Abstract
Human adenoviruses (HAdVs) are highly contagious pathogens causing acute respiratory disease (ARD), among other illnesses. Of the ARD genotypes, HAdV-7 presents with more severe morbidity and higher mortality than the others. We report the isolation and identification of a genome type HAdV-7d (DG01_2011) from a recent outbreak in Southern China. Genome sequencing, phylogenetic analysis, and restriction endonuclease analysis (REA) comparisons with past pathogens indicate HAdV-7d has re-emerged in Southern China after an absence of twenty-one years. Recombination analysis reveals this genome differs from the 1950s-era prototype and vaccine strains by a lateral gene transfer, substituting the coding region for the L1 52/55 kDa DNA packaging protein from HAdV-16. DG01_2011 descends from both a strain circulating in Southwestern China (2010) and a strain from Shaanxi causing a fatality and outbreak (Northwestern China; 2009). Due to the higher morbidity and mortality rates associated with HAdV-7, the surveillance, identification, and characterization of these strains in population-dense China by REA and/or whole genome sequencing are strongly indicated. With these accurate identifications of specific HAdV types and an epidemiological database of regional HAdV pathogens, along with the HAdV genome stability noted across time and space, the development, availability, and deployment of appropriate vaccines are needed.
Human adenoviruses (HAdVs) are highly contagious pathogens that are associated with a wide spectrum of human illnesses involving the respiratory, ocular, gastrointestinal, and genitourinary systems1,2, and a metabolic disorder (obesity)2. Of the several respiratory disease-associated HAdV pathogens, HAdV-4 and HAdV-7 are among the most commonly reported and associated with febrile respiratory disease, in particular, acute respiratory disease (ARD)2,3. These two genotypes4, particularly HAdV-7, circulate globally and frequently in civilian populations3,5,6,7. They are of such public health concerns that specific vaccines have been developed and deployed against them at two different time periods by the U.S. military8,9. These successful vaccine periods provided an unintentional experiment reinforcing the effectiveness of vaccines in public health10. In between the two periods of highly effective vaccine deployments, the inexplicable suspension of the vaccination program resulted in a resurgence of ARD cases to prevaccine-era levels10. This demonstrates the effectiveness and supports the development, availability, and deployment of vaccines against the HAdVs that affect certain populations routinely and predictably, particularly those under stressed and high-density conditions. Two important considerations for this public health program are required, along with vaccines: 1) the ability to monitor, and to identify circulating HAdV types and 2) the presence of molecular data quantifying and characterizing genome changes that may occur during viral evolution, particularly at the antigenic epitopes, e.g., the hexon hypervariable L1/L2 regions11,12,13,14.
HAdV-7 is of particular concern as it is associated often with illnesses presenting with more severe and higher levels of morbidity than other respiratory HAdV pathogens, and also may result in higher levels of fatalities3,15,16,17,18,19. As an example, a higher mortality rate was reported for children infected by HAdV-7 in China during 1958–1990 and also in Korea (Seoul) during 1990–2000, with mortality rates of 18%, caused by HAdV-7 genome types 7d and 7l, as compared with 3.6% noted for HAdV-3 infections15. HAdV strains compete against each other in host populations, with the presumably more robust ones replacing the previously dominant circulating strains. In a survey of 166 HAdV strains from the U.S. and Eastern Ontario/Canada (1966–2000), two genomic variants, that were previously absent in the population, were identified: HAdV-7d2 comprised 28% of the HAdVs reported and HAdV-7h comprised 2%3. HAdV-7d2 was first identified in the U.S. in 1993 and subsequently spread further in the U.S. and into Eastern Ontario3. It was concluded that both genomic variants represented “recent introduction[s]” from “previously geographically restricted areas… herald[ing] a shift in predominant [genome type] circulating in the [U.S.]”3. The origins of HAdV-7d, presumably the parent of HAdV-7d2, was noted as China, where it circulated from 1958 to 1990, having “replaced HAdV-7b as the predominant circulating virus”3,7 over a period of 10 years (1980–1990)20. HAdV-7d, in particular HAdV-7d2, from China subsequently spread “beyond [its] formerly geographically restricted regions”, e.g., to South Korea (1995–1997)15 and also Japan, Israel and the U.S. and Canada3,21,22. As another example of the severity of type 7 HAdV pathogens, the genomic variant HAdV-7h also produced more severe symptoms, including higher fatality levels, when it emerged and circulated in South America in the 1990s23.
Novel and re-emergent strains of highly contagious HAdV pathogens identified within mainland China are of public health concerns to the global community, and vice versa. Therefore, we call attention to and report the apparent re-emergence of HAdV-7d, after an approximately nineteen years absence in mainland China (1990 to 2009)20,24 and twenty-one years in Southern China (2011), with the isolation, identification, and analysis of the genome sequence of an adenovirus from a child afflicted with ARD during an outbreak in a primary school located in Dongguan of the Guangdong province in Southern China. This genome is nearly identical to two other recently characterized genomes, one isolated from Shaanxi Province in Northwest China (2009)24 and the other from Chongqing in Southwestern China (2010) (unpublished; JX625134). Given the caveat that the viruses may have been circulating earlier but had not been identified properly nor reported, the re-emergence of this genomic variant of HAdV-7 has potentially serious consequences in China, and globally, if it follows a similar trajectory as the earlier HAd-7d and HAdV-7d2 genome types that emerged in other countries and caused higher morbidity and mortality rates3,7,15.
Results
Adenovirus identification and genome annotation
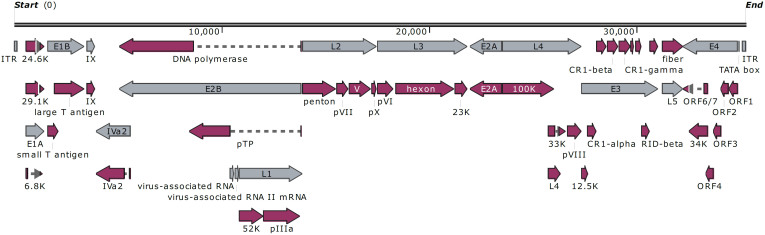
All 11 specimens collected were amplified “PCR-positive” for adenovirus and identified as HAdV-7 by type-specific PCR analysis. Of these, two, isolated from hospitalized and presumably more severe cases, produced visible CPE upon culturing. They were archived as DG01_2011 and DG02_2011. Sequence analysis revealed identical hexons, which were identified as HAdV type 7 by BLASTn analysis. The genome from DG01_2011 was then sequenced, assembled, annotated, and analyzed. Figure 1 presents the genomic organization and transcription map of DG01_2011. This genome contains 35,240 bp with a GC content of 51.08%. A total of 39 coding sequences were identified. These genome data, noted formally as “human adenovirus 7 strain CHN/DG01/2011/7[P7H7F7]” and in this report as “DG01_2011”, were deposited in GenBank (accession number KC440171).
Figure 1. Genomic organization and transcription map of human adenovirus 7d strain CHN/DG01/2011/7[P7H7F7]″ (DG01_2011).
The grey arrows indicate early, intermediate, and late transcription units; the red indicates coding regions. Arrows reflect the direction of the coding regions.
Genome type determination of HAdV-7 strains DG01_2011, 0901HZ/ShX/2009, and CQ1198_2010
The genome type of DG01_2011 was determined by comparing its in silico REA profiles with other HAdV-7 genome types reported in the literature3,7,11,15,20. Although seemingly antiquated in comparison to genome sequencing, REA profiles are still useful for comparisons with unsequenced but previously reported genome types and strains, and also as rapid and less-expensive alternatives for large-scale characterizations of viruses given a correct reference strain. Using the genome type denomination of Li, et al.20, DG01_2011 is identified as HAdV-7d, evidenced by the REA patterns and identical with the first reported HAdV-7d25, as shown in Figure 2. The REA patterns generated from DG01_2011, 0901HZ/ShX/2009, which caused acute bronchitis and pneumonia in an ARD outbreak comprising 70 cases amongst young children in the Shaanxi Province in 2009, including one fatality24, and CQ1198_2010, which was associated with an epidemic in Chongqing, Southwestern China (Ni, K., et al., unpublished; JX625134), are identical to each other and also identical with those of HAdV-7d reported earlier in Israel (1998) and Japan (1999)26,27. The REA patterns of CQ1198_2010 in this study provide evidence to amend the less-descriptive designation of “mutant HAdV-7d2” noted by Ni, K., et al. in the GenBank entry for CQ1198_2010 (JX625134). For reference, the in silico REA profile for the prototype Gomen HAdV-7 is provided; the REA patterns for these recent isolates differ clearly from the prototype, as shown in Figure 2. HAdV-7 prototype is the correct reference genome as three REA profiles, BclI, SalI, and XhoI, showed identical patterns and complement the REA patterns that differ, along with sequence similarities across the genome.
Figure 2. Identification of genome types and comparison of three recent HAdV-7d genomes in China.
The in-silico restriction endonuclease analysis (REA) pattern analysis of the HAdV-7d sequenced genomes from recently circulating strains DG01_2011, 0901HZ/ShX/2009, and CHN_CQ1198_2010 were generated using the Vector NTI 11.5 software (Invitrogen Corp.; San Diego). REA allows for comparisons with historical strains that were not sequenced but were characterized by REA patterns and descriptions. GenBank-available genome sequences of strains Gomen- the type 7 prototype (lane A; AY594255), DG01_2011 (lane B; KC440171), 0901HZ/ShX/2009 (lanes C; JF800905), and CHN_CQ1198_2010 (lane D; JX625134) were analyzed with BamHI, BclI, Bgll, BglII, BstEII, HindIII, HpaI, SmaI, EcoRI, SalI, XbaI, and XhoI, respectively, as described by Li, et al. (Li et al. J Med Virol, 1996, 49(3):170–177). M: molecular weight markers corresponding to a 1 kb DNA Ladder are provided for reference. All three recent outbreak genomes correspond to the genome type of HAdV-7d, and have identical REA profiles to each other, divergent from the HAdV-7 prototype (1952).
Phylogenetic analysis of hexon genes and whole genomes confirms the genome types
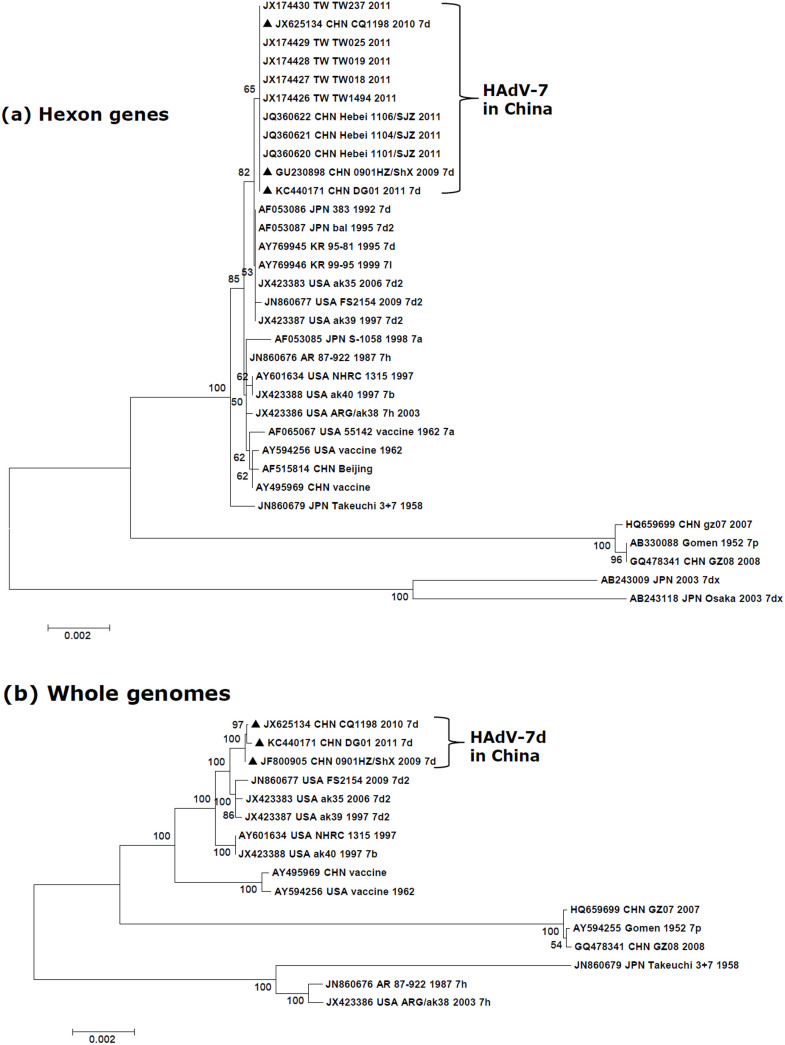
Phylogenetic analysis of 33 archived HAdV-7 hexon genes showed that DG01_2011 has an origin common to strains 0901HZ/ShX/2009, CQ1198_2010, Hebei_SJZ_2011, and TW_2011. These hexons form a subclade that is on the same branch with another subclade containing several non-China isolates, including HAdV-7d2 from the U.S., as shown in Figure 3A. The bootstrap value of 65 indicates the hexons from the China genomes are highly similar to each, but are separate from the U.S. HAdV-7d2 subclade (bootstrap value 82). Furthermore, the phylogenetic analysis of 16 available HAdV-7 whole genomes revealed DG01_2011, 0901HZ/ShX/2009, and CQ1198_2010 forming a subclade comprising HAdV-7d, and confirming the close relationships with each other, reaffirming a common lineage (Figure 3B) that is distinct from the HAdV-7d2 strains of the U.S.A. (bootstrap value 100). All of the genome types form subclades that are separate from the clade containing the prototype 7 (Gomen; 1952), with HAdV-7h forming a separate subclade in the genome phylogenetic analysis in contrast to the hexon gene phylogenetic analysis.
Figure 3. Phylogenetic analysis of HAdV-7d strain DG01_2011 isolated from Dongguan, Southern China (2011).
Nucleotide sequences of the archived HAdV-7 hexon genes (A) and whole genomes (B) are accessible from GenBank. These include five hexon gene sequences from Taiwan (TW; JX174426-JX174430); three from Hebei (JQ360620-JQ360622); one from Chongqing (JX625134); one from Shaanxi Province (JF800905); and one, this report, from Dongguan (DG) (KC440171). Additional HAdV-7 sequences from other global isolates were also analyzed. For reference, taxon names include the corresponding GenBank accession number, country of isolation, strain name, year of isolation (if available), and genome type (if available). Boot-strapped, neighbor-joining trees with 1,000 replicates were constructed using the MEGA 5.1.0 software (http://www.megasoftware.net) and by applying default parameters, with a maximum-composite-likelihood method. Bootstrap numbers shown at the nodes indicate the percentages of 1,000 replications producing the clade. A bootstrap value of 80 indicates robustness and confidence in the branching. The scale bar indicates units of nucleotide substitutions per site. Sequences from strains DG01_2011, 0901HZ/ShX/2009, and CQ1198_2010 are noted for reference (▴).
Comparative genomic analysis and single nucleotide differences of HAdV-7 strains causing ARD outbreaks in China
Comparative genomics analysis showed DG01_2011 has near genome identity with an earlier HAdV-7 isolate, 0901HZ/ShX/2009 (99.97%) and also with CQ1198_2010 (99.96%). Comparative genomics analysis documented seven single nucleotide substitution and one single base insertion differences between the DG01_2011 and 0901HZ/ShX/2009 genomes. Of these, two single nucleotide substitutions were localized in the ITRs and one non-synonymous substitution each was located in the DNA polymerase, penton base, and 34 kDa protein coding sequences (Table 1). One synonymous nucleotide substitution each was present in the 100-kDa hexon assembly-associated protein and Virus-Associated (VA) RNA II. The single nucleotide insertion was in a non-coding region of DG01_2011. There were three single nucleotide substitutions in coding sequences and seven base deletion differences in the ITRs between CQ1198_2010 and 0901HZ/ShX/2009 genomes. One synonymous substitution (C to T) was located in hexon assembly-associated protein (A254A) and the other two non-synonymous substitutions G to C and G to T were located in DNA polymerase (S55C) and 34 kDa protein (P87Q), respectively. The nucleotide deletions in ITRs of CQ1198_2010 may be sequencing errors given that the left ITR was not identical with the right ITR, or may represent recent mutations. If exclusive of ITR differences, there were only three single nucleotide substitutions between the CQ1198_2010 and 0901HZ/ShX/2009 genomes (99.99%). For strain DG01_2011, it had a higher genome identity with CQ1198_2010 (99.98%) than 0901HZ/ShX/2009 (99.97%) if exclusive of ITR difference. There were only four single nucleotide substitutions and one single nucleotide insertion in non-coding region between both genomes, which led to three non-synonymous substitutions in DNA polymerase (D1039E, S55C) and penton base gene (V239A), respectively.
Table 1. Comparative genomics analysis of three respiratory pathogens from recent China outbreaks: HAdV-7d strains DG01_2011, 0901HZ/ShX/2009, and CQ1198_2010. Nucleic acid and amino acid sequence changes are noted, along with their genome locations and coding consequences. The reference genome is 0901HZ/ShX/2009. *ITR: inverted terminal repeat. “-“: no change or not applicable.
| Nucleotide change | Amino acid substitution (DG01_2011/CQ1198_2010) | |||||
|---|---|---|---|---|---|---|
| Region | Position | Gene | DG01_2011 | CQ1198_2010 | Non-synonymous | Synonymous |
| ITR* | 1-6 | - | - | Deletion of CATCAT | -/- | -/- |
| ITR | 58 | - | T→C | - | -/- | -/- |
| Non-coding Region | 518 | - | Insertion of T | - | -/- | -/- |
| E2B | 5517 | DNA polymerase | G→C | - | D→E/- | -/- |
| 8470 | - | G→C | -/S→C | -/- | ||
| L1 | 10830 | Virus-Associated RNA II | G→A | - | -/- | -/- |
| L2 | 14594 | Penton base | T→C | - | V→A/- | -/- |
| L4 | 24298 | Hexon assembly -associated protein | C→T | C→T | -/- | A→A/A→A |
| E4 | 33155 | 34 kDa protein | G→T | G→T | P→Q/P→Q | -/- |
| ITR | 35182 | - | A→G | - | -/- | -/- |
| ITR | 35239 | - | - | Deletion of G | -/- | -/- |
Nucleotide substitution rates and selection pressures for HAdV-7d strain DG01_2011 major capsid protein genes
The selective pressures at the protein level for the three HAdV-7 capsid protein genes, hexon, penton base and fiber, were examined by comparing synonymous and non-synonymous mutations. All three genes have Ka/Ks ratios of less than 1 (Table 2). This is in accordance with the hypothesis that organismal evolution is dominated by negative selection, i.e., ones removing mutations harmful to fitness28. Specifically, both hexon and penton base genes have less non-synonymous substitutions per site, which leads to the low ratios of Ka/Ks. Although the non-synonymous substitutions and Ka/Ks ratio of the fiber gene is also low, it is relatively higher than for the hexon and penton base genes. This may indicate that the fiber gene has less negative selection pressures, likely due to tissue tropism being determined and constrained by the fiber gene. Overall, the majority of mutations are synonymous and do not affect the integrity of the hexon, penton base, and fiber proteins.
Table 2. Nucleotide substitution rates and selection pressures for the HAdV-7d strain DG01_2011 major capsid protein genes. The mean numbers of non-synonymous (Ka) and synonymous (Ks) substitutions per site from between sequences were noted and Ka/Ks ratios were calculated. This selection pressure analysis was conducted using the Nei-Gojobori model, and included nucleotide sequences from 33 hexon, 57 fiber, and 19 penton base genes of HAdV-7 available from GenBank. All positions containing gaps were eliminated automatically. Evolutionary analyses were performed with MEGA 5.1.0 (http://www.megasoftware.net).
| Gene | Length (bp) | No. of Sequences | Mean Non-synonymous Substitutions/site | Mean Synonymous Substitutions/site | Ka/Ks |
|---|---|---|---|---|---|
| Hexon | 2805 | 33 | 0.003 | 0.043 | 0.069 |
| Penton base | 1635 | 19 | 0.003 | 0.025 | 0.12 |
| Fiber | 978 | 57 | 0.013 | 0.053 | 0.245 |
Genome recombination analysis of HAdV-7d
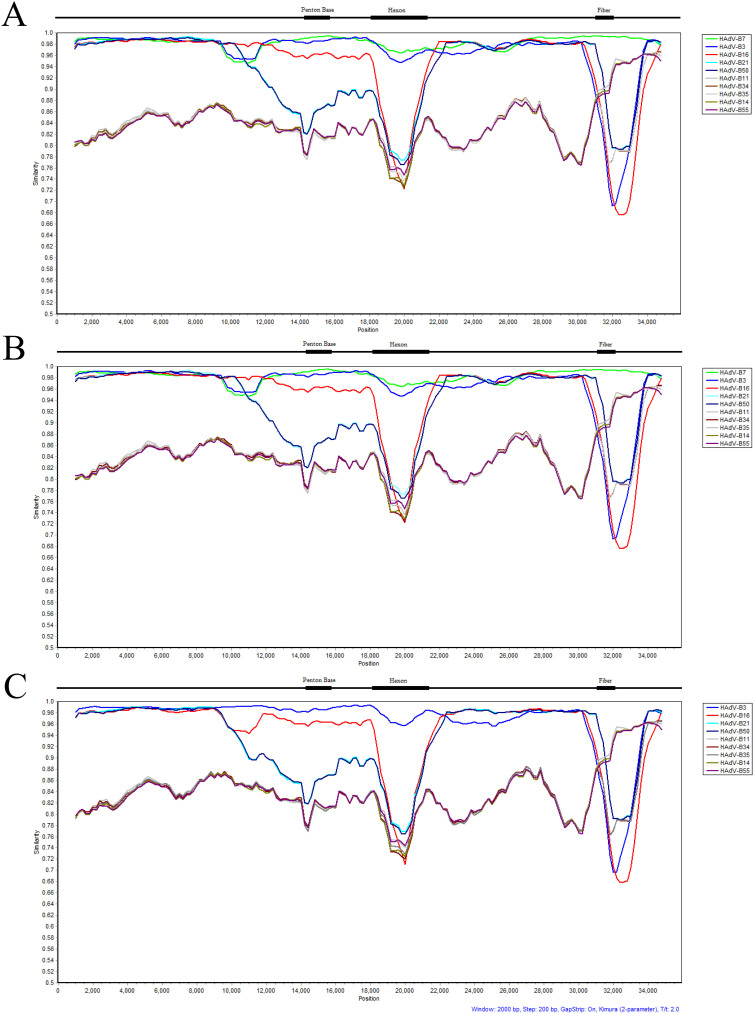
Genome recombination analysis using Simplot software29 reveals a lateral transfer of a small portion of the genome upstream of the penton base gene. This recombination contains the entire L1 52/55 kDa gene from HAdV-16 into HAdV-7d, as shown in Figure 4A. Its importance remains to be revealed. The gene transfer is also found in the genomes from the earlier strains CQ1198_2010 (Southwestern China; 2010; unpublished) and 0901HZ/ShX/2009 (Northwestern China; 2009)24, respectively, shown in Figure 4B, but not found in the prototype Gomen HAdV-7 genome, as displayed in Figure 4C.
Figure 4. Genome recombination analysis.
The genomes of HAdV-B7d strain DG_2011 (Guangdong Province, China; 2011), CQ1198_2010 (Southwestern China; 2010, unpublished) and 0901HZ/ShX/2009 (Northwestern China; 2009) were analyzed for sequence recombination events along with HAdV species B genomes using the software Simplot (http://sray.med.som.jhmi.edu/SCRoftware/simplot/). For the recombination analysis, MAFFT software was used first to align the sequences using default parameters (http://mafft.cbrc.jp/alignment/server/). Default parameter settings for the Simplot software were used for analyzing the whole genomes, with the following input: window size (2000 nucleotides [nt]), step size (200 nt), replicates used (n 100), gap stripping (on), distance model (Kimura), and tree model (neighbor-joining). Genome nucleotide positions are noted along the x-axis, and the sequence similarities are indicated along the y-axis. For reference, select genome landmarks of the three major capsid protein genes are noted above each graph. Simplot analysis of the genome of strain DG_2011 (A) shows a lateral transfer of a small portion of the genome upstream of the penton base gene from HAdV-B16. This is also present in the genomes of HAdV-7d strains CQ1198_2010 and 0901HZ/ShX/2009 (B). Surprisingly, this recombination is not found in the genome from the prototype virus from which these strains descend, Gomen HAdV-B7 (Fort Ord, U.S.A.; 1955) (C). Colors: green, HAdV-B7p (AY594255); blue, HAdV-B3 (AY599834.1); magenta, HAdV-B16 (AY601636.1); light blue, HAdV-B21 (AY601633.1); dark blue, HAdV-B50 (AY737798.1); grey, HAdV-B11 (AY163756.1); brown, HAdV-B34 (AY737797.1); light grey, HAdV-B35 (AY271307.1); dark green HAdV-B14 (AY803294.1); and dark violet HAdV-B55 (FJ643676.1).
Discussion
Among the two HAdV species B respiratory pathogens most frequently associated with ARD outbreaks globally, HAdV-7 is reported to cause a higher mortality rate than HAdV-3 in one long-term survey (1958–1990)20, as well as in a recent shorter term survey of adenoviral pneumonia cases in Beijing (2009–2011)19. Genome type HAdV-7d apparently originated and circulated in China from 1958–1990, becoming the predominant strain during the period of 1980–199020. It was also the prevalent genome type found in Korea during two outbreaks in 1995–1996 and 2001–2002, accounting for 98–100% all of the type 7 HAdV strains assayed30. Interestingly, despite reports of global circulation, HAdV-7, and in particular HAdV-7d, epidemics had not been reported in mainland China from 1990 to 2009. In 2009, HAdV-7 was identified as the respiratory pathogen in an outbreak that included a fatality in Shaanxi24 and also in a 2010 outbreak in Chongqing (unpublished), signaling a reemergence. Thorough characterization of these pathogens is evidenced by the availability of two genome sequences (JF800905 and JX625134), both of which are further identified as the HAdV-7d genome type in this report, and shown to be nearly identical to this report of an isolate from a 2011 ARD outbreak in Guangdong Province (strain DG01_2011) by comparative genomics and, in particular, in silico REA pattern analysis, as presented in Figure 2.
Although not ideal and largely replaced by whole genome sequencing, REA patterns can still provide rapid and relatively inexpensive characterizations of the genomes of large number of pathogens in an outbreak31,32,33,34,35,36. For HAdV comparisons, the caveat is to use the correct reference genome; for example, HAdV-55 contains a partial hexon gene from HAdV-11, comprising approximately only 2.6% of the length of the genome, in a chassis of HAdV-14, comprising approximately 97.4% of the length of the genome37. Using the genome of HAdV-11 as a reference yields meaningless patterns that are subject to researcher-biased interpretations and leads to erroneous conclusions that HAdV-55 is a genome type of HAdV-11. Using the HAdV-14 genome as a reference provides a closer approximation of the genome identities4,38. However, the recombination event revealed by whole genome sequencing, with the conflicting “Trojan Horse” renal pathogen epitope observed with ARD symptoms, indicates this was a novel and emergent pathogen4,37,39,40,41,42. In contrast, for HAdV-7d, the prototype HAdV-7 genome provides the correct reference: three REA patterns are identical (BclI, SalI, and XhoI); four are obviously different (BamHI, BglI, HpaI, and SmaI); and four are highly similar with a few differences in the band patterns (BglII, BstEII, HindIII, EcoRI, and XbaI), shown in Figure 2.
The major advantages of REA comparisons are the value and abundance of earlier molecular epidemiology studies, prior to the genome sequencing era, presenting REA data, and, in many cases, relating particular genome types to clinical, epidemiological, and pathogenicity observations. All of these historical strains are physically lost and no longer available for further genomic or laboratory characterization. In essence, however, the value and knowledge of the outbreaks, pathogens, and researchers of the past are not entirely lost if genomes of current pathogenic strains of interest may be compared with published REA patterns of past pathogens, as demonstrated in the genome type identities presented in this report.
Whole genome characterization of HAdV provides a higher-resolution perspective of understanding this pathogen, which may or may not lead to better public health strategies and measures to prevent outbreaks. As noted for two species B ARD pathogens, HAdV-4 and HAdV-7, “restricted use” but effective vaccines can be and are deployed currently in the U.S. military to prevent ARD outbreaks8,9,10. However, even if there were no viable strategy to manage HAdV outbreaks, knowing the genome type, either by REA or by whole genome sequencing, allows an understanding of the epidemiology, including potential morbidity and mortality profiles, of the circulating pathogens.
As discussed earlier, genome types may have different pathogenicity, infectivity, and virulence profiles; for example, a higher mortality rate was reported for children infected by genome types 7d and 7l in Korea, with mortality rates of 18%, compared to 3.6% for HAdV-3 infections15. Another genome type, HAdV-7h, also resulted in more severe symptoms, including fatalities in South America23. For their molecular epidemiological studies of HAdV-7, Wadell and colleagues presented numerous REA patterns generated with restriction endonucleases (BamHI, BclI, Bgll, BglII, BstEII, HindIII, HpaI, SmaI, EcoRI, SalI, XbaI, and XhoI), parsing HAdV-7 isolates from various regions and across many years to divide them into more than 20 genome types15,25,43,44,45.
Adenoviruses contain relatively stable double-stranded DNA genomes14,46,47. There are seven single base substitutions and a one-base insertion between strains DG01_2011 and 0901HZ/ShX/2009, which led to three non-synonymous substitutions in the DNA polymerase, penton base, and 34 kDa protein coding sequences. Interesting, there are only three single base substitutions between strains CQ1198_2010 and 0901HZ/ShX/2009, exclusive of the nucleotide deletions in ITRs of CQ1198_2010 which may be due to possible sequencing errors. The high genome percent identity between strains CQ1198_2010 and 0901HZ/ShX/2009 and the adjacent locations of Chongqing and Shaanxi (394 kilometers apart) where strains CQ1198_2010 and 0901HZ/ShX/2009 were isolated indicate strain 0901HZ/ShX/2009 may be the origin of strain CQ1198_2010. Strain DG01_2011 has a higher genome identity with CQ1198_2010 than 0901HZ/ShX/2009, which also supports the hypothesis that strain CQ1198_2010 is be the ancestor of strain DG01_2011.
Although HAdV genomes appear stable in terms of single base changes, as expected for double stranded DNA viruses and as observed in pairs of HAdV genomes examined to date, e.g., the prototype versus circulating strains of HAdV-3 and -5, separated by approximately fifty years46,47, less common but biologically and clinically significant larger genome changes are observed either as a single, small recombination event, such as the lateral transfer of the renal pathogen epsilon epitope (HAdV-11) providing a “Trojan Horse” effect to the recombinant HAdV-55, an emergent acute respiratory disease (ARD) pathogen in a putatively immune naive host population37, or as multiple and larger recombination events, such as the lateral transfer of the non-pathogen epsilon epitope (HAdV-D22) along with multiple other sequences to an emergent recombinant resulting in the highly contagious ocular pathogen causing epidemic keratoconjunctivitis (EKC), HAdV-D5348. Additionally, the presence of the epsilon epitope of a nonpathogenic type, HAdV-19, found in several recently reported emergent recombinant EKC pathogens, HAdV-64, support the hypothesis that recombination amongst HAdVs is an important mechanism driving the molecular evolution and genesis of HAdV pathogens49. In both of these latter examples, newly emergent HAdV pathogens have the “serotype” of nonpathogens but are potent, significant, and highly contagious human pathogens.
Recombination appears to play another novel and major role in the molecular evolution of HAdVs and genesis of human pathogens. Recent reports of HAdV genomes containing genome segments, including near-entire genomes, derived from simian adenoviruses (SAdVs) indicate zoonosis is an avenue of lateral gene transfer. Thus, nonhuman primates may be a wellspring of emergent human pathogens50,51, and vice versa52.
A novel third type of lateral gene transfer is revealed in this newly reported genome of HAdV-7d strain DG01_2011, that of a “moderate-sized” single whole gene recombination. This serendipitous insight into the molecular evolution of these respiratory pathogens from HAdV species B demonstrates the genomes of individual HAdV types, such as type 7, contain changes revealed only by high-resolution genome sequences and may be important in the context of HAdV molecular evolution, viral fitness, origins and bases of clinical and pathogenicity differences, and account for emergent and re-emergent pathogens. BLAST analysis reveals the recombinant region to encode the entire L1 52/55 kDa gene of HAdV-B16 with flanking non-coding sequences. The BLAST scores indicate the first highly similar sequence, aside from several type 7 sequences, is that from the HAdV-B16 prototype (Max. score 2710, Total score 2710, Query cover 100%, E-value 0.0, Ident. 96%) and a HAdV-B16 recombinant (2687, 2687, 100%, 0.0. 96%), with additional homologous and highly similar sequences found in HAdV-B50 (2436, 2436, 100%, 0.0, 93%) and HAdV-B21 strains (2422, 2422, 100%, 0.0, 93%). This encodes a DNA-binding protein that is expressed in both the early and late stages of infection, suggesting it could play multiple roles in the adenoviral life cycle. The L1 52/55 kDa protein interacts with the IVa2 protein and is an essential protein that is absolutely required for DNA packaging as well53,54. Effects of this particular moderate-sized recombination from HAdV-B16 into the HAdV-B7 genome chassis and the resultant emergent pathogen are unknown pending wet-bench investigations and additional clinical reports. The HAdV-7 prototype strain (AY594255)55 analyzed is also known as the Gomen strain, which was isolated as a clinical specimen from a throat washing of a U.S. military recruit with pharyngitis56. This strain is nearly contemporaneous with the Greider strain (AY594256)57, aka HAdV-7a, which was used to develop the vaccine strain14,58. Although there are minor genome differences, e.g., point mutations, between the prototype and the vaccine strains, pairwise genome dot blot analysis (PipMaker) indicated no recombination events14.
These observations strongly support and validate the recent paradigm change of using the genome data along with biological and clinical profile changes to recognize, characterize, type, and name novel HAdVs rather than relying solely on the epsilon and/or gamma epitopes4,59, determined either by serology or imputed by limited DNA sequencing, in the past.
With the exception of sporadic HAdV-7 infections reported in children in Guangzhou (2011)60 and the three recent outbreaks, the apparent absence of type 7 ARD pathogen circulating in the population of Southern China before 2011 leads to a concern that the dense city populations in China are now immunogenically naïve with respect to HAdV-7. In Northern China, recently, 312 isolates were typed as HAdV-7 by PCR and sequencing of hexon genes from 848 HAdV-positive specimens during 2003–2012; HAV-7 was associated with most of the severe lower respiratory HAdV infections18. Coupled with increased opportunities for travel, a “Perfect Storm” for present and near-future outbreaks of the apparently more severe disease-causing HAdV-7d strains is foreboding.
In Chongqing (Southwestern China), 92 (48.17%) cases involving HAdV-7 were identified from children presenting with ARD during 2009–2012 by hexon sequencing40. Recently, there were two ARD outbreaks caused by HAdV-7, both of which occurred in military training camps, one in Shaanxi Province (Northwest China) from February to March of 201261, the other in Wuhan (Central China) from January to February of 201362. In the former outbreak, a total of 176 patients were sampled, with all of the patients being males, with ages between 16–34 years61. In the latter, 440 patients aged between 17–22 years were reported as afflicted with ARD62. In Taiwan, there was a large community outbreak of HAdV-7 in 201116. In this instance, an abrupt increase in percentage of HAdV-7 infections occurred, from 0.3% in 2008–2010 to 10% in 201116. The hexon nucleotide sequences of five HAdV-7 isolates collected in Taiwan were identical to the sequence of HAdV-7 strain 0901HZ/ShX/200916, which was also identical to DG01_2011. In the context of the data in this report, these “Taiwan” hexon genes formed the same subclade with strains 0901HZ/ShX/2009, CQ1198_2010 and DG01_2011 (Fig. 3A). Given that only the hexon genes were sequenced, the exact genome types of these strains in the two outbreaks remain unknown. However, the possibility of a HAdV-7d genome type circulating is foreboding. Further data, including complete genome sequencing and in silico REA, are important to confirm this possibility.
In the interest of global public health, with these recent outbreaks and the identification of nearly identical contemporary HAdV-7d genome types, we strongly urge molecular surveillance and genotyping of newly isolated HAdV strains in China by whole genome sequencing and/or in silico REA. Additionally, the newly-redeveloped vaccines, which are now only accessible to the U.S. military9, should be made available to the civilian “at-risk” public to prevent “preventable” highly contagious outbreaks involving HAdVs associated with high morbidity rates and fatalities5,7,15,16,17,18,19,20,21,22,23,24,25,26,27,30,43,45,60,61,63. In particular, the vaccine against HAdV-7 is urgently needed in China, due to the apparent decades-absence of circulating HAdV-7, which presumably resulted in a corresponding lower level of herd immunity in today's population. Given the higher severity of diseases and fatality rates caused by HAdV-7, especially HAdV-7d, extensive surveillance and corresponding molecular investigation, including genotyping, genome typing, and genome sequencing, should be carried out when confronting outbreaks of HAdV pathogens in the high-density populations of China to protect the public and the global community.
Methods
Specimen collection and handling
During February 27 to March 6 of 2011, twenty-three primary school children under the age of 12 (Dongguan; Guangdong Province) presented with flu-like symptoms, including fever, pharyngalgia, and coughing as well as other indications of ARD. Two were hospitalized with severe symptoms. Eleven throat swab specimens were collected into 2-ml viral transport media; transported at 2°C–8°C; and preserved at −80°C for virus isolation and nucleic acids extraction. This study protocol was approved by the institutional ethics committee of the Center for Disease Control and Prevention of Guangdong Province (Guangdong CDC) and was carried out in accordance with the approved guidelines. The guardians of all under-aged participants gave signed informed consent for participation in the study. Data records of the samples and sample collection are de-identified and completely anonymous.
Detection of respiratory pathogens
Total nucleic acids were extracted from the specimens using the QIAamp minElute virus spin kit (Qiagen; Hombrechtikon, Germany). Human adenovirus, respiratory syncytial virus, influenza virus A and B, parainfluenza virus types 1–3, human rhinovirus, human metapneumovirus, and human coronavirus OC43 and 229E were detected by real-time PCR as described earlier63. For HAdV identification, type-specific primers were used to characterize the type by PCR, as described in an earlier report60.
Adenovirus isolation and genomic DNA extraction
Adenovirus-positive throat swab specimens, identified by PCR analysis, were inoculated into A549 cell cultures, and grown in Dulbecco's minimum essential medium supplemented with 100 IU penicillin ml−1, 100 mg streptomycin ml−1, and 2% (v/v) fetal calf serum, at an atmosphere of 5% (v/v) carbon dioxide. Cytopathic effect (CPE) was monitored for at least ten days. Viral genomic DNA was extracted from infected cells for genomic analysis, as described by Le, et al.64.
Genome sequencing and annotation
The genome of HAdV strain DG01_2011 was sequenced using a Sanger chemistry-based, primer-walking method by PCR-amplification, with overlapping regions sequenced39,65. Both 5′- and 3′-ends (including both inverted terminal repeats) were sequenced directly by primers Ad7-LTRS1A (5′-GCCTCTTGACGGAACTCG-3′) and Ad14-LTRS2 (5′-GGTCCCTCTAAATACACATACA-3′), respectively, using genomic DNA as template; this ensured the accurate determination of the end sequences39,65. The sequence data, collected with an ABI 3730 Genetic Analyzer, provided an average genome coverage of 3- to 5-fold, with both strands represented. Gaps and ambiguous sequences were PCR-amplified using different primers and resequenced. These sequencing ladders were assembled with the SeqMan Pro software 7.0.1 (DNASTAR, Inc.; Madison, WI. USA). Nucleotide and amino acid sequences were aligned with CLUSTAL and BLAST software. The genome sequence was annotated based on the previous annotation of HAdV-7 prototype strain (Gomen)55 and deposited into GenBank with the accession number KC440171.
In silico restriction endonuclease analysis (REA)
The specific adenovirus genome type was determined using in silico REA analysis of the whole-genome sequences in accordance with the in vitro protocol described by Li, et al20. This was performed using the software Vector NTI Advance 11.5 (Invitrogen Corp.; San Diego, CA. USA). Twelve restriction enzymes were used for this analysis, as performed by Li, et al.20: BamHI, BclI, Bgll, BglII, BstEII, HindIII, HpaI, SmaI, EcoRI, SalI, XbaI, and XhoI.
Phylogenetic analyses of HAdV-7 hexon genes and the whole genome sequences. The Molecular Evolutionary Genetics Analysis (MEGA) version 5.1.0 software was used for phylogenetic analyses of the HAdV-7 hexon genes and the whole genomes, with additional sequences retrieved from GenBank database, as described previously66,67. Neighbor-joining phylogenetic trees with 1,000 boot-strap replicates were constructed using a maximum-composite-likelihood method with default parameters. Bootstrap numbers shown at the nodes indicate the percentages of 1,000 replications producing the clade, with a value of 80 noted as robust and significant.
Archived HAdV-7 genome sequences from GenBank were used for phylogenetic analysis. These are as follows (for reference, the names include the corresponding GenBank accession number, country of isolation, strain name, year of isolation (if available), and genome type (if available)): AY594255_Gomen_1952_7p, JX625134_CHN_CQ1198_2010_7d, JF800905_CHN_0901HZ/ShX_2009_7d, JX423388_USA_ak40_1997_7b, JX423386_USA_ARG/ak38_2003_7h, JX423387_USA_ak39_1997_7d2, JX423383_USA_ak35_2006_7d2, JN860677_USA_FS2154_2009_7d2, JN860679_JPN_Takeuchi_3+7_1958, JN860676_AR_87-922_1987_7h, GQ478341_CHN_GZ08_2008, HQ659699_CHN_GZ07_2007, AY594256_USA_vaccine_1962, AY495969_CHN_vaccine, AY601634_USA_NHRC_1315_1997, and KC440171_CHN_DG01_2011_7d.
The HAdV-7 hexon complete sequences used for these analyses are as follows: AB330088_Gomen_1952_7p, JN860679_JPN_Takeuchi_3+7_1958, AF065067_USA_55142_vaccine_1962_7a, AY594256_USA_vaccine_1962, AF515814_CHN_Beijing, AY495969_CHN_vaccine, JN860676_AR_87-922_1987_7h, AF053086_JPN_383_1992_7d, AF053087_JPN_bal_1995_7d2, AY769945_KR_95-81_1995_7d, JX423387_USA_ak39_1997_7d2, JX423388_USA_ak40_1997_7b, AY601634_USA_NHRC_1315_1997, AF053085_JPN_S-1058_1998_7a, AY769946_KR_99-95_1999_7l, AB243009_JPN_2003_7dx, AB243118_JPN_Osaka_2003_7dx, JX423386_USA_ARG/ak38_2003_7h, JX423383_USA_ak35_2006_7d2, HQ659699_CHN_gz07_2007, GQ478341_CHN_GZ08_2008, GU230898_CHN_0901HZ/ShX_2009_7d, JN860677_USA_FS2154_2009_7d2, JX625134_CHN_CQ1198_2010_7d, JQ360620_CHN_Hebei_1101/SJZ_2011, JQ360621_CHN_Hebei_1104/SJZ_2011, JQ360622_CHN_Hebei_1106/SJZ_2011, JX174426_TW_TW1494_2011, JX174427_TW_TW018_2011, JX174428_TW_TW019_2011, JX174429_TW_TW025_2011, JX174430_TW_TW237_2011, and KC440171_CHN_DG01_2011_7d.
Genome recombination analysis
The genomes of HAdV-B7d strains DG01_2011 (Guangdong Province, China; 2011), CQ1198_2010 (Southwestern China; 2010, unpublished), and 0901HZ/ShX/2009 (Northwestern China; 2009)24, along with the prototype Gomen genome were analyzed for sequence recombination events using the software tool Simplot (http://sray.med.som.jhmi.edu/SCRoftware/simplot/)29. For the recombination analysis, MAFFT software was used first to align the HAdV-B species sequences using default parameters (http://mafft.cbrc.jp/alignment/server/). Default parameter settings for the Simplot software were used for analyzing the whole genomes, along with the following input: window size (2000 nucleotides [nt]), step size (200 nt), replicates used (n 100), gap stripping (on), distance model (Kimura), and tree model (neighbor-joining). The following genomic sequences of HAdV-B members were used: HAdV-B7p (AY594255), HAdV-B3 (AY599834), HAdV-B16 (AY601636), HAdV-B21 (AY601633), HAdV-B50 (AY737798), HAdV-B11 (AY163756), HAdV-B34 (AY737797), HAdV-B35 (AY271307), HAdV-B14 (AY803294), and HAdV-B55 (FJ643676).
Substitution Rate analysis of the hexon, penton base and fiber genes in HAdV-7
The numbers of non-synonymous (Ka) and synonymous (Ks) substitutions per site from between sequences were noted and the Ka/Ks ratios were calculated. This HAdV-7 analysis was conducted using the Nei-Gojobori model68, and included nucleotide sequences from 33 hexon genes, 57 fiber genes, and 19 penton base genes available from GenBank. All positions containing gaps and missing data were eliminated automatically. Evolutionary analyses were performed with MEGA 5.1.066.
The HAdV-7 complete hexon, penton base and fiber gene sequences available in GenBank were achieved for analysis. The HAdV-7 complete hexon gene sequences used for this analysis are same with previous those in phylogenetic analysis. The following HAdV-7 complete fiber gene sequences were used: AY495969, AY594255, AY594256, AY601634, GQ478341, HQ659699, JF800905, JN860677, JX625134, GQ265864, GQ265865, GQ265866, GQ265867, GQ265868, GQ265869, GQ265871, GQ265872, GQ265873, HM057190, JQ410438, JQ410439, JQ410440, JQ410441, JQ410442, JQ410443, JX174431, JX174432, JX174433, JX174434, JX174435, KC456126, KC456127, KC456128, KC456129, KC456130, KC456132, KC456133, KC456134, KC456135, KC456136, KC456137, KC456138, KC456139, KC456140, KC456141, KC456142, AC_000018, KJ195467, KF268117, KF268125, KF268134, KF268135, JX423383, JX423386, JX423387, JX423388, and JX625134. The HAdV-7 complete penton base gene sequences used in this analysis: AY495969, AY594255, AY594256, AY601634, GQ478341, HQ659699, JN860677, JX625134, AC_000018, AD001675, KF268117, KF268125, KF268134, KF268135, JX423383, JX423386, JX423387, JX423388, and JX625134.
Author Contributions
Q.Z. and D.S. conceived and designed experiments. S.Z., C.W., C.K., J.S., S.D., L.Zou, J.Z., Z.C., S.J., Z.Z., J.Z., X.Wan, X.Wu, W.Z., L.Zhu, D.S. and Q.Z. performed the experiments and analyzed the data. S.Z., C.W., D.S. and Q.Z. wrote the manuscript. All authors reviewed the manuscript.
Acknowledgments
This work was supported by funding from the National Natural Science Foundation of China (31100133 and 31370199) and the Natural Science Foundation of Guangdong Province (S2012010009261), along with Special Grants from the Guangzhou Pearl River Young Talents of Science and Technology (2013J2200052) and the Excellent Young Teacher Training Plan of Guangdong Province (Yq2013039), and the Guangzhou Healthcare Collaborative Innovation Major Project (201400000002). D. Seto thanks the Office of the Provost at George Mason University for funding a Sabbatical Study Leave (2013) and Professor James Chodosh, his research group, and the Massachusetts Eye and Ear Infirmary at Harvard Medical School (Boston, MA) for providing a stimulating intellectual environment (2013). S.D. and D.S. dedicate this work to their friend, colleague, and collaborator Jason Seto (1964–2014).
References
- Hierholzer J. C. in Diagnostic Procedures for Viral, Rickettsial, and Chlamydial Infections (eds Lennette, E. H., Lennette, D. A. & Lennette, E. T.) 169–188 (American Public Health Association, 1995). [Google Scholar]
- Lion T. Adenovirus Infections in Immunocompetent and Immunocompromised Patients. Clin. Microbiol. Rev. 27, 441–462 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman D. D. et al. Molecular Epidemiology of Adenovirus Type 7 in the United States, 1966–2000. Emerg. Infect. Dis. 8, 269–277 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto D., Jones M. S., Dyer D. W. & Chodosh J. Characterizing, typing, and naming human adenovirus type 55 in the era of whole genome data. J. Clin. Virol. 58, 741–742 (2013). [DOI] [PubMed] [Google Scholar]
- Kajon A. & Wadell G. Genome analysis of South American adenovirus strains of serotype 7 collected over a 7-year period. J. Clin. Microbiol. 32, 2321–2323 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadell G. & Varsanyi T. M. Demonstration of three different subtypes of adenovirus type 7 by DNA restriction site mapping. Infect. Immun. 21, 238–246 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadell G. et al. Molecular epidemiology of adenoviruses: global distribution of adenovirus 7 genome types. J. Clin. Microbiol. 21, 403–408 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock R. M., Ludwig W., Heubner R. J., Cate T. R. & Chu L. W. Immunization by selective infection with type 4 adenovirus grown in human diploid tissue cultures. I. Safety and lack of oncogenicity and tests for potency in volunteers. JAMA 195, 445–452 (1966). [PubMed] [Google Scholar]
- Hoke C. H. Jr & Snyder C. E. Jr History of the restoration of adenovirus type 4 and type 7 vaccine, live oral (Adenovirus Vaccine) in the context of the Department of Defense acquisition system. Vaccine 31, 1623–1632 (2013). [DOI] [PubMed] [Google Scholar]
- Russell K. L. et al. Vaccine-preventable adenoviral respiratory illness in US military recruits, 1999–2004. Vaccine 24, 2835–2842 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford-Miksza L. K., Nang R. N. & Schnurr D. P. Strain variation in adenovirus serotypes 4 and 7a causing acute respiratory disease. J. Clin. Microbiol. 37, 1107–1112 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford-Miksza L. & Schnurr D. P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70, 1836–1844 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X. H. et al. Structure-based high-throughput epitope analysis of hexon proteins in B and C species human adenoviruses (HAdVs). PLoS ONE 7, e32938 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A. et al. Genomic and bioinformatics analyses of HAdV-4vac and HAdV-7vac, two human adenovirus (HAdV) strains that constituted original prophylaxis against HAdV-related acute respiratory disease, a reemerging epidemic disease. J. Clin. Microbiol. 43, 3083–3094 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J. et al. Genome type analysis of adenovirus types 3 and 7 isolated during successive outbreaks of lower respiratory tract infections in children. J. Clin. Microbiol. 41, 4594–4599 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou T. P. et al. Community outbreak of adenovirus, Taiwan, 2011. Emerg. Infect. Dis. 18, 1825–1832 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. Y. et al. Lower respiratory tract infections due to adenovirus in hospitalized Korean children: epidemiology, clinical features, and prognosis. Clin. Infect. Dis. 32, 1423–1429 (2001). [DOI] [PubMed] [Google Scholar]
- Deng J. et al. Identification and typing of adenovirus from acute respiratory infections in pediatric patients in Beijing from 2003 to 2012. Bing Du Xue Bao 29, 615–620 (2013). [PubMed] [Google Scholar]
- Hu F. H. et al. Clinical analysis of 47 children with types 3 and 7 adenovirus pneumonia. Zhonghua Yi Xue Za Zhi 92, 3393–3397 (2012). [PubMed] [Google Scholar]
- Li Q.-G., Zheng Q.-J., Liu Y.-H. & Wadell G. Molecular epidemiology of adenovirus types 3 and 7 isolated from children with pneumonia in Beijing. J. Med. Virol. 49, 170–177 (1996). [DOI] [PubMed] [Google Scholar]
- Noda M. et al. Molecular and epidemiological analyses of human adenovirus type 7 strains isolated from the 1995 nationwide outbreak in Japan. J. Clin. Microbiol. 40, 140–145 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanaiem H. et al. An outbreak of adenovirus type 7 in a residential facility for severely disabled children. Pediatr. Infect. Dis. J. 30, 948–952 (2011). [DOI] [PubMed] [Google Scholar]
- Murtagh P., Cerqueiro C., Halac A., Avila M. & Kajon A. Adenovirus type 7h respiratory infections: a report of 29 cases of acute lower respiratory disease. Acta Paediatr. 82, 557–561 (1993). [DOI] [PubMed] [Google Scholar]
- Tang L., Wang L., Tan X. & Xu W. Adenovirus serotype 7 associated with a severe lower respiratory tract disease outbreak in infants in Shaanxi Province, China. Virol J 8, 10.1186/1743-1422X-1188-1123 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. G. & Wadell G. Analysis of 15 different genome types of adenovirus type 7 isolated on five continents. J. Virol. 60, 331–335 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashido M. et al. Molecular and serological characterization of adenovirus genome type 7h isolated in Japan. Epidemiol. Infect. 122, 281–286 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azar R., Varsano N., Mileguir F. & Mendelson E. Molecular epidemiology of adenovirus type 7 in Israel: identification of two new genome types, Ad7k and Ad7d2. J. Med. Virol. 54, 291–299 (1998). [DOI] [PubMed] [Google Scholar]
- Ostrow S. L., Barshir R., DeGregori J., Yeger-Lotem E. & Hershberg R. Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes. Plos Genetics 10, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole K. S. et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73, 152–160 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E. H. et al. Ten-year analysis of adenovirus type 7 molecular epidemiology in Korea, 1995–2004: implication of fiber diversity. J. Clin. Virol. 35, 388–393 (2006). [DOI] [PubMed] [Google Scholar]
- Jin X. H. et al. Genome variability of human adenovirus type 8 causing epidemic keratoconjunctivitis during 1986–2003 in Japan. Mol. Vis. 17, 3121–3127 (2011). [PMC free article] [PubMed] [Google Scholar]
- Selvaraju S. B., Kovac M., Dickson L. M., Kajon A. E. & Selvarangan R. Molecular epidemiology and clinical presentation of human adenovirus infections in Kansas City children. J. Clin. Virol. 51, 126–131 (2011). [DOI] [PubMed] [Google Scholar]
- Marinheiro J. C. et al. A naturally occurring human adenovirus type 7 variant with a 1743 bp deletion in the E3 cassette. J. Gen. Virol. 92, 2399–2404 (2011). [DOI] [PubMed] [Google Scholar]
- Adhikary A. K., Banik U., Okabe N. & Fujimoto T. Molecular characterization of human adenovirus type 8 (HAdV-8), including a novel genome type detected in Japan. Jpn J Infect Dis 64, 493–498 (2011). [PubMed] [Google Scholar]
- Adhikary A. K., Hanaoka N. & Fujimoto T. Simple and Cost-Effective Restriction Endonuclease Analysis of Human Adenoviruses. Biomed. Res. Int. 2014, 363790 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary A. K., Ushijima H. & Fujimoto T. Human adenovirus type 8 genome typing. J. Med. Microbiol. 61, 1491–1503 (2012). [DOI] [PubMed] [Google Scholar]
- Walsh M. P. et al. Computational analysis identifies human adenovirus type 55 as a re-emergent acute respiratory disease pathogen. J. Clin. Microbiol. 48, 991–993 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajon A. E. et al. Molecular and serological characterization of species B2 adenovirus strains isolated from children hospitalized with acute respiratory disease in Buenos Aires, Argentina. J. Clin. Virol. 58, 4–10 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang Q., Seto D., Cao B., Zhao S. & Wan C. Genome Sequence of Human Adenovirus Type 55, a Re-Emergent Acute Respiratory Disease Pathogen in China. J. Virol. 86, 12441–12442 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q. B. et al. Epidemiology of human adenovirus and molecular characterization of human adenovirus 55 in China, 2009–2012. Influenza Other Respir Viruses 8, 302–308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. et al. Genomic analyses of recombinant adenovirus type 11a in China. J. Clin. Microbiol. 47, 3082–3090 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. et al. Outbreak of acute respiratory disease in China caused by B2 species of adenovirus type 11. J. Clin. Microbiol. 47, 697–703 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Yamaoka K., Noda M. & Ogino T. Genome types of adenovirus type 7 isolated in Hiroshima City. J. Med. Virol. 69, 215–219 (2003). [DOI] [PubMed] [Google Scholar]
- Wadell G. Molecular epidemiology of human adenoviruses. Curr. Top. Microbiol. Immunol. 110, 191–220 (1984). [DOI] [PubMed] [Google Scholar]
- Larranaga C., Kajon A., Villagra E. & Avendano L. F. Adenovirus surveillance on children hospitalized for acute lower respiratory infections in Chile (1988–1996). J. Med. Virol. 60, 342–346 (2000). [PubMed] [Google Scholar]
- Mahadevan P., Seto J., Tibbetts C. & Seto D. Natural variants of human adenovirus type 3 provide evidence for relative genome stability across time and geographic space. Virology 397, 113–118 (2010). [DOI] [PubMed] [Google Scholar]
- Seto J., Walsh M. P., Metzgar D. & Seto D. Computational analysis of adenovirus serotype 5 (HAdV-C5) from an HAdV coinfection shows genome stability after 45 years of circulation. Virology 404, 180–186 (2010). [DOI] [PubMed] [Google Scholar]
- Walsh M. P. et al. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS ONE 4, e5635 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. Analysis of human adenovirus type 19 associated with epidemic keratoconjunctivitis and its reclassification as adenovirus type 64. Invest. Ophthalmol. Vis. Sci. 53, 2804–2811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan S. et al. Computational analysis of four human adenovirus type 4 genomes reveals molecular evolution through two interspecies recombination events. Virology 443, 197–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A. et al. Genomic and bioinformatics analysis of HAdV-4, a human adenovirus causing acute respiratory disease: implications for gene therapy and vaccine vector development. J. Virol. 79, 2559–2572 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghan S. et al. Simian adenovirus type 35 has a recombinant genome comprising human and simian adenovirus sequences, which predicts its potential emergence as a human respiratory pathogen. Virology 447, 265–273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K. E. & Imperiale M. J. Encapsidation of viral DNA requires the adenovirus L1 52/55-kilodalton protein. J. Virol. 72, 7860–7870 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K. E., Lutz P. & Imperiale M. J. Interaction of the adenovirus L1 52/55-kilodalton protein with the IVa2 gene product during infection. J. Virol. 70, 6463–6467 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkayastha A., Su J., Carlisle S., Tibbetts C. & Seto D. Genomic and bioinformatics analysis of HAdV-7, a human adenovirus of species B1 that causes acute respiratory disease: implications for vector development in human gene therapy. Virology 332, 114–129 (2005). [DOI] [PubMed] [Google Scholar]
- Berge T. O., England B., Mauris C., Shuey H. E. & Lennette E. H. Etiology of acute respiratory disease among service personnel at Fort Ord, California. Am J Hyg 62, 283–294 (1955). [DOI] [PubMed] [Google Scholar]
- Gundelfinger B. F., Hantover M. J., Bell J. A., Loosli C. G. & Rowe W. P. Evaluation of a trivalent adenovirus vaccine for prevention of acute respiratory disease in naval recruits. Am J Hyg 68, 156–168 (1958). [DOI] [PubMed] [Google Scholar]
- Top F. H. Jr, Buescher E. L., Bancroft W. H. & Russell P. K. Immunization with live types 7 and 4 adenovirus vaccines. II. Antibody response and protective effect against acute respiratory disease due to adenovirus type 7. J. Infect. Dis. 124, 155–160 (1971). [DOI] [PubMed] [Google Scholar]
- Seto D., Chodosh J., Brister J. R., Jones M. S. & Community M. o. t. A. R. Using the Whole-Genome Sequence To Characterize and Name Human Adenoviruses. J. Virol. 85, 5701–5702 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G. et al. Identification and typing of respiratory adenoviruses in Guangzhou, Southern China using a rapid and simple method. Virol. Sin. 28, 103–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P. et al. An Outbreak of Acute Respiratory Disease Caused by “Human Adenovirus Type 7” in a Military Training Camp in Shaanxi, China. Microbiol. Immunol. 57, 553–560 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C., Zhang T., Qu L., Hu H. & Liu H. Epidemiological investigation of an outbreak of acute respiratory infection caused by adenovirus type 7. Medical Journal of National Defending forces in Southwest China 24, 497–499 (2014). [Google Scholar]
- Zou L. R. et al. Etiology survey on virus of acute respiratory infection in Guangzhou from 2006 to 2009. Zhonghua yu fang yi xue za zhi [Chinese journal of preventive medicine] 45, 825–829 (2011). [DOI] [PubMed] [Google Scholar]
- Le C. T., Gray G. C. & Poddar S. K. A modified rapid method of nucleic acid isolation from suspension of matured virus: applied in restriction analysis of DNA from an adenovirus prototype strain and a patient isolate. J. Med. Microbiol. 50, 571–574 (2001). [DOI] [PubMed] [Google Scholar]
- Zhang Q. et al. Genome Sequence of the First Human Adenovirus Type 14 Isolated in China. J. Virol. 86, 7019–7020 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. et al. Parental LTRs Are Important in a Construct of a Stable and Efficient Replication-Competent Infectious Molecular Clone of HIV-1 CRF08_BC. PLoS ONE 7, e31233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. & Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3, 418–426 (1986). [DOI] [PubMed] [Google Scholar]