Abstract
Genetic alterations and etiology of thymic epithelial tumors (TETs) are largely unknown, hampering the development of effective targeted therapies for patients with TETs. Here TETs of advanced-stage patients enrolled in a clinical trial of molecularly-guided targeted therapies were employed for targeted sequencing of 197 cancer-associated genes. Comparative sequence analysis of 78 TET/blood paired samples obtained from 47 thymic carcinoma (TC) and 31 thymoma patients revealed a total of 86 somatic non-synonymous sequence variations across 39 different genes in 33 (42%) TETs. TCs (62%; 29/47) showed higher incidence of somatic non-synonymous mutations than thymomas (13%; 4/31; p < 0.0001). TP53 was the most frequently mutated gene in TETs (n = 13; 17%), especially in TCs (26%), and was associated with a poorer overall survival (p < 0.0001). Genes in histone modification [BAP1 (n = 6; 13%), SETD2 (n = 5; 11%), ASXL1 (n = 2; 4%)], chromatin remodeling [SMARCA4 (n = 2; 4%)], and DNA methylation [DNMT3A (n = 3; 7%), TET2 (n = 2; 4%), WT1 (n = 2; 4%)] pathways were recurrently mutated in TCs, but not in thymomas. Our results suggest a potential disruption of epigenetic homeostasis in TCs, and a substantial difference in genetic makeup between TCs and thymomas. Further investigation is warranted into the roles of epigenetic dysregulation in TC development and its potential for targeted therapy.
Thymic epithelial tumors (TETs) are rare neoplasms arising from the epithelial cells of the thymus with an incidence of 0.13 per 100,000 person/year in the United States1. According to the 2004 World Health Organization (WHO) classification, TETs are divided into thymomas (A, A/B, B1, B2, B3 subtypes) and thymic carcinomas (TCs) based on the tumor cell morphology, atypia, and on the extent of the thymocyte component2. The diagnosis of TETs by and large relies on histology supported by immunohistochemistry3.
Although thymomas can be relatively indolent, with a 10-year survival of almost 100% for type A thymoma, TCs are much more aggressive with 5-year survival rate of only 50%3. Surgical resection is the cornerstone treatment for operable tumors. Besides histotype, prognosis is determined by the stage at the diagnosis and completeness of removal in resectable cases. The treatment of advanced TETs however is limited to chemotherapy which usually is not curative3.
Targeted therapies against “actionable” genetic alterations in individual tumors have yielded impressive results in several types of cancers4. Despite a few case reports of dramatic responses to single agent-targeted therapies5,6,7,8, the development of a rational targeted therapy for TETs has been hampered by insufficient knowledge of the genetic alterations of these tumors. Although the next generation sequencing (NGS) approach has shed light on the cancer genome of several common cancer types9, efforts on mapping the genetic makeup of rare tumors such as TETs has lagged behind, and the knowledge of TETs at the genomic level is limited to the sporadic reports of TP53 and KIT mutations3,10.
Large scale efforts to catalogue genetic mutations in cancer such as the cancer genome atlas (TCGA) have employed samples obtained from resection of the primary tumors11. However, solid tumors are heterogeneous and that advanced tumors have dynamically evolved at the genetic level12. By whole genome and transcriptome sequencing, we previously demonstrated that common cancer-associated gene mutations were rare in an aggressive B3 thymoma13. Very recently, we identified a unique somatic missense mutation in the GTF2I gene that is prevalent in indolent early stage TETs and associated with good prognosis14. Genomic studies of advanced-stage TETs have not been performed yet.
In this study, we profiled somatic genetic variations in 78 advanced-stage TET samples with the aid of targeted-capture sequencing of a panel of 197 cancer-related genes (Table S1). Advanced TCs exhibited a higher incidence of somatic non-synonymous mutations than advanced stage thymomas. Moreover, we observed recurrent somatic mutations of epigenetic regulatory genes in TCs but not in thymomas. To our knowledge, this study represents the largest series of advanced-stage TETs which have been characterized by targeted sequencing.
Results
Patient characteristics
A total of 82 TET patients were enrolled in the molecular profiling study from March 2011 to December 2012. Out of the 82 patients, tumor specimens and paired blood samples were obtained from 78 patients, including 31 thymomas and 47 TCs (Table 1), and were processed for sequence analysis. According to the Masaoka staging system2, 4 patients had stage III, 24 stage IVA and 50 stage IVB disease (Table 1). All patients were not eligible for surgery or radiation therapy.
Table 1. Patient characteristics and mutation status.
| Cases | w/somatic mutations | w/o somatic mutations | p-value | mt epi | wt epi | p-value | mt TP53 | wt TP53 | p-value | mt BAP1 | wt BAP1 | p-value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | 78 | |||||||||||||
| Median Age (range), years | 53 (20–80) | |||||||||||||
| Gender | Male | 43 | 22 | 21 | 0.108 | 14 | 29 | 0.305 | 10 | 33 | 0.084 | 3 | 40 | 0.793 |
| Female | 35 | 11 | 24 | 7 | 28 | 3 | 32 | 3 | 32 | |||||
| Race | Caucasian | 62 | 26 | 36 | 0.775* | 16 | 46 | 0.532 | 12 | 50 | 0.443 | 5 | 57 | 1.000 |
| Asian | 8 | 3 | 5 | 2 | 6 | 0 | 8 | 1 | 7 | |||||
| Black | 7 | 4 | 3 | 3 | 4 | 1 | 6 | 0 | 7 | |||||
| Hispanian | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | |||||
| Stage | III | 4 | 1 | 3 | 0.031** | 1 | 3 | 0.197 | 0 | 4 | 0.119 | 0 | 4 | 0.553 |
| IV A | 24 | 6 | 18 | 4 | 20 | 2 | 22 | 0 | 24 | |||||
| IV B | 50 | 26 | 24 | 16 | 34 | 11 | 39 | 6 | 44 | |||||
| WHO histotype | AB | 3 | 1 | 2 | 0.0001*** | 1 | 2 | 0.005 | 0 | 3 | 0.010 | 0 | 3 | 0.038 |
| B1 | 2 | 1 | 1 | 1 | 1 | 0 | 2 | 0 | 2 | |||||
| B2 | 17 | 1 | 16 | 1 | 16 | 1 | 16 | 0 | 17 | |||||
| B3 | 6 | 1 | 5 | 0 | 6 | 0 | 6 | 0 | 6 | |||||
| NOS | 3 | 0 | 3 | 0 | 3 | 0 | 3 | 0 | 3 | |||||
| TC | 42 | 28 | 14 | 18 | 24 | 11 | 31 | 6 | 36 | |||||
| TNEC | 5 | 1 | 4 | 0 | 5 | 1 | 4 | 0 | 5 | |||||
| Biopsy site | Primary | 34 | 13 | 21 | 0.645 | 9 | 25 | 1.000 | 5 | 29 | 0.766 | 3 | 31 | 1.000 |
| Metastais | 44 | 20 | 24 | 12 | 32 | 8 | 36 | 3 | 41 | |||||
| Autoimmune/other paraneoplastic diseases# | Autoimmne | 10 | 1 | 9 | 0.005**** | 1 | 9 | 0.031 | 0 | 10 | 0.037 | 1 | 9 | 0.752 |
| Non-autoimmune | 7 | 1 | 6 | 0 | 7 | 0 | 7 | 0 | 7 | |||||
| No | 61 | 31 | 30 | 20 | 41 | 13 | 48 | 5 | 56 |
Age was calculated from birth to date of diagnosis.
*P-values were calculated by Chi-Square test of Caucasian + Hispanian vs Black + Asian.
**P-values were calculated by Chi-Square test of III + IVA vs IVB.
***P-values were calculated by Chi-Square test of AB + B1 + B2 + B3 + thymoma NOS vs TC + TNEC.
****P-values were calculated by Chi-Square test of autoimmune + nonautoimmune diseases vs no paraneoplastic diseases.
#see Table 2 for details.
NOS: thymoma not otherwise specified; mt, wt epi: mutant, wild-type epigenetic regulatory genes.
Exome capture sequencing and somatic mutation analysis
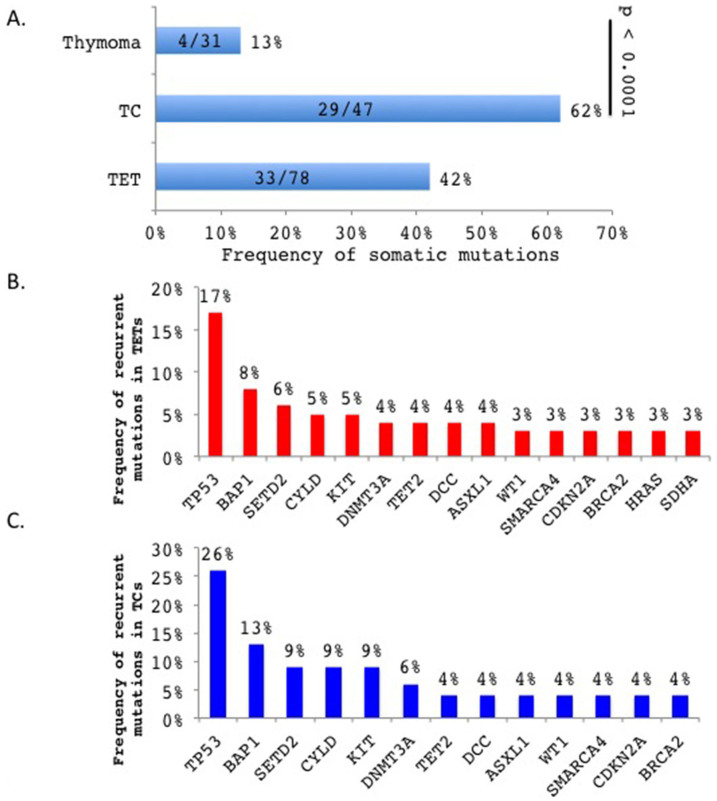
We identified a total of 86 somatic variations affecting 39 genes (Table S2) in 33 of the 78 (42%) TETs with significantly higher incidence in TCs (29/47; 62%) than thymomas (4/31; 13%) (p < 0.0001; Figure 1A). As the 197 cancer-gene panel was selected independently of the mutations found in our recent exome sequencing study of TETs14, the results are in line with our previous data and further confirm that somatic mutations are more prevalent in TCs than thymomas14. Forty-eight variants, including 41 of the 64 recurrent variants, were randomly chosen for validation by Sanger sequencing using gene-specific primers and 46 (96%) variants were confirmed (Table S2). Among the 86 variants, 62 were single nucleotide variations (49 missense mutations, 4 splice site mutations and 9 nonsense mutations) derived from 31 genes in 34 patients, and 24 were indels (17 frameshift and 7 inframe indels) from 15 genes in 17 patients (Table S2). Thirty-seven (43%) of those somatic variations were previously reported in COSMIC and 12 in dbSNP137 and ESP6500 polymorphism databases (Table S2). The latter were all tumor-associated SNPs. None of the 86 somatic mutations were identical. The tumor that carried the highest number of mutations was a TC with 13 mutations affecting 12 genes (Table S2), which was probably due to the presence of mutations of the DNA repair genes MLH1 and XRCC115,16.
Figure 1.
(A). TCs exhibited higher incidence of somatic mutations than thymomas (p < 0.0001, Chi-square test). The numbers in the bars indicate the number of cases with somatic mutations out of total number of cases analyzed. (B). Frequency of recurrent mutations in TETs. (C). Frequency of recurrent mutations in TCs.
Genes recurrently mutated in at least two cases included ASXL1, BAP1, BRCA2, CDKN2A, CYLD, DCC, DNMT3A, HRAS, KIT, SDHA, SETD2, SMARCA4, TET2, TP53 and WT1 (Figure 1B). Among the 15 recurrently mutated genes in TETs, 13 occurred in TCs (Figure 1C) but none in thymomas (Table S2). Similarly, recurrent mutations of BAP1, CDKN2A, CYLD and TP53 were also found in TCs of our recent study14. To examine the potential impact of the mutations on gene function, we employed SIFT and Polyphen2 algorithms17,18 and found that 51 of 58 SIFT- and Polyphen2-predictable somatic variations (88%) are likely to affect the gene functions based on at least one of the two prediction models (Table S2). Consistent with previous reports10,19, TP53 mutations were found in 17% (13/78) of TETs, 26% (12/47) being in TCs and 3% (1/31) in thymomas (Chi square test, p = 0.0097), and KIT mutations in 9% (4/47) TCs and none in thymomas (p = 0.1471) (Figure 1B and 1C). A KIT L576P mutation identified in a TC (Table S2) was previously reported in melanomas that responded to imatinib treatment20. Another noticeable recurrently mutated gene was CYLD deubiquitinase, which functions as a negative regulator of NFKB pathway21,22. Four TCs (9%, 4/47) carried CYLD mutations and no CYLD mutation was detected in the 31 sequenced thymomas. This observation is in line with our previous work in independent samples where we found 3 of 16 (19%) TCs harboring CYLD mutations, and none in 38 sequenced thymomas14. Combined together, a total of 9 CYLD mutations (3 frameshift, 1 nonsense, 1 splice-site error and 4 missenses) were found in 11% (7 of 63) TCs and none in 69 thymomas (Fisher's exact test, p < 0.001). A detailed summary of the CYLD mutations is illustrated in Table S3.
Recurrent mutations of epigenetic regulatory genes in TCs
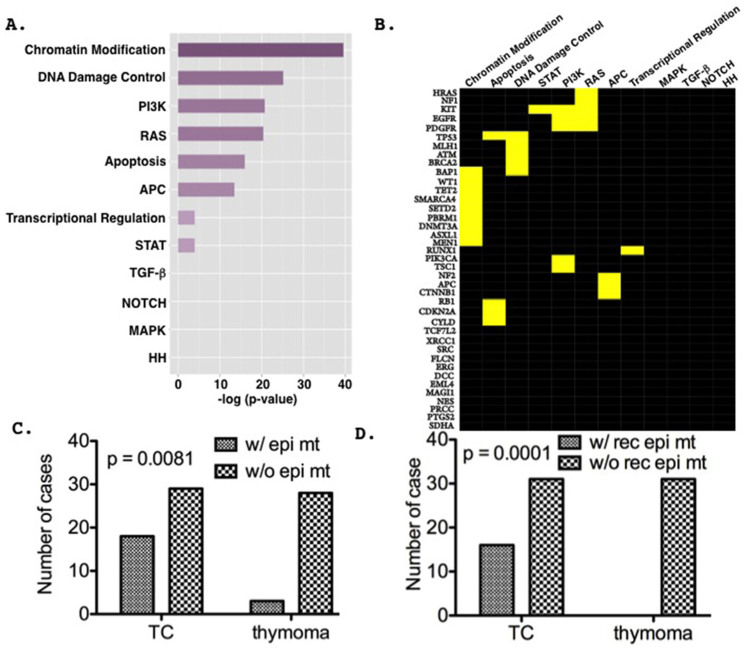
Based on the recurrent mutation and truncation frequencies at the same amino acid positions described in cancer, ~140 driver genes were identified and classified into 12 core pathways23. Gene enrichment analysis revealed that out of the 39 mutated genes identified in this study, 27 were significantly enriched in 8 of the 12 defined core-pathways (Figure 2A; Tables S4 and S5), among which the chromatin modification pathway showed the highest enrichment score (Fisher's exact tests, p < 0.0001, Figure 2B and Table S5). However, the results need to be interpreted with caution as 197 genes were pre-selected based on their association with cancers and thus the 12 core pathways may be over-represented in the panel. Intriguingly, 9 out of the 39 mutated genes identified in this study encode epigenetic regulatory proteins (chromatin remodeling, histone modification and DNA methylation) and were found mutated with significantly higher incidence in TCs (18/47, 38%) than thymomas (3/31, 10%) (p = 0.0081, Fisher's exact test; Figure 2C), within which 7 were recurrently mutated in TCs (16/47) but not in thymomas (0/31) (Fisher's exact tests; p = 0.0001; Figure 2D).
Figure 2. Mutations of epigenetic regulatory genes in chromatin modification pathway are significantly enriched in TCs.
(A). Bar plot shows the hierarchical order of the predefined core pathway enrichment23 of the mutated genes based on their enrichment scores (−log [p-value]). (B). Heat map of the mutated genes within the core pathways. Yellow bars represent genes that are significantly enriched in the corresponding pathways. (C). Epigenetic gene mutations are more prevalent in TCs than in thymomas (p = 0.0053, Chi-square test). (D). Recurrent epigenetic gene mutations in TC but not in thymomas.
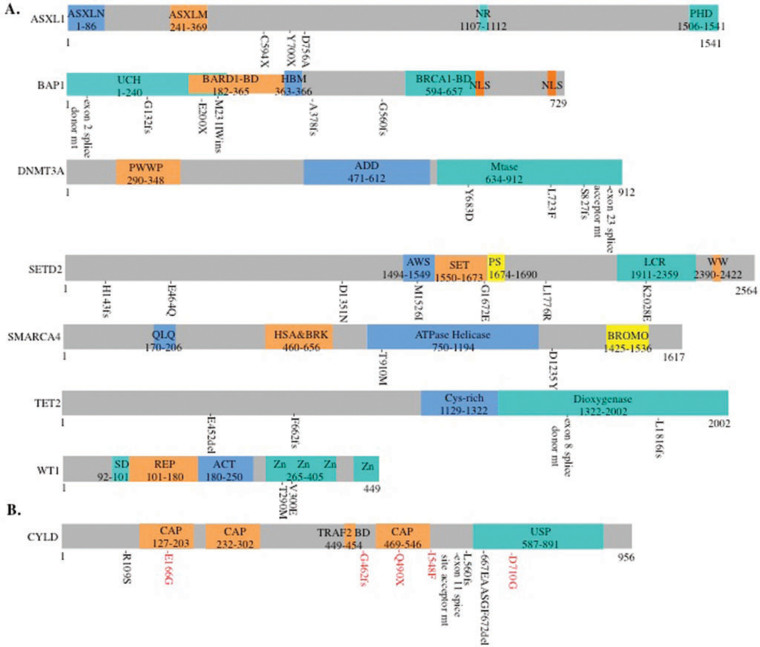
The mutated epigenetic regulatory genes that directly affect histone modification and chromatin remodeling include genes encoding histone deubiquitinase BAP124,25, polycomb chromatin binding protein ASXL126, histone H3K36 trimethyltransferase SETD227, and ATPase of the SWI-SNF complex SMARCA428 (Figure 3A and Table S2). Six TCs (13%) showed BAP1 mutations (Table S2) of which three were frameshift indels (A378fs, G560fs, G132fs), one nonsense (E200*), one exon 2 splice site error and one inframe insertion (M231IWins). All are likely to be deleterious to the protein except M231IWins mutation. Similar loss-of-function mutations of BAP1 have been documented in renal clear cell carcinoma24. Loss-of-function germline mutations of BAP1 also predispose to uveal and cutaneous melanomas, mesothelioma, clear cell renal cancer and atpical cutaneous melanocytic proliferation29. Two ASXL1 nonsense mutations (C594* and Y700*) were identified in two TCs and one missense (D756A) in a B2 thymoma (Figure 3A and Tables S2). A nonsense mutation of ASXL1 has been recently reported in a cytogenetically normal B3 thymoma30. Recurrent somatic ASXL1 mutations also occur in myelodysplastic syndrome, myeloproliferative neoplasms, and acute myeloid leukemia, and are associated with adverse outcome31. Another frequently mutated histone modification gene in TETs was the SETD2 tumor suppressor gene, encoding the sole H3K36 trimethyltransferase in humans27. Six SEDT2 missense mutations (G1672E, M1526I, E464Q, D1351N, K2028E and L1776R) and one frameshift deletion (H143 fs) were found in four TCs and one B2 thymoma (Figure 3A and Table S2). In clear cell renal carcinoma and pediatric glioma, SETD2 loss-of-function mutations have been observed32,33. Whether the SETD2 mutations, the majority of which (6/7) are missense in TETs, may disrupt its tumor suppressor function remains to be investigated.
Figure 3.
(A). Amino acid positions of recurrently mutated genes relative to the domains of the epigenetic regulatory gene-encoded proteins. ASXL1: ASXN, conserved domain at the N-terminus; ASXM, conserved domain in the middle part; NR, nuclear receptor/PHD, plant homeodomain. BAP1: UCH, Ubiquitin C-terminal hydrolase; HBM, HCF-binding motif (NHNY sequence); NLS, Nuclear localization signal. DNMT3A: PWWP, a proline-tryptophan-tryptophan-proline domain; ADD, an ATRX-DNMT3-DNMT3L-type zinc finger domain; Mtase, a methyltransferase domain. SETD2: AWS, AWS domain; SET, SET domain; PS, post-SET domain; LCR, low-charge region; WW, WW domain. SMARCA4: QLQ, Gln, Leu, Gln motifi; HSA&BRK, domain associated with helicase, SANT, transcription and chromo domain helicases; ATPase Helicase, DEAD-like helicase, helicase C terminal domain; Bromo, bromodomain. TET2: Cys-rich, CXXC Cysteine rich domain; Dioxygenase, dioxygenase domain. WT1: SD, 10-AA suppression domain; REP, repression domain; Act, activation domain; Zn, Zinc finger. (B). Somatic mutation positions relative to the domains of CYLD protein. CAP, CAP-Gly domain; TRAF2, TRAF2 binding site; CAP 469–546, NEMO binding site; USP, ubiquitin specific protease domain. CYLD somatic mutations identified in this study are marked in black, and mutations identified in our recent study14 are in red. X, stop codon; del, deletion; ins, insertion; fs, frameshift.
Another group of the mutated epigenetic regulatory genes includes DNA methylation genes cytosine dioxymethyltransferase TET2, de-novo cytosine methyltransferase DNMT3A34 and WT1, positive regulator of DNMT3A35. Four potential loss-of-function mutations of TET2 (E452 del, L1816fs, F662fs and exon 8 splice donor site error) were found in two TCs and one AB thymoma (Figure 3A and Table S2). TET2 inactivation mutations have been associated with lymphoid and myeloid malignancies36. Three TCs showed four DNMT3A mutations including one TC with S827fs and Y683D mutations, two other TCs with L723F and exon 23 splice acceptor site mutations respectively (Figure 3A and Table S2). All DNMT3A mutations occurred in the catalytic domain (Figure 3A), which may result in impairment of enzymatic activity37. Similar DNMT3A mutations with high frequency spreading around the catalytic domain were also found in T-cell lymphomas, acute myeloid leukemia and recently a B2 thymoma30,34,38.
Effect of mutations on overall survival and response to targeted agents
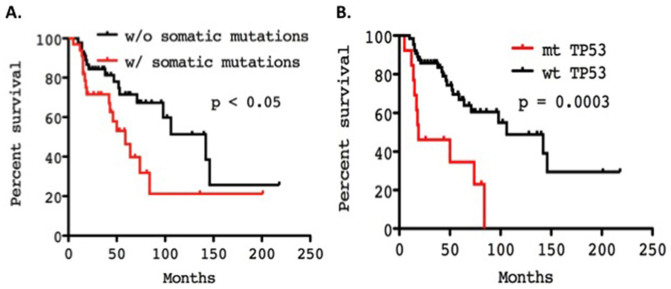
Although all patients with thymoma enrolled in this study had advanced-stage disease (stage IVA/IVB) (Table 2), the mutation frequency among thymomas was significantly lower than that observed in TCs. TET patients with somatic mutations exhibited a worse overall survival than patients without mutations (median survival 59 months vs 142 months; Log-rank Mantel-Cox, p < 0.05, 95%CI: 1.902 to 2.912; Figure 4A). Interestingly patients with tumors harboring mutant TP53 displayed poorer overall survival than those with wild-type TP53 (median survival 19 months vs. 106 months; Log-rank Mantel-Cox Test, p = 0.0003; 95% CI of ratio 2.234–22.39; Figure 4B). However, no difference was observed in survival of TET patients with and without epigenetic regulatory gene mutations (p = 0.194).
Table 2. Patient history.
| Patient ID | Sub- type | Stage | Sex | Age | Survival (mo) | Race | Autoimmune/other disease | Biopsy Site | BEL | PACB | PHA | IMC-A12 | Sutent | Epig MT | TP53 MT | # of Somatic MT Genes |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1010109 | TC | IV a | M | 60 | 201 | Black | none | Pre-sternal mass | SD | ASXL1 C594* | 2 | |||||
| 1010168 | B1 | IV a | F | 41 | 46 | Caucasian | None | Pleural mass | SD | ASXL1 D756A | 2 | |||||
| 1010153 | TC | IV b | M | 64 | 19 | Caucasian | None | Left Lung | PD | SD | ASXL1 Y700*, DNMT2A S827fs, DNMT3A Y683D | C135* | 3 | |||
| 1010380 | TC | IV b | F | 45 | 19 | Caucasian | None | Brain | BAP1 G132fs | 2 | ||||||
| 1010144 | TC | IV b | M | 39 | 51 | Asian | Sarcoidosis | Mediastinal Mass | BAP1 A378fs | 2 | ||||||
| 1010358 | TC | IV b | M | 59 | 16 | Caucasian | None | RLL lung | SD | BAP1 E200* | 1 | |||||
| 1010089 | TC | IV b | M | 51 | 26 | Caucasian | None | Anterior mediastinal mass | SD | SD | SD | BAP1 Ex3 splice donor; PBRM1 L1494fs | R248Q | 4 | ||
| 1010294 | TC | IV b | F | 64 | 59 | Caucasian | None | Thymus | BAP1 G560fs | 2 | ||||||
| 1010304 | TC | Iv b | F | 62 | 16 | Caucasian | none | Lung, RML | SD | BAP1 M231IWins | 1 | |||||
| 1010201 | TC | IV b | M | 40 | 31 | Caucasian | None | Right Pleural | SD | PR | DNMT3A Ex23 splice acceptor | 1 | ||||
| 1010257 | TC | III | M | 67 | 53 | Caucasian | None | Liver | DNMT3A L723F, SMARCA4 D1235Y | 2 | ||||||
| 1010112 | TC | IV b | M | 69 | 42 | Caucasian | None | Mediastinal Mass | PD | MEN1 S84fs | 1 | |||||
| 1010137 | TC | IV b | M | 55 | 81 | Caucasian | None | Medialstinal Mass | SD | SETD2 (M1526I, E464Q, D1351N), SMARCA4 T910M | R282P | 3 | ||||
| 1010075 | B2 | IV a | F | 41 | 15 | Caucasian | None | Soft Tissue; Thymus | PR | SETD2 G1672E | R267W | 10 | ||||
| 1010024 | TC | IV b | M | 69 | 18 | Caucasian | none | Lymph node | SD | SETD2 H143 fs | P27S | 2 | ||||
| 1010148 | TC | IV b | M | 67 | 50 | Caucasian | None | Thymus/Liver | SETD2 K2028E | C176Y | 3 | |||||
| 1010018 | AB | IV a | M | 30 | 43 | Asian | None | Mediastinal Mass | SD | TET2 E452 del | 2 | |||||
| 1010217 | TC | IV b | M | 59 | 17 | Caucasian | None | Mediastinal Mass | SD | SD | TET2 F662fs, SETD2 L1776R | M237I | 4 | |||
| 1010103 | TC | IV b | M | 62 | 15 | Caucasian | None | Liver | TET2 L1816fs, TET2 Ex8 splice donor | 2 | ||||||
| 1010336 | TC | IV b | F | 57 | 69 | Black | None | Paraspinal mass | SD | WT1 T290M | 1 | |||||
| 1010174 | TC | IV b | F | 69 | 44 | Black | None | Liver | WT1 V300E | Q331* | 3 | |||||
| 1010010 | TNEC | IV b | M | 24 | 38 | Caucasian | Cushings disease | Mediatinal mass | SD | |||||||
| 1010016 | TC | IV b | F | 39 | 14 | Caucasian | None | Liver | M133K | 3 | ||||||
| 1010019 | TNEC | IV b | M | 51 | 64 | Caucasian | Cushings disease | Lymph Node | SD | 1 | ||||||
| 1010038 | B2 | IV b | F | 43 | 146 | Caucasian | PRCA | Liver | SD | |||||||
| 1010049 | B2 | IV a | M | 46 | 63 | Caucasian | None | Anterior Mediastinal | SD | SD | SD | |||||
| 1010050 | TC | IV b | F | 50 | 96 | Caucasian | None | Pleura and pericardial mass | SD | PR | ||||||
| 1010054 | TC | IV a | F | 42 | 36 | Caucasian | None | thymic resection | PD | |||||||
| 1010056 | TC | IV b | F | 53 | 58 | Caucasian | None | Supraclavicular lymph node | PR | SD | SD | 2 | ||||
| 1010059 | AB | IV b | M | 44 | 106 | Caucasian | None | Left pleural mass | PD | SD | ||||||
| 1010060 | NOS | IV a | F | 69 | 71 | Caucasian | PRCA | Lung | SD | |||||||
| 1010061 | NOS | IV a | F | 64 | 86 | Black | None | Thymoma resection | SD | |||||||
| 1010073 | TC | IV b | M | 61 | 18 | Caucasian | None | Pleural effusion | ||||||||
| 1010077 | B1 | IV a | M | 49 | 31 | Caucasian | None | Thymus mediastinal mass | PR | |||||||
| 1010085 | B2 | IV b | M | 38 | 98 | Asian | Infections- PCP | Pleura | SD | |||||||
| 1010086 | B2 | IV a | F | 61 | 52 | Asian | PRCA | Pleura | ||||||||
| 1010097 | B2 | IV b | M | 66 | 142 | Black | None | Right lung | SD | |||||||
| 1010098 | B3 | IV a | F | 60 | 75 | Caucasian | None | Thymoma resection | ||||||||
| 1010101 | B2 | IV a | M | 37 | 47 | Caucasian | Ulcerative colitis | Right Pleural Mass | PR | SD | ||||||
| 1010108 | B3 | IV a | F | 30 | 28 | Caucasian | None | Mediastinal mass | ||||||||
| 1010120 | B2 | IV a | F | 56 | 146 | Caucasian | PRCA | Retroperitoneum | SD | |||||||
| 1010134 | TC | IV b | M | 38 | 14 | Asian | None | Thymus | PD | |||||||
| 1010138 | B3 | IV a | F | 44 | 35 | Caucasian | None | Left Pleura | SD | SD | SD | |||||
| 1010152 | TC | Iv b | M | 59 | 74 | Caucasian | None | Ant Mediastinum | SD | Y234C | 1 | |||||
| 1010154 | TC | IV b | F | 76 | 14 | Caucasian | None | Mediastinal Mass | SD | |||||||
| 1010178 | TNEC | IV b | M | 27 | 10 | Hispanic | None | Left para-mediastinum | ||||||||
| 1010184 | B3 | IV a | M | 60 | 136 | Caucasian | None | Thymus | PD | PR | SD | 1 | ||||
| 1010196 | B2 | IV b | M | 69 | 140 | Caucasian | None | Mediastinal Mass | SD | |||||||
| 1010198 | TC | Iv b | M | 69 | 84 | Caucasian | None | Kidney | G361fs | 12 | ||||||
| 1010200 | TC | IV b | M | 50 | 37 | Caucasian | none | Liver | SD | SD | ||||||
| 1010204 | TC | IV b | F | 72 | 22 | Black | None | Thymectomy, RUL (lobe) | SD | |||||||
| 1010212 | AB | IV a | F | 63 | 64 | Asian | None | Soft Tissue, Right chest | PR | |||||||
| 1010219 | NOS | IV a | F | 53 | 218 | Caucasian | None | Thymus, pericardium & LUL | PR | |||||||
| 1010224 | TC | III | M | 47 | 16 | Caucasian | None | Thymus | 1 | |||||||
| 1010232 | TC | IV b | M | 73 | 34 | Black | None | RLL visceral pleura implant | SD | 1 | ||||||
| 1010239 | B2 | IV b | F | 65 | 96 | Caucasian | MAI | Right lung | ||||||||
| 1010242 | TC | IV a | M | 35 | 16 | Caucasian | None | Pleura | ||||||||
| 1010253 | TC | IV a | F | 80 | 28 | Caucasian | None | Ant mediastinal mass | SD | SD | ||||||
| 1010256 | TC | IV a | M | 47 | 40 | Caucasian | None | Ant mediastinal mass | SD | |||||||
| 1010261 | TNEC | III | M | 20 | 53 | Caucasian | Cushing's disease | Thymic Mass | ||||||||
| 1010264 | B2 | IV b | F | 31 | 59 | Caucasian | Uveitis | Ant Mediastinum | SD | |||||||
| 1010268 | TC | IV b | M | 65 | 12 | Caucasian | None | Pleura | PR | PD | R282W | 1 | ||||
| 1010281 | B2 | IVa | F | 58 | 80 | Caucasian | Myasthenia Gravis; Ulcerative Colitis | Lymph Node | NE | |||||||
| 1010286 | TC | IV a | F | 22 | 53 | Caucasian | None | Left Axilla mass | ||||||||
| 1010292 | B2 | IV b | M | 49 | 19 | Caucasian | None | Thymus | SD | |||||||
| 1010310 | TC | IV b | F | 72 | 21 | Caucasian | None | Liver | PR | |||||||
| 1010318 | B2 | IV b | M | 40 | 102 | Caucasian | None | Diaphragm | SD | |||||||
| 1010323 | B2 | IV b | F | 62 | 73 | Caucasian | Nephrotic syndrome- minimal change disease | Thymic Mass | CR | |||||||
| 1010327 | B3 | IV b | M | 58 | 128 | Caucasian | None | Mediastinal Mass | SD | |||||||
| 1010346 | TC | IV b | F | 74 | 25 | Caucasian | None | Thymus | SD | |||||||
| 1010347 | B3 | IV b | M | 33 | 21 | Asian | None | Thymus | ||||||||
| 1010348 | B2 | IV b | F | 74 | 44 | Caucasian | PRCA | Thymus | SD | |||||||
| 1010363 | TC | IV b | F | 62 | 15 | Caucasian | None | Pleural Biopsy | 2 | |||||||
| 1010376 | TC | IV b | F | 64 | 20 | Asian | None | Anterior mediastinal mass | 1 | |||||||
| 1010379 | TC | III | M | 40 | 17 | Caucasian | None | Thymus | ||||||||
| 900006335 | B2 | IV b | M | 67 | 62 | Caucasian | Polycythemia vera | Pleural effusion | PD | |||||||
| 1010290 | TNEC | IV b | F | 36 | 80 | Caucasian | Cushings disease | Breast, left lateral core | ||||||||
| 1010298 | TC | IV a | M | 45 | 5 | Caucasian | None | RUL | PD | PD | P87fs | 1 |
PR, partial response; CR, complete response; PD, progressive disease; SD, stable disease; NE, not evaluable; Ex, exon; del, deletion; ins, insertion;
*, stop codon; fs, frameshift; Epig, epigenetic gene;
MT, mutation.
TNEC: Thymic neuroendocrine carcinoma; TC: thymic carcinoma; NOS: thymoma not otherwise specified.
PACB: Platinum, Adriamycin and Cyclophosphamide with Belinostat.
IMC-A12 (cixutumamab): IGF-1R antibody.
Bel, Belinostat (PXD101): HDAC inhbitor.
Sutent (Sunitinib): multi-targeted receptor tyrosine kinase inhibitor.
PHA (PHA-848125AC, milciclib): CDK2/TRKA inhibitor.
MAI: Mycobacterium Avium-Intracellulare.
PRCA: Pure red cell aplasia.
PCP: Pneumocystis Pneumonia.
Figure 4.
(A). Overall survival of TET patients with and without somatic mutations. (B). Overall survival of TET patients with and without TP53 mutations. Survival curves were generated with Kaplan-Meier method and the differences evaluated by the Log-rank (Mantel-Cox) test. mt, mutant; wt, wild-type.
A total of 56 patients were enrolled in phase II studies evaluating the following investigational agents (Table 2): (1) the multi-kinase inhibitor, sunitinib5, (2) the CDK2/TRKA inhibitor, milciclib (PHA-848125AC)6, (3) the histone deacetylase (HDAC) inhibitor, belinostat in combination with cisplatin, doxorubicin and cyclophosphamide (PACB)39, (4) a monoclonal antibody targeting the insulin-like growth factor-1 receptor (IGF-1R), cixutumumab7 and (5) a trial evaluating single-agent belinostat therapy8. Six TET patients with epigenetic gene mutations received PACB treatment and all patients showed stable disease except a patient with a SETD2 (G1672E)-mutant B2 thymoma who achieved a partial response (Table 2). In contrast, among 13 TET patients without epigenetic gene mutations who received PACB treatment, five showed partial response and one complete response. Nevertheless the response to PACB was not significantly different between patients with and without epigenetic gene mutations (p = 0.12). Moreover, there was no significant difference in the tumor response to the above-mentioned targeted therapies between patients with and without somatic gene mutations identified in this study (Table 2), although the number of patients enrolled in each of these studies is too small to draw firm conclusions5,6,7,8,39.
Discussion
Our report represents the largest study of somatic mutations in patients with advanced-stage TETs and aggressive histological types (i.e. 95% stage IVA/IVB and 60% TC). In addition to the previously reported TP53 and KIT mutations10,19, here we showed that chromatin remodeling, histone modification, and DNA methylation genes are frequently mutated in advanced-stage TCs. Although none of the epigenetic regulatory genes showed high mutation frequency, altogether they were present in 38% of TCs or 27% of TETs. Our finding is not without precedent. Recent NGS projects revealed high frequency of clusters of epigenetic regulatory gene mutations in kidney carcinoma, metastasizing uveal melanomas and pediatric high grade glioma, medulloblastoma, and T-lineage acute lymphoblastic leukaemia26,32,33,34. Considering that not all the epigenetic regulatory genes were included in our 197 cancer-associated gene panel, we might have underestimated the mutation frequencies of epigenetic regulatory genes in TCs. Whole exome sequencing of more patient samples will be required to clarify the frequency of the epigenetic regulatory gene mutations in TETs, especially in TCs.
The seven epigenetic regulatory genes (BAP1, ASXL1, SETD2, SMARCA4, DNMT3A, TET2, and WT1) have been previously categorized as drivers when mutated in other cancers23 and were found recurrently mutated only in TCs but not thymomas. SIFT and Polyphen2 algorithms predicted the damaging nature of most of the mutations in these seven genes (Table S2), implicating the potential importance of these mutations in TCs. The most prevalent epigenetic gene mutation was in the BAP1 tumor suppressor gene, that has histone deubiquitinase activity25. Five (frameshift, splice-site and nonsense mutations) of the six identified mutations are likely to be deleterious to the protein40. BAP1 mutation in clear cell renal carcinoma correlated with high tumor grade and worse prognosis41. All BAP1 mutations affected stage IVB TCs. However, given the relatively small sample size and variability of therapies, we have not been able to establish the prognostic values of BAP1 mutations in TCs. BAP1 interacts with ASXL1 polycomb chromatin binding protein forming a polycomb repressive deubiquitinase complex that regulates histone H2A ubiquitination26. Loss of BAP1 and ASXL1 functions are reported to compromise their tumor suppressor activities in cancer26. ASXL1 and BAP1 mutations were mutually exclusive and together accounted for 17% of TCs.
Mutations of de-novo cytosine methyltransferase DNMT3A have been reported in T-cell lymphomas which are often accompanied with mutations of TET2 cytosine dioxymethyltransferase, an enzyme acting at an intermediate step toward methylcytosine demethylation34. Unlike T-cell lymphomas, TET2 and DNMT3A mutations were mutually exclusive in TETs, indicating that the two genes may be involved in deregulation of cytosine methylation and demethylation processes independently in thymic tumors.
As the mutated epigenetic regulatory genes identified in this study function at different levels of epigenetic regulations (chromatin remodeling, histone modification and DNA methylation), it is not surprising that patients with epigenetic regulatory gene mutations may not respond to chemotherapy and HDAC inhibitor combination therapy (Table 2). Understanding how and whether the mutated epigenetic regulatory genes may play a role in thymic epithelial cell transformation will help to tailor specific drugs to tumors with specific epigenetic alterations, as piloted in non-small cell lung cancer and leukemia studies42,43. Future work requires the determination of the mutation impact on the functions of the encoded proteins and the epigenetic landscape of the tumor cells.
TP53 mutations correlated with poorer patient survival, reflecting the high mutation frequency in stage IVB TCs. Similar findings have been published for other more common solid tumors44. However the prognostic role of mutant P53 remains controversial, probably mainly because most studies are based only on immunohistochemistry analysis45.
CYLD activates NFKB signaling through deubiquitination of NFKB upstream regulators ubiquitylated by cIAP ubiquitin E3 ligase, and inhibition of cIAP by cIAP antagonist blocks NFKB activation21,22. Five of the 9 mutations in TETs were framshift, nonsense and splice-site error (Figure 3B), which are probably non-functional, in line with the tumor suppressor role of CYLD22. Anti-inflammation drugs aspirin and prostaglandin A1 were used in a phase I trial to block NFKB signaling in cylindromatosis patients with CYLD mutations46,47. Whether similar treatment strategies may be applicable to TCs remains to be evaluated.
In conclusion, our study shows that chromatin remodeling, histone modification and DNA methylation regulatory genes are frequently mutated in TCs, and that the genetic architecture of thymoma is different from that of TCs. Further investigation on the role of epigenetic dysregulation in TC development and the potential for targeted therapy is warranted.
Methods
Patients
The study was approved by the institutional ethics committee at the National Cancer Institute (NCI) and performed according to institutional guidelines. All participants provided written informed consent. Patients with advanced-stage TETs who were referred to NCI and were eligible for a pilot study of molecular profiling in thoracic cancers (Clinical trial.gov ID: NCT01306045) were included. This was a large trial in which patients with advanced chest tumors (lung cancers and TETs) were molecularly profiled and treatment allocated depending on predefined molecular alterations in tumors48. Eligibility criteria included: patients with advanced disease that was not amenable to radical surgery or radiation, Eastern Cooperative Oncology Group (ECOG) performance status of 0–2 and availability of tumor tissue material (either archival tissues or new biopsies). At the same time there were several Phase II trials available at NCI for patients with advanced TETs and patients were enrolled on those studies if no predefined targetable molecular abnormality was found. Clinical data on these Phase II trials have been previously reported5,6,7,8,39. Stages were defined according to the Masaoka staging system and histotypes according to the WHO 2004 classification2,
DNA extraction from formalin-fixed paraffin-embedded block sections
Formalin-fixed paraffin-embedded (FFPE) slides prepared from fresh biopsies or archival material were stained with hematoxylin and eosin (H&E) and evaluated by a pathologist (KJK) to confirm the tumor diagnosis and select regions rich (>80%) in tumor cells. DNA was extracted from sections of enriched epithelial tumor cells by macro-dissection from unstained FFPE blocks using DNeasy Blood & Tissue kit (Qiagen, Valencia, CA) with an extended proteinase K digestion over 24 hrs at 65°C. Control DNAs were prepared from paired patients' peripheral blood using Agencourt Genfind v2 kit (Beckman Coulter, Brea, CA). All patients gave their written informed consent to this study that was conducted in agreement with the principles of the declaration of Helsinki and approved by the ethical institutional review board of the NIH clinical center (ClinicalTrial.gov ID#: NCT01306045).
Massively parallel sequencing of 197 cancer-related genes
A panel of 197 cancer-associated genes (Table S1) were selected for targeted exome capture based on their known frequency of mutations in most common tumor types, in particular: (1) variations were reported in the Catalogue of Somatic Mutations in Cancer (COSMIC) Cancer Gene Census or (2) were known or drivers of solid tumors not yet listed in the COSMIC census at the time this reagent was designed or (3) were in pathways under investigation within NCI for other projects. Massively parallel sequencing was performed for high depth sequencing on MiSeq sequencers (Illumina, San Diego, CA). Briefly, DNAs were extracted from clinical specimens, and were fragmented by sonication. Indexed DNA libraries were prepared by 3 consecutive steps including end-repair, A-tailing and adapter ligation to the DNA fragments. In subsequent PCR amplification steps, primers containing a flow cell attachment site (P5), sequencing primer sites for index read (Index SP) and application read 2 (Rd2 SP), unique 6 bp indices (Index) and a second flow cell attachment site (P7), were incorporated. The indexed libraries were then pooled in groups of up to 12, target enriched by Sure Select custom targeted-capture kit (Agilent) according to vendor protocol, and proceeded with paired-end (2 × 150 bp) sequencing using the Miseq V2 kit (15 M, 300 cycles). The design of the 197 genes covers about 96.3% of 9946 exons, which are equal to 682,932 bps. In the 156 sequenced samples (78 TET/blood pairs), the average sequence coverage was 125X (range 27X–420X), with 130X (27X–420X) for TETs and 121X (33X–300X) for normal blood controls.
Workflow for sequence read processing, somatic mutation call and annotation
Data processing and variant calling procedure mainly followed the Best Practices workflow recommended by the Broad Institute (http://www.broadinstitute.org/gatk/guide/best-practices). Briefly, the raw sequencing reads were mapped to human genome version 19 by Burrows-Wheeler Aligner49, followed by local realignment using the GATK suite from the Broad Institute and duplicated reads were marked by Picard tools (http://picard.sourceforge.net). Somatic variant calling was performed on sequencing reads of matched tumor-normal samples by the Strelka somatic variant caller50, and germline variant calling was done with the UnifiedGenotyper from the Broad Institute. Mutiple annotation databases and open source packages were used to annotate and predict the effects of variants, including SnpEff51, dbNSFP52, dbSNP 137 (NCBI), ESP6500 (NHLBI Exome Sequencing Project), and COSMIC database. SIFT scores range from 0 to 1, and scores < 0.05 suggest that the amino acid change is damaging (sift.jcvi.org/www/SIFT_help.html). PolyPhen-2 scores > 0.85 are interpreted as probably damaging (genetics.bwh.harvard.edu/pph2/dokuwiki/overview#prediction).
The following filtering criteria were used for somatic variation calls: (1) SNVs and indels in tumors were considered somatic if they were completely absent in the paired blood samples; (2) Only SNVs and indels of >15 reads with allelic fraction of >15% were reported; (3) MAPQ score of <20 were excluded for variant count; (4) Somatic SNVs and indels were reported only when they were within the open reading frames and splice sites; (5) Synonymous variations, non-coding region mutations outside splice-sites, and single nucleotide polymorphisms (SNPs) that have not been reported to be disease-related were filtered out; (6) All the identified somatic SNVs and indels were validated visually using Integrative Genomics Viewer (IGV, Broad Institute).
Validation of somatic mutations by Sanger sequencing
Somatic mutations identified in the study were randomly selected for validation by dideoxynucleotide Sanger sequencing on tumor/blood paired samples. Primers were designed using Primer 3.0 or Oligo 6.0 software (Molecular Biology Insights, Cascade, CO), and blast-searched against the whole human genome sequence (NCBI build 37) for primer specificity. DNAs were PCR-amplified with gene specific primers that were flanked by M13F and M13R sequences at 5′ ends for sequencing (sequences of primers are available upon request).
Statistical analysis
Pathway enrichment analysis of mutated genes was performed using Fisher's exact or Chi square tests to quantify the association between the somatic mutations identified in this study and the driver gene-enriched 12 core-pathways recently described by Vogelstein et al23. Briefly the representation of each core pathway genes within the mutated genes identified in our study was compared with the representation of the core pathway genes within all the reported cancer-associated genes in the COSMIC database by Fisher's exact test to calculate the enrichment scores (−log [p-value]). We also calculated q-values from the empirical p-values by the bootstrap estimation method53,54. We considered q-values of ≤0.05 as an acceptable proportion of false positives in combination with p-value of ≤0.05. All the data were processed and analyzed using the statistical computing software R with specific add-on packages55.
Correlations between patient and tumor characteristics and molecular results were assessed using Fisher's exact test. Survival curves were generated with Kaplan-Meier method, and differences evaluated using the Log-rank (Mantel-Cox) test. Overall survival was calculated from the time of initial diagnosis to death or censored to the time at which the patient was last known to be alive. All calculations were performed using PRISM version 5.0b (GraphPad Software Inc., La Jolla, CA).
Author Contributions
Y.W., P.S.M. and G.G. designed, managed the project and wrote the manuscript; Y.W., C.L., Y.Z., I.P., X.Z., P.S.M. and G.G. performed data analysis; C.L., J.K.K. and T.P. performed sequence assays; A.T., A.R., B.M. and G.G. provided samples and collected clinical data. All authors reviewed the paper.
Supplementary Material
Supplemental Information
Acknowledgments
The study was supported by NIH/NCI intramural research program and by Lombardi Cancer Center, Georgetown University.
References
- Engels E. A. Epidemiology of thymoma and associated malignancies. J Thorac Oncol 5, S260–265 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis W. D., Brambilla E., Muller-Hermelink H. K. & Harris C. C. Pathology and genetics: Tumors of the lung, pleura, thymus and heart. in World Health Organization Classification of Tumours (series eds. Kleihues, P. & Sobin, L. H.) 145–247 (IARC Press, Lyon, 2004). [Google Scholar]
- Kelly R. J., Petrini I., Rajan A., Wang Y. & Giaccone G. Thymic Malignancies: From Clinical Management to Targeted Therapies. J Clin Oncol 29, 4820–4827 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macconaill L. E. & Garraway L. A. Clinical implications of the cancer genome. J Clin Oncol 28, 5219–5228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A. et al. Phase II trial of sunitinib in patients with thymic epithelial tumors (TET). J Clin Oncol 32:5s, suppl; abstr 7525–7525 (2014). [Google Scholar]
- Besse B. et al. A phase II study of milciclib (PHA-848125AC) in patients (pts) with thymic carcinoma (TC). J Clin Oncol 32:5s, Suppl; abstr 7526–7526 (2014). [Google Scholar]
- Rajan A. et al. Cixutumumab for patients with recurrent or refractory advanced thymic epithelial tumours: a multicentre, open-label, phase 2 trial. Lancet Oncol 15, 191–200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaccone G. et al. Phase II study of belinostat in patients with recurrent or refractory advanced thymic epithelial tumors. J Clin Oncol 29, 2052–2059 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M., Gabriel S. & Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11, 685–696 (2010). [DOI] [PubMed] [Google Scholar]
- Girard N. Thymic tumors: relevant molecular data in the clinic. J Thorac Oncol 5, S291–295 (2010). [DOI] [PubMed] [Google Scholar]
- Alexandrov L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell R. A., McGranahan N., Bartek J. & Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013). [DOI] [PubMed] [Google Scholar]
- Petrini I. et al. Whole genome and transcriptome sequencing of a B3 thymoma. PloS One 8, e60572 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrini I. et al. A specific missense mutation in GTF2i occurs at high frequency in thymic epithelial tumors. Nat Genet 46, 844–849 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilar E. & Gruber S. B. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 7, 153–162 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P. A. Holding damaged DNA together. Nat Struct Biol 6, 805–806 (1999). [DOI] [PubMed] [Google Scholar]
- Kumar P., Henikoff S. & Ng P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4, 1073–1081 (2009). [DOI] [PubMed] [Google Scholar]
- Adzhubei I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateyama H. et al. p53 protein expression and p53 gene mutation in thymic epithelial tumors. An immunohistochemical and DNA sequencing study. Am J Clin Pathol 104, 375–381 (1995). [DOI] [PubMed] [Google Scholar]
- Beadling C. et al. KIT gene mutations and copy number in melanoma subtypes. Clin Cancer Res 14, 6821–6828 (2008). [DOI] [PubMed] [Google Scholar]
- Blake P. W. & Toro J. R. Update of cylindromatosis gene (CYLD) mutations in Brooke-Spiegler syndrome: novel insights into the role of deubiquitination in cell signaling. Hum Mutat 30, 1025–1036 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S. C. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ 17, 25–34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B. et al. Cancer genome landscapes. Science 339, 1546–1558 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena-Llopis S. et al. BAP1 loss defines a new class of renal cell carcinoma. Nat Genet 44, 751–759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone M. et al. BAP1 and cancer. Nat Rev Cancer 13, 153–159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Wahab O. & Dey A. The ASXL-BAP1 axis: new factors in myelopoiesis, cancer and epigenetics. Leukemia 27, 10–15 (2013). [DOI] [PubMed] [Google Scholar]
- Schmidt C. K. & Jackson S. P. On your mark, get SET(D2), go! H3K36me3 primes DNA mismatch repair. Cell 153, 513–515 (2013). [DOI] [PubMed] [Google Scholar]
- Trotter K. W. & Archer T. K. The BRG1 transcriptional coregulator. Nucl Recept Signal 6, e004 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fouchardiere A. et al. Germline BAP1 mutations predispose also to multiple basal cell carcinomas. Clin Genet, 10.1111/cge.12472 (2014). [DOI] [PubMed] [Google Scholar]
- Belani R. et al. ASXL1 and DNMT3A mutation in a cytogenetically normal B3 thymoma. Oncogenesis 3, e111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsi-Boyer V., Brecqueville M., Devillier R., Murati A., Mozziconacci M. J. & Birnbaum D. Mutations in ASXL1 are associated with poor prognosis across the spectrum of malignant myeloid diseases. J Hematol & Oncol 5, 12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgliesh G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huether R. et al. The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat Commun 5, 3630 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couronne L., Bastard C. & Bernard O. A. TET2 and DNMT3A mutations in human T-cell lymphoma. New Engl J Med 366, 95–96 (2012). [DOI] [PubMed] [Google Scholar]
- Szemes M. et al. Control of epigenetic states by WT1 via regulation of de novo DNA methyltransferase 3A. Hum Mol Genet 22, 74–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solary E., Bernard O. A., Tefferi A., Fuks F. & Vainchenker W. The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia 28, 485–496 (2014). [DOI] [PubMed] [Google Scholar]
- Yan X. J. et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet 43, 309–315 (2011). [DOI] [PubMed] [Google Scholar]
- Ley T. J. et al. DNMT3A mutations in acute myeloid leukemia. New Engl J Med 363, 2424–2433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A. et al. A phase (Ph) I/II study of belinostat (Bel) in combination with cisplatin, doxorubicin, and cyclophosphamide (PAC) in the first-line treatment of advanced or recurrent thymic malignancies. J Clin Oncol 30, suppl; abstr 7103–7103 (2012). [Google Scholar]
- Nicholson P. et al. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol life Sci 67, 677–700 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur P. et al. Effects on survival of BAP1 and PBRM1 mutations in sporadic clear-cell renal-cell carcinoma: a retrospective analysis with independent validation. Lancet Oncol 14, 159–167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens R. A. et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 1, 598–607 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff T. & Armstrong S. A. Recent progress toward epigenetic therapies: the example of mixed lineage leukemia. Blood 121, 4847–4853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi A. & Kuwano H. TP53 mutations in nonsmall cell lung cancer. J Biomed Biotechnol 2011, 583929 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoccianti C. et al. Prognostic value of TP53, KRAS and EGFR mutations in nonsmall cell lung cancer: the EUELC cohort. ERJ 40, 177–184 (2012). [DOI] [PubMed] [Google Scholar]
- Brummelkamp T. R., Nijman S. M., Dirac A. M. & Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424, 797–801 (2003). [DOI] [PubMed] [Google Scholar]
- Pfeiffer N. Topical aspirin promising against rare inherited skin cancer. Oncol Times 25, 42–45 (2003). [Google Scholar]
- Giaccone G. et al. Custom (Molecular Profiling and Targeted Therapy for Advanced Non-Small Cell Lung Cancer, Small Cell Lung Cancer, and Thymic Malignancies) trial. J Clin Oncol 31, suppl; abstr 7513–7513 (2013). [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C. T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- Cingolani P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Jian X. & Boerwinkle E. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum Mutat 34, E2393–2402 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, Series B 57, 289–300 (1995). [Google Scholar]
- Storey J. D. & Tibshirani R. Statistical significance for genomewide studies. Pro Natl Acad Sci USA 100, 9440–9445 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R. & Gentleman R. R. A language for data analysis and graphics. J Comut Graph Stat 5, 299–314 (1996). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Information