the redox behavior of the protein. P Natl Acad Sci USA. 1996;93:12240–12244. doi: 10.1073/pnas.93.22.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khare SD, Ding F, Dokholyan NV. Folding of Cu, Zn superoxide dismutase and familial amyotrophic lateral sclerosis. J Mol Biol. 2003;334:515–525. doi: 10.1016/j.jmb.2003.09.069. [DOI] [PubMed] [Google Scholar]
- 18.Ding F, Furukawa Y, Nukina N, Dokholyan NV. Local Unfolding of Cu, Zn Superoxide Dismutase Monomer Determines the Morphology of Fibrillar Aggregates. J Mol Biol. 2012;421:548–560. doi: 10.1016/j.jmb.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sparta M, Shirvanyants D, Ding F, Dokholyan NV, Alexandrova AN. Hybrid Dynamics Simulation Engine for Metalloproteins. Biophys J. 2012;103:767–776. doi: 10.1016/j.bpj.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bard AJ, Faulkner LR. ELECTROCHEMICAL METHODS Fundamentals and Applications. 2. JOHN WILEY & SONS, INC; 2001. [Google Scholar]
- 21.Kurtz HA. Lumo Energies and Negative Electron-Affinities. J Chem Educ. 1984;61:580–581. [Google Scholar]
- 22.Khan SUM, Kainthla RC, Bockris JO. The Redox Potential and the Fermi Level in Solution. J Phys Chem-Us. 1987;91:5974–5977. [Google Scholar]
- 23.Zhang G, Musgrave CB. Comparison of DFT methods for molecular orbital eigenvalue calculations. J Phys Chem A. 2007;111:1554–1561. doi: 10.1021/jp061633o. [DOI] [PubMed] [Google Scholar]
- 24.Zhan CG, Nichols JA, Dixon DA. Ionization potential, electron affinity, electronegativity, hardness, and electron excitation energy: Molecular properties from density functional theory orbital energies. J Phys Chem A. 2003;107:4184–4195. [Google Scholar]
- 25.Perdew JP, Levy M. Comment on “Significance of the highest occupied Kohn-Sham eigenvalue”. Phys Rev B. 1997;56:16021–16028. [Google Scholar]
- 26.Perdew JP. Density Functional Theory and the Band-Gap Problem. Int J Quant Chem. 1985:497–523. [Google Scholar]
- 27.Koopmans T. The classification of wave functions and eigen-values to the single electrons of an atom. Physica. 1934;1:104–113. [Google Scholar]
- 28.Martyshkin DV, Mirov SB, Zhuang YX, Crow JP, Ermilov V, Beckman JS. Fluorescence assay for monitoring Zn-deficient superoxide dismutase in vitro. Spectrochim Acta A. 2003;59:3165–3175. doi: 10.1016/s1386-1425(03)00135-5. [DOI] [PubMed] [Google Scholar]
- 29.Sparta M, Valdez CE, Alexandrova AN. Metal-Dependent Activity of Fe and Ni Acireductone Dioxygenases: How Two Electrons Reroute the Catalytic Pathway. J Mol Biol. 2013;425:3007–3018. doi: 10.1016/j.jmb.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Valdez CE, Sparta M, Alexandrova AN. The Role of the Flexible L43-S54 Protein Loop in the CcrA Metallo-beta-lactamase in Binding Structurally Dissimilar beta-Lactam Antibiotics. J Chem Theory Comput. 2013;9:730–737. doi: 10.1021/ct300712j. [DOI] [PubMed] [Google Scholar]
- 31.Sparta M, Alexandrova AN. How Metal Substitution Affects the Enzymatic Activity of Catechol-O-Methyltransferase. Plos One. 2012:7. doi: 10.1371/journal.pone.0047172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valdez CE, Alexandrova AN. Why Urease Is a Di-Nickel Enzyme whereas the CcrA beta-Lactamase Is a Di-Zinc Enzyme. J Phys Chem B. 2012;116:10649–10656. doi: 10.1021/jp302771n. [DOI] [PubMed] [Google Scholar]
- 33.Dirac PAM. Quantum mechanics of many-electron systems. P R Soc Lond a-Conta. 1929;123:714–733. [Google Scholar]
- 34.Slater JC. A Simplification of the Hartree-Fock Method. Phys Rev. 1951;81:385–390. [Google Scholar]
- 35.Perdew JP, Wang Y. Accurate and Simple Analytic Representation of the Electron-Gas Correlation-Energy. Phys Rev B. 1992;45:13244–13249. doi: 10.1103/physrevb.45.13244. [DOI] [PubMed] [Google Scholar]
- 36.Tao JM, Perdew JP, Staroverov VN, Scuseria GE. Climbing the density functional ladder: Nonempirical meta-generalized gradient approximation designed for molecules and solids. Phys Rev Lett. 2003:91. doi: 10.1103/PhysRevLett.91.146401. [DOI] [PubMed] [Google Scholar]
- 37.Sierka M, Hogekamp A, Ahlrichs R. Fast evaluation of the Coulomb potential for electron densities using multipole accelerated resolution of identity approximation. J Chem Phys. 2003;118:9136–9148. [Google Scholar]
- 38.Skylaris CK, Gagliardi L, Handy NC, Ioannou AG, Spencer S, Willetts A. On the resolution of identity Coulomb energy approximation in density functional theory. J Mol Struc-Theochem. 2000;501:229–239. [Google Scholar]
- 39.Von Arnim M, Ahlrichs R. Performance of parallel TURBOMOLE for density functional calculations. J Comput Chem. 1998;19:1746–1757. [Google Scholar]
- 40.Grimme S, Antony J, Ehrlich S, Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J Chem Phys. 2010:132. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 41.Schafer A, Horn H, Ahlrichs R. Fully Optimized Contracted Gaussian-Basis Sets for Atoms Li to Kr. J Chem Phys. 1992;97:2571–2577. [Google Scholar]
- 42.Weigend F, Ahlrichs R. Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy. Phys Chem Chem Phys. 2005;7:3297–3305. doi: 10.1039/b508541a. [DOI] [PubMed] [Google Scholar]
- 43.Ahlrichs R, Bar M, Haser M, Horn H, Kolmel C. Electronic-Structure Calculations on Workstation Computers - the Program System Turbomole. Chem Phys Lett. 1989;162:165–169. [Google Scholar]
- 44.Dokholyan NV, Buldyrev SV, Stanley HE, Shakhnovich EI. Discrete molecular dynamics studies of the folding of a protein-like model. Fold Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- 45.Ding F, Tsao D, Nie HF, Dokholyan NV. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reed AE, Weinstock RB, Weinhold F. Natural-Population Analysis. J Chem Phys. 1985;83:735–746. [Google Scholar]
- 47.Vosko SH, Wilk L, Nusair M. Accurate Spin-Dependent Electron Liquid Correlation Energies for Local Spin-Density Calculations - a Critical Analysis. Can J Phys. 1980;58:1200–1211. [Google Scholar]
- 48.Becke AD. Density-Functional Exchange-Energy Approximation with Correct Asymptotic-Behavior. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 49.Lee CT, Yang WT, Parr RG. Development of the Colle-Salvetti Correlation-Energy Formula into a Functional of the Electron-Density. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 50.Becke AD. Density-Functional Thermochemistry. 3 The Role of Exact Exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 51.Grimme S. Semiempirical hybrid density functional with perturbative second-order correlation. J Chem Phys. 2006:124. doi: 10.1063/1.2148954. [DOI] [PubMed] [Google Scholar]
- 52.Tsipis AC. DFT flavor of coordination chemistry. Coordination Chem Rev. 2014;272:1–29. [Google Scholar]
- 53.Klamt A, Schuurmann G. Cosmo - a New Approach to Dielectric Screening in Solvents with Explicit Expressions for the Screening Energy and Its Gradient. J Chem Soc Perk Trans. 1993;25:799–805. [Google Scholar]
- 54.Antosiewicz J, McCammon JA, Gilson MK. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994;238:415. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- 55.Pople JA, Nesbet RK. Self-Consistent Orbitals for Radicals. J Chem Phys. 1954;22:571–572. [Google Scholar]
- 56.Sabel CE, Neureuther JM, Siemann S. A spectrophotometric method for the determination of zinc, copper, and cobalt ions in metalloproteins using Zincon. Anal Biochem. 2010;397:218–226. doi: 10.1016/j.ab.2009.10.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.