Abstract
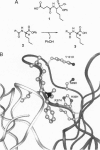
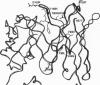
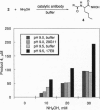
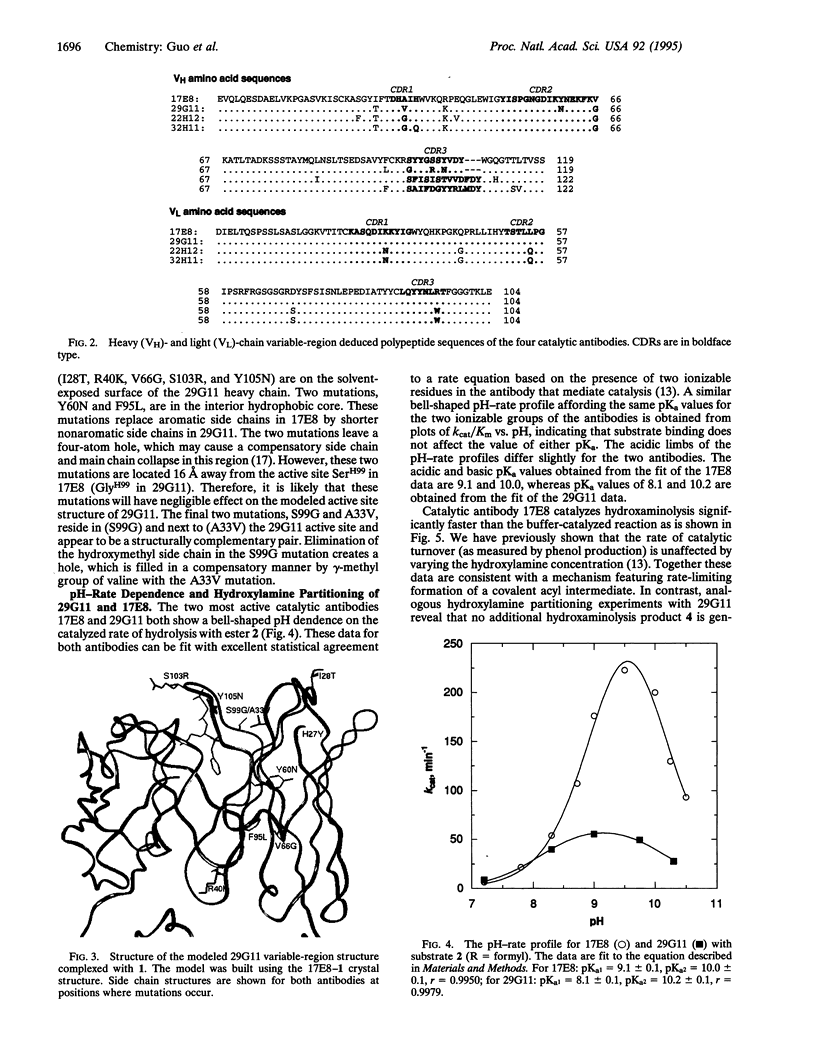
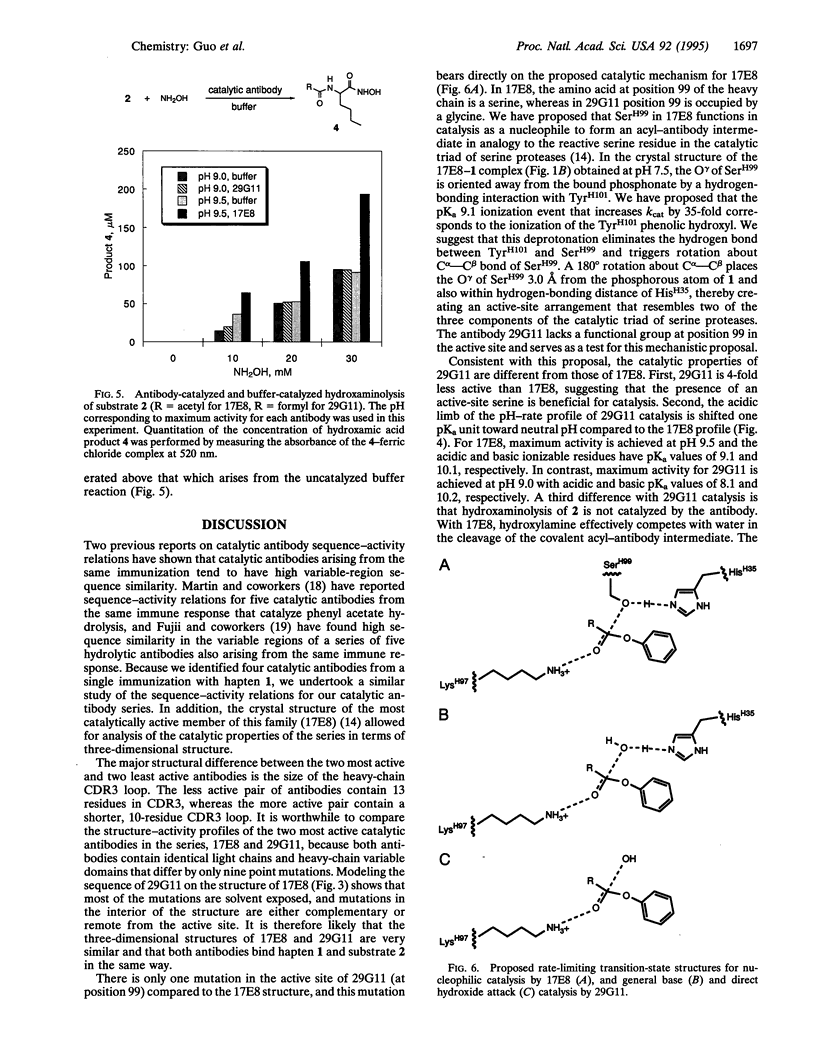
The variable-region peptide sequence and steady-state kinetic behavior are compared for a family of catalytic antibodies that arose from the same immune response to a transition-state analog. The crystal structure of the most catalytically active member of the family (17E8) has been solved to 2.5 A resolution and shows that the antibody active site contains a SerH99-HisH35 (H = heavy chain) catalytic dyad analogous to the Ser-His-Asp catalytic triad of serine proteases. The variable-region peptide sequence of the next most active antibody (29G11) differs from that of 17E8 by nine heavy-chain point mutations, and results from computer modeling suggest that the three-dimensional structure of 29G11 is similar to that of 17E8. In addition, 29G11 is an efficient catalytic antibody; it possesses 26% of the hydrolytic activity of 17E8. There is one active-site mutation in 29G11 compared to 17E8; position 99 of the heavy chain of 29G11 contains a glycine residue in place of the nucleophilic serine at this position in 17E8. Consistent with this mutation, results from pH-rate studies and hydroxylamine partitioning experiments indicate that in contrast to the catalytic mechanism of 17E8, the mechanism of 29G11-catalyzed esterolysis does not feature nucleophilic catalysis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angeles T. S., Smith R. G., Darsley M. J., Sugasawara R., Sanchez R. I., Kenten J., Schultz P. G., Martin M. T. Isoabzymes: structurally and mechanistically similar catalytic antibodies from the same immunization. Biochemistry. 1993 Nov 16;32(45):12128–12135. doi: 10.1021/bi00096a025. [DOI] [PubMed] [Google Scholar]
- DIXON M. The determination of enzyme inhibitor constants. Biochem J. 1953 Aug;55(1):170–171. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson A. E., Baase W. A., Zhang X. J., Heinz D. W., Blaber M., Baldwin E. P., Matthews B. W. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992 Jan 10;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- Golinelli-Pimpaneau B., Gigant B., Bizebard T., Navaza J., Saludjian P., Zemel R., Tawfik D. S., Eshhar Z., Green B. S., Knossow M. Crystal structure of a catalytic antibody Fab with esterase-like activity. Structure. 1994 Mar 15;2(3):175–183. doi: 10.1016/s0969-2126(00)00019-8. [DOI] [PubMed] [Google Scholar]
- Haynes M. R., Stura E. A., Hilvert D., Wilson I. A. Routes to catalysis: structure of a catalytic antibody and comparison with its natural counterpart. Science. 1994 Feb 4;263(5147):646–652. doi: 10.1126/science.8303271. [DOI] [PubMed] [Google Scholar]
- Hilvert D., Hill K. W. Antibody catalysis of concerted, carbon-carbon bond-forming reactions. Methods Enzymol. 1991;203:352–369. doi: 10.1016/0076-6879(91)03020-h. [DOI] [PubMed] [Google Scholar]
- Lerner R. A., Benkovic S. J., Schultz P. G. At the crossroads of chemistry and immunology: catalytic antibodies. Science. 1991 May 3;252(5006):659–667. doi: 10.1126/science.2024118. [DOI] [PubMed] [Google Scholar]
- Miyashita H., Hara T., Tanimura R., Tanaka F., Kikuchi M., Fujii I. A common ancestry for multiple catalytic antibodies generated against a single transition-state analog. Proc Natl Acad Sci U S A. 1994 Jun 21;91(13):6045–6049. doi: 10.1073/pnas.91.13.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis D. L., Cheah E., Cygler M., Dijkstra B., Frolow F., Franken S. M., Harel M., Remington S. J., Silman I., Schrag J. The alpha/beta hydrolase fold. Protein Eng. 1992 Apr;5(3):197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- Pollack S. J., Jacobs J. W., Schultz P. G. Selective chemical catalysis by an antibody. Science. 1986 Dec 19;234(4783):1570–1573. doi: 10.1126/science.3787262. [DOI] [PubMed] [Google Scholar]
- Roberts V. A., Stewart J., Benkovic S. J., Getzoff E. D. Catalytic antibody model and mutagenesis implicate arginine in transition-state stabilization. J Mol Biol. 1994 Jan 21;235(3):1098–1116. doi: 10.1006/jmbi.1994.1060. [DOI] [PubMed] [Google Scholar]
- Stewart J. D., Krebs J. F., Siuzdak G., Berdis A. J., Smithrud D. B., Benkovic S. J. Dissection of an antibody-catalyzed reaction. Proc Natl Acad Sci U S A. 1994 Aug 2;91(16):7404–7409. doi: 10.1073/pnas.91.16.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J. D., Roberts V. A., Thomas N. R., Getzoff E. D., Benkovic S. J. Site-directed mutagenesis of a catalytic antibody: an arginine and a histidine residue play key roles. Biochemistry. 1994 Mar 1;33(8):1994–2003. doi: 10.1021/bi00174a004. [DOI] [PubMed] [Google Scholar]
- Tramontano A., Janda K. D., Lerner R. A. Catalytic antibodies. Science. 1986 Dec 19;234(4783):1566–1570. doi: 10.1126/science.3787261. [DOI] [PubMed] [Google Scholar]
- Zhou G. W., Guo J., Huang W., Fletterick R. J., Scanlan T. S. Crystal structure of a catalytic antibody with a serine protease active site. Science. 1994 Aug 19;265(5175):1059–1064. doi: 10.1126/science.8066444. [DOI] [PubMed] [Google Scholar]