Abstract
Academic radiology is poised to play an important role in the development and implementation of quantitative imaging (QI) tools. This manuscript, drafted by the Association of University Radiologists (AUR) Radiology Research Alliance (RRA) Quantitative Imaging Task Force, reviews current issues in QI biomarker research. We discuss motivations for advancing QI, define key terms, present a framework for QI biomarker research, and outline challenges in QI biomarker development. We conclude by describing where QI research and development is currently taking place and discussing the paramount role of academic radiology in this rapidly evolving field.
Keywords: radiology, quantitative imaging, biomarker development
Medical imaging has evolved dramatically since the first Roentgenogram nearly 125 years ago (1). Modern techniques including ultrasound, computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) now provide an unprecedented level of spatial detail and functional information (2). As medical imaging has progressed, older analog techniques have been steadily replaced with newer digital methods of image acquisition, processing, archiving, and display. This evolution has occurred in parallel with advancements in our understanding of the molecular underpinnings of disease and the rise of a more statistical and evidence-based approach to diagnosis and treatment. Medical imaging is now poised to leverage quantitative techniques in support of a wide range of clinical and research goals (3, 4).
In a broad sense, quantitative imaging (QI) refers to the extraction and use of numerical/statistical features from medical images (see Box 1 for definitions of key terms). As a research field, QI includes the development, standardization, optimization, and application of anatomical, functional, and molecular imaging acquisition protocols, data analyses, display methods, and reporting structures, as well as the validation of QI results against relevant biological and clinical data (5, 6). The QI concept is closely tied to that of a biomarker, defined as a characteristic that is objectively measured and evaluated as an indicator of a normal biologic process, pathologic process, or response to a therapeutic intervention (7). A QI biomarker is therefore an objectively measured characteristic, derived from a medical image, that can be correlated with anatomically and physiologically relevant parameters including disease presence, disease severity, disease characterization (particularly on a molecular level), predicted disease course (both with and without treatment), and treatment response. The Quantitative Imaging Biomarkers Alliance (QIBA), organized by the Radiological Society of North America (RSNA), has formally defined a QI biomarker as “an objective characteristic derived from an in vivo image measured on a ratio or interval scale as indicators [sic] of normal biological processes, pathogenic processes, or a response to a therapeutic intervention.” This definition’s emphasis on ratio or interval variables would imply that tumor volumes or PET standardized uptake values (SUVs) would be considered QI biomarkers, because the difference or ratio between two values is meaningful, whereas ordinal variables such as Breast Imaging Reporting and Data System (BIRADS) assessment categories would not. This strict definition is meant to guide QI research toward biomarkers that may be assessed and compared with robust statistical calculations including frequency distributions, medians, means, standard deviations, and standard errors of the mean (8).
Box 1. Definitions related to quantitative imaging biomarker development.
Analytical validation – Demonstration of the accuracy, precision, and feasibility of biomarker measurement
Biomarker – A characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or responses to a therapeutic intervention
Predictive biomarker – A biomarker intended to forecast disease course in the presence of a specific treatment
Prognostic biomarker – A biomarker intended to forecast disease course in the absence of treatment
Qualification – Demonstration that a biomarker is associated with a clinical endpoint
Quantitative imaging – The extraction and use of numerical/statistical features from medical images
Quantitative imaging biomarker (modified QIBA definition) – An objective characteristic derived from an in vivo image measured on a ratio or interval scale as an indicator of a normal biological process, a pathogenic process, or a response to a therapeutic intervention (8)
Repeatability – The agreement between successive measurements made under the same conditions
Reproducibility – The agreement between successive measurements made with varying conditions, such as location or operator
Surrogate endpoint – A biomarker intended to substitute for a clinical endpoint
Utilization – Assessment of biomarker performance in the specific context of its proposed use
This manuscript, drafted by the Association of University Radiologists (AUR) Radiology Research Alliance (RRA) Quantitative Imaging Task Force, addresses issues related to QI biomarker research and development. A separate manuscript from our Task Force outlines current clinical applications of QI (9). In this article, we describe motivations for QI biomarker development and discuss challenges for QI research using a three-part framework. We then provide an overview of where QI research and development is currently taking place. We conclude by discussing the particular role of academic radiology in advancing QI. Sections of this manuscript were derived from individual mini-scoping studies based on focused research questions (10).
Motivations for QI biomarker development
The promise of QI lies in the potential for increased precision and standardization of image interpretation, in both the research and clinical settings. Potential gains from the growth of QI include increased diagnostic accuracy; decreased variability and subjectivity of image analysis; increased automation of data reporting; more robust association of imaging findings with other biological and clinical parameters, including rigorous statistical correlations between quantitative datasets; and the opportunity for large-scale attempts to link phenotypic imaging patterns with genomic profiles (11). The development of QI is being driven in large part by the environment of evidence-based medicine, in which diagnoses across the clinical spectrum are reinforced with quantitative data (12, 13).
Perhaps the greatest demand for QI at present is from cancer clinical trials, where quantitative measurements of tumor response are used to determine the efficacy of investigational treatments. Imaging-based response assessment guidelines such as the Response Evaluation Criteria in Solid Tumors (RECIST) (14) have been used for decades and have been successfully validated against long-term patient outcomes in certain settings (15, 16). However, in the era of targeted agents that may promote tumor stability rather than tumor regression (17-21), the oncologic imaging community has embarked on developing novel imaging biomarkers to identify and interrogate underlying molecular and functional changes in tissue, with the premise that these measurements will provide earlier and/or more accurate response assessment than tumor size changes (Fig 1) (22). Validated QI biomarkers reporting on different elements of tumor status may enhance drug development by establishing proof of concept for investigational agents, by facilitating selection of candidate agents for promotion to later stage testing, and by determining patient subgroups in which the likelihood of drug response is higher (23, 24). QI biomarkers may also be useful for clinical care by offering the ability to stratify patients to the most appropriate treatments and by promoting earlier identification of patients with a poor response to a particular regimen (25).
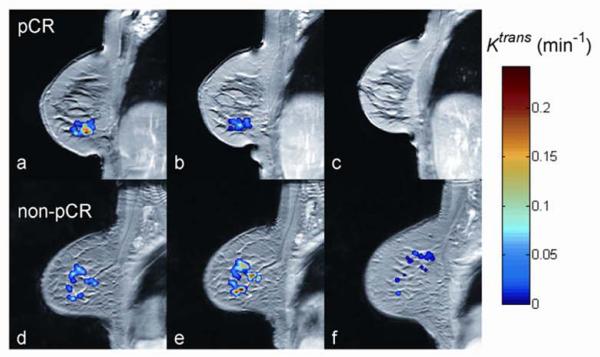
Fig. 1.
Dynamic contrast-enhanced MRI (DCE-MRI) as a quantitative imaging (QI) technique for assessing breast cancer response to neoadjuvant therapy (color overlay = tumor). The top row illustrates an early reduction in the quantitative DCE-MRI parameter Ktrans in a patient who had a documented pathological complete response (pCR) at surgery (A: prior to therapy, B: after one cycle of neoadjuvant therapy, C: at the conclusion of neoadjuvant therapy). The bottom row illustrates an early increase in Ktrans in a patient who had residual disease (non-pCR) at surgery (D: prior to therapy, E: after one cycle of neoadjuvant therapy, F: at the conclusion of neoadjuvant therapy. (Image courtesy of Lisa Li, Ph.D., Vanderbilt University)
Imaging researchers are responding to the demand for QI biomarkers by advancing a broad array of quantitative techniques across a wide spectrum of clinical and research indications (24, 26-35). The common denominator linking all of these efforts is the drive toward producing standardized, unbiased, and precise imaging data in support of the larger medical research and clinical enterprise. This endeavor involves particular research challenges, as presented in the next section.
Challenges in QI biomarker development
Rigorous evaluation is required before a QI biomarker can be safely and sensibly adopted (36). This section describes this evaluation process and presents key challenges in QI biomarker development. We have organized our discussion using a framework from the Institute of Medicine (IOM) that considers biomarker evaluation in three parts: (1) analytical validation, (2) qualification, and (3) utilization (37).
Analytical validation
Analytical validation involves demonstration of the accuracy, precision, and feasibility of biomarker measurement. If a QI biomarker cannot be reliably measured, it will have little or no use as an indicator for a biological process or clinical outcome. The process of analytical validation includes generating data on limits of detection, limits of quantification, and reference normal values (23). It also includes assessing both repeatability (i.e., the agreement between successive measurements made under the same conditions) and reproducibility (i.e., the agreement between successive measurements made with varying conditions such as location or operator) (8, 38, 39), with both specified by appropriate statistical parameters including the kappa (or weighted kappa), the intra-class correlation coefficient, or the confidence interval of the mean (Fig. 2) (40-50).
Fig. 2.
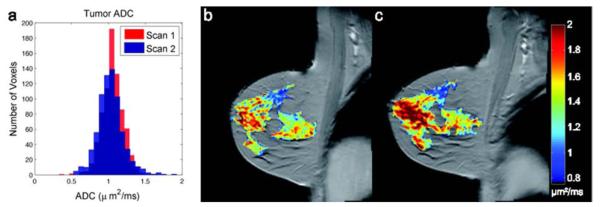
Repeatability of apparent diffusion coefficient (ADC) measurements from diffusion-weighted MRI (DW-MRI) in a breast cancer patient (color overlay = tumor). Panel A shows the distributions of the ADC values from the tumor obtained on two separate scans within one week of each other. Panels B (visit 1) and C (visit 2) show the spatial variations at the voxel level. The mean with 95% confidence intervals for the two visits were 1.06 +/− 0.01 mm2/ms and 1.03 +/− 0.01 mm2/ms, respectively. The lack of overlap in the confidence intervals, despite the apparent similarity in the histograms, illustrates the importance of analytical validation studies to establish ranges of measurement error before deploying quantitative techniques to interrogate changes in underlying biology. (Image courtesy of Lori Arlinghaus, Ph.D, Vanderbilt University.)
Validation studies are also used to generate preliminary reporting standards for QI biomarkers (51). Evaluations of technical performance and measurement error provide the foundation for establishing whether biomarkers should be reported as continuous variables or categorical (e.g., mild, moderate or severe dysfunction of the left ventricle, as assessed by cardiac MRI with quantification of ejection fraction), and also provide data to inform selection of rational cutoff values.
It is important to note that QI techniques typically rest on a foundation of image processing steps used to generate the values for subsequent biomarker definition. These initial steps present their own research challenges. Examples of challenges at the image processing stage include validating automated feature generation in absence of a reliable ground truth or plausible simulation model, and achieving accurate data while minimizing radiation dose.
Qualification
Qualification involves demonstrating that a biomarker is associated with a clinical endpoint. The qualification process establishes the ability of a QI biomarker to serve as a measurable indicator of a biological process, pathologic process, or response to an intervention (31, 52-55). This critical step in biomarker evaluation provides the basis for biomarker adoption in clinical and research applications, as well as for consideration of biomarker data by regulatory authorities as evidence of drug and device efficacy (51).(51). Qualification is fundamentally a statistical challenge, one with important methodological requirements that must be taken into account in the design of biomarker studies (38, 39).
For a prognostic biomarker, i.e., a biomarker intended to forecast disease course in the absence of treatment, a correspondence must be shown between the biomarker and the outcome of interest. Once a relationship has been established in an initial derivation cohort, it must be confirmed independently in an entirely different set of patients (validation cohort) to prove that the initial correspondence was neither due to chance nor the result of overfitting a statistical model to the derivation cohort dataset (56). Initial relationships can be demonstrated through small retrospective studies, but more robust biomarker qualification requires testing in multiple patient cohorts and preferably within a randomized or prospective clinical trial (57). If test performance is standardized rigorously, the biomarker’s ability to predict clinical outcomes can be tested across multiple centers with varying scanners and viewing platforms. Qualification of prognostic biomarkers must also evaluate biomarker performance as a function of time; even if a strong correspondence is established early in a disease between a biomarker and a clinical outcome, comprehensive biomarker qualification must also examine whether the strength of that correspondence wanes over time as the disease progresses.
For a predictive biomarker, i.e., a biomarker intended to forecast disease course in the presence of a specific treatment, the statistical challenges are greater (58, 59). The same general principle still applies (i.e., establishing an initial relationship between biomarker and outcome and then confirming that relationship in an independent validation cohort), but the analysis requires data from patients with both high and low biomarker levels. Different clinical trial designs exist for analyzing treatment effects in patients stratified by biomarker status. Given the challenges of performing prospective, randomized studies, retrospective analyses of completed trials may be an important source of evidence for predictive biomarker qualification.
One of the greatest statistical challenges for biomarker qualification is in establishing a biomarker as a surrogate endpoint, i.e., a valid substitute for a clinical endpoint. Only a small subset of biomarkers ever meets criteria for surrogacy. In order to qualify as a surrogate for a clinical endpoint, not only must the biomarker forecast the clinical outcome without reference to a specific intervention (“‘individual level” surrogacy), but also the effect of treatment on the biomarker must correlate closely with the effect of treatment on the clinical outcome (“trial level” surrogacy). Generally, individual-level surrogacy is established using standard correlation coefficients, while trial-level surrogacy can be established only through meta-analysis of multiple randomized trials (60). A major challenge for the validation of surrogate endpoints is the need for separate qualification of surrogate endpoints in the setting of different treatments; if a biomarker is qualified as a surrogate endpoint with one treatment, it cannot be assumed that it automatically qualifies as a surrogate for a novel treatment with a different mechanism of action (59).
It should be noted that there is not always consensus on the appropriate clinical endpoint against which a biomarker should be qualified. In oncology clinical trials, for example, many observers have commented on the difficulties in using overall survival (OS) as the gold standard clinical endpoint in tumors for which several lines of treatment are available, leading to adoption in many trials of progression-free survival (PFS) as the primary clinical endpoint (61-64). Considerations such as these demand that we develop a pragmatic approach to biomarker qualification based not only on statistics but also incorporating elements of biological plausibility and practical usefulness (65).
Utilization
Utilization involves the assessment of biomarker performance in the specific context of its proposed use. This important step in biomarker evaluation asks whether the available evidence from validation and qualification provides sufficient support for the intended use of the biomarker (37). Different research and clinical settings may have distinct requirements and performance thresholds for incorporating a QI biomarker. For example, the U.S. Food and Drug Administration (FDA) may require a higher level of qualification when using biomarker data in support of drug approval than required by a pharmaceutical company when using biomarker data to prioritize compounds in its development pipeline (23).
In the clinical environment, contextual consideration of biomarker utilization allows for a holistic evaluation of a biomarker’s usefulness for decision making. For example, even if a QI biomarker is well correlated with clinical response to a drug agent, it may not demonstrate important drug side effects or toxicities (66), which in turn may imply the need for additional information beyond biomarker status (including qualitative information from imaging) for patient management. Proper consideration of QI utilization in the clinical setting must also address the possibility of assigning too much importance to statistical results and too little to clinical intuition and empirical judgment (67). A comprehensive evaluation of QI biomarker utilization would ideally consider the long-term effects of biomarker use on patient outcomes, notwithstanding the well-known difficulties in separating and measuring the effects of diagnostic imaging on improving patient health (68).
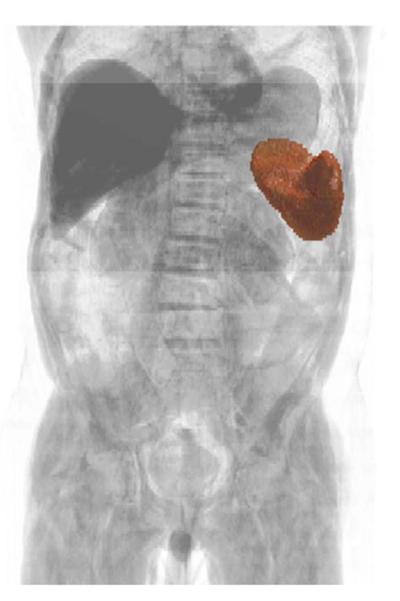
Comprehensive evaluation of QI biomarker utilization also addresses practical issues around biomarker incorporation into routine workflows. If biomarker data cannot be extracted and reported efficiently and at a reasonable cost, there is little likelihood of biomarker translation and adoption into standard-of-care clinical practice (69). Critical imperatives for QI biomarker research therefore include development of semi-automated and fully automated methods of data extraction (Fig. 3) (70), refinement of software tools for importing biomarker data into structured reports (71, 72), development of tools to facilitate QI biomarker tracking over time (73), integration of these tools with existing PACS and other radiology information systems, and investigations into the time-efficiency and cost-effectiveness of QI biomarker reporting (74, 75). Integration of QI biomarker archives with other clinical databases will likely assume greater importance with the anticipated transformation of radiology from a transactional to an information management business (76). Finally, implementing QI biomarker reporting in clinical practice may require dedicated insurance reimbursement to cover the costs of equipment upgrades, phantoms, software, image processing and interpretation, altered workflow, and ongoing quality assurance. These changes to reimbursement are difficult to achieve, and are likely to occur only following recommendations by expert panels and incorporation of QI biomarkers into clinical practice guidelines (77).
Fig. 3.
Volume-rendered CT of the abdomen and pelvis with overlaid 3-D surface rendering of the spleen, segmented by a fully automated multi-atlas content labeling algorithm. This technology is under investigation as a means of efficiently and accurately extracting spleen volume data for biomarker analyses. (Image courtesy of Zhoubing Xu, Ph.D. graduate student, Vanderbilt University.)
Where QI research is taking place
Conceptualization of QI biomarker research requires an appreciation of the developmental needs for QI and also consideration of the most appropriate environments for conducting QI investigations. The validation-qualification-utilization framework establishes an ambitious agenda for QI research that requires engagement from multiple stakeholders with different skill sets and end objectives.
Government funding for QI biomarker development exists through several arms of the National Institutes of Health (NIH), mostly notably the National Cancer Institute (NCI). The NCI’s Imaging Response Assessment Teams (IRAT) were an initial effort by the NCI to advance QI biomarkers for assessing therapy response, to increase the use of QI biomarkers in clinical trials, and to strengthen collaborations between basic imaging scientists and clinical oncology investigators. The NCI now encourages QI biomarker development principally through its Quantitative Imaging Network (QIN), which currently includes 17 “centers of imaging excellence” throughout the U.S. (78).
QI biomarker development is also taking place within a number of partnerships and consortia. The Quantitative Imaging Biomarkers Alliance (QIBA), established in 2007 by the RSNA, brings together imaging scientists, radiologists, and industry stakeholders for the advancement and use of QI biomarkers in both research and clinical practice. As of May 2014, QIBA has released publicly reviewed profiles for DCE-MRI quantification, CT tumor volume change, and FDG-PET/CT as an imaging biomarker for measuring response to cancer therapy (79). Meanwhile, the American College of Radiology Imaging Network (which recently merged with the Eastern Cooperative Oncology Group as ECOG-ACRIN) facilitates collaboration in QI clinical trials by academic and community radiologists as well as public and private stakeholders.
Private industry is also involved with QI biomarker development. Pharmaceutical companies are a source of funding for QI biomarker investigations, especially those designed to establish proof-of-concept for compounds in early-stage clinical testing (80). Several small companies are now marketing either stand-alone or plug-in software solutions for automated lesion measurement tracking (e.g., Mint Lesion, Mint Medical, Heidelberg, Germany; Median Lesion Management Solutions, Median Technologies, Valbonne, France; MimVista Software Inc., Cleveland, OH), and several large PACS vendors now offer biomarker tracking and reporting packages for various applications.
As QI biomarker research efforts move across the validation-qualification-utilization spectrum, deployment within clinical trials assumes greater importance (2). It is important to note that in many clinical trials, QI biomarkers are not the primary focus of the trial itself, but are rather deployed as tools to facilitate or accelerate a larger study objective (e.g., demonstrating efficacy of an investigational drug agent). The use of QI biomarkers confers several potential advantages within a clinical trial, including the possibility of populating the trial with biomarker-selected patients who have a higher likelihood of a positive therapeutic response; the opportunity to measure response earlier, more accurately, and/or less invasively than with other methods; and the potential ability to decrease overall study duration and cost by reducing both sample size and patient follow-up requirements (31, 36, 54). Clinical trials also provide a cost-effective environment for conducting QI research because QI correlative studies can be attached to trials with a broader funding appeal. However, conducting QI research within the confines of a larger clinical trial has important limitations, including the “two-variable” problem, i.e., the inherent difficulty in testing an exploratory biomarker and an investigational drug simultaneously (81), and the reluctance of trial sponsors to pay for additional imaging beyond standard-of-care scans.
The role of academic radiology in QI biomarker research
Academic radiology occupies a crucial role at the interface of basic imaging science and clinical research and is the proving ground for the eventual translation of novel techniques and approaches into routine practice. As such, academic radiology is poised to play a unique role in the development and dissemination of QI methods. Specific roles for academic radiologists include partnering with basic science colleagues to ensure that biomarker efforts are directed toward clinically relevant objectives; coordinating interdisciplinary collaboration between basic science and clinical researchers; designing and participating in analytical validation and qualification studies; and spearheading efforts to establish the potential advantages and appropriate utilization of QI biomarkers. Additional partnership opportunities include working with clinical colleagues from other disciplines to incorporate QI biomarkers into standardized diagnostic and therapeutic algorithms; working with informatics professionals to develop and test technology solutions for efficient QI biomarker extraction, reporting, and management; working with industry stakeholders to promote standardization of biomarker acquisition across vendor platforms; and working with collaborative groups and government agencies to establish data registries and new funding opportunities.
Challenges for academic radiology in QI biomarker research include prioritizing and focusing among a wide set of important objectives, avoiding redundant efforts given a broad array of stakeholders, and staying grounded with respect to basic tenets of standardization and quality assurance while pursuing higher-level technology evaluation. The latter challenge is especially relevant given the recent heightened interest in QI. It is crucial to address variability in methods before attempting to qualify QI biomarkers for broad clinical use (82). Finally, QI biomarker research output from academic radiology is currently hampered by the lack of training among radiologists in advanced clinical research techniques (83); the academic radiology community is addressing this deficiency through programs such as the AUR GE-Radiology Research Academic Fellowship (GERRAF) program, but additional initiatives would be beneficial.
Conclusion
Researchers and clinicians from across the biomedical spectrum are increasingly demanding QI biomarkers for incorporation into algorithmic decision making. The imaging community is responding to this demand by developing QI biomarkers in numerous modalities across a broad set of functional areas. QI biomarker development requires painstaking evaluation with sequential attention to analytical validation, qualification, and utilization of novel techniques and metrics. Academic radiology is poised to play a significant role in these efforts, especially in framing research questions and facilitating translation of emerging techniques from the laboratory into practice.
Acknowledgements
The authors gratefully acknowledge Lisa Li, Ph.D., and Lori Arlinghaus, Ph.D., and Zhoubing Xu for contributing figures to this manuscript.
Funding acknowledgements: AUR GE Radiology Research Academic Fellowship (RGA), P30 CA068485 (RGA), P50 CA098131 (RGA), NCI U01CA142565 (TEY), RSNA ESCH1319 (RMS), AHRQ HHSA290201200007I (RMS), T32 EB001631 (JPY)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Richard G. Abramson, Department of Radiology and Radiological Sciences Vanderbilt University 1161 21st Ave. S, CCC-1121 MCN Nashville, TN 37232-2675 (615)322-6759 Fax (615) 322-3764 richard.abramson@vanderbilt.edu.
Kirsteen R. Burton, Dept. of Medical Imaging and Institute of Health Policy, Management and Evaluation University of Toronto 263 McCaul Street, 4th Floor Toronto, ON M5T1W7 (416) 978-6801 kirsteen.burton@utoronto.ca.
John-Paul J. Yu, Department of Radiology and Biomedical Imaging University of California, San Francisco 505 Parnassus Ave., M-391 Box 0628 San Francisco, CA 94143-0628 jp.yu@ucsf.edu.
Ernest M. Scalzetti, Department of Radiology SUNY Upstate Medical University 750 E. Adams St. Syracuse NY 13210 scalzete@upstate.edu.
Thomas E. Yankeelov, Institute of Imaging Science Vanderbilt University 1161 21st Ave. S, AA-1105 MCN Nashville, TN 37232-2310 thomas.e.yankeelov@vanderbilt.edu.
Andrew B. Rosenkrantz, Department of Radiology NYU Langone Medical Center 550 First Avenue New York, NY 10016 (212) 263-0232 fax: (212) 263-6634 Andrew.Rosenkrantz@nyumc.org.
Mishal Mendiratta-Lala, Abdominal and Cross-sectional Interventional Radiology Henry Ford Hospital 2799 West Grand Blvd. Detroit, MI 48202 (313) 461-1648 mishall@rad.hfh.edu.
Brian J. Bartholmai, Chair, Division of Radiology Informatics Mayo Clinic Rochester, MN Phone 507-284-4292 FAX: 507-284-8996 Bartholmai.Brian@mayo.edu.
Dhakshinamoorthy Ganeshan, Department of Abdominal Imaging University of Texas MD Anderson Cancer Center Houston, TX 77030 713-792-2486 Fax: 713-745-1151 dganeshan@mdanderson.org.
Leon Lenchik, Department of Radiology Wake Forest School of Medicine Medical Center Boulevard Winston-Salem, NC 27157 Phone: 336-716-4316 Fax: 336-716-1278 llenchik@wakehealth.edu.
Rathan M. Subramaniam, Russell H Morgan Department of Radiology and Radiological Sciences Johns Hopkins School of Medicine Department of Health Policy and Management Johns Hopkins Bloomberg School of Public Health Johns Hopkins University Baltimore, MD.
References
- 1.DiSantis DJ. Early American Radiology: the pioneer years. AJR Am J Roentgenol. 1986;147(4):850–3. doi: 10.2214/ajr.147.4.850. [DOI] [PubMed] [Google Scholar]
- 2.Buckler AJ, Bresolin L, Dunnick NR, Sullivan DC, Group A collaborative enterprise for multi-stakeholder participation in the advancement of quantitative imaging. Radiology. 2011;258(3):906–14. doi: 10.1148/radiol.10100799. [DOI] [PubMed] [Google Scholar]
- 3.Buckler AJ, Paik D, Ouellette M, Danagoulian J, Wernsing G, Suzek BE. A novel knowledge representation framework for the statistical validation of quantitative imaging biomarkers. J Digit Imaging. 2013;26(4):614–29. doi: 10.1007/s10278-013-9598-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaffer FA, Weissleder R. Molecular imaging in the clinical arena. JAMA : the journal of the American Medical Association. 2005;293(7):855–62. doi: 10.1001/jama.293.7.855. [DOI] [PubMed] [Google Scholar]
- 5.Buckler AJ, Bresolin L, Dunnick NR, et al. Quantitative imaging test approval and biomarker qualification: interrelated but distinct activities. Radiology. 2011;259(3):875–84. doi: 10.1148/radiol.10100800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer CR, Armato SG, Fenimore CP, et al. Quantitative imaging to assess tumor response to therapy: common themes of measurement, truth data, and error sources. Translational oncology. 2009;2(4):198–210. doi: 10.1593/tlo.09208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biomarkers Definitions Working G Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clinical pharmacology and therapeutics. 2001;69(3):89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 8.Kessler LG, Barnhart HX, Buckler AJ, et al. The emerging science of quantitative imaging biomarkers terminology and definitions for scientific studies and regulatory submissions. Statistical methods in medical research. 2014 doi: 10.1177/0962280214537333. [DOI] [PubMed] [Google Scholar]
- 9.Rosenkrantz A. E. Clinical utility of quantitative imaging. Acad Radiol. doi: 10.1016/j.acra.2014.08.011. al. (accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arksey H, O’Malley L. Scoping studies: towards a methodological framework. International Journal of Social Research Methodology. 2005;8:19–32. [Google Scholar]
- 11.Sullivan DC. Imaging as a quantitative science. Radiology. 2008;248(2):328–32. doi: 10.1148/radiol.2482080242. [DOI] [PubMed] [Google Scholar]
- 12.Boone JM. Radiological interpretation 2020: toward quantitative image assessment. Med Phys. 2007;34(11):4173–9. doi: 10.1118/1.2789501. [DOI] [PubMed] [Google Scholar]
- 13.Choong MK, Tsafnat G. The implications of biomarker evidence for systematic reviews. BMC medical research methodology. 2012;12:176. doi: 10.1186/1471-2288-12-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) European journal of cancer. 2009;45(2):228–47. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Vidaurre T, Wilkerson J, Simon R, Bates SE, Fojo T. Stable disease is not preferentially observed with targeted therapies and as currently defined has limited value in drug development. Cancer J. 2009;15(5):366–73. doi: 10.1097/PPO.0b013e3181b9d37b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buyse M, Thirion P, Carlson RW, Burzykowski T, Molenberghs G, Piedbois P. Relation between tumour response to first-line chemotherapy and survival in advanced colorectal cancer: a meta-analysis. Meta-Analysis Group in Cancer. Lancet. 2000;356(9227):373–8. doi: 10.1016/s0140-6736(00)02528-9. [DOI] [PubMed] [Google Scholar]
- 17.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. The New England journal of medicine. 2001;344(11):783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 18.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. The New England journal of medicine. 2004;350(23):2335–42. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 19.Johnson DH, Fehrenbacher L, Novotny WF, et al. Randomized phase II trial comparing bevacizumab plus carboplatin and paclitaxel with carboplatin and paclitaxel alone in previously untreated locally advanced or metastatic non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2004;22(11):2184–91. doi: 10.1200/JCO.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 20.Jubb AM, Oates AJ, Holden S, Koeppen H. Predicting benefit from anti-angiogenic agents in malignancy. Nat Rev Cancer. 2006;6(8):626–35. doi: 10.1038/nrc1946. [DOI] [PubMed] [Google Scholar]
- 21.Workman P, Aboagye EO, Chung YL, et al. Minimally invasive pharmacokinetic and pharmacodynamic technologies in hypothesis-testing clinical trials of innovative therapies. Journal of the National Cancer Institute. 2006;98(9):580–98. doi: 10.1093/jnci/djj162. [DOI] [PubMed] [Google Scholar]
- 22.Desar IM, van Herpen CM, van Laarhoven HW, Barentsz JO, Oyen WJ, van der Graaf WT. Beyond RECIST: molecular and functional imaging techniques for evaluation of response to targeted therapy. Cancer treatment reviews. 2009;35(4):309–21. doi: 10.1016/j.ctrv.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 23.Abramson RG, Yankeelov TE. Imaging biomarkers and surrogate endpoints in oncology clinical trials. In: Luna A, Vilanova JC, Hygino Da, Cruz LC, Rossi SE, editors. Functional Imaging in Oncology. Springer; Heidelberg: 2014. [Google Scholar]
- 24.O’Connor JP, Jackson A, Asselin MC, Buckley DL, Parker GJ, Jayson GC. Quantitative imaging biomarkers in the clinical development of targeted therapeutics: current and future perspectives. Lancet Oncol. 2008;9(8):766–76. doi: 10.1016/S1470-2045(08)70196-7. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Donas J, Rodriguez-Antona C, Jonasch E. Molecular markers to predict response to therapy. Semin Oncol. 2013;40(4):444–58. doi: 10.1053/j.seminoncol.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Bhooshan N, Giger ML, Jansen SA, Li H, Lan L, Newstead GM. Cancerous breast lesions on dynamic contrast-enhanced MR images: computerized characterization for image-based prognostic markers. Radiology. 2010;254(3):680–90. doi: 10.1148/radiol.09090838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuleihan GE, Testa MA, Angell JE, Porrino N, Leboff MS. Reproducibility of DXA absorptiometry: a model for bone loss estimates. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 1995;10(7):1004–14. doi: 10.1002/jbmr.5650100704. [DOI] [PubMed] [Google Scholar]
- 28.Hua X, Lee S, Yanovsky I, et al. Optimizing power to track brain degeneration in Alzheimer’s disease and mild cognitive impairment with tensor-based morphometry: an ADNI study of 515 subjects. Neuroimage. 2009;48(4):668–81. doi: 10.1016/j.neuroimage.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Klaveren RJ, Oudkerk M, Prokop M, et al. Management of lung nodules detected by volume CT scanning. The New England journal of medicine. 2009;361(23):2221–9. doi: 10.1056/NEJMoa0906085. [DOI] [PubMed] [Google Scholar]
- 30.Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: Evolving Considerations for PET response criteria in solid tumors. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2009;50(Suppl 1):122S–50S. doi: 10.2967/jnumed.108.057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wintermark M, Albers GW, Alexandrov AV, et al. Acute stroke imaging research roadmap. AJNR American journal of neuroradiology. 2008;29(5):e23–30. doi: 10.1161/STROKEAHA.107.512319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merckel LG, Verkooijen HM, Peters NH, et al. The added diagnostic value of dynamic contrast-enhanced MRI at 3.0 T in nonpalpable breast lesions. PloS one. 2014;9(4):e94233. doi: 10.1371/journal.pone.0094233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Liu H, Sun J, et al. Varying correlation between 18F-fluorodeoxyglucose positron emission tomography and dynamic contrast-enhanced MRI in carotid atherosclerosis: implications for plaque inflammation. Stroke; a journal of cerebral circulation. 2014;45(6):1842–5. doi: 10.1161/STROKEAHA.114.005147. [DOI] [PubMed] [Google Scholar]
- 34.Kickingereder P, Sahm F, Wiestler B, et al. Evaluation of microvascular permeability with dynamic contrast-enhanced MRI for the differentiation of primary CNS lymphoma and glioblastoma: radiologic-pathologic correlation. AJNR American journal of neuroradiology. 2014;35(8):1503–8. doi: 10.3174/ajnr.A3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim I, Noh WC, Park J, et al. The combination of FDG PET and dynamic contrast-enhanced MRI improves the prediction of disease-free survival in patients with advanced breast cancer after the first cycle of neoadjuvant chemotherapy. Eur J Nucl Med Mol Imaging. 2014 doi: 10.1007/s00259-014-2797-4. [DOI] [PubMed] [Google Scholar]
- 36.Kurland BF, Gerstner ER, Mountz JM, et al. Promise and pitfalls of quantitative imaging in oncology clinical trials. Magnetic resonance imaging. 2012;30(9):1301–12. doi: 10.1016/j.mri.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Institute of Medicine . Evaluation of Biomarkers and Surrogate Endpoints in Chronic Disease. National Academies Press; Washington, DC: 2010. [PubMed] [Google Scholar]
- 38.Obuchowski NA, Reeves AP, Huang EP, et al. Quantitative imaging biomarkers: A review of statistical methods for computer algorithm comparisons. Statistical methods in medical research. 2014 doi: 10.1177/0962280214537390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raunig DL, McShane LM, Pennello G, et al. Quantitative imaging biomarkers: A review of statistical methods for technical performance assessment. Statistical methods in medical research. 2014 doi: 10.1177/0962280214537344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Padhani AR, Hayes C, Landau S, Leach MO. Reproducibility of quantitative dynamic MRI of normal human tissues. NMR Biomed. 2002;15(2):143–53. doi: 10.1002/nbm.732. [DOI] [PubMed] [Google Scholar]
- 41.Yankeelov TE, DeBusk LM, Billheimer DD, et al. Repeatability of a reference region model for analysis of murine DCE-MRI data at 7T. Journal of magnetic resonance imaging : JMRI. 2006;24(5):1140–7. doi: 10.1002/jmri.20729. [DOI] [PubMed] [Google Scholar]
- 42.Barnes SL, Whisenant JG, Loveless ME, Ayers GD, Yankeelov TE. Assessing the reproducibility of dynamic contrast enhanced magnetic resonance imaging in a murine model of breast cancer. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2013;69(6):1721–34. doi: 10.1002/mrm.24422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dula AN, Arlinghaus LR, Dortch RD, et al. Amide proton transfer imaging of the breast at 3 T: Establishing reproducibility and possible feasibility assessing chemotherapy response. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2012 doi: 10.1002/mrm.24450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alonzi R, Taylor NJ, Stirling JJ, et al. Reproducibility and correlation between quantitative and semiquantitative dynamic and intrinsic susceptibility-weighted MRI parameters in the benign and malignant human prostate. Journal of magnetic resonance imaging : JMRI. 2010;32(1):155–64. doi: 10.1002/jmri.22215. [DOI] [PubMed] [Google Scholar]
- 45.Whisenant JG, Peterson TE, Fluckiger JU, Tantawy MN, Ayers GD, Yankeelov TE. Reproducibility of Static and Dynamic (18)F-FDG, (18)F-FLT, and (18)F-FMISO MicroPET Studies in a Murine Model of HER2+ Breast Cancer. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2012 doi: 10.1007/s11307-012-0564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tseng JR, Dandekar M, Subbarayan M, et al. Reproducibility of 3′-deoxy-3′-(18)F-fluorothymidine microPET studies in tumor xenografts in mice. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2005;46(11):1851–7. [PMC free article] [PubMed] [Google Scholar]
- 47.Dandekar M, Tseng JR, Gambhir SS. Reproducibility of 18F-FDG microPET studies in mouse tumor xenografts. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2007;48(4):602–7. doi: 10.2967/jnumed.106.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sim J, Wright CC. The kappa statistic in reliability studies: use, interpretation, and sample size requirements. Physical therapy. 2005;85(3):257–68. [PubMed] [Google Scholar]
- 49.Beaton DE, Katz JN, Fossel AH, Wright JG, Tarasuk V, Bombardier C. Measuring the whole or the parts? Validity, reliability, and responsiveness of the Disabilities of the Arm, Shoulder and Hand outcome measure in different regions of the upper extremity. Journal of hand therapy : official journal of the American Society of Hand Therapists. 2001;14(2):128–46. [PubMed] [Google Scholar]
- 50.Bruton A, Conway JH, Holgate ST. Reliability: what is it, and how is it measured? Physiotherapy. 2000;86(2):94–9. [Google Scholar]
- 51.Katz R. Biomarkers and surrogate markers: an FDA perspective. NeuroRx : the journal of the American Society for Experimental NeuroTherapeutics. 2004;1(2):189–95. doi: 10.1602/neurorx.1.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bruns A, Kunnecke B, Risterucci C, Moreau JL, von Kienlin M. Validation of cerebral blood perfusion imaging as a modality for quantitative pharmacological MRI in rats. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2009;61(6):1451–8. doi: 10.1002/mrm.21779. [DOI] [PubMed] [Google Scholar]
- 53.Yoo AJ, Sheth KN, Kimberly WT, et al. Validating imaging biomarkers of cerebral edema in patients with severe ischemic stroke. Journal of stroke and cerebrovascular diseases : the official journal of National Stroke Association. 2013;22(6):742–9. doi: 10.1016/j.jstrokecerebrovasdis.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wintermark M, Albers GW, Broderick JP, et al. Acute Stroke Imaging Research Roadmap II. Stroke; a journal of cerebral circulation. 2013;44(9):2628–39. doi: 10.1161/STROKEAHA.113.002015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huynh TJ, Flaherty ML, Gladstone DJ, et al. Multicenter accuracy and interobserver agreement of spot sign identification in acute intracerebral hemorrhage. Stroke; a journal of cerebral circulation. 2014;45(1):107–12. doi: 10.1161/STROKEAHA.113.002502. [DOI] [PubMed] [Google Scholar]
- 56.Buyse M, Michiels S, Sargent DJ, Grothey A, Matheson A, de Gramont A. Integrating biomarkers in clinical trials. Expert Rev Mol Diagn. 2011;11(2):171–82. doi: 10.1586/erm.10.120. [DOI] [PubMed] [Google Scholar]
- 57.Puntmann VO. How-to guide on biomarkers: biomarker definitions, validation and applications with examples from cardiovascular disease. Postgraduate medical journal. 2009;85(1008):538–45. doi: 10.1136/pgmj.2008.073759. [DOI] [PubMed] [Google Scholar]
- 58.Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: theoretical considerations and practical challenges. J Clin Oncol. 2009;27(24):4027–34. doi: 10.1200/JCO.2009.22.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buyse M, Sargent DJ, Grothey A, Matheson A, de Gramont A. Biomarkers and surrogate end points--the challenge of statistical validation. Nature reviews Clinical oncology. 2010;7(6):309–17. doi: 10.1038/nrclinonc.2010.43. [DOI] [PubMed] [Google Scholar]
- 60.Sargent DJ, Rubinstein L, Schwartz L, et al. Validation of novel imaging methodologies for use as cancer clinical trial end-points. European journal of cancer. 2009;45(2):290–9. doi: 10.1016/j.ejca.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sargent DJ, Hayes DF. Assessing the measure of a new drug: is survival the only thing that matters? Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26(12):1922–3. doi: 10.1200/JCO.2007.14.8064. [DOI] [PubMed] [Google Scholar]
- 62.Hoering A, Leblanc M, Crowley JJ. Randomized phase III clinical trial designs for targeted agents. Clin Cancer Res. 2008;14(14):4358–67. doi: 10.1158/1078-0432.CCR-08-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Freidlin B, McShane LM, Korn EL. Randomized clinical trials with biomarkers: design issues. J Natl Cancer Inst. 2010;102(3):152–60. doi: 10.1093/jnci/djp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Villaruz LC, Socinski MA. The clinical viewpoint: definitions, limitations of RECIST, practical considerations of measurement. Clin Cancer Res. 2013;19(10):2629–36. doi: 10.1158/1078-0432.CCR-12-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Green E, Yothers G, Sargent DJ. Surrogate endpoint validation: statistical elegance versus clinical relevance. Statistical methods in medical research. 2008;17(5):477–86. doi: 10.1177/0962280207081863. [DOI] [PubMed] [Google Scholar]
- 66.Smith JJ, Sorensen AG, Thrall JH. Biomarkers in imaging: realizing radiology’s future. Radiology. 2003;227(3):633–8. doi: 10.1148/radiol.2273020518. [DOI] [PubMed] [Google Scholar]
- 67.Jha S. The allure of quantification. Magnetic resonance imaging. 2013;31(6):1035–6. doi: 10.1016/j.mri.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Mackenzie R, Dixon AK. Measuring the effects of imaging: an evaluative framework. Clin Radiol. 1995;50(8):513–8. doi: 10.1016/s0009-9260(05)83184-8. [DOI] [PubMed] [Google Scholar]
- 69.Abramson RG, Su PF, Shyr Y. Quantitative metrics in clinical radiology reporting: a snapshot perspective from a single mixed academic-community practice. Magnetic resonance imaging. 2012 doi: 10.1016/j.mri.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Lorenzo D, Francis S, Narayanan S, Arnold DL, Collins DL. Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal. 2013;17(1):1–18. doi: 10.1016/j.media.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 71.Pomar-Nadal A, Perez-Castillo C, Alberich-Bayarri A, Garcia-Marti G, Sanz Requena R, Marti-Bonmati L. [Integrating information about imaging biomarkers into structured radiology reports] Radiologia. 2013;55(3):188–94. doi: 10.1016/j.rx.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 72.Travis AR, Sevenster M, Ganesh R, Peters JF, Chang PJ. Preferences for structured reporting of measurement data: an institutional survey of medical oncologists, oncology registrars, and radiologists. Academic radiology. 2014;21(6):785–96. doi: 10.1016/j.acra.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 73.Abajian AC, Levy M, Rubin DL. Informatics in radiology: improving clinical work flow through an AIM database: a sample web-based lesion tracking application. Radiographics : a review publication of the Radiological Society of North America, Inc. 2012;32(5):1543–52. doi: 10.1148/rg.325115752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bresnahan BW. Economic evaluation in radiology: reviewing the literature and examples in oncology. Academic radiology. 2010;17(9):1090–5. doi: 10.1016/j.acra.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 75.Rubin DL, Willrett D, O’Connor MJ, Hage C, Kurtz C, Moreira DA. Automated tracking of quantitative assessments of tumor burden in clinical trials. Translational oncology. 2014 doi: 10.1593/tlo.13796. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Enzmann DR. Radiology’s value chain. Radiology. 2012;263(1):243–52. doi: 10.1148/radiol.12110227. [DOI] [PubMed] [Google Scholar]
- 77.Institute of Medicine . Developing biomarker-based tools for cancer screening, diagnosis and treatment: workshop summary. National Academies Press; Washington, DC: 2007. [Google Scholar]
- 78.Clarke LP, Nordstrom RJ, Zhang H, et al. The Quantitative Imaging Network: NCI’s Historical Perspective and Planned Goals. Translational oncology. 2014;7(1):1–4. doi: 10.1593/tlo.13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Radiological Society of North America [Accessed 5/20/14];QIBA Protocols and Profiles. Available at: https://www.rsna.org/QIBA_Protocols_and_Profiles.aspx.
- 80.Chan E, Arlinghaus LR, Cardin DB, et al. Gastrointestinal Cancers Symposium. San Francisco, CA: 2013. Phase I trial of chemoradiation with capecitabine and vorinostat in pancreatic cancer (scientific abstract) [Google Scholar]
- 81.Stadler WM. Radiological Society of North America (RSNA) Chicago, IL: 2010. Clinical needs/values of imaging biomarkers (oral presentation) [Google Scholar]
- 82.Doot RK, Thompson T, Greer BE, et al. Early experiences in establishing a regional quantitative imaging network for PET/CT clinical trials. Magnetic resonance imaging. 2012;30(9):1291–300. doi: 10.1016/j.mri.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krishnaraj A, Weinreb JC, Ellenbogen PH, Allen B, Jr., Norbash A, Kazerooni EA. The Future of Imaging Biomarkers in Radiologic Practice: Proceedings of the Thirteenth Annual ACR Forum. Journal of the American College of Radiology : JACR. 2014;11(1):20–3. doi: 10.1016/j.jacr.2013.08.017. [DOI] [PubMed] [Google Scholar]