Mutations in transcription factor 4 (TCF4) have recently been described in myeloid dysplastic syndromes (MDS) and acute myeloid leukemia (AML). We analyzed the impact of TCF4 mRNA expression on clinical outcome in AML patients (n=525). Patients with high TCF4 expression (TCF4high, defined as the 25% highest TCF4 expressors) had a significantly worse overall survival (OS) and event-free survival (EFS) than patients with lower TCF4 expression (TCF4low) (5-year OS 18% vs. 44%, P<0.0001; 5-year EFS 15% vs. 34%, P<0.0001, respectively). This was confirmed in an independent cohort (n=436). Multivariate analysis showed that TCF4high is an independent prognostic factor for OS and EFS in the whole cohort and in patients carrying a normal karyotype.
Importantly, TCF4high patients benefited most from an allogeneic hematopoietic cell transplantation (HCT), compared to an autologous HCT or additional chemotherapy (CT) (5-year OS 39%, 8%, 10%, P<0.0001; 5-year EFS 31%, 0%, 10%, P=0.001, respectively), while TCF4low patients seemed to benefit most from an autologous HCT, compared to allogeneic HCT or additional CT (5-year OS: 61%, 45%, 39% P=0.002; 5-year EFS: 42%, 32%, 34%, P=0.102, respectively).
We demonstrate that high expression of TCF4 is an independent adverse prognostic factor in AML that could guide treatment decisions.
TCF4 plays a role in a variety of developmental processes, including hematopoiesis. TCF4 is part of the basic helix-loop-helix (bHLH) class 1 family, also called E-proteins. These E-proteins recognize an E-box DNA binding site (CANNTG), which are present in a variety of tissue-specific enhancers.1,2 Recently, Papaemmanuil and colleagues reported mutations in TCF4 in MDS patients.3 A total of 9 mutations were found in 7 of the 738 (0.9%) sequenced MDS patients. The TCF4 mutations were found in various MDS subclasses. Mutations in TCF4 have also been reported for AML cases (0.5%)4 and were associated with a poor prognosis,5 suggesting a potential role of TCF4 in the pathogenesis of these myeloid malignancies. Here we report that TCF4 mRNA expression levels are an independent prognostic factor in AML patients.
TCF4 expression values measured using Affymetrix HGU133 plus 2.0 arrays were derived from a database which contains a cohort of 525 AML patients treated according to HOVON protocols (AML -04, -04A, -29, -32, -42, -43; available at http://www.hovon.nl).6 Both bone marrow aspirates or peripheral-blood samples (at the time of diagnosis) have been analyzed. Blasts and mononuclear cells were purified by Ficoll–Hypaque (Nygaard) centrifugation and cryopreserved. The AML samples contained 80–100% blast cells after thawing, regardless of the blast count at diagnosis. To determine the TCF4 expression, an average of 5 probe sets (which bind at different locations of the gene) were used. The microarray expression data were confirmed by qPCR (Online Supplementary Figure S1). In addition, the TCF4 expression levels of healthy CD34+ control cells (hCD34+; n=11) and mononuclear cell fractions derived from normal bone marrow (NBM; n=5) were available. A second, independent cohort of 436 AML patients was used for validation.7 Patients were divided into genetic risk groups according to the European LeukemiaNet (ELN) guidelines.8
In the studied cohort of 525 AML patients, TCF4 is differentially expressed in AML blasts compared to NBM and hCD34+ (Figure 1A). To study the impact of TCF4 expression levels on survival, the cohort was divided on the basis of differences in expression levels; expression below or above the median, tertiles, quartiles, quintiles, sixtiles and septiles. In all these cohorts, univariate analysis showed that high expression of TCF4 was associated with poor outcome. The highest expressors of TCF4 showed a more than 2-fold shorter 5-year OS than the lowest expressors (Online Supplementary Figure S2). Since we found that TCF4 expression is not normally distributed and because approximately 25% of the patients showed a much higher expression (Figure 1B), a distribution of the cohort based on the highest 25% (TCF4high) and the lowest 75% TCF4 expression (TCF4low) was used for further analysis. Characteristics of the patients in the TCF4low and TCF4high groups are described in Online Supplementary Table S1. TCF4high patients more often had high-risk cytogenetic abnormalities (P<0.0001), FLT3-ITD (P<0.0001) and their morphology more frequently corresponded with M0 or M1 FAB-subgroups (P<0.0001). TCF4low patients were more likely to have biallelic CEBPA mutations (P=0.011). No associations between TCF4 expression and age, sex, white blood cell (WBC) count, or other cytogenetic or molecular abnormalities could be identified.
Figure 1.
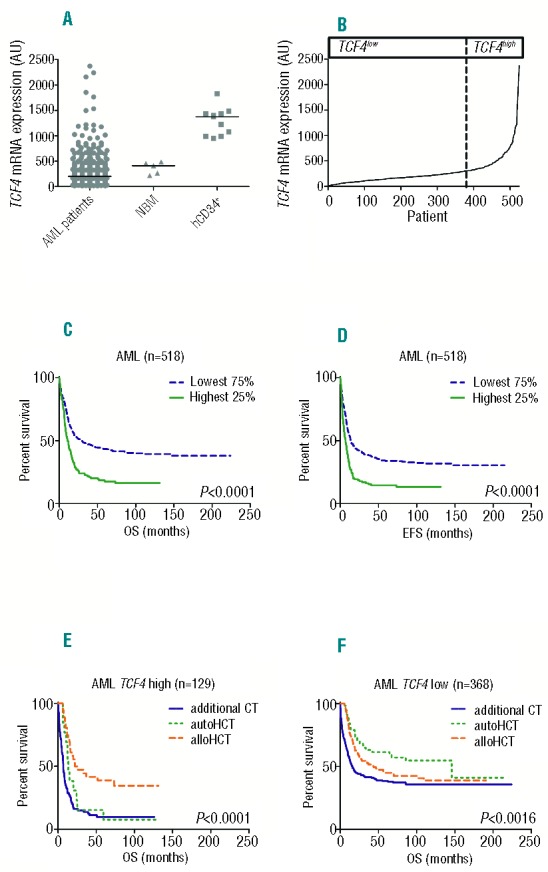
TCF4 expression and survival curves in the first cohort. (A) Expression of TCF4 in AML patients (n=525), NBM (n=5) and hCD34+ (n=11). (B) TCF4 expression ranked from lowest to highest expression (n=525). (C) Overall survival (OS) curves for AML patients with available follow-up data (n=518) stratified by TCF4high (n=129) and TCF4low (n=389). (D) Same for event-free survival (EFS). (E) OS curves for 7CF4high AML patients with available follow up and consolidation treatment data (n=129) stratified for conditioning with alloHCT (n=36), autoHCT (n=13) or additional CT (n=80). (F) OS curves for TCF4low AML patients with available follow up and consolidation treatment data (n=386) stratified for conditioning with alloHCT (n=99), autoHCT (n=57) or additional CT (n=212).
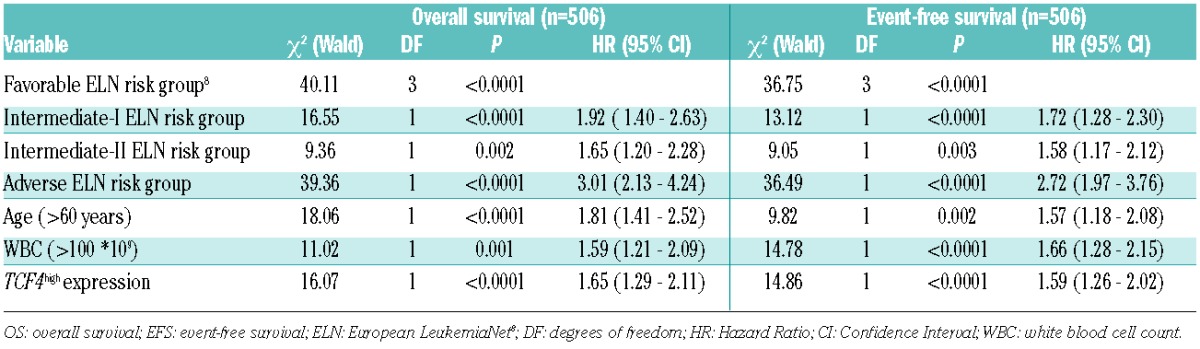
Survival analysis according to the Kaplan-Meier method showed that TCF4high patients had a worse survival than patients classified as TCF4low (5-year OS 18% vs. 44%, P<0.0001; 5-year EFS 15% vs. 34%, P<0.0001, respectively) (Figure 1C and D). We confirmed the impact of TCF4 expression levels on survival in the second cohort of 436 AML patients7 (OS: P=0.001; EFS: P<0.0001) (Online Supplementary Figure S3). In the multivariate Cox regression analysis, patients classified as TCF4high had a significantly higher risk of death (HR 1.7, CI: 1.3–2.1; P<0.0001), relapse or not obtaining a CR than TCF4low patients (HR 1.6, CI: 1.3–2.0; P<0.0001) (Table 1A). In addition, multivariate Cox regression analysis revealed TCF4 expression, as a continuous variable per 100 arbitrary units (AU), was a significant predictor of OS and EFS (HR 1.04, CI: 1.01–1.07, P=0.024; HR 1.05, CI: 1.02–1.08, P=0.002, respectively) (Online Supplementary Table S2A). When selecting for AML patients with a normal karyotype, TCF4high patients again showed a worse OS and EFS than TCF4low patients (5-year OS 21% vs. 41%, P<0.0001; 5-year EFS 18% vs. 33%, P<0.0001, respectively) (Online Supplementary Figure S4). In the multivariate Cox regression analysis of normal karyotype AML patients, TCF4 expression is also an independent predictor of survival (OS: HR 1.7, CI: 1.2–2.5, P=0.003 EFS: HR 1.7, CI: 1.2–2.4, P=0.005) (Online Supplementary Table S2B). Also as a continuous variable, TCF4 expression remained an independent prognostic factor in this cohort (OS: HR 1.07 (per 100 AU), CI: 1.02–1.13, P=0.004; EFS: HR 1.08 (per 100 AU), CI: 1.03–1.13, P=0.003) (Online Supplementary Table S2C).
Table 1.
Multivariate Cox’s regression survival analysis. Factors predicting overall survival and event-free survival in acute myeloid leukemia patients of the first cohort with available complete data of all cytogenetic and molecular parameters (n=506).
Interestingly, survival analysis according to the Kaplan-Meier method showed that TCF4high and TCF4low patients of the first cohort demonstrated a different survival benefit depending on the consolidation treatment they received, i.e, an additional cycle of chemotherapy (CT), autologous or allogeneic hematopoietic cell transplantation (autoHCT, alloHCT, respectively) (OS: Figure 1E and F; EFS: Online Supplementary Figure S5). TCF4high patients who received alloHCT showed a superior survival compared to TCF4high patients who received autoHCT or who received additional CT (5-year OS 39%, 8%, 10%, P<0.0001; 5-year EFS 31%, 0%, 10%, P=0.001, respectively). In contrast, patients classified as TCF4low showed a trend towards significant superior survival after autoHCT, compared to TCF4low patients who received alloHCT or additional CT (5-year OS: 61%, 45%, 39% P=0.002; 5-year EFS: 42%, 32%, 34%, P=0.102, respectively). Moreover, this difference in outcome, depending on type of consolidation treatment between the TCF4high and the TCF4low patients, was confirmed in multivariate Cox regression analysis (Online Supplementary Table S3). In the second cohort, only 7 patients in the TCF4high group received autoHCT, hampering validation of our observations in this subgroup. Nevertheless, also in this cohort, consolidation treatment with alloHCT (n=44) resulted in significantly better OS for TCF4high patients compared to TCF4high patients who received additional chemotherapy (n=58) (5-year OS 41% vs. 8%, respectively; P<0.0001). Furthermore, in this cohort TCF4low patients who received autoHCT (n=52) showed a superior OS compared with those patients who received alloHCT (n=86) or additional CT (n=186) (5-year OS 61%, 48% vs. 26%, respectively; P<0.0001), confirming the observations from the first cohort.
The biological role of TCF4 is poorly understood,2 and contrasting observations are described in the literature. For example, enforced expression of members of the bHLH class A family, including TCF4, suppresses colony-forming efficiency of various cell lines due to upregulation of p21, p15 and p16, suggesting that these bHLH proteins act as negative regulators of cell growth.9 In contrast, Tcf4 expression appeared increased in rat-E1A-immortalized RK3E cells following β-catenin induced neoplastic transformation and aberrant expression of Tcf4 promoted neoplastic transformation of RK3E cells.10 These different observations might be explained by differences in cellular context, or by the different transcript variants of TCF4,11–14 which could affect the function of the protein.10 Possibly, TCF4 can either stimulate or inhibit cell growth, depending on its environment, which might indicate that an aberrant expression is not only a prognostic marker, but also a pathological feature. This would be in line with the report of mutations in TCF4 in MDS and AML.3,4
TCF4 has also been reported to be highly expressed in hematopoietic stem cells (HSC) and to show a decreased expression in committed progenitors.15 Since the frequency of TCF4 mutations is relatively low (0.5% in AML), obviously not all patients with high expression of TCF4 can have mutated TCF4. Interestingly, in MLL-AF9-mediated transformation of progenitor cells, TCF4 has been shown to be up-regulated.15 In the first cohort, patients with high TCF4 expression are significantly more classified in the M0 or M1 FAB-subgroups than TCF4low patients, suggesting that the leukemic cells of the TCF4high patients derive from more immature cells. In addition, TCF4 expression of patients in the TCF4high group is comparable to the level of TCF4 expression of hCD34+ cells. Furthermore, when looking at the CD34 mRNA expression in the first cohort, 73.3% of the TCF4high patients show a high CD34 expression (above the median), compared to 42.1% of the TCF4low patients. When including CD34 expression in the multivariate Cox regression analysis, CD34 expression is an independent prognostic factor in OS and EFS; nevertheless TCF4 expression also remains an independent prognostic factor (data not shown).
Our observations report on the prognostic relevance of the level of TCF4 expression in AML and demonstrate that high TCF4 expression is associated with a worse survival. In addition, the TCF4 expression levels seem to provide additional information in the response to treatment. Before considering TCF4 expression levels in clinical decision-making, additional validation studies, also to define optimal cut-off levels, are needed. Further mechanistic studies are warranted on the role of TCF4 in myeloid diseases.
Acknowledgments
the authors would like to thank Dr. P. Valk for providing cDNA.
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Ellenberger T, Fass D, Arnaud M, Harrison SC. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Genes Dev. 1994;8(8):970–80. [DOI] [PubMed] [Google Scholar]
- 2.Massari ME, Murre C. MINIREVIEW Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol Cell Biol. 2000;20(2):429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makishima H, LaFramboise T, Przychodzen JP, Yoshida K, Ruffalo M, Gómez-Seguí I, et al. Clinical “MUTATOME” Of Myelodysplastic Syndrome; Comparison To Primary Acute Myelogenous Leukemia. ASH Annual Meeting Abstract. Blood. 2013;Abstract 518. [Google Scholar]
- 6.Wouters BJ, Löwenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R, et al. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood. 2009;113(13):3088–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kharas MG, Lengner CJ, Al-Shahrour F, Bullinger L, Ball B, Zaidi S, et al. Musashi-2 regulates normal hematopoiesis and promotes aggressive myeloid leukemia. Nat Med. 2010;16(8):903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Döhner H, Estey EH, Amadori S, Appelbaum FR, Büchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–74. [DOI] [PubMed] [Google Scholar]
- 9.Pagliuca A, Gallo P, De Luca P, Lania L. Class A helix-loop-helix proteins are positive regulators of several cyclin-dependent kinase inhibitors’ promoter activity and negatively affect cell growth. Cancer Res. 2000;60(5):1376–82. [PubMed] [Google Scholar]
- 10.Kolligs FT, Nieman MT, Winer I, Hu G, Van Mater D, Feng Y, et al. ITF-2, a downstream target of the Wnt/TCF pathway, is activated in human cancers with beta-catenin defects and promotes neoplastic transformation. Cancer Cell. 2002;1(2):145–55. [DOI] [PubMed] [Google Scholar]
- 11.Corneliussen B, Thornell A, Hallberg B, Grundström T. Helix-loop-helix transcriptional activators bind to a sequence in glucocorticoid response elements of retrovirus enhancers. J Virol. 1991;65(11):6084–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990;247(4941):467–70. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Ray SK, Yang XQ, Luntz-Leybman V, Chiu IM. A splice variant of E2-2 basic helix-loop-helix protein represses the brain-specific fibroblast growth factor 1 promoter through the binding to an imperfect E-box. J Biol Chem. 1998;273(30):19269–76. [DOI] [PubMed] [Google Scholar]
- 14.Skerjanc IS, Truong J, Filion P, McBurney MW. A splice variant of the ITF-2 transcript encodes a transcription factor that inhibits MyoD activity. J Biol Chem. 1996;271(7):3555–61. [DOI] [PubMed] [Google Scholar]
- 15.Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442(7104):818–22. [DOI] [PubMed] [Google Scholar]


