Abstract
Pmel17 is an important protein for pigmentation in human skin and eyes. Proteolytic fragments from Pmel17 form fibrils upon which melanin is deposited in melanosomes. The repeat domain (RPT) derived from Pmel17 only forms fibrils under acidic melanosomal conditions. Here, we examined the effects of lipids on RPT aggregation to explore whether intramelanosomal vesicles can facilitate fibrillogenesis. Using transmission electron microscopy, circular dichroism, and fluorescence spectroscopy, fibril formation was monitored at the ultrastructural, secondary conformational and local level, respectively. Phospholipid vesicles and lysophospholipid (lysolipid) micelles were employed as membrane mimics. The surfactant-like lysolipids are particularly pertinent due to their high content in melanosomal membranes. Interestingly, RPT aggregation kinetics were influenced only by lysolipid-containing phospholipid vesicles. While both vesicles containing either anionic lysophosphatidylglycerol (LPG) or zwitterionic lysophosphatidylcholine (LPC) stimulate aggregation, LPG exerted a greater effect on reducing the apparent nucleation time. A detailed comparison showed distinct behaviors of LPG versus LPC monomers and micelles plausibly originating from their headgroup hydrogen bonding capabilities. Acceleration and retardation of aggregation were observed for LPG monomers and micelles, respectively. Because a specific interaction between LPG and RPT was identified by intrinsic W423 fluorescence and induced α-helical structure, it is inferred that binding of LPG near the C-terminal amyloid core initiates intermolecular association, whereas stabilization of α-helical conformation inhibits β-sheet formation. Contrastingly, LPC promotes RPT aggregation at both submicellar and micellar concentrations via non-specific binding with undetectable secondary structural change. Our findings suggest that protein-lysolipid interactions within melanosomes may regulate amyloid formation in vivo.
Keywords: melanosome, protein-lipid interaction, tryptophan, TEM, lysolipids
INTRODUCTION
Amyloids are insoluble fibrous protein aggregates with characteristic cross β-sheet structures, widely known for their involvement in neurodegenerative disorders such as Parkinson’s, Alzheimer’s, and prion diseases [1–3]. However, discovery of functional (beneficial) amyloids from bacteria to humans [4–9] implores us to question the molecular basis for distinct cellular outcomes. Factors that initiate amyloid formation as well as those that alter mechanisms of fibril assembly are the topic of intense research; of particular interest is what species are cytotoxic and how they are formed.
Mounting evidence connects amyloid formation and lipid membrane binding propensities to disease etiology. Membranes have been shown to modulate both protein structure and aggregation kinetics of pathological amyloidogenic polypeptides such as α-synuclein (α-syn) (Parkinson’s), amyloid-β (Aβ) peptides (Alzheimer’s) and amylin (type-II diabetes) [10–16]. To begin to elucidate distinguishing features of a functional amyloid, it is pertinent to study how functional amyloid formation is influenced by a membranous environment. Here, we report on the effects of membrane lipids on aggregation kinetics and fibrillar structures of the repeat domain (RPT) of the human functional amyloid, Pmel17 [17].
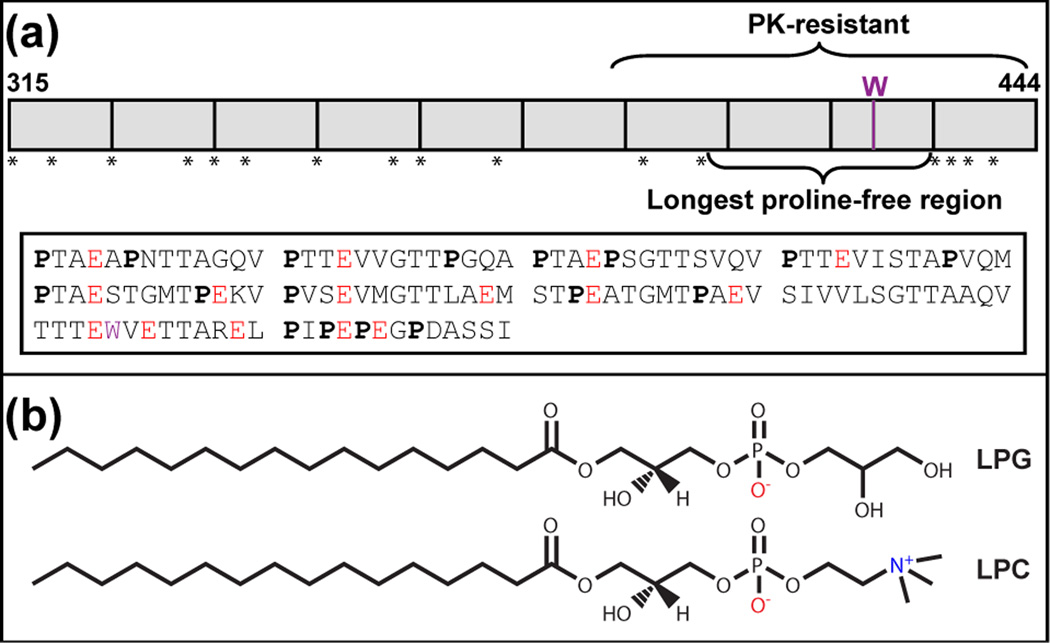
Fibrils composed of proteolytically processed Pmel17 domains including RPT (residues 315–444) [18] serve as structural scaffolds for melanin deposition in melanosomes, acidic organelles found in the skin and eyes [19]. In addition to playing a structural role, fibrils are proposed to sequester toxic intermediates formed during melanin biosynthesis [20, 21]. Failed or improper fibril formation can cause hypopigmentation in animals [22]. RPT is a critical component of melanosome fibrils [18]. Composed of 10 imperfect amino acid repeats rich in proline, serine, threonine, and glutamic acid residues, RPT contains only one native tryptophan, W423 (Fig. 1a). Both data from the limited proteinase K digestion of RPT fibrils [23] and solution nuclear magnetic resonance (NMR) spectroscopy [24] indicate that the amyloidogenic core resides in the C-terminus. This region also contains the longest proline-free stretch (403– 431, Fig. 1a).
Fig. 1.
(a) Top: Schematic drawing of the repeat domain (RPT) of human Pmel17. RPT (residues 315–444) is composed of 10 imperfect repeats (shown as boxes) rich in proline, indicated by stars. The only tryptophan (W) is denoted in purple. The proposed amyloid core (proteinase K (PK)-resistant region) and the longest proline-free stretch are as indicated. Bottom: Amino acid sequence of RPT. Prolines (P) are in bold, glutamic acids (E) are in red, and tryptophan (W) is in purple. (b) Chemical structures of lysophosphatidylglycerol (LPG), and lysophosphatidylcholine (LPC).
We have previously determined a critical pH regime for RPT fibril formation that closely matches the mildly acidic pH (5 ± 0.5) of early melanosomes [25, 26] with rapid aggregation (~minutes) observed at pH 4, the pH of stage I melanosomes [27]. Remarkably, fibril morphology is highly pH responsive and can be transformed in situ as the pH is varied with fibril dissolution occurring at pH 6 and beyond [24]. Most recently, our mutational study shows that a single C-terminal glutamic acid, E422, is predominantly responsible for this pH behavior; neutralization of the negative charge at E422 accelerates fibril formation and increases fibril stability by a full pH unit [23]. This highly reversible aggregation/disaggregation process under physiological pH is a unique property of RPT and we speculate that this is a potential way for keeping these amyloids benign if they were to escape melanosomes into the cytosol [27]. Moreover, it may be plausible to recycle amyloid fibrils via this mechanism. Notably, it also has been suggested that the storage of peptide hormones in pituitary secretory granules [28] and the formation of RNA granules [29] involve reversible polymerization of amyloid-like structures.
Studies have shown that Pmel17 trafficking and its subsequent proteolytic processing in melanosomes is important for melanin formation [30]; however, the precise series of events and players involved in initiating and propagating fibril formation remain to be defined. Electron tomographic analyses of early stage melanosomes suggest that fibril formation is initiated on intralumenal membrane vesicles (ILVs) [31]. Moreover, Marks and coworkers speculate that the RPT domain may interact with ILVs and facilitate amyloid formation [32]. In this study, we sought to determine the effects of membrane lipids on RPT fibril formation, including vesicles and micelles formed from phospholipids and lysophospholipids (lysolipids), respectively. Lysolipids are particularly interesting due to their high content in melanosomal membranes (> 10% [33, 34], see Fig. 1b for chemical structures) as compared to plasma membranes (< 2% [35]). With a combination of biophysical techniques, including circular dichroism (CD) and tryptophan fluorescence spectroscopy, dynamic light scattering (DLS) as well as transmission electron microscopy (TEM), mechanistic insights were gained for the modulation of RPT amyloid formation by two specific lysolipids, negatively-charged lysophosphatidylglycerol (LPG) and zwitterionic lysophosphatidylcholine (LPC).
RESULTS AND DISCUSSION
Effect of Lysolipid-containing Phospholipid Vesicles on RPT Aggregation
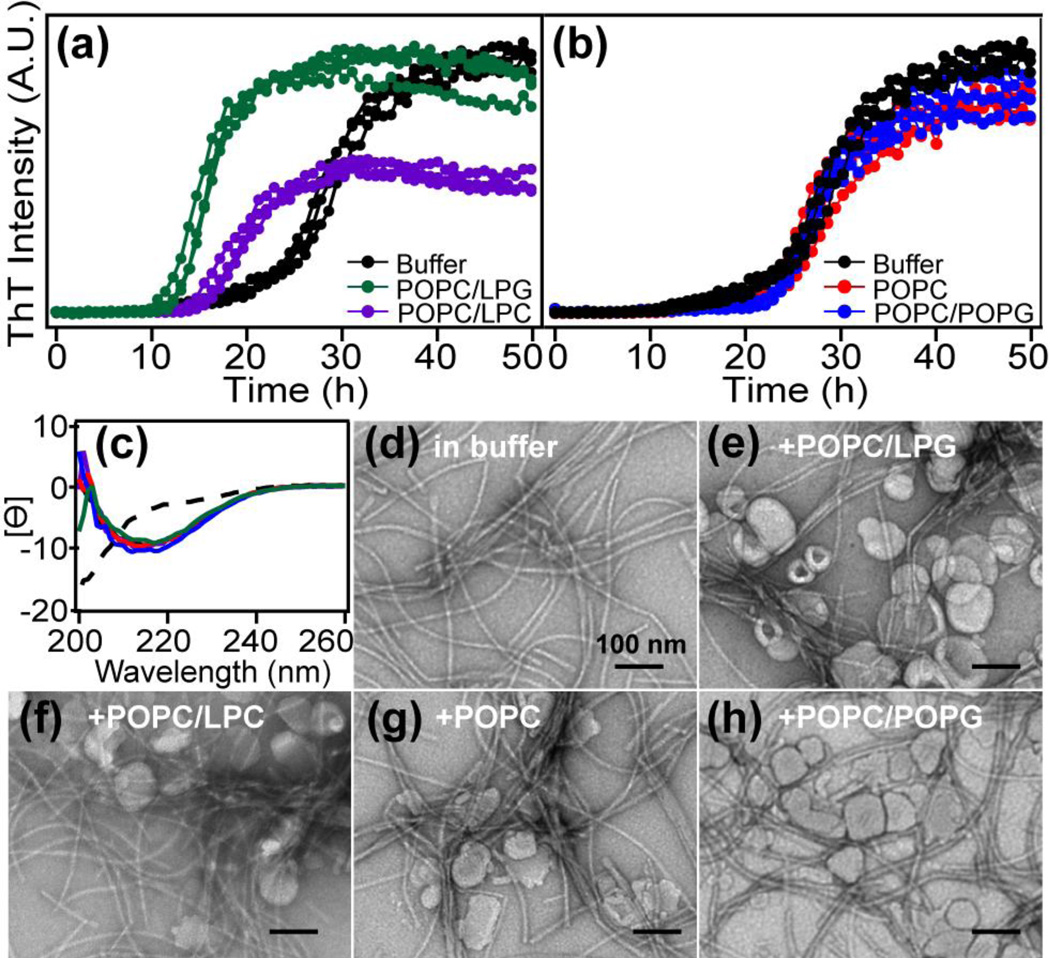
RPT forms fibrils in vitro under mildly acidic conditions (pH 4.5–5.5) [27]. To determine the effects of lipid membranes on RPT fibril formation, aggregation kinetics at pH 5 were monitored by thioflavin T (ThT) fluorescence in the absence and presence of small unilamellar vesicles (SUVs, average diameter ~80 nm). Vesicles were composed of phosphatidylcholine (POPC, 90% molar ratio), the most abundant phospholipid in biological membranes, and different lysolipids (10% molar ratio: LPG and LPC Fig. 2a). In buffer, RPT exhibits sigmoidal aggregation kinetics typical for amyloid formation with distinct lag, growth, and stationary phases. Upon adding lysolipid-containing vesicles (1.5 mM, lipid-to-protein molar ratio (L/P = 50)), faster RPT aggregation kinetics are observed with LPG exerting the greatest influence compared to LPC (Fig. 2a). With LPG, the time it takes to reach half maximal signal (t½) decreases from 28 to 15 h. In all cases, lag phases are hastened (LPG < LPC) with comparable growth rates. Interestingly, vesicles composed of only phospholipids (either POPC or POPC/POPG) have little to no effect on RPT aggregation (Fig. 2b), suggesting that the observed stimulated aggregation is lysolipid-specific and not solely a factor of headgroup chemical composition. Using CD spectroscopy, all samples exhibit negative maxima at ~218 nm characteristic of β-sheets after aggregation (Fig. 2c). TEM images of resulting RPT filaments show no morphological differences between amyloids formed under various lipid conditions (Fig. 2d–h).
Fig. 2.
Effects of lipid composition on RPT amyloid formation. (a) RPT aggregation kinetics monitored by ThT in the absence and presence of POPC vesicles containing 10% (molar ratio) lysolipids (n=3). (b) RPT aggregation kinetics monitored by ThT in the absence and presence of phospholipid vesicles. Compositions are as shown. POPC/POPG vesicles contain 10% (molar ratio) POPG (n=3). (c) CD spectra of RPT at the end of aggregation experiments. Mean residue ellipticity ([Ɵ] (deg cm2 dmol−1) × 10−3) is reported with lipid compositions colored as in (a) and (b). The dashed line is spectrum for monomeric RPT before aggregation. (d–h) Representative TEM images at the end of aggregation experiments are shown with compositions indicated. Scale bar is 100 nm.
Effects of Lysolipid Monomers and Micelles on RPT Fibril Formation
Because the presence of lysolipids in vesicles exerted strong influence on RPT aggregation kinetics, we tested whether lysolipids alone could stimulate fibril assembly. Specifically, we compared LPG versus LPC to test the effect of headgroup (Fig. 1b). Unlike phospholipids which contain two chains and form vesicles (bilayers) in solution, lysolipids have only one acyl chain and are more structurally related to surfactants/detergents. Like detergents, lysolipids form micelles when their concentration is above the critical micelle concentration (CMC).
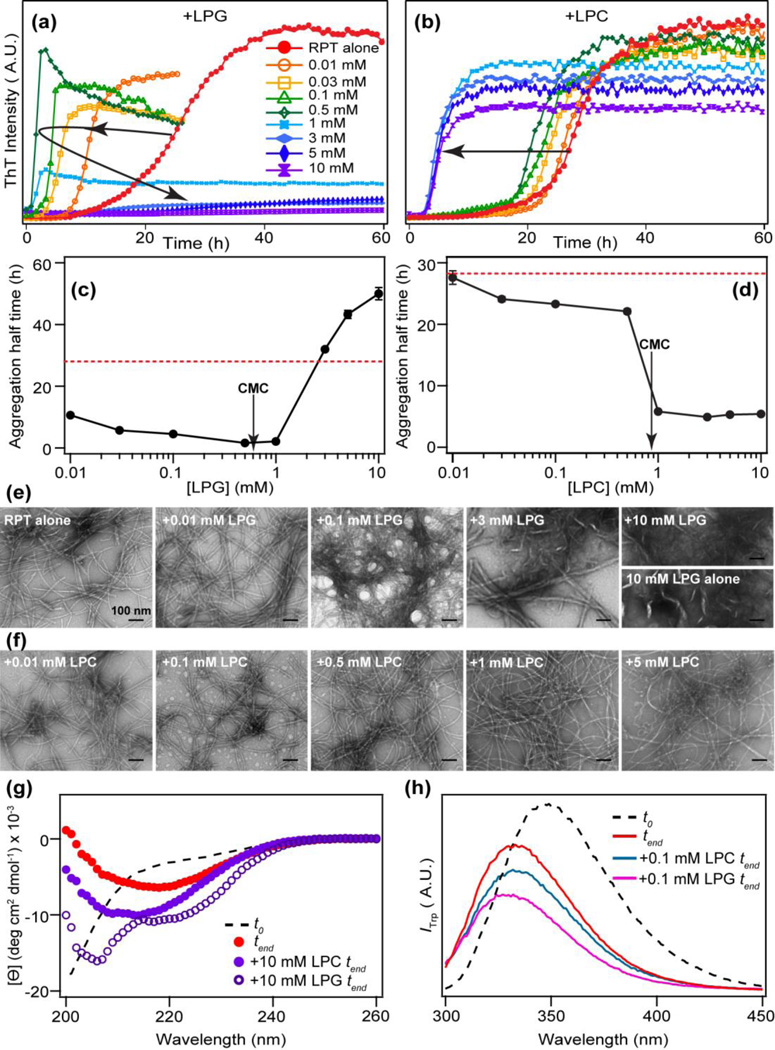
We monitored the effects of both monomeric and micellar forms of lysolipids on RPT aggregation kinetics by ThT fluorescence (Fig. 3a–b). Values of t½ are plotted as a function of lipid concentration (Fig. 3c–d). The presence of LPG monomers (<1 mM) dramatically shortens the lag time and enhances the growth rate compared to that of RPT alone in buffer; t½ changes from ~28 to 3.5 h with the greatest change occurring at a LPG-to-protein ratio ~15–30 (Fig. 3c). Contrastingly, LPG micelles (CMC ~0.6 mM [36, 37]) appear to slow down RPT fibril formation as indicated by the negligible ThT response (Fig. 3a). For LPC, RPT aggregation is stimulated at both submicellar and micellar concentrations with shorter lag times exhibited by micelles (Fig. 3d). ThT intensities at the stationary phase seem inversely correlated with the concentration of LPC micelles.
Fig. 3.
Effects of lysolipids on RPT aggregation kinetics and fibril morphology. RPT aggregation kinetics were monitored by ThT in the presence of different concentrations of LPG (a) and LPC (b). Each reported kinetic curve is an average of at least 3 wells from one plate. Lysolipid concentrations are as indicated. Aggregation half time (t1/2) vs. lipid concentration is shown in (c) for LPG and (d) for LPC, respectively. For reference, t1/2 for RPT alone in buffer is also shown as a red dashed line. Critical micelle concentrations (CMC) are indicated with arrows. Errors bars are standard deviations (n ≥ 2). Representative TEM images are shown for RPT aggregated in the presence of LPG (e) and LPC (f). Lipid concentration of each sample is shown. The image of 10 mM LPG alone is shown for comparison. Scale bar is 100 nm. (g) CD spectra of RPT in the presence of 10 mM LPC and LPG compared to RPT alone at the end of aggregation experiments (tend). (h) W423 fluorescence in the presence of 0.1 mM LPG and LPC at the end of aggregation experiments (tend). Black dashed line in (g) and (h) is RPT in solution before aggregation (t0).
Morphology of RPT fibrils were visualized with TEM (Fig. 3e–f). Long, unbranched filaments are observed for RPT alone and in the presence of LPG and LPC monomers (Fig. 3e–f), whereas filaments formed in the presence of increasing amounts of LPG monomers seem to have more clustered appearance. Fewer filaments are found in the presence of LPG micelles, consistent with the minimal ThT response. Fibrils formed in the presence of LPC micelles show consistently thinner diameter (~ 7 nm) compared to those formed in buffer or with LPG (~11 nm) (Fig. 3e).
Characterization of RPT Fibrils Formed in the Presence of Lysolipids
RPT monomers in solution exhibit characteristic CD spectral feature for unfolded proteins with a negative maximum at ~198 nm (Fig. 3g, dashed line) which is in agreement with disorder propensity prediction program such as VL3H [38] that above 80% of the sequence is unstructured. Upon aggregation, RPT fibrils show a negative maximum around 218 nm, typical of β-sheets (Fig. 3g, red line). In the presence of high concentration of lysolipids (10 mM), the secondary structural content of RPT aggregates appears to be somewhat altered. RPT shows increased signal intensity at 222 nm, which indicates the presence of α-helices in the presence of LPG (Fig. 3g, open circles) However, in the case of LPC, β-sheet content is increased with its negative maximum slightly shifted (215 nm, solid circles), which could be due to the presence of more fibrils and/or fibrillar structures with greater β-sheet character.
W423 resides inside the proposed amyloid core region (Fig. 1a) and has been successfully used as a site-specific structural probe in previous RPT studies [23, 24, 27]. To delineate any local structural differences within fibrils formed in the presence of LPG versus LPC, W423 fluorescence was measured in the absence and presence of 0.1 mM LPG and LPC, concentration at which sufficient amounts of fibrils were observed by TEM. RPT monomers shows an emission peak (λmax) at 350 nm (Fig. 3h, dashed line), indicating a water-exposed tryptophan. After aggregation, W423 blue shifts (λmax = 335 nm, Fig. 3h, red line), consistent with a more hydrophobic environment as it is incorporated into fibrils. While the presence of LPC does not change λmax (Fig. 3h, blue line), W423 exhibits a slightly greater blue shift (λmax = 330 nm, Fig. 3h, magenta line) after aggregation in the presence of LPG, hinting at subtle molecular differences within the RPT fibril core.
pH Sensitivity of RPT Fibrils Formed in the Presence of Lysolipids
In contrast to pathological amyloids, which are notoriously resistant to the solution environment once they are formed, RPT amyloid formation is reversible and fibrils fully dissolve at neutral pH. Thus, we next evaluated both the pH dependence of fibril formation and whether these amyloids formed in the presence of either LPG or LPC retain their unique property of dissolution at neutral pH. No fibrils were formed at pH 7 in the presence of LPG or LPC, demonstrating that the presence of lysolipids does not promote RPT fibril formation at neutral pH (data not shown).
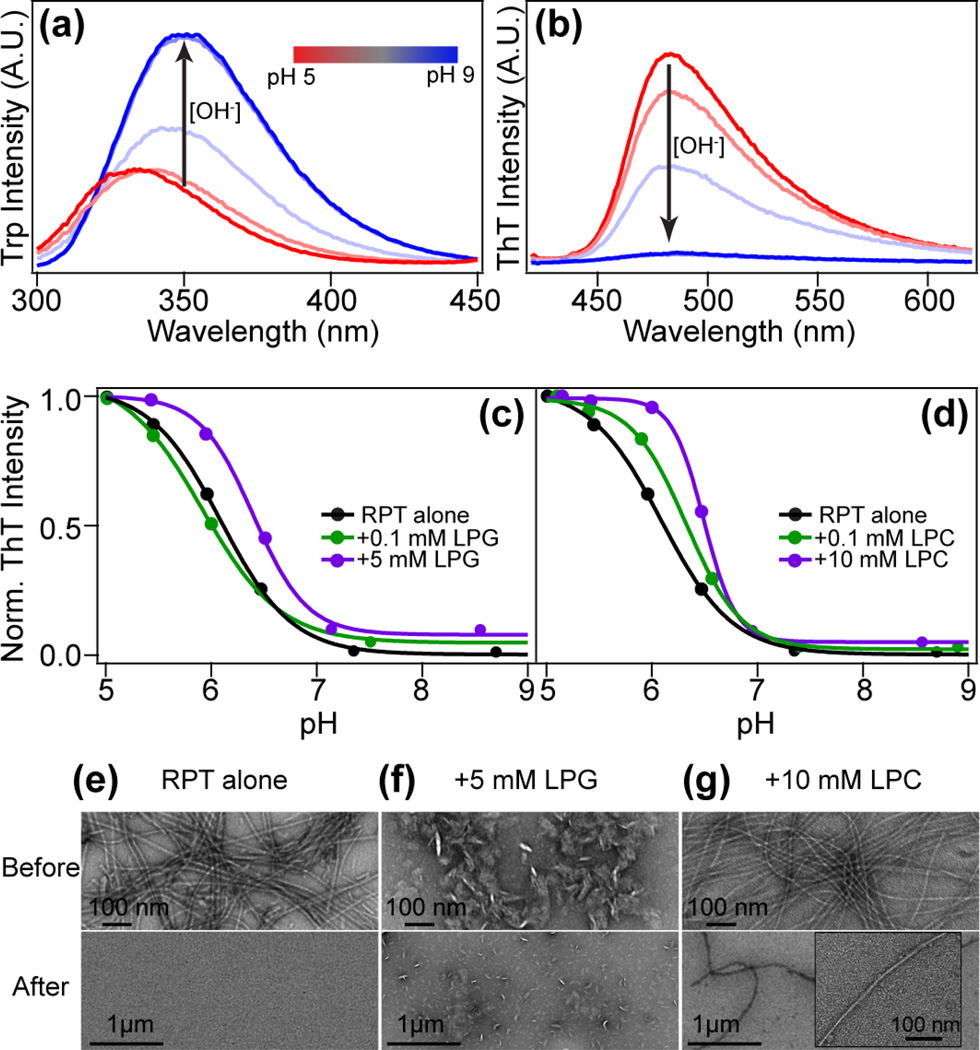
To test the reversibility of RPT fibril formation, dissolution experiments were performed on fibrils formed at pH 5 with different concentrations of lysolipids. The process was monitored by both W423 (Fig. 4a) and ThT (Fig. 4b) fluorescence. As the solution pH is increased by titrating trace amounts of concentrated NaOH, W423 emission red-shifts and increases in intensity, indicating that the sidechain is becoming more solvent-exposed and released from a self-quenched state [27]. In accord, ThT fluorescence decreases as the fibrils disassemble at higher pH. Fibrils formed in the presence of lysolipids show similar trends (Fig.4c and 4d). Intriguingly, fibrils formed in the presence of micelles show slightly later ThT descending profiles, especially in the case of LPC micelles where filaments appear to be stable up to pH 6 (Fig. 4d).
Fig. 4.
Dissolution of RPT fibrils formed in the presence of lysolipids. (a) W423 spectral changes of RPT filaments formed at pH 5 in the presence of 0.1 mM LPG as the solution pH is increased (pH 5–9). (b) Corresponding ThT spectral changes of RPT filaments formed at pH 5 in the presence of 0.1 mM LPG as the solution pH is increased (pH 5–9). Values of pH are presented as a color bar from red to blue. Fibril dissolving profile with LPG (c) and LPC (d) is plotted as normalized ThT intensity vs. solution pH. Dissolution of RPT fibrils formed in the absence of lysolipids (RPT alone) is also shown for comparison. (e)–(g) Corresponding TEM images of before (top) and after (bottom) dissolution experiments. A zoomed-in fibril image is shown as an inset (panel g bottom). Scale bars are as shown.
TEM images were taken before (Fig. 4e–g top) and after dissolution (Fig. 4e–g bottom) to corroborate the fluorescence data. RPT fibrils formed in buffer alone or with submicellar concentrations of lysolipids dissolve completely by the end of the pH titration (pH ~9) (Fig. 4e). In contrast, a few long fibrils decorated with LPC micelles are observed at pH 9 (for a magnified view see Fig. 4g bottom), suggesting enhanced stability, consistent with ThT results. While fibrils do dissolve in the presence of LPG micelles, the reduced level of fibrils formed to begin with and the presence of lipid micelle structures (Fig. 3e) make it difficult to evaluate whether some fibrils remain at the end of titration (Fig. 4f). CD measurements also confirmed the loss of β-sheet structure as the pH is increased from 5 to 9 (data not shown). Nevertheless, these experiments show that the presence of lysolipids does not change the highly reversible and pH dependent RPT fibril formation [24], even though lysolipids substantially alter its aggregation kinetics.
Lysolipid–RPT Interactions Probed by Trp Fluorescence and CD Spectroscopy
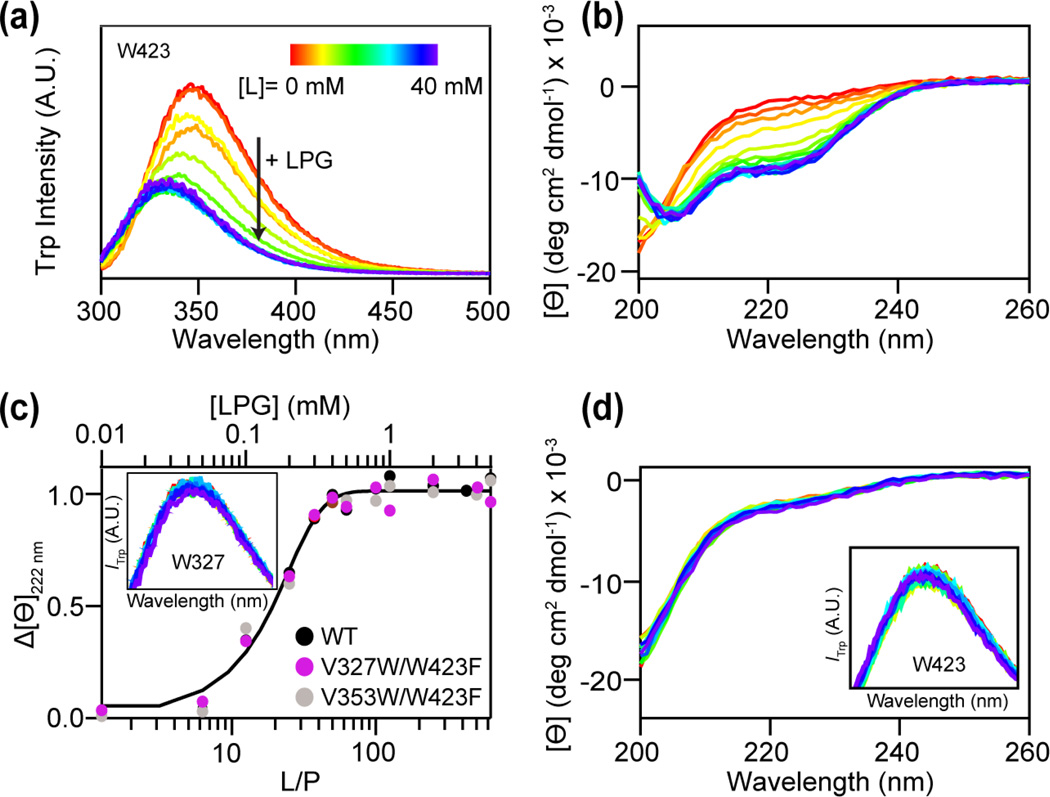
We sought to elucidate whether specific lysolipid-protein interaction leads to conformational changes of the polypeptide, thus altering RPT aggregation propensity. Trp fluorescence and CD spectroscopy were employed as site-specific and secondary structural probes, respectively. First, intrinsic W423 fluorescence was measured as a function of LPG concentration at pH 5 (Fig. 5a). A blue shift (Δλmax ~10 nm) along with a quantum yield decrease is observed as increasing amounts of LPG are added to the protein, indicating LPG binding. Consistently, CD spectroscopic data suggest that as LPG is introduced to RPT, the highly disordered polypeptide develops α-helical features (Fig. 5b). The negative maximum at 222 nm saturates at ~1 mM LPG with a calculated maximal helicity ~27%. A binding curve is generated by plotting the mean residue ellipticity (Θ) at 222 nm vs. LPG concentration (top x-axis) or lipid-to-protein (L/P) ratio (bottom x-axis, Fig. 5c). Because RPT aggregation propensity is exquisitely pH sensitive [27, 39, 40], we also ascertained that the interaction between LPG and RPT is pH dependent. Similar LPG titrations were performed at pH 7, no changes from W423 fluorescence or CD spectra are observed, indicating that the measured interaction is selective for acidic solution conditions (data not shown). Unexpectedly, neither W423 nor CD spectral changes were observed for RPT with varying concentrations of LPC (Fig. 5d), suggesting that LPC interacts with RPT differently than that of LPG.
Fig. 5.
RPT interaction with lysolipids probed by W423 and CD spectroscopy. (a) W423 fluorescence in the presence of increasing LPG (0–40 mM). (b) Secondary structure changes of RPT with increasing concentrations of LPG measured by CD spectroscopy. (c) Representative plot of the mean residue ellipticity (Ɵ) at 222 nm versus lipid-to-protein ratio (L/P, bottom x-axis) and LPG concentration (top x-axis) for WT (black), V327W/W423F (magenta), and V353W/W423F (light grey). A sigmoid curve is shown as a guide. Inset: Fluorescence spectra of V327W/W423F in the presence of increasing LPG (0–40 mM). (d) CD spectra of WT RPT in the presence of increasing LPC (0–40 mM). Inset: W423 fluorescence in the presence of LPC. All measurements were performed with RPT monomers at 25 °C.
Two additional N-terminal Trp variants (V327W/W423F (repeat 1) and V353W/W423F (repeat 3)) were generated to examine the binding specificity of LPG. Each variant contains a single Trp (Val-to-Trp) whereas the native W423 was mutated to a phenylalanine. These two sites were chosen due to the reported solid-state NMR data which suggested potential involvement of N-terminal repeats in segmental polymorphs of RPT fibrils [41]. While nearly identical CD binding curves confirmed minimal perturbations from mutations (Fig. 5c), neither W327 nor W353 emission was sensitive to the presence of LPG (See Fig. 5c inset for representative W327 spectra, W353 data not shown). Clearly, LPG binds in close proximity to W423, near the RPT C-terminus.
Molecular Dimensions of RPT in the Presence of Lysolipids
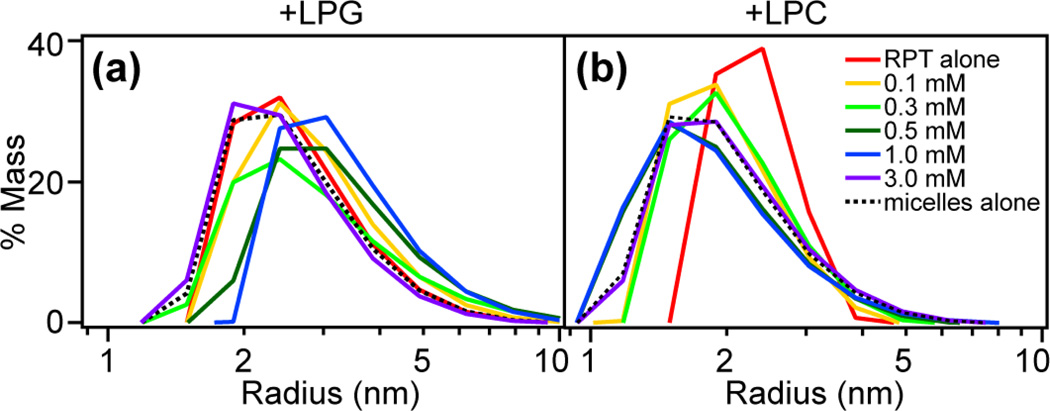
Dynamic light scattering (DLS) was used to examine the molecular dimensions (state of aggregation) of RPT upon adding lysolipids. Unstructured RPT monomers show an average hydrodynamic radius (<r>) around 2.5 nm in solution. Interestingly, RPT increases in size as LPG is added as shown by the shift of the size distributions to <r>~3.5 nm (Fig. 6a). We rationalize that larger LPG–protein particles are formed as a result of the specific interaction with the C-terminal region (Fig. 5a). In contrast to LPG, RPT monomer peak gradually shifts to the left upon adding LPC, corresponding to proteins with a smaller size (Fig. 6b). Eventually, it is indistinguishable from that of LPC micelles measured alone with <r> ~ 1.2 nm, suggesting that RPT polypeptides undergo compaction while remaining unstructured in the presence of LPC (Fig. 5d).
Fig. 6.
Molecular size of RPT in the presence of lysolipids. DLS experiments were performed to monitor the particle size of RPT in solutions with varying concentrations (0–3 mM) of LPG (a) and LPC (b), respectively. Shown are size distributions based on regularization fits. Size distributions for lysolipid micelles measured alone are shown as black dashed lines.
Mechanistic Insights of RPT Aggregation in the Presence of Lysolipids
RPT is highly negatively charged due to the presence of multiple carboxylic acid residues (15 Glu and 1 Asp) with only two basic sidechains (Fig. 1a). As a result, RPT is unlikely to interact with the phosphate group on LPG due to electrostatic repulsion. However, its glycerol headgroup can function as both a hydrogen bond donor and an acceptor; therefore, we propose that a hydrogen bond network is formed between RPT and LPG monomers, proximate to repeat 9 as suggested by W423 fluorescence (Fig. 5a). Hydrogen bond formation would reduce the electrostatic repulsion within the polypeptides and bring them closer, e.g. by sharing a micelle-like structure with LPG as proposed by Giehm et al. for sodium dodecyl sulfate and α-syn [42] (Fig. 7a top). Moreover, since charge neutralization of E422 in repeat 9 has been shown to be crucial in RPT fibril initiation [23], it is possible that the molecular basis for stimulating aggregation is due to a specific interaction between LPG and E422. Hydrogen bonding with Ser and Thr neighbors in repeats 8 and 9 may also be occurring, resulting in a nucleation event and hence faster kinetics. This hypothesis is supported by the observation of larger particles in DLS measurements as LPG is added to RPT. The optimal aggregation condition for RPT is ~15–30 LPG per RPT molecule; although it may be irrelevant, it is surprisingly similar to what has been reported for SDS/α-syn (~12–30) [42].
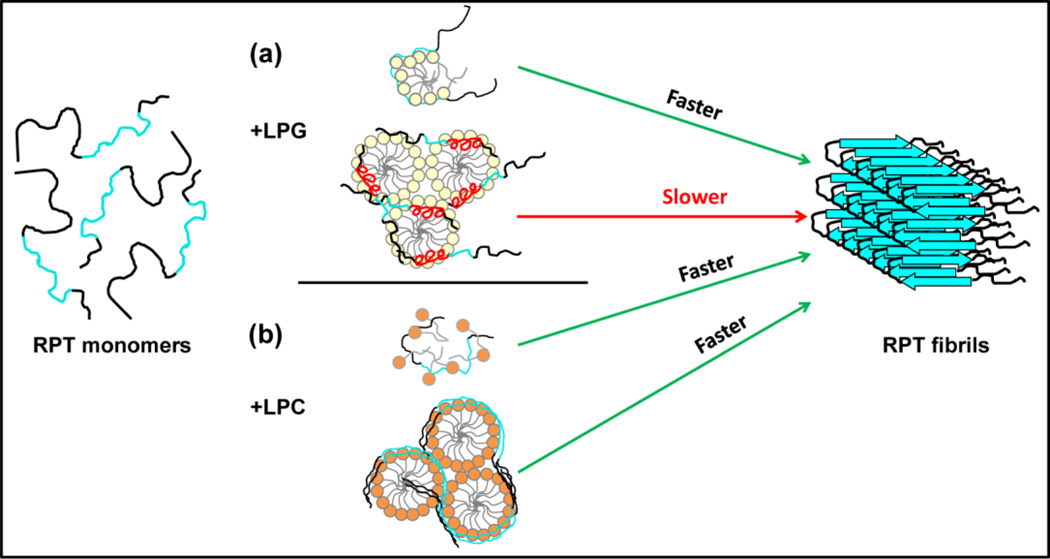
Fig. 7.
Proposed mechanism for lysolipid-modulated RPT aggregation. RPT molecules are depicted with the amyloidogenic region colored cyan. (a) RPT aggregation in the presence of LPG. LPG monomers (top) stimulate RPT aggregation by specifically binding to the C-terminus, forming protein-lipid micelle-like structures likely through hydrogen bonding, and promoting intermolecular interaction between RPT molecules. In contrast, LPG micelles (bottom) induce RPT to develop α-helices (red) possibly within the amyloid core region (repeats 8–9), preventing β-sheet formation and inhibiting aggregation. (b) RPT aggregation in the presence of LPC. The non-specific binding of LPC monomers (top) exposes the hydrophobic regions of RPT, which facilitates protein-protein association, leading to faster aggregation. The unfolded RPT molecules accumulate on the surface of LPC micelles (bottom), functioning as a protein-concentrating platform and enhancing aggregation kinetics.
The amino acid sequence of RPT also contains many prolines, which are well-known helix breakers due to their conformational rigidity. However, there is a proline-free C-terminal stretch of 29 residues (403–431), constituting ~ 20% of the protein length (Fig. 1a) which also encompasses essential residues (VSIVVL, 405–410) required for amyloid formation [23]. Due to the presence of hydrophobic (Val and Trp) and hydrophilic residues (Thr and Ser), it is plausible for this region to form an amphipathic helix. Indeed, a predicted helical region is within this stretch [43, 44] and a comparable helical content percentage increase (Δ~20%) is measured in the presence of LPG. Therefore, we propose that the helical region is formed from this crucial stretch of residues, 403–431. Under this assertion, the presence of an α-helix inside the amyloid core would then retard β-sheet structure formation, leading to fewer RPT fibrils in the presence of LPG micelles, and would inhibit aggregation completely at high LPG concentrations (≥ 10 mM) (Fig. 7a bottom).
Unlike the glycerol headgroup of LPG, the choline moiety of LPC cannot form intermolecular hydrogen bonds with RPT and would likely work differently from the abovementioned mechanism. For the zwitterionic LPC, we propose that it stimulates RPT aggregation similarly to the reported case for the non-charged surfactant OG and Aβ [45, 46]. The presence of amphipathic LPC molecules would promote the exposure of hydrophobic parts of RPT monomers in solution [47–49], thereby facilitating oligomerization and initiating fibril formation. On the other hand, the surface of LPC micelles functions as a nucleation site by increasing local RPT concentration through non-specific electrostatic interaction (Fig. 7b), resulting in even faster aggregation with shorter lag times compared to LPC at submicellar concentration.
Concluding Remarks
RPT is an intrinsically disordered protein (IDP, [50, 51]) that forms amyloid. We have, for the first time, characterized the interaction between a human functional amyloid and membrane lipids, including phospholipids and lysolipids. Negatively charged LPG has dual and opposing effects on RPT aggregation: monomers accelerate whereas micelles retard kinetics. For the zwitterionic LPC, both monomers and micelles stimulate fibril formation, with micelles exerting a stronger effect. Lysolipids are structurally closer to surfactants than to phospholipids. Numerous studies have shown that amphipathic surfactants including sodium dodecyl sulfate (SDS) and fatty acids affect the conformation and fibril formation process of pathological amyloidogenic proteins such as Aβ and α-syn [42, 45, 48, 52–56].
Lysolipids are inverted cone-shaped due to their large headgroup and relatively small acyl chain, a structure that contributes to positive spontaneous curvature [57]. As a result, it is not surprising to see a higher lysolipid content in melanosomes [33] because these structural features of lysolipids would favor the formation of highly curved small ILVs and stabilize the ellipsoid-shaped melanosomes. Therefore, it supports the proposal that the fibril initiation occurs on the highly curved ILV membrane surface [31]. Moreover, the preference of lysolipids for positive spontaneous curvature has been shown to promote membrane fusion by minimizing the bending energy [58–62]. We conjecture that a high concentration of lysolipids is needed in ILVs to aid their merging with melanosomal membranes [31].
Because intermediate species (oligomers) en-route to fibrils are proposed to be the most potent cytotoxic agents, one way for melanosomes to cope with the adverse effect of amyloid formation is through the spatial and temporal regulation of aggregation. By using different lysolipids and their relative distribution and concentrations on ILVs, both the location and the speed of fibrillation can be controlled. With faster aggregation kinetics, over-population of oligomers may also be circumvented as well as the prompt sequestration of melanin and its associated toxic intermediates on the fibrils may be achieved. Taken together, our data suggest that lysolipids may play a key role in modulating Pmel17 fibril formation and possibly be involved in melanosome maturation.
MATERIALS AND METHODS
Chemicals
Phospholipids and lysolipids: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (POPG), 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) (LPG), and 1-lauroyl-2-hydroxy-sn-glycero-3-phosphocholine (LPC) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Thioflavin T (ThT) and buffers, sodium acetate trihydrate (NaOAc) and 3-(N-morpholino) propanesulfonic acid (MOPS), were purchased from Sigma-Aldrich (St. Louis, MO). Guanidinium hydrochloride was purchased from MP Biomedicals (Solon, OH).
Protein Expression and Purification
His-tagged wild-type RPT was expressed in E. Coli BL21(DE3) RIPL (Agilent Technologies) and purified as previously described with minor modifications [39]. Transformed cells were grown in a 20-L fermentor using modified LB media (10 g/L tryptone, 5 g/L yeast extract, 5 g/L NaCl, 14.2 g/L Na2HPO4, 3.5 g/L glucose, 0.5 g/L MgSO4·7H2O, and 100 mg/L ampicilin) and induced with 1 mM isopropyl β-D-1-thiogalactopyranoside at 30 °C when the optical density at 600 nm reached around 3 (Protein Expression Facility). N-terminal Trp mutants (V327W/W423F; V353W/W423F) were generated using QuickChange Mutagenesis Kit (Agilent Technologies Inc., CA) and verified by DNA sequencing. Lysis buffer (pH 7.5) contained 6 M guanidinium hydrochloride, 100 mM NaCl and 100 mM Na2HPO4. A washing step (10× column volume of 30 mM imidazole) was added before eluting RPT (200 mM imidazole) from Ni-NTA agarose (Qiagen). Protein collected after elution was desalted into 20 mM Tris, pH 8.0 buffer using a HiPrep desalting 26/10 column (GE Healthcare). Protein purity was examined by SDS-PAGE visualized either with silver or Coomassie blue staining. Anionic exchange chromatography (MonoQ, GE Healthcare) using a linear NaCl gradient (0 – 300 mM) was used for further purification if necessary. Protein concentrations were determined using a molar extinction coefficient estimated on the basis of amino-acid content: ε280 nm = 5,500 M−1 cm−1. Protein molecular weights were confirmed by electrospray ionization mass spectrometry (Biochemistry Core Facility). Purified protein was flash-frozen in liquid N2 and stored at −80 °C until use.
Lipid and Vesicle Preparation
Lipids (~30 mM) were stored in chloroform/methanol (2:1) stock solutions. Lipid films were prepared by drying appropriate amounts of the stock solution in a stream of dry nitrogen followed by vacuum desiccation for at least 2 h to ensure removal of organic solvent. To rehydrate the lipid films, appropriate buffer solutions were added (20 mM NaOAc, 100 mM NaCl, pH 5 and 20 mM MOPS, 100 mM NaCl, pH 7) and vortexed at least 3 times for 60 s. The multilamellar vesicle solution was extruded 30 times through a 50-nm diameter polycarbonate membrane (Whatman, GE Healthcare) using an extrusion kit (Avanti Polar Lipids, Inc.). Vesicle sizes (average hydrodynamic radius ~ 40 nm) were determined by dynamic light scattering using Dynapro NanoStar (Wyatt). Each measurement contained ten 5-s acquisitions and globular proteins were used as the model for calculating the radius with regularization fit. Aqueous lysolipid solutions were prepared from 200 mM stock solutions by dissolving dry powder in working buffers. CMC determined using DLS under our solution conditions are 0.6 mM and 0.8 mM for LPG and LPC, respectively. All buffers were filtered through a 0.22 µm filter (Millipore) before use.
Fluorescence and Circular Dichroism Spectroscopy
Trp was excited at 280 nm and emission was collected from 300 – 500 nm with integration time of 0.3 s in 1 cm quartz cuvettes. ThT was excited at 400 nm and emission was recorded from 450 – 600 nm with integration time of 0.25 s in 1 cm quartz cuvettes. Slits for excitation and emission were 1 and 2 nm, respectively. Fluorescence was recorded with a Fluorolog 3 spectrofluorometer (Horiba Jobin Yvon) equipped with a temperature-controlled sample holder. CD measurements (198 to 260 nm, 1 nm data pitch, continuous scanning with 1 nm bandwidth, 100 nm/min and 3 accumulations) with 8 µM RPT were carried out in 1 mm quartz cuvettes using a Jasco J-715 spectropolarimeter (Jasco Analytical Instruments, Biophysics Core). All measurements were performed at 25 °C. Data were analyzed using IgorPro 6.22 (Wavemetrics). The mean residue ellipticity is calculated using the equation: [Θ]=(100θ)/(Cln), where θ is the measured CD signal in mdeg, C is the protein concentration in mM, l is the path length in cm, and n is the number of amino acids. The percent helicity (100fhelix) is calculated using the equation [63]: fhelix=([Θ]222 nm− [Θ]coil)/([Θ]helix− [Θ]coil), where [Θ]coil = 640 − 45T, and [Θ]helix= −40,000(1 − 2.5/n)+100T (T is the temperature in °C and n is the number of amino acid residues).
Aggregation Kinetics and Fibril Dissolution
Purified RPT was exchanged into working buffer (pH 5 buffer: 20 mM NaOAc, 100 mM NaCl; pH 7 buffer: 20 mM MOPS, 100 mM NaCl) using a PD-10 column (GE Healthcare) and filtered through YM-100 filters (Millipore) to remove any preformed aggregates immediately prior to aggregation. Final solutions contained 30 µM RPT and specified lipids/vesicles. Aggregation experiments were performed in a sealed 96-well flat bottom plates (Corning Costar, 200 µL per well and a 2-mm sterile, glass bead was added to facilitate shaking) at 37 °C with continuous orbital shaking (1 mm, ca. 87.6 rpm) using a microplate reader (Tecan Infinite M200 Pro). Lipid dependent trends were comparable and reproducible though variability in the absolute lag times was observed. ThT was excited at 415 nm and emission was recorded at 480 nm as a function of time.
Increments of 1 M NaOH (<10 µL) were added to pre-aggregated RPT samples (600 µL) to increase pH from pH 5 to 9. The solution pH was monitored with a micro-pH probe (Mettler Toledo). At each pH, following a 15 min equilibration time at RT, ThT and W423 fluorescence were measured as described above (dilution effects were negligible <1%).
Transmission Electronic Microscopy
TEM was performed using a JEOL JEM 1200EX transmission electron microscope (accelerating voltage 80 keV) equipped with an AMT XR-60 digital camera (Electron Microscopy Core). Some sample grids (400-mesh formvar and carbon coated copper, Electron Microscopy Sciences) were glow discharged using an EMScope TB500 (Emscope Laboratories) to increase surface hydrophilicity prior to deposition of protein sample (3 µL). This treatment increases the amount of deposited material. After sample deposition (1 min) and wicking off, good background contrast was obtained by applying an additional wash step with deionized water (3 µL). Immediately, the samples were stained with 2–3 drops of 1% (w/v) aqueous uranyl acetate solution. Excess solution was absorbed with filter paper and air dried.
Highlights.
Investigated the effect of lipids on a functional amyloid RPT aggregation
Lysolipids-containing phospholipid vesicles accelerate RPT fibril formation
LPC stimulates RPT aggregation at both submicellar and micellar concentrations
LPG modulates RPT aggregation with both stimulatory and inhibitory effects
Lysolipids may play an important role in melanosomal fibril formation and maturation
ACKNOWLEDGMENTS
Supported by the Intramural Research Program at the National Institutes of Health, National Heart, Lung, and Blood Institute. We thank M. Daniels, P. Connelly (EM Core Facility), Y. He (Protein Expression Facility), D.-Y. Lee (Biochemistry Core Facility) and G. Piszczek (Biophysics Core Facility) for technical assistance and use of equipment. We acknowledge R. P. McGlinchey for performing Trp variant PCR and helpful discussions and S.K. Hess for manuscript proof reading.
Abbreviations
- α-syn
α-synuclein
- Aβ
amyloid-β
- RPT
repeat domain
- ThT
thioflavin T
- CD
circular dichroism
- TEM
transmission electron microscopy
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol)
- LPG
lysophosphatidylglycerol
- LPC
lysophosphatidylcholine
- DLS
dynamic light scattering
- CMC
critical micelle concentration
- OG
octyl β-D-glucopyranoside
- SDS
sodium dodecyl sulfate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Shewmaker F, McGlinchey RP, Wickner RB. Structural insights into functional and pathological amyloid. J. Biol. Chem. 2011;286:16533–16540. doi: 10.1074/jbc.R111.227108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Jahn TR, Radford SE. Folding versus aggregation: Polypeptide conformations on competing pathways. Arch. Biochem. Biophys. 2008;469:100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenwald J, Riek R. Biology of amyloid: Structure, function, and regulation. Structure. 2010;18:1244–1260. doi: 10.1016/j.str.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid - from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH. Amyloid fibrils of the HET-s(218–289) prion form a β solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 7.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGlinchey RP, Yap TL, Lee JC. The yin and yang of amyloid: insights from α-synuclein and repeat domain of Pmel17. Phys. Chem. Chem. Phys. 2011;13:20066–20075. doi: 10.1039/c1cp21376h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balguerie A, Dos Reis S, Ritter C, Chaignepain S, Coulary-Salin B, Forge V, Bathany K, Lascu I, Schmitter JM, Riek R, Saupe SJ. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams TL, Serpell LC. Membrane and surface interactions of Alzheimer's Aβ peptide - insights into the mechanism of cytotoxicity. FEBS J. 2011;278:3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- 11.Torres-Bugeau CM, Borsarelli CD, Minahk CJ, Chehin RN. The key role of membranes in amyloid formation from a biophysical perspective. Curr. Protein Pept. Sci. 2011;12:166–180. doi: 10.2174/138920311795860197. [DOI] [PubMed] [Google Scholar]
- 12.Gorbenko G, Trusova V. Protein aggregation in a membrane environment. In: Donev R, editor. Adv. Protein Chem. Struct. Biol. Vol. 84. 2011. pp. 113–142. [DOI] [PubMed] [Google Scholar]
- 13.Pfefferkorn CM, Jiang Z, Lee JC. Biophysics of α-synuclein membrane interactions. BBA Biomembr. 2012;1818:162–171. doi: 10.1016/j.bbamem.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang Z, de Messieres M, Lee JC. Membrane remodeling by α-synuclein and effects on amyloid formation. J. Am. Chem. Soc. 2013;135:15970–15973. doi: 10.1021/ja405993r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abedini A, Raleigh DP. A critical assessment of the role of helical intermediates in amyloid formation by natively unfolded proteins and polypeptides. Protein Eng. Des. Sel. 2009;22:453–459. doi: 10.1093/protein/gzp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brender JR, Salamekh S, Ramamoorthy A. Membrane disruption and early events in the aggregation of the diabetes related peptide IAPP from a molecular perspective. Acc. Chem. Res. 2012;45:454–462. doi: 10.1021/ar200189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:100–107. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoashi T, Muller J, Vieira WD, Rouzaud F, Kikuchi K, Tamaki K, Hearing VJ. The repeat domain of the melanosomal matrix protein PMEL17/GP100 is required for the formation of organellar fibers. J. Biol. Chem. 2006;281:21198–21208. doi: 10.1074/jbc.M601643200. [DOI] [PubMed] [Google Scholar]
- 19.Raposo G, Marks MS. Melanosomes-dark organelles enlighten endosomal membrane transport. Nat. Rev. Mol. Cell Biol. 2007;8:786–797. doi: 10.1038/nrm2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee ZH, Hou L, Moellmann G, Kuklinska E, Antol K, Fraser M, Halaban R, Kwon BS. Characterization and subcellular localization of human Pmel 17/silver, a 110-kDa (pre)melanosomal membrane protein associated with 5,6,-dihydroxyindole-2-carboxylic acid (DHICA) converting activity. J. Invest. Dermatol. 1996;106:605–610. doi: 10.1111/1523-1747.ep12345163. [DOI] [PubMed] [Google Scholar]
- 21.Chakraborty AK, Platt JT, Kim KK, Kwon BS, Bennet DC, Pawelek JM. Polymerization of 5,6-dihydroxyindole-2-carboxylic acid to melanin by the Pmel 17 silver locus protein. Eur. J. Biochem. 1996;236:180–188. doi: 10.1111/j.1432-1033.1996.t01-1-00180.x. [DOI] [PubMed] [Google Scholar]
- 22.Theos AC, Truschel ST, Raposo G, Marks MS. The silver locus product Pmel17/gp100/Silv/ME20: controversial in name and in function. Pigment Cell Res. 2005;18:322–336. doi: 10.1111/j.1600-0749.2005.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGlinchey RP, Jiang Z, Lee JC. Molecular origin of pH-dependent fibril formation of a functional amyloid. ChemBioChem. 2014;15:1569–1572. doi: 10.1002/cbic.201402074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGlinchey RP, Gruschus JM, Nagy A, Lee JC. Probing fibril dissolution of the repeat domain of a functional amyloid, Pmel17, on the microscopic and residue level. Biochemistry. 2011;50:10567–10569. doi: 10.1021/bi201578h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brilliant MH. The mouse p (pink-eyed dilution) and human P genes, oculocutaneous albinism type 2 (OCA2), and melanosomal pH. Pigment Cell Res. 2001;14:86–93. doi: 10.1034/j.1600-0749.2001.140203.x. [DOI] [PubMed] [Google Scholar]
- 26.Puri N, Gardner JM, Brilliant MH. Aberrant pH of melanosomes in pink-eyed dilution (p) mutant melanocytes. J. Invest. Dermatol. 2000;115:607–613. doi: 10.1046/j.1523-1747.2000.00108.x. [DOI] [PubMed] [Google Scholar]
- 27.Pfefferkorn CM, McGlinchey RP, Lee JC. Effects of pH on aggregation kinetics of the repeat domain of a functional amyloid, Pmel17. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21447–21452. doi: 10.1073/pnas.1006424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KPR, Simon R, Schubert D, Eisenberg D, Rivier J, Sawchenko P, Vale W, Riek R. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato M, Han TNW, Xie SH, Shi K, Du XL, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei JM, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, Eisenberg D, Tycko R, McKnight SL. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leonhardt RM, Vigneron N, Rahner C, van den Eynde BJ, Cresswell P. Endoplasmic reticulum export, subcellular distribution, and fibril formation by Pmel17 require an intact N-terminal domain junction. J. Biol. Chem. 2010;285:16166–16183. doi: 10.1074/jbc.M109.097725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurbain I, Geerts WJC, Boudier T, Marco S, Verkleij AJ, Marks MS, Raposo G. Electron tomography of early melanosomes: Implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19726–19731. doi: 10.1073/pnas.0803488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theos AC, Truschel ST, Tenza D, Hurbain I, Harper DC, Berson JF, Thomas PC, Raposo G, Marks MS. A lumenal domain-dependent pathway for sorting to intralumenal vesicles of multivesicular endosomes involved in organelle morphogenesis. Dev. Cell. 2006;10:343–354. doi: 10.1016/j.devcel.2006.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jimbow M, Kanoh H, Jimbow K. Characterization of biochemical-properties of melanosomes for structural and functional differentiation: Analysis of the compositions of lipids and proteins in melanosomes and their subfractions. J. Invest. Dermatol. 1982;79:97–102. doi: 10.1111/1523-1747.ep12500034. [DOI] [PubMed] [Google Scholar]
- 34.Ward WC, Simon JD. The differing embryonic origins of retinal and uveal (iris/ciliary body and choroid) melanosomes are mirrored by their phospholipid composition. Pigment Cell Res. 2007;20:61–69. doi: 10.1111/j.1600-0749.2006.00357.x. [DOI] [PubMed] [Google Scholar]
- 35.Carruthers A, Melchior DL. Effects of lipid environment on membrane transport: the human erythrocyte sugar transport protein/lipid bilayer system. Annu. Rev. Physiol. 1988;50:257–271. doi: 10.1146/annurev.ph.50.030188.001353. [DOI] [PubMed] [Google Scholar]
- 36.Stafford RE, Fanni T, Dennis EA. Interfacial properties and critical micelle concentration of lysophospholipids. Biochemistry. 1989;28:5113–5120. doi: 10.1021/bi00438a031. [DOI] [PubMed] [Google Scholar]
- 37.Chou JJ, Baber JL, Bax A. Characterization of phospholipid mixed micelles by translational diffusion. J. Biomol. NMR. 2004;29:299–308. doi: 10.1023/B:JNMR.0000032560.43738.6a. [DOI] [PubMed] [Google Scholar]
- 38.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinform. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McGlinchey RP, Shewmaker F, McPhie P, Monterroso B, Thurber K, Wickner RB. The repeat domain of the melanosome fibril protein Pmel17 forms the amyloid core promoting melanin synthesis. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13731–13736. doi: 10.1073/pnas.0906509106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGlinchey RP, Shewmaker F, Hu KN, McPhie P, Tycko R, Wickner RB. Repeat domains of melanosome matrix protein Pmel17 orthologs form amyloid fibrils at the acidic melanosomal pH. J. Biol. Chem. 2011;286:8385–8393. doi: 10.1074/jbc.M110.197152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu KN, McGlinchey RP, Wickner RB, Tycko R. Segmental polymorphism in a functional amyloid. Biophys. J. 2011;101:2242–2250. doi: 10.1016/j.bpj.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giehm L, Oliveira CLP, Christiansen G, Pedersen JS, Otzen DE. SDS-induced fibrillation of α-synuclein: An alternative fibrillation pathway. J. Mol. Biol. 2010;401:115–133. doi: 10.1016/j.jmb.2010.05.060. [DOI] [PubMed] [Google Scholar]
- 43.Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Honigschmid P, Schafferhans A, Roos M, Bernhofer M, Richter L, Ashkenazy H, Punta M, Schlessinger A, Bromberg Y, Schneider R, Vriend G, Sander C, Ben-Tal N, Rost B. PredictProtein-an open resource for online prediction of protein structural and functional features. Nucleic Acids Res. 2014;42:W337–W343. doi: 10.1093/nar/gku366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raghava GPS. Protein secondary structure prediction using nearest neighbor and neural network approach. CASP4. 2000:75–76. [Google Scholar]
- 45.Rocha S, Loureiro JA, Brezesinski G, Pereira MD. Peptide-surfactant interactions: Consequences for the amyloid-β structure. Biochem. Biophys. Res. Commun. 2012;420:136–140. doi: 10.1016/j.bbrc.2012.02.129. [DOI] [PubMed] [Google Scholar]
- 46.Loureiro JA, Rocha S, Pereira MD. Charged surfactants induce a non-fibrillar aggregation pathway of amyloid-β peptide. J. Pept. Sci. 2013;19:581–587. doi: 10.1002/psc.2535. [DOI] [PubMed] [Google Scholar]
- 47.Otzen DE, Sehgal P, Westh P. α-Lactalbumin is unfolded by all classes of surfactants but by different mechanisms. J. Colloid Interface Sci. 2009;329:273–283. doi: 10.1016/j.jcis.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 48.Friedman R, Caflisch A. Surfactant effects on amyloid aggregation kinetics. J. Mol. Biol. 2011;414:303–312. doi: 10.1016/j.jmb.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Otzen DE. Protein-surfactant interactions: A tale of many states. BBA:Proteins Proteom. 2011;1814:562–591. doi: 10.1016/j.bbapap.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 50.Dunker AK, Babu MM, Barbar E, Blackledge M, Bondos SE, Dosztányi Z, Dyson HJ, Forman-Kay J, Fuxreiter M, Gsponer J, Han K-H, Jones DT, Longhi S, Metallo SJ, Nishikawa K, Nussinov R, Obradovic Z, Pappu RV, Rost B, Selenko P, Subramaniam V, Sussman JL, Tompa P, Uversky VN. What's in a name? Why these proteins are intrinsically disordered. Intrinsically Disordered Proteins. 2013;1:0–4. doi: 10.4161/idp.24157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: Introducing the D2 concept. Annu. Rev. Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 52.Tian JH, Sethi A, Anunciado D, Vu DM, Gnanakaran S. Characterization of a disordered protein during micellation: Interactions of α-synuclein with sodium dodecyl sulfate. J. Phys. Chem. B. 2012;116:4417–4424. doi: 10.1021/jp210339f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sabate R, Estelrich J. Stimulatory and inhibitory effects of alkyl bromide surfactants on β-amyloid fibrillogenesis. Langmuir. 2005;21:6944–6949. doi: 10.1021/la050472x. [DOI] [PubMed] [Google Scholar]
- 54.Sureshbabu N, Kirubagaran R, Jayakumar R. Surfactant-induced conformational transition of amyloid β-peptide. Eur. Biophys. J. Biophy. 2009;38:355–367. doi: 10.1007/s00249-008-0379-8. [DOI] [PubMed] [Google Scholar]
- 55.Abelein A, Kaspersen JD, Nielsen SB, Jensen GV, Christiansen G, Pedersen JS, Danielsson J, Otzen DE, Graslund A. Formation of dynamic soluble surfactant-induced amyloid β peptide aggregation intermediates. J. Biol. Chem. 2013;288:23518–23528. doi: 10.1074/jbc.M113.470450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dahse K, Garvey M, Kovermann M, Vogel A, Balbach J, Fandrich M, Fahr A. DHPC strongly affects the structure and oligomerization propensity of Alzheimer's Aβ(1–40) peptide. J. Mol. Biol. 2010;403:643–659. doi: 10.1016/j.jmb.2010.09.021. [DOI] [PubMed] [Google Scholar]
- 57.Fuller N, Rand RP. The influence of lysolipids on the spontaneous curvature and bending elasticity of phospholipid membranes. Biophys. J. 2001;81:243–254. doi: 10.1016/S0006-3495(01)75695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chizmadzhev YA. Membrane fusion. Biochemistry(Moscow) 2012;6:152–158. [Google Scholar]
- 59.Elamrani K, Blume A. Incorporation kinetics of lysolecithin into lecithin vesicles, kinetics of lysolecithin-induced vesicle fusion. Biochemistry. 1982;21:521–526. doi: 10.1021/bi00532a017. [DOI] [PubMed] [Google Scholar]
- 60.Poole AR, Howell JI, Lucy JA. Lysolecithin and cell fusion. Nature. 1970;227:810. doi: 10.1038/227810a0. [DOI] [PubMed] [Google Scholar]
- 61.Croce CM, Sawicki W, Kritchev D, Koprowsk H. Induction of homokaryocyte, heterokaryocyte and hybrid formation by lysolecithin. Exp. Cell Res. 1971;67:427. doi: 10.1016/0014-4827(71)90428-9. [DOI] [PubMed] [Google Scholar]
- 62.Chernomordik LV, Melikyan GB, Chizmadzhev YA. Biomembrane fusion: A new concept derived from model studies using two interacting planar lipid bilayers. BBA Biomembr. 1987;906:309–352. doi: 10.1016/0304-4157(87)90016-5. [DOI] [PubMed] [Google Scholar]
- 63.Chen YH, Yang JT, Chau KH. Determination of helix and β-form of proteins in aqueous-solution by circular dichroism. Biochemistry. 1974;13:3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]