Abstract
Several studies have demonstrated that 3D culture systems influence human embryonic stem cell (hESC) phenotypes and fate choices. However, the effect that these microenvironmental changes have on signaling pathways governing hESC behaviors is not well understood. Here, we have used a 3D microwell array to investigate differences in activation of developmental pathways between 2D and 3D cultures of both undifferentiated hESCs and hESCs undergoing initial differentiation in embryoid bodies (EBs). We observed increased induction into mesoderm and endoderm and differences in expression of genes from multiple signaling pathways that regulate development, including Wnt/β-catenin, TGF-β superfamily, Notch and FGF during EB-mediated differentiation, in 3D microwells as compared with the 2D substrates. In undifferentiated hESCs, we also observed differences in epithelial-mesenchymal transition phenotypes and the TGFβ/BMP pathway between cultures in 3D and 2D. These results illustrate that 3D culture influences multiple pathways that may regulate the differentiation trajectories of hESCs.
Introduction
Human embryonic stem cells (hESCs) have the potential to differentiate into over 200 diversely functioning cell types found in the human body1–3. The many processes involved in mammalian development, such as survival, self-renewal and differentiation, are regulated by the microenvironment, which is the coordinated spatial and temporal presentation of molecular, structural, mechanical and hydrodynamic cues4. In order to understand and control hESC behavior for downstream applications such as drug screening, tissue engineering, and regenerative medicine applications, in vitro studies that provide mechanistic understanding of how the physiologically relevant 3D microenvironment regulates hESC fates will be necessary.
To this end, various materials-based culture platforms have been engineered to systematically study how microenvironmental contributions, such as substrate stiffness, extra-cellular matrix (ECM) proteins, scaffolding and mechanical stimuli, affect ESC behavior5–10. 3D microwell cell culture platforms have been utilized to modulate cell-cell contact, intercellular and autoregulatory signaling that occur in hESCs and during differentiation, particularly in embryoid-body (EB) based differentiation11. Some studies have demonstrated that pro-pluripotency autoregulation can be accentuated in various configurations of 3D microwell culture12–14. In addition, many studies have demonstrated that microwell culture affects differentiation toward multiple cell types including hematopoietic lineages15, myeloid and erythroid cells16 and cardiomyocytes17–19.
However, very little is yet known regarding the molecular mechanisms by which 3D microwell culture drive the significant differences in cell fate observed. A study by Azarin et al. showed that hESCs cultured in microwells exhibited less nuclear β-catenin when compared to hESCs cultured on 2D substrates20. However, upon enzymatic removal of hESC colonies from microwells and EB differentiation, significantly higher canonical Wnt signaling was observed in EBs made from microwell-cultured hESCs. The cells that demonstrated higher levels of Wnt signaling activity exhibited greater expression of genes associated with the primitive streak, mesoderm and cardiac lineage. In another study by Hwang et al, EBs derived from murine ESCs cultured in microwell sizes that favored cardiogenesis over endothelial cell differentiation expressed non-canonical Wnt11 at a greater level than Wnt5a21. Upon siRNA knock down of Wnt5a, endothelial vessel sprouting was ablated while cardiac markers became highly expressed. Exogenous Wnt5a addition increased the expression of endothelial markers and the sprouting of endothelial vessel structures, though with no significant change to cardiac markers. Finally, a study by Bauwens et al described a cellular mechanism by which microwell-derived EBs of different sizes influenced differentiation potential22. They posited that EB size specified the outer surface area-to-volume ratio, which determined the amount of cardiac-promoting extra-embryonic endoderm cells in the EB, thus promoting cardiomyogenesis. Much has yet to be identified regarding the milieu of molecular mechanism by which morphology of ESC colonies regulates developmental signaling pathways and affects lineage commitment.
Therefore, using a microwell system previously established in our lab13, we sought to identify major signaling pathways modulated by 3D microwell culture of hESCs in the undifferentiated state and also during subsequent EB differentiation. In undifferentiated hESCs cultured in 3D versus 2D, we observed differences in epithelial-mesenchymal transition (EMT) and the TGFβ/BMP pathway. However, comparing 3D EB-mediated differentiation of aggregated hESCs cultured from 3D microwells versus 2D substrates, we observed increased induction into mesoderm and endoderm, and differences in expression of genes from multiple signaling pathways that regulate development, including Wnt/β-catenin, TGF-β superfamily, Notch and FGF.
Materials & Methods
Microwell fabrication
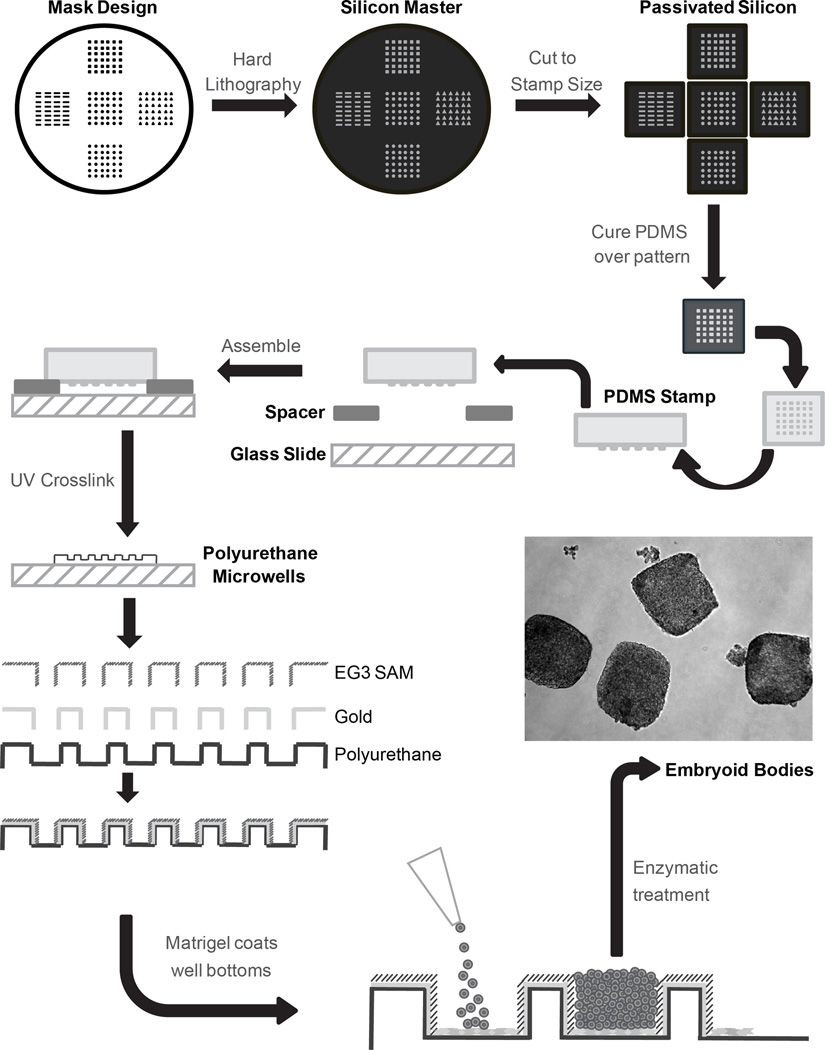
Microwells were prepared in three major steps as previously described13. First, silicon master molds with desired microwell patterns were made using hard lithography techniques. The cuboidal microwells had lateral dimensions of 300 µm and a depth of 120 µm. These master molds were fluorinated with (tridecafluoro-1,1,2,2,-tetrahydrooctyl)-1-trichlorosilane vapor to passivate the surface and minimize fouling. Second, via soft-lithography, polydimethylsiloxane (PDMS) stamps were formed with the inverted pattern from the silicon masters. Finally, these PDMS stamps were elevated over glass microscope slides with 250 µm spacers. In the space between, polyurethane prepolymer was fed, distributed via capillary action and crosslinked under UV light yielding patterned microwells.
Preparation of microwells for cell seeding
On the areas surrounding the wells, microwells were coated with an 80 Å titanium layer followed by a 200 Å gold layer via e-beam evaporation. After sterilization with UV and 100% ethanol, microwell slides were immersed in tri(ethylene glycol)-terminated (EG3) alkanethiol self-assembling monolayer (SAM) solution overnight. This SAM functionalized only on the surfaces coated with gold while the EG3 functional group prevented protein adsorption and subsequent cell attachment. Thus, of the approximate 250 mm2 total surface area of the microwell array, about 45 mm2 are available for protein adsorption. After washing in ethanol, functionalized microwells were coated with growth factor reduced Matrigel for at least 1 hour at 37°C. To seed the microwells, cells were singularized via Accutase (Innovative Cell Technologies), resuspended in CM/F+ at 1×106 cells/100 µL/microwell slide, and aliquoted to the top of each microwell array. The cell solution was maintained on top of arrays for at least 15 min to allow cells to settle into microwells before adding 2 ml/well of CM/F+. A flowchart of microwell fabrication and cell seeding is represented in Fig 1.
Figure 1.
Schematic of 3D microwell design, manufacturing and preparation for cell culture. Silicon master molds were fabricated via hard lithography. The master molds were passivated in order to pattern PDMS stamps. The reusable stamps were then elevated over glass slides with spacers. Polyurethane pre-polymer filled the space and was UV-crosslinked to form polyurethane microwells. E-beam evaporation was then used to coat the areas outside of the wells with a thin layer of gold. Finally, a tri(ethylene glycol)-terminated alkanethiol self-assembled monolayer (EG3) was assembled on the gold surface. This confers protein resistance on all but the bottom of the microwells. Cells were seeded into the microwells and allowed to grow for 6 days before enzymatic detachment and EB formation to initiate differentiation. The phase contrast inset shows suspended cell aggregates immediately after microwell colonies were detached.
hESC culture and embryoid body differentiation
H9 hESCs were cultured on tissue culture polystyrene (TCPS) coated with growth factor reduced Matrigel (BD Bioscience) for at least 1 hour at 37°C. Unconditioned medium (UM/F−) composed of DMEM/F12 culture medium (Invitrogen) containing 20% KnockOut Serum Replacer (Invitrogen), 0.1 mM MEM non-essential amino acids (Invitrogen), 1 mM L-glutamine (Invitrogen), and 0.1 mM β-mercaptoethanol (Sigma) was conditioned on irradiated mouse embryonic fibroblasts (MEFs) for 24 hours and supplemented with 4 ng/mL bFGF (Invitrogen), which resulted in the culture medium CM/F+.
For embryoid body differentiation studies, hESCs were cultured on TCPS or in microwells for 6 days in CM/F+. To initiate differentiation, colonies were detached from the Matrigel matrix at day 6 of culture using 1 mg/mL dispase (Invitrogen) and placed in suspension in UM/F− in Corning 3741 ultra-low attachment plates. Following 24 hours in UM/F−, the EBs were maintained in suspension for 4 more days in EB20 medium: DMEME/F12 containing 20% fetal bovine serum (Invitrogen), 0.1 mM non-essential amino acids, 1 mM L-glutamine and 0.1 mM β-mercaptoethanol.
RT-PCR and Quantitative RT-PCR
For RNA extraction, cells were dissociated with Versene (Invitrogen), Accutase (Innovative Cell Technologies), TrypLE (Invitrogen) or 0.25% Trypsin-EDTA (Invitrogen). Total RNA was extracted using an RNeasy MiniKit (Qiagen) according to the manufacturer’s instructions. And treated with DNase I (Invitrogen). cDNA was generated from 1 µg of RNA using Omniscript reverse transcriptase (Qiagen) and Oligo-dT(20) primers (Invitrogen). End-point PCR was performed with GoTaq Green Master Mix (Promega) and then subjected to 2% agarose gel electrophoresis and imaged using ethidium bromide. Amplicon band intensity was semi-quantified using ImageJ with GAPDH and ACTB used as endogenous controls. Quantitative RT-PCR (qPCR) was performed using iQ SYBR Green Supermix (Bio-Rad) on an iCycler (Bio-Rad).
Western Blot Analysis
Cells were lysed in M-PER Mammalian Protein Extraction Reagent (Pierce) in the presence of Halt Protease and Phosphatase Inhibitor Cocktail (Pierce). Proteins were separated by 10% Tris-Glycine SDS-PAGE under denaturing conditions and transferred to a nitrocellulose membrane. After blocking with 5% BSA or Milk in TBS + 0.1% Tween-20, the membrane was labeled with primary antibody overnight at 4°C. The membrane was then washed and incubated with a horse-radish peroxidase-conjugated secondary antibody for 1 hour at room temperature or overnight at 4°C. Protein levels were detected via a SuperSignal West Pico Chemiluminescent Substrate (Pierce). Equal protein loading was confirmed via β-Actin and GAPDH levels. Band intensity was quantified using Image Lab Software (BIO-RAD).
Flow Cytometry
Cells were dissociated into single cells and then fixed with 1% paraformaldehyde for >20 minutes at room temperature and permeabilized in ice-cold 90% methanol for at least 30 minutes. Primary antibodies were incubated overnight in PBS plus 0.1% Triton X-100 and 0.5% BSA. Data were collected on a FACS Caliber flow cytometer (Beckton Dickinson) and analyzed using FlowJo.
Statistics
Data are presented as mean ± standard deviation (SD) and p-values were determined using an unpaired Student’s t-test.
Results
Microwell culture increased induction into mesendoderm in embryoid bodies
As previously demonstrated18, the differentiation of hESCs into cardiomyocytes can be enhanced by culturing the hESCs in 3D microwells system prior to inducing differentiation in embryoid bodies. Differentiation to functional, contracting cardiomyocytes did not occur until at least 9 days after hESC colony removal from microwell culture. However, since the direct effects from microwell culture end upon EB formation, we expect to observe differences in differentiation trajectory at much earlier developmental stages, including exit from pluripotency and germ lineage specification. The earliest steps in cardiogenesis involve the induction of pluripotent hESCs into mesendoderm and precardiac mesoderm, which are specified during gastrulation in vivo.
To ascertain whether microwell culture influences induction into mesendoderm during early EB differentiation, hESCs were seeded into Matrigel-coated, 3D microwells. The protein-resistant SAM prevented cell attachment on all but the bottom of the microwells. hESCs were also seeded onto Matrigel-coated TCPS as a control representing traditional 2D culture of ESCs. After 6 days of culture in CM/F+, hESCs were enzymatically removed and cultured in suspension to form EBs. Cells were harvested for PCR, flow cytometry and/or Western Blot analysis immediately prior to suspension culture and after selected number of days in suspension culture. A schematic representation of the general experimental plan is shown in Fig 2.
Figure 2.
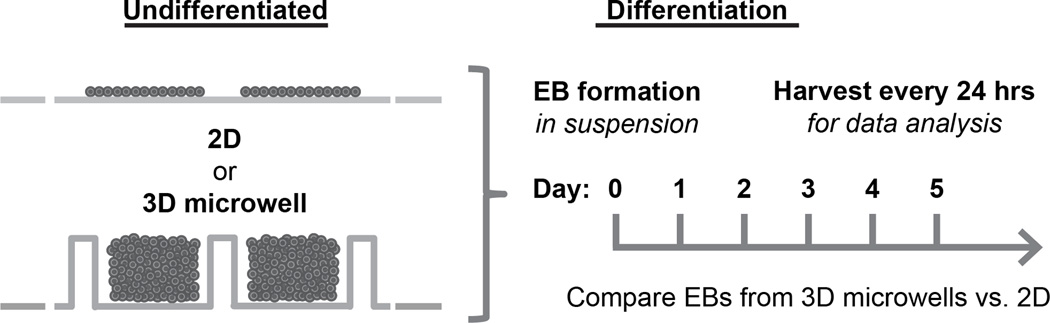
Schematic representation of the experimental plan. Undifferentiated hESCs were cultured either on 2D TCPS or in 3D microwells. After 6 days in culture as undifferentiated cells, colonies were enzymatically detached and placed in suspension to initiate EB formation and differentiation. EBs were collected immediately after removal from the substrate and every 24 hours following for data analysis to compare differences between EBs generated from 2D and 3D microwell culture of undifferentiated hESCs.
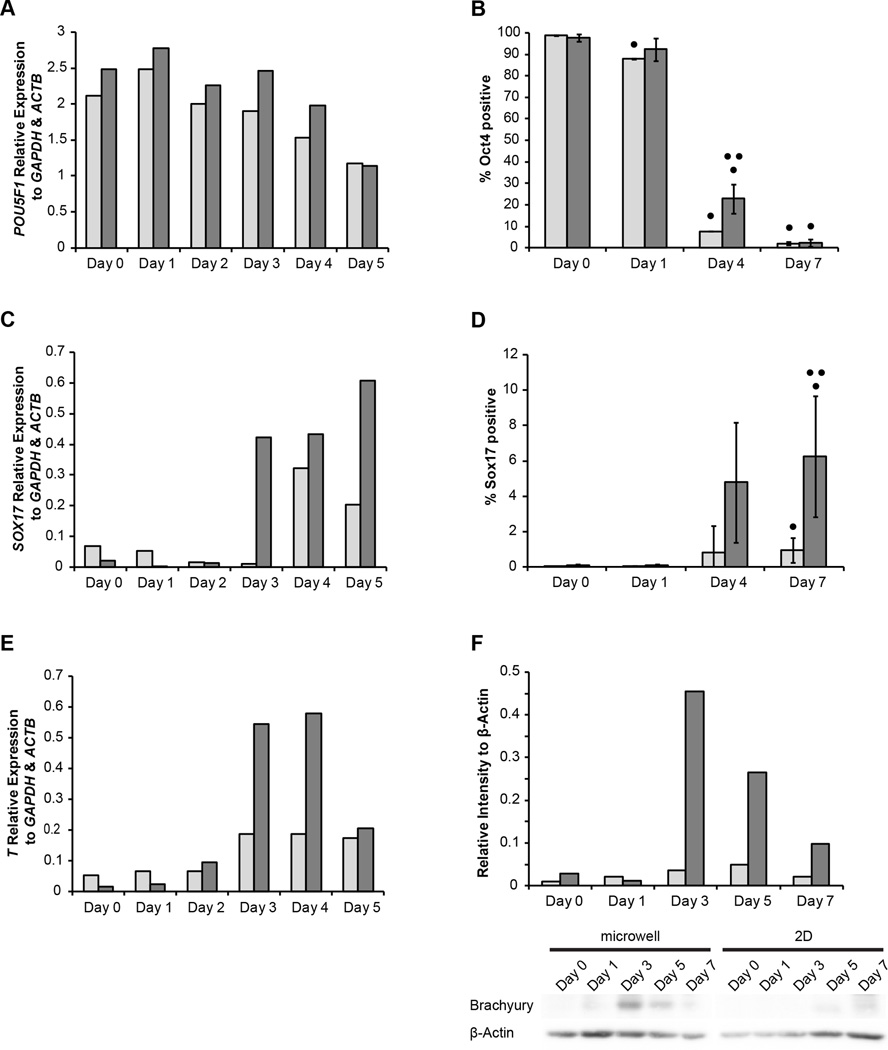
The expression of POU5F1 (Oct4) decreased at similar rates in EBs made from hESCs cultured on 2D TCPS and in 3D microwells, though the EBs from microwell-cultured hESCs continued to express more POU5F1 through day 4 of EB formation (Fig 3A). The percentage of Oct4 positive cells was nearly zero by the seventh day of EB formation from both 2D TCPS and 3D microwell culture, though EBs from microwell cultured hESCs exhibited a higher percent of Oct4 positive cells at day 4 of EB formation (Fig 3B).
Figure 3.
(A) RT-PCR quantification of POU5F1 expression in EBs 0–5 days after formation from hESCs cultured on Matrigel-coated TCPS (2D, light) and 3D microwells (dark) relative to expression of GAPDH and ACTB. (B) Flow cytometry for percentage Oct4-positive cells of EBs days 0, 1, 4 and 7 after formation. (C) RT-PCR quantification of SOX17 expression, relative to expression of GAPDH and ACTB, in EBs 0–5 days after formation from hESCs cultured on either 2D TCPS or in 3D microwells. (D) Flow cytometry for percentage Sox17-positive cells in EBs days 0, 1, 4 and 7 after formation. (E) RT-PCR quantification of T expression in EBs days 0–5 after formation from hESCs cultured on either 2D TCPS or in 3D microwells. (F) A representative western blot of Brachyury in EBs from hESCs cultured from either 2D TCPS or 3D microwells is shown with quantification of western blot intensities of Brachyury relative to housekeeping β-actin. (• indicates p<0.05 compared to Day 0, •• indicated p<0.05 compared to same-day 2D control)
We observed significant differences in expression of the transcription factors that control induction into the germ layers associated with cardiogenesis. The expression of SOX17, a marker of embryonic definitive endoderm, was detected by the third day of formation of EBs from microwell-cultured hESCs, which was about a day before detection of SOX17 in EBs made from 2D cultured hESCs. SOX17 expression continued to rise through the fourth day of EB differentiation (Fig 3C). This correlated with a higher percentage of cells expressing Sox17 protein in the EBs from microwell-cultured hESCs as compared to EBs made from 2D culture (Fig 3D). Importantly, there was a strong peak in expression of T gene (Brachyury), which encodes a transcription factor that controls mesendoderm induction early after loss of pluripotency23, at days 3 and 4 in EBs made from microwell-cultured hESCs, but no such peak in T expression in EBs made from hESCs cultured on 2D TCPS (Fig 3E). A similar peak at days 3 and 4 was observed in the expression of additional mesendoderm markers GSC24 and MIXL125 (Fig 4). A corresponding peak in the expression of Brachyury protein was also observed at day 3 (Fig 3F). This profile suggests EBs from hESC-cultured microwells favor mesendoderm lineages.
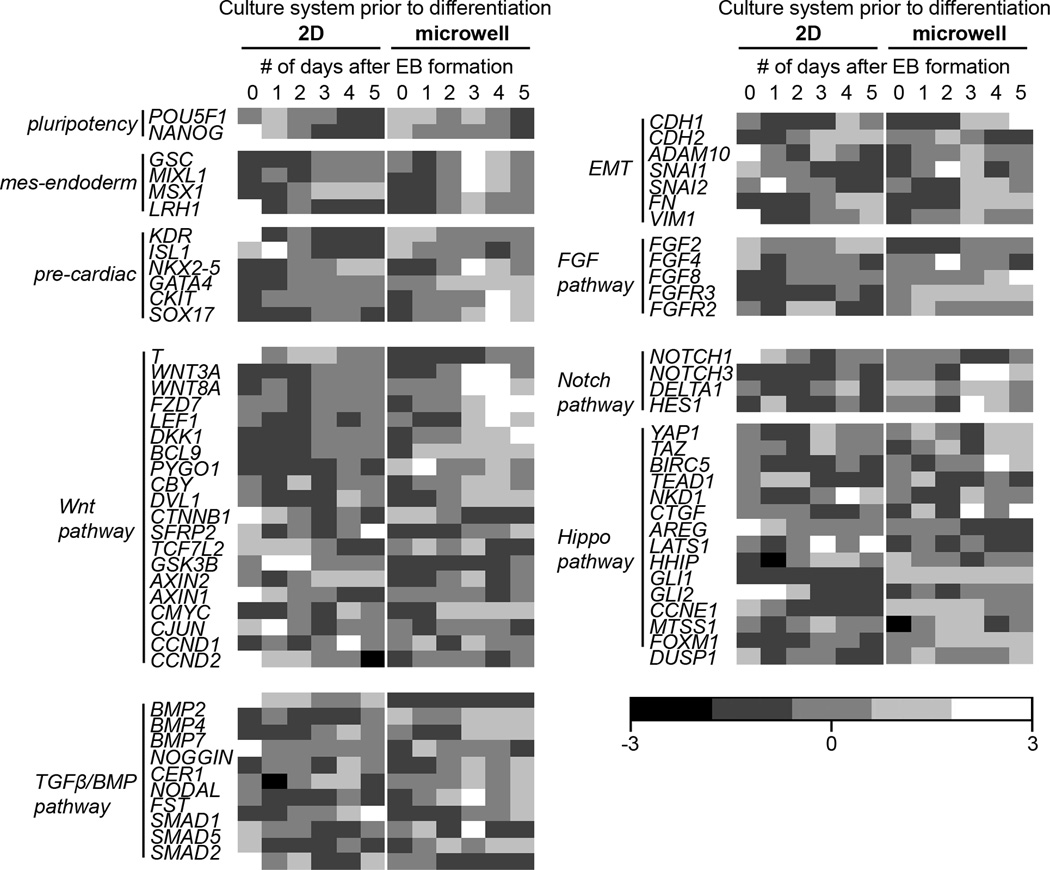
Figure 4.
Gene expression heat maps in EBs from 2D TCPS-cultured hESCs and 3D microwell-cultured hESCs. RT-PCR quantification of gene expression, relative to GAPDH and ACTB, in EBs 0–5 days after formation from hESCs cultured on 2D TCPS or in 3D microwells. RT-PCR analysis of pluripotent, mesendoderm, pre-cardiac, FGF pathway, Wnt pathway, Notch pathway, Hippo pathway and TGFβ/BMP pathway was performed. Values along each row (representing a single gene) were normalized so that the mean equals 0 and the standard deviation equals 1. Lighter shades of gray indicate higher (>0) expression and darker shades of gray indicate lower (<0) expression.
Though the enhancement in definitive cardiomyocyte specification from microwell culture was not observed until 1.5 weeks after the start of EB differentiation via functional beating or flow cytometry for cardiomyocyte markers18, the earliest markers of mesendoderm specification, an event in cardiac development, were observed as early as 3 days after EB formation. Thus, microwell culture influences the differentiation trajectory that correlate with the differences in cardiomyocyte differentiation observed at later times.
Differences in gene expression of multiple signaling pathways in EBs from microwell and 2D culture
In order to broaden our understanding of the potential differences that occur during the differentiation of EBs made from hESCs grown in 3D microwells or on 2D TCPS, expression of genes related to signaling pathways and cellular transitions involved in hESC differentiation was surveyed. These include the Wnt/β-catenin pathway, TGF-β superfamily, Notch pathway, FGF signaling and epithelial-to-mesenchymal transition (EMT). Though this analysis was not comprehensive, it illustrated major differences in signaling pathway gene expression in hESCs cultured in 2D and 3D when subjected to differentiation in EBs.
In order to identify differences in gene expression in EBs, hESCs were seeded into Matrigel-coated microwells or on 2D plates as previously described. After 6 days of expansion, hESCs were enzymatically removed and cultured in suspension to form EBs. Cells were harvested for RNA purification and RT-PCR immediately prior to suspension culture and every day for 5 days. The amplified products were run on an agarose gel and the intensities were quantified to the intensities of housekeeping genes GAPDH and ACTB. Finally, the quantified intensities of each gene were standardized in Matlab across each of the biological samples and are presented as a heatmap (Fig 4).
As expected, there was a strong upregulation of genes associated with the mesendoderm lineage and early cardiogenesis, including ISL1, GATA4 and NKX2-5, at day 3 of differentiation in EBs formed from microwell-cultured hESCs. Moreover, there was also an upregulation of the genes responsive to Wnt signaling, such as WNT3A, WNT8A and LEF1, at day 3 of differentiation in EBs formed from microwell-cultured hESCs. These two trends had very similar expression profiles. Interestingly, some target genes of Wnt signaling, including DKK1 and BCL9, were upregulated in EBs from microwell-cultured hESCs even before the third day of differentiation.
Genes associated with the Notch pathway, FGF pathway and TGF-β superfamily were also generally upregulated in EBs derived from microwell-cultured hESCs. Genes in the Notch pathway such as NOTCH1 and DELTA1 were upregulated in EBs made from microwell-cultured hESCs by day 3 of differentiation, while NOTCH3 was upregulated throughout differentiation. Similarly, FGF2 and FGF8, genes in the FGF pathway, were upregulated in EBs from microwell-cultured hESCs by the first day of differentiation. Representative genes from the TGF-β superfamily, such as BMP2, BMP7, NOGGIN and NODAL, were also upregulated in EBs from microwell-cultured hESCs when compared to those from TCPS-cultured hESCs.
Differences in EMT phenotypes and TGFβ/BMP pathway activation in hESCs in 3D and 2D culture
While a general upregulation of Wnt, FGF, TGFβ and Notch family genes was observed during EB-based differentiation of hESCs cultured in microwells, there was also modular expression of some developmentally important signaling pathway genes between undifferentiated hESCs cultured in 3D microwells or on 2D TCPS. Such differences included genes related to epithelial and mesenchymal phenotypes and members of the TGFβ/BMP signaling pathway. Thus, differences in gene expression were supplemented with analysis of protein expression and activation.
To study differences in phenotypes of undifferentiated hESCs, cells were seeded into Matrigel-coated 3D microwells or 2D TCPS substrates. After 3 or 6 days of culture in CM/F+, hESCs were harvested for PCR, flow cytometry and/or western blot analysis.
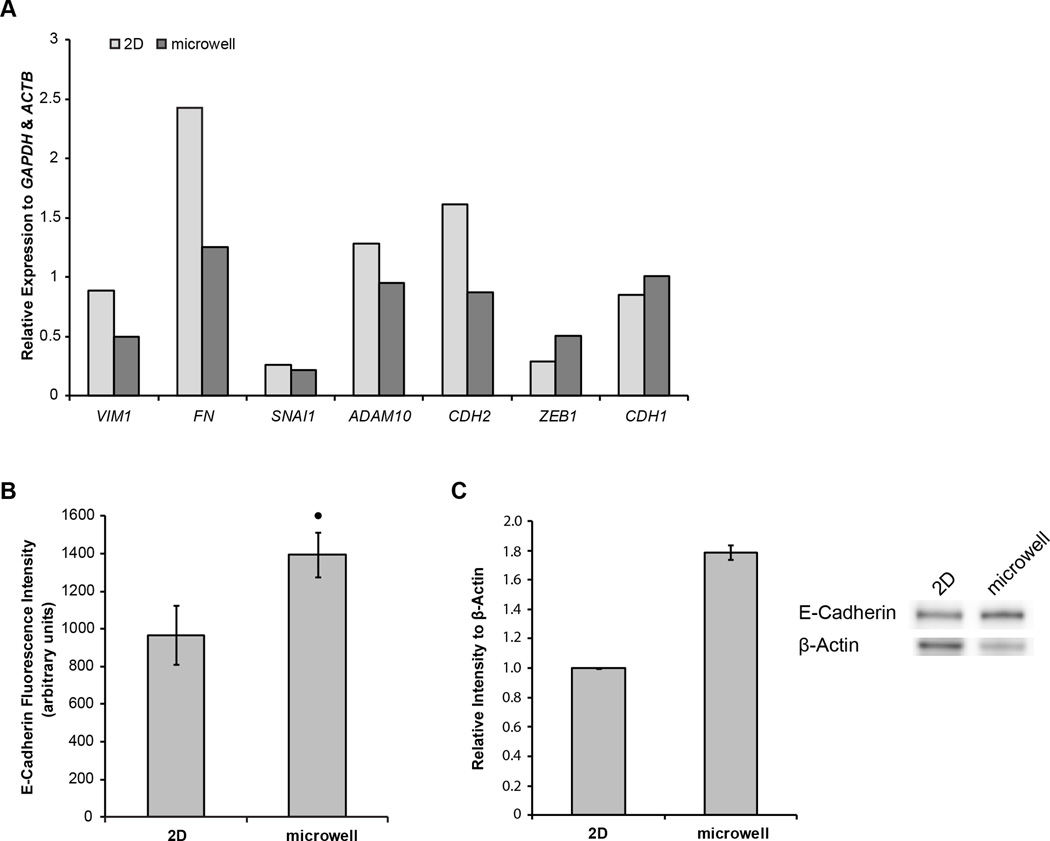
First, cells cultured in microwells exhibited reduced expression of markers associated with mesenchymal-like cells and the epithelial-mesenchymal transition (EMT), such as FN and CDH2 (Fig 5A). Conversely, a marker of epithelial-like cells, CDH1, was slightly upregulated. This trend suggests that the cells cultured in microwells exhibit a more epithelial phenotype, possibly due to increased cell-cell contact from neighboring cells in 3D culture. CDH1 encodes for E-cadherin, the protein necessary for the establishment and maintenance of cell-cell contact in adherens junctions. Thus, by measuring the amount of E-cadherin per cell, the degree of functional cell-cell contact per cell can also be determined. Therefore, the intensity of E-cadherin expression per cell was quantified using flow cytometry. The average fluorescence intensity of E-cadherin per cell was ~50% greater in cells cultured in 3D microwells than cells cultured on 2D TCPS (Fig 5B). The elevated expression of E-cadherin in hESCs cultured in microwells vs. TCPS was confirmed via western blot analysis (Fig 5C).
Figure 5.
(A) Quantification of representative EMT-related gene expression agarose gel bands, relative to GAPDH and ACTB, in undifferentiated hESCs cultured for 6 days on 2D TCPS (light) and in 3D microwells (dark) (B) Average intensity of E-cadherin fluorescence per cell, determined by flow cytometry, in undifferentiated hESCs cultured on 2D TCPS and in 3D microwells. (C) Quantification of western blot intensities of E-cadherin relative to β-actin in biological replicates of hESCs cultured for 6 days on 2D TCPS or in 3D microwells. Sample western blots representative of data are shown (• indicates p<0.05 compared to same-day 2D control)
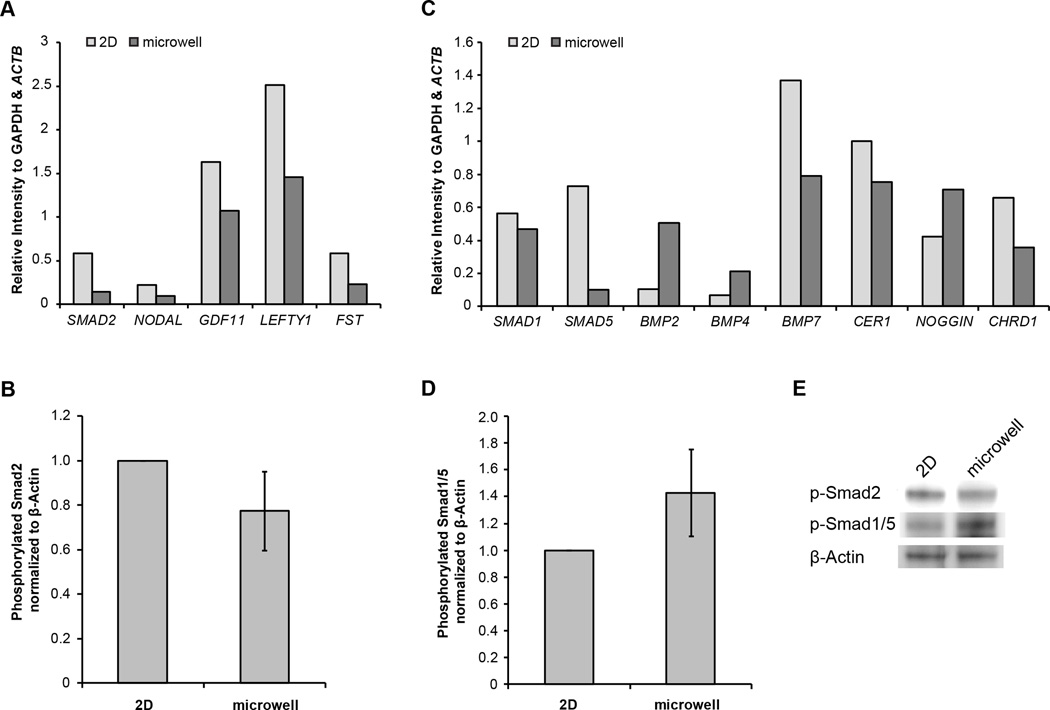
Another developmental signaling pathway regulated by microwell culture is the TGF-β superfamily. While the TGF-β superfamily interactome is extensive, most signaling channels through the Smad family of intracellular transcriptional mediators. In the Smad family, receptor-regulated Smads (R-Smads) are phosphorylated by receptor kinases that respond to specific TGF-β superfamily ligands26. For example, Smad2 and 3 are phosphorylated by TGFβ/Activin/Nodal receptors and Smad1, 5 and 8 are phosphorylated by BMP receptors. Once phosphorylated, these R-Smads complex with a single co-Smad, Smad4, and translocate into the nucleus to regulate gene expression. Thus, R-Smad phosphorylation can serve as an indicator of TGFβ/Activin/Nodal or BMP signaling.
NODAL and GDF11, genes related to TGFβ/Activin/Nodal signaling,, exhibited reduced expression in hESCs in microwells as compared hESCs on TCPS (Fig 6A). To determine whether the downregulation in expression of these genes correlated with less Smad2 signaling, the protein level of phosphorylated Smad2 was measured via western blot. The relative protein level of phosphorylated Smad2 to β-actin was found to be lower in hESCs cultured in microwells as compared to those on a 2D substrate (Fig 6B). Together, the gene expression and phosphorylated Smad2/3 levels suggest that the cells cultured in 3D microwells compared to 2D surfaces may transduce less signaling through the TGFβ/Activin/Nodal pathway.
Figure 6.
(A) Quantification of representative TGFβ/Activin/Nodal pathway gene expression agarose gel bands, normalized to GAPDH and ACTB expression in undifferentiated hESCs cultured for 6 days on 2D TCPS (light) or in 3D microwells (dark) (B) Quantification of western blot intensities of phosphorylated Smad2 normalized to β-actin in biological triplicates of protein extracts obtained from undifferentiated hESCs cultured for 6 days on 2D TCPS or in 3D microwells. Quantification of intensities was normalized to 2D samples. (C) Quantification of representative BMP pathway gene expression agarose gel bands, relative to GAPDH and ACTB, in undifferentiated hESCs cultured for 6 days on 2D TCPS (light) and in 3D microwells (dark) (D) Quantification of western blot intensities of phosphorylated Smad1/5 normalized to β-actin in biological triplicates of protein extracts obtained from undifferentiated hESCs cultured for 6 days on 2D TCPS or in 3D microwells. Quantification of intensities was normalized to 2D samples. (E) Sample western blots representative of data in panels (B) and (D).
The expression of genes associated with the BMP branch of the TGF-β superfamily did not provide clear evidence of activation or repression in 3D vs. 2D culture (Fig 6C). In hESCs cultured in microwells, there was greater expression of some BMP ligands, such as BMP2 and BMP4 and less expression of other BMP ligands, such as BMP7. The expression of the BMP ligand inhibitor NOGGIN was upregulated in microwells, but expression of the inhibitor CHRD1 was diminished. However, the cells in microwells exhibited a greater ratio of phosphorylated Smad 1/5 than those in 2D culture (Fig 6D), suggesting elevated BMP associated Smad signal transduction in 3D compared to 2D.
Discussion
In this study, we utilized a microwell system to observe how major signaling pathways in hESCs are affected by culturing within a 3D microenvironment while in the undifferentiated state and also during subsequent EB differentiation. Based upon previous work18,20, we hypothesized that the gene targets of the canonical Wnt signaling pathway would be upregulated in the EBs made from microwells. Moreover, we hypothesized that many of the markers for the earliest stages of cardiogenesis would also be upregulated in the EBs made from microwells. Not only did we see an upregulation of multiple direct gene targets of canonical Wnt signaling, we also observed a corresponding upregulation in mesendoderm genes (T, GSC, MIXL1, SOX17) and early cardiac mesoderm genes (ISL1, NKX2-5). Moreover, we observed a higher percentage of Brachyury and Sox17-expressing cells in EBs formed from microwells. Sox17, an endodermal transcription factor, has been shown to be indispensable for cardiac myogenesis27. The T gene that encodes Brachyury is a direct gene target of Wnt signaling in ESCs28. Furthermore, our group has also demonstrated that by modulating the Wnt signaling pathway alone, a high yield and purity of cardiomyocytes can be generated from ESCs29,30. Thus, information garnered from the study of the effects from 3D culture is pertinent to understanding the signaling pathways important during cardiogenesis.
Several studies have demonstrated that the proper balance and timing of TGFβ/Activin/Nodal and BMP signaling can induce cardiogenesis in hESCs in vitro31–34. Moreover, these pathways are known to regulate gastrulation, primitive streak formation and mesendoderm specification35. Accordingly, our gene expression data demonstrate that TGFβ signaling pathways are influenced by 3D culture prior the start of differentiation and upregulated during EB differentiation of microwell-cultured hESCs. Activin/Nodal and BMP signaling is induced by activating Wnt pathway via GSK3β-inhibitors during directed cardiomyocyte differentiation29. However, during EB based differentiation from microwell-cultured hESCs, it is unclear how the interplay and crosstalk between these developmental signaling pathways operates. Also of note, immediately after differentiation began, we noticed an upregulation in FGF-related genes such as FGF4 and FGF8. FGF4 synergizes with BMP signaling to induce cardiogenesis36 and both FGF4 and 8 are known to be essential during primitive streak establishment and mesoderm formation37.
Interestingly, genes associated with the Notch pathway were upregulated in EBs made from microwell-cultured hESCs compared to TCPS-cultured hESCs. In prior ESC differentiation studies, activation of the Notch pathway during early differentiation was associated with neural commitment38,39 and is not believed to directly specify cardiac mesoderm during early stages of mesoderm formation in the primitive streak40. However, the context of Notch signaling may direct hESCs to alternative fates, such as early endocardium specification, which then provides coordinating cues to the developing cardiac tissue via lateral inhibition41. Alternatively, microwell culture may encourage differentiation down multiple lineages or promote the activation of juxtacrine signaling due to increased cell-cell contact. However, the heterogeneity of EB-based differentiation makes studying which signaling pathways are most important in directing any specific cell fate difficult. Batch-to-batch variability exists among experiments and the composition of individual EBs within any single experiment is variable. The population-level overview of gene expression presented here highlights the fact that there are substantial differences in the signaling and differentiation trajectories between EBs made from hESCs cultured in 3D microwells and on 2D TCPS. The clearest difference was that genes associated with primitive streak, mesendoderm and cardiac differentiation were upregulated in microwell-cultured hESCs and the resulting EBs.
These differences between EBs generated from 2D substrates and 3D microwells were likely influenced by the effects of 3D culture and also the tighter control over EB size distribution that microwells confer. A previous study demonstrated that that the distribution of EB diameters generated from microwells could be described by Gaussian distributions tightly centered at the dimensions of the microwell confinement18. In contrast, since hESC colony size was not constrained in 2D culture, EBs derived from hESCs cultured on 2D substrates were more broadly size distributed and not well described by a simple Gaussian distribution. Studies utilizing both mouse and human ESCs that were aggregated into EBs within different sized microwells have also indicated that EBs formed from microwell culture can affect lineage progression. Park et al. reported higher expression of ectoderm-associated genes and proteins in EBs made in smaller microwells (100 µm), while the EBs from larger microwells favored mesoderm and endoderm gene expression (500 µm)42. Nguyen et al. observed that EBs made from microwells around 100 µm preferentially upregulated endoderm and ectoderm markers when compared to EBs from smaller microwells43. Bauwens et al. controlled EB size by changing hESC colony size in 2D using circular extracellular matrix islands of varying diameters44. Their results indicated that aggregate size alone can play an important role in differentiation trajectories. Our recent study and others have also demonstrated that controlling EB size by changing 3D microwell confinement dimensions influences cardiac differentiation18,22. However, the entire influence of 3D culture cannot be restricted to the effects of EB size distribution control. Mohr et al. generated EBs from 3D microwells of dimensions ranging from 100 to 500 µm, which spans the range of EB sizes generated from unconstrained 2D culture18. In terms of cardiomyocyte differentiation efficiency quantified via flow cytometry and PCR for cardiomyocyte markers and % beating EBs for functioning cardiomyocytes, EBs generated from all microwell sizes outperformed those from 2D culture. These data suggest that the size differences in EBs generated from 2D substrates and 3D microwell culture cannot entirely account for the observed differences in cell signaling and differentiation fates.
In addition to observing substantial differences in gene expression during EB differentiation, we detected multiple molecular differences in undifferentiated hESCs cultured in 3D and 2D platforms. Culturing hESCs in a 3D microenvironment increased functional cell-cell contact and modulated developmental signaling pathways including TGFβ/Activin/Nodal and BMP signaling, and Wnt/β-catenin signaling. We observed that hESCs cultured in microwells, as compared to 2D, exhibited a lower ratio of phosphorylated Smad2 and a higher ratio of phosphorylated Smad1/5. This suggests less Smad signal transduction through the TGFβ/Activin/Nodal branch and more through the BMP branch. In hESCs, TGFβ/Activin/Nodal regulates pluripotency while BMP signaling induces differentiation46,47. While this may suggest that hESCs cultured in microwells may be more primed for differentiation than those on 2D, the percentage of Oct4 positive cells remained the same, as indicated in Fig 3B. Importantly, the BMP antagonist NOGGIN was more highly expressed in microwell-cultured hESCs. Noggin cooperates with Fgf2 to maintain pluripotency by inhibiting BMP signaling in hESCs48. In addition, we demonstrated that the amount of E-cadherin per cell was significantly higher in hESCs from microwells than from 2D. This trend was further expounded upon in a study by Azarin et al20, which demonstrated that the E-cadherin surface density was 5-fold higher in cells cultured in microwells. Moreover, Azarin et al demonstrated that changing the dimensions of the 3D microwells can modulate E-cadherin expression, further suggesting a link between the 3D culture conditions and molecular changes like cell-cell contact. In a separate study, Nakazawa et al. also linked the expression level of E-cadherin in mouse ESCs to microwell dimension49. E-cadherin has been shown to function as both a regulator of pluripotent signaling pathways as well as improve hESC derivation50,51. Thus, the changes in other major signaling pathways, such as TGFβ/BMP and Wnt, may at least in part result from the increased cell-cell contact in the 3D microwell culture to change the hESC cellular context. This may serve as a means to bias the cells towards the mesendoderm lineages during differentiation. Nonetheless, it is intriguing that by simply culturing hESCs in a 3D microenvironment without additional changes to media or ECM, multiple pathways can be affected.
Furthermore, it is still unclear by what specific mechanisms these differences arise. The 3D microenvironment can influence not only cell-cell contact but also cell-ECM contact, mechanical interactions between cells, cytoskeletal tension, and cell morphology. Due to the many changes a cell can experience when cultured in 3D, it is reasonable that multiple mechanisms synergize to produce the range of molecular and signaling differences observed. However, it is also possible that one primary pathway is responsible for regulating the molecular differences observed during 3D culture and directly activates other pathways.
The collective data from this study illustrate that 2D and 3D culture of hESC leads to distinct developmental signaling early in differentiation and may potentially provide molecular targets for studying pathways by which the 3D microenvironment tranduces signals that prime hESCs for differentiation and influence desired differentiation outcomes. Not only may these molecular targets be exploited to engineer cell fate decisions in 2D differentiation processes, understanding how microenvironmental contributions from the intercellular and autoregulatory signaling that occur in 3D culture of hESCs and their derivatives can aid in the transition to 3D downstream applications, such as tissue engineering and regenerative medicine.
Supplementary Material
Supplementary Table 1. Numerical values from which the Figure 4 heat maps were generated. RT-PCR quantification of gene expression, relative to GAPDH and ACTB expression, in EBs 0–5 days after formation from hESCs cultured on 2D TCPS or in 3D microwells. RT-PCR analysis of pluripotent, mesendoderm, pre-cardiac, FGF pathway, Wnt pathway, Notch pathway, Hippo pathway and TGFβ/BMP pathway gene expression was performed. Values along each row (representing a single gene) were normalized so that the mean equals 0 and the standard deviation equals 1.
Acknowledgements
This work was supported by NIH grant R01 EB007534 and NSF grant EFRI 0735903. The authors would like to thank Amritava Das in assistance with Matlab and the WiCell Research Institute for providing cells and reagents. We also thank Prof. Bird for his inspiration and wish him a happy 90th birthday.
References
- 1.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19(3):193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 3.Greenow K, Clarke AR. Controlling the stem cell compartment and regeneration in vivo: the role of pluripotency pathways. Physiol Rev. 2012;92(1):75–99. doi: 10.1152/physrev.00040.2010. [DOI] [PubMed] [Google Scholar]
- 4.Pampaloni F, Reynaud EG, Stelzer EHK. The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol. 2007;8(10):839–845. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 5.Kraehenbuehl TP, Langer R, Ferreira LS. Three-dimensional biomaterials for the study of human pluripotent stem cells. Nat Methods. 2011;8(9):731–736. doi: 10.1038/nmeth.1671. [DOI] [PubMed] [Google Scholar]
- 6.Burdick JA, Vunjak-Novakovic G. Engineered microenvironments for controlled stem cell differentiation. Tissue Eng Part A. 2009;15(2):205–219. doi: 10.1089/ten.tea.2008.0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bratt-Leal AM, Carpenedo RL, McDevitt TC. Engineering the embryoid body microenvironment to direct embryonic stem cell differentiation. Biotechnol Prog. 2009;25(1):43–51. doi: 10.1002/btpr.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keung AJ, Kumar S, Schaffer DV. Presentation counts: microenvironmental regulation of stem cells by biophysical and material cues. Annu Rev Cell Dev Biol. 2010;26:533–556. doi: 10.1146/annurev-cellbio-100109-104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keung AJ, Healy KE, Kumar S, Schaffer DV. Biophysics and dynamics of natural and engineered stem cell microenvironments. Wiley Interdiscip Rev Syst Biol Med. 2010;2(1):49–64. doi: 10.1002/wsbm.46. [DOI] [PubMed] [Google Scholar]
- 10.Peerani R, Zandstra PW. Enabling stem cell therapies through synthetic stem cell-niche engineering. J Clin Invest. 2010;120(1):60–70. doi: 10.1172/JCI41158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsiao C, Palecek SP. Microwell Regulation of Pluripotent Stem Cell Self-Renewal and Differentiation. Bionanoscience. 2012;2(4):266–276. doi: 10.1007/s12668-012-0050-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khademhosseini A, Ferreira L, Blumling J, Yeh J, Karp JM, Fukuda J, Langer R. Co-culture of human embryonic stem cells with murine embryonic fibroblasts on microwell-patterned substrates. Biomaterials. 2006;27(36):5968–5977. doi: 10.1016/j.biomaterials.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 13.Mohr JC, de Pablo JJ, Palecek SP. 3-D microwell culture of human embryonic stem cells. Biomaterials. 2006;27(36):6032–6042. doi: 10.1016/j.biomaterials.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 14.Sakai Y, Yoshida S, Yoshiura Y, Mori R, Tamura T, Yahiro K, Mori H, Kanemura Y, Yamasaki M, Nakazawa K. Effect of microwell chip structure on cell microsphere production of various animal cells. J Biosci Bioeng. 2010;110(2):223–229. doi: 10.1016/j.jbiosc.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 15.Hong S-H, Werbowetski-Ogilvie T, Ramos-Mejia V, Lee JB, Bhatia M. Multiparameter comparisons of embryoid body differentiation toward human stem cell applications. Stem Cell Res. 2010;5(2):120–130. doi: 10.1016/j.scr.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Ng ES, Davis RP, Azzola L, Stanley EG, Elefanty AG. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106(5):1601–1603. doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- 17.Burridge PW, Anderson D, Priddle H, Barbadillo Muñoz MD, Chamberlain S, Allegrucci C, Young LE, Denning C. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25(4):929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- 18.Mohr JC, Zhang J, Azarin SM, Soerens AG, de Pablo JJ, Thomson Ja, Lyons GE, Palecek SP, Kamp TJ. The microwell control of embryoid body size in order to regulate cardiac differentiation of human embryonic stem cells. Biomaterials. 2010;31(7):1885–1893. doi: 10.1016/j.biomaterials.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burridge PW, Thompson S, Millrod Ma, Weinberg S, Yuan X, Peters A, Mahairaki V, Koliatsos VE, Tung L, Zambidis ET. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6(4):e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Azarin SM, Lian X, Larson Ea, Popelka HM, de Pablo JJ, Palecek SP. Modulation of Wnt/β-catenin signaling in human embryonic stem cells using a 3-D microwell array. Biomaterials. 2012;33(7):2041–2049. doi: 10.1016/j.biomaterials.2011.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang Y-S, Chung BG, Ortmann D, Hattori N, Moeller H-C, Khademhosseini A. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11. Proc Natl Acad Sci U S A. 2009;106(40):16978–16983. doi: 10.1073/pnas.0905550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauwens CL, Song H, Thavandiran N, Ungrin M, Massé S, Nanthakumar K, Seguin C, Zandstra PW. Geometric control of cardiomyogenic induction in human pluripotent stem cells. Tissue Eng Part A. 2011;17(15–16):1901–1909. doi: 10.1089/ten.TEA.2010.0563. [DOI] [PubMed] [Google Scholar]
- 23.Kimelman D, Griffin KJ. Mesoderm induction: a postmodern view. Cell. 1998;94(4):419–421. doi: 10.1016/s0092-8674(00)81582-2. [DOI] [PubMed] [Google Scholar]
- 24.Tada S, Era T, Furusawa C, Sakurai H, Nishikawa S, Kinoshita M, Nakao K, Chiba T, Nishikawa S-I. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132(19):4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- 25.Pereira La, Wong MS, Mei Lim S, Stanley EG, Elefanty AG. The Mix family of homeobox genes--key regulators of mesendoderm formation during vertebrate development. Dev Biol. 2012;367(2):163–177. doi: 10.1016/j.ydbio.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 26.Heldin C-H, Landström M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176. doi: 10.1016/j.ceb.2009.01.021. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Asakura M, Inoue H, Nakamura T, Sano M, Niu Z, Chen M, Schwartz RJ, Schneider MD. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104(10):3859–3864. doi: 10.1073/pnas.0609100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnold SJ, Stappert J, Bauer a, Kispert a, Herrmann BG, Kemler R. Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech Dev. 2000;91(1–2):249–258. doi: 10.1016/s0925-4773(99)00309-3. [DOI] [PubMed] [Google Scholar]
- 29.Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, Palecek SP. PNAS Plus: Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci. 2012:1–10. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013;8(1):162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laflamme Ma, Chen KY, Naumova AV, Muskheli V, Fugate Ja, Dupras SK, Reinecke H, Xu C, Hassanipour M, Police S, O’Sullivan C, Collins L, Chen Y, Minami E, Gill Ea, Ueno S, Yuan C, Gold J, Murry CE. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25(9):1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 32.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, Henckaerts E, Bonham K, Abbott GW, Linden RM, Field LJ, Keller GM. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453(7194):524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 33.Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Zhang J, Klos M, Wilson GF, Herman AM, Lian X, Raval KK, Barron MR, Hou L, Soerens AG, Yu J, Palecek SP, Lyons GE, Thomson Ja, Herron TJ, Jalife J, Kamp TJ. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ Res. 2012;111(9):1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beyer TA, Narimatsu M, Weiss A, David L, Wrana JL. The TGFβ superfamily in stem cell biology and early mammalian embryonic development. Biochim Biophys Acta. 2013;1830(2):2268–2279. doi: 10.1016/j.bbagen.2012.08.025. [DOI] [PubMed] [Google Scholar]
- 36.Lough J, Barron M, Brogley M, Sugi Y, Bolender DL, Zhu X. Combined BMP-2 and FGF-4, but neither factor alone, induces cardiogenesis in non-precardiac embryonic mesoderm. Dev Biol. 1996;178(1):198–202. doi: 10.1006/dbio.1996.0211. [DOI] [PubMed] [Google Scholar]
- 37.Böttcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26(1):63–77. doi: 10.1210/er.2003-0040. [DOI] [PubMed] [Google Scholar]
- 38.Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4(5):e121. doi: 10.1371/journal.pbio.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nemir M, Croquelois A, Pedrazzini T, Radtke F. Induction of cardiogenesis in embryonic stem cells via downregulation of Notch1 signaling. Circ Res. 2006;98(12):1471–1478. doi: 10.1161/01.RES.0000226497.52052.2a. [DOI] [PubMed] [Google Scholar]
- 40.Niessen K, Karsan A. Notch signaling in cardiac development. Circ Res. 2008;102(10):1169–1181. doi: 10.1161/CIRCRESAHA.108.174318. [DOI] [PubMed] [Google Scholar]
- 41.De la Pompa JL, Epstein Ja. Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev Cell. 2012;22(2):244–254. doi: 10.1016/j.devcel.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park J, Cho CH, Parashurama N, Li Y, Berthiaume F, Toner M, Tilles AW, Yarmush ML. Microfabrication-based modulation of embryonic stem cell differentiation. Lab Chip. 2007;7(8):1018–1028. doi: 10.1039/b704739h. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen D, Sa S, Pegan JD, Rich B, Xiang G, McCloskey KE, Manilay JO, Khine M. Tunable shrink-induced honeycomb microwell arrays for uniform embryoid bodies. Lab Chip. 2009;9(23):3338–3344. doi: 10.1039/b914091c. [DOI] [PubMed] [Google Scholar]
- 44.Bauwens CL, Peerani R, Niebruegge S, Woodhouse Ka, Kumacheva E, Husain M, Zandstra PW. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26(9):2300–2310. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 45.Massagué J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103(2):295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 46.Dalton S. Signaling networks in human pluripotent stem cells. Curr Opin Cell Biol. 2013;25(2):241–246. doi: 10.1016/j.ceb.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li W, Ding S. Human pluripotent stem cells: decoding the naïve state. Sci Transl Med. 2011;3(76):76ps10. doi: 10.1126/scitranslmed.3000996. [DOI] [PubMed] [Google Scholar]
- 48.Wang G, Zhang H, Zhao Y, Li J, Cai J, Wang P, Meng S, Feng J, Miao C, Ding M, Li D, Deng H. Noggin and bFGF cooperate to maintain the pluripotency of human embryonic stem cells in the absence of feeder layers. Biochem Biophys Res Commun. 2005;330(3):934–942. doi: 10.1016/j.bbrc.2005.03.058. [DOI] [PubMed] [Google Scholar]
- 49.Nakazawa K, Yoshiura Y, Koga H, Sakai Y. Characterization of mouse embryoid bodies cultured on microwell chips with different well sizes. J Biosci Bioeng. 2013;xx(xx):1–6. doi: 10.1016/j.jbiosc.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 50.Soncin F, Ward CM. The Function of E-Cadherin in Stem Cell Pluripotency and Self-Renewal. Genes (Basel) 2011;2(1):229–259. doi: 10.3390/genes2010229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redmer T, Diecke S, Grigoryan T, Quiroga-Negreira A, Birchmeier W, Besser D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12(7):720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Numerical values from which the Figure 4 heat maps were generated. RT-PCR quantification of gene expression, relative to GAPDH and ACTB expression, in EBs 0–5 days after formation from hESCs cultured on 2D TCPS or in 3D microwells. RT-PCR analysis of pluripotent, mesendoderm, pre-cardiac, FGF pathway, Wnt pathway, Notch pathway, Hippo pathway and TGFβ/BMP pathway gene expression was performed. Values along each row (representing a single gene) were normalized so that the mean equals 0 and the standard deviation equals 1.