Abstract
Rigid attachment of microtubules (MTs) to glass cover slip surfaces is a prerequisite for a variety of microscopy experiments in which MTs are used as substrates for MT-associated proteins, such as the molecular motors kinesin and cytoplasmic dynein. We present an MT-surface coupling protocol in which aminosilanized glass is formylated using the cross-linker glutaraldehyde, fluorescence-labeled MTs are covalently attached, and the surface is passivated with highly pure beta-casein. The technique presented here yields rigid MT immobilization while simultaneously blocking the remaining glass surface against nonspecific binding by polystyrene optical trapping microspheres. This surface chemistry is straightforward and relatively cheap and uses a minimum of specialized equipment or hazardous reagents. These methods provide a foundation for a variety of optical tweezers experiments with MT-associated molecular motors and may also be useful in other assays requiring surface-immobilized proteins.
Keywords: Microtubules, Protein immobilization, Plasma cleaning, Silanization, Aminosilane functionalization, Chemical cross-linking, Glutaraldehyde, Fluorescence labeling, Single-molecule assays, Microtubule motor proteins
1 Introduction
In vitro, single-molecule microscopy studies of molecular motor proteins can provide valuable insights into how they generate forces essential to mitosis and other critical cellular functions. Single-molecule studies of microtubule (MT) motors and other MT-associated proteins (MAPs) frequently employ surface-immobilized MTs. While axonemes (bundles of MTs and other associated proteins) have been used frequently for this purpose [1-5] and have the advantage of adsorbing strongly to untreated glass, they contain other proteins in addition to tubulin, and they are not the natural substrates for cytosolic molecular motors. Moreover, the large diameter of axonemes (~160 nm) may be undesirable in certain assays. Therefore, MTs are generally preferable, and so a reliable method for fixing them to a glass surface is of utility.
Rigid MT attachment is important in order to achieve high-quality measurements: unwanted motion makes it impossible to precisely track the positions of MT-bound MAPs and introduces compliance to the motor-MT system that complicates interpretation of forces measured by optical tweezers and other methods. Unfortunately, MTs adsorb very poorly to untreated glass. Any MTs that do bind tend to bow and agitate on the surface and often detach when a force is applied. Therefore, specialized methods of MT attachment to glass are required. For optical trapping, it is also required that the remaining surface be well passivated (usually using a blocking agent such as bovine serum albumin (BSA), casein, or polyethylene glycol (PEG) derivatives), so that the trapped microspheres (“beads”) do not bind to the glass via nonspecific adsorption. This is important because attractive bead-surface interactions could, for example, impair molecular motor movement and/or give false indications of MAP-MT binding.
We sought a method to eliminate essentially all nonspecific microsphere-surface interactions. A variety of techniques have been reported in the literature for immobilizing MTs on glass. We experimented with several approaches, such as coating glass with nonspecifically adsorbed poly-l-lysine [6-9], antibodies [10], rigor kinesins (both G234A [11, 12] and T93N [13, 14] mutants), or BSA-biotin (with streptavidin and biotinylated MTs) [15, 16]. We also experimented with silane-PEG-N-hydroxysuccinimide (silane-PEG-NHS) reagents. In our hands, several of these techniques were effective at either immobilizing MTs or preventing trapping beads from binding to the surface but did not accomplish both simultaneously (see Note 1). For example, although rigor kinesin is very effective at binding MTs (we use this technique in other single-molecule assays in our laboratory), microspheres frequently bind irreversibly to the kinesin-coated surface, even in the presence of blocking agents.
Aminosilanization (coating of surfaces, typically containing free hydroxyl groups, with aminated alkoxysilane molecules) has been used for years to promote binding of a variety of biological materials to glass (see, e.g., [17] and references therein), including MTs [18-20]. Reported aminosilanization conditions and protocols vary widely, and optimization of this surface chemistry is still an active area of research [21-26]. However, we found that, when used in combination with chemical cross-linking [27-31] and appropriate surface blocking [20], aminosilanized cover slips perform exceptionally well, both in terms of binding MTs and limiting microsphere adhesion. Certain parameters, such as incubation times and blocking reagents, are critical to achieving good results with MTs. Therefore, we thought it would be of general benefit to provide the protocols we established, based on current literature and optimization in the laboratory. The procedures given here are cheap and easy and do not require hazardous cleaning reagents. MTs attach rigidly to the glass and support robust movement by a variety of MT-associated molecular motors. Trapping beads, whether coated in protein or not, show no binding (even transiently) to the blocked cover slip surface when pushed into or dragged against the glass (even after days in the chamber at room temperature).
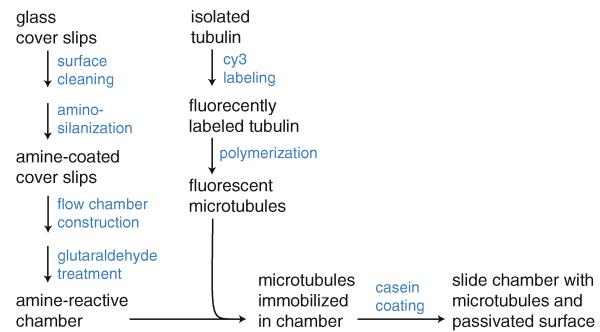
The method works as follows. First, the glass is prepared through a series of cleaning procedures that remove contaminants and hydroxylate the surface. Next, the surface is aminosilanized, thereby coating the glass with positively charged amine groups that attract MTs (due to the strongly negative MT surface charge). The treated glass cover slip is then used to assemble a microscopy flow chamber. This flow chamber is treated with glutaraldehyde and extensively washed, leaving a mixture of surface amines and reactive formyl groups on the glass. Fluorescent tubulin is then prepared and polymerized into MTs. The MTs are bound to the surface, attracted initially by the surface charge, and then covalently coupled to the glass via reaction between exposed lysyl residues on the MT and the surface-bound formyl groups. Finally, the remaining surface is blocked with β-casein, which physically adsorbs to the glass, thereby passivating it. The casein also reacts with any remaining formyl groups to inactivate them. Figure 1 summarizes how the individual steps contribute to the overall procedure. The reader is encouraged to consult each of the Notes, which contain important practical information, as well as explanations of theoretical principles.
Fig. 1.
Protocol scheme. Each pathway summarizes the major steps in preparing a slide chamber with MTs attached and the surface passivated against nonspecific microsphere binding
The procedures presented here yield a flow chamber suitable for a number of optical tweezers studies involving MAP function, such as our accompanying protocol for measurement of kinesin force generation (see Chapter 10). These methods are also likely to be useful in other single-molecule studies of MAP-MT interactions and other assays requiring immobilization of proteins on glass surfaces.
2 Materials
2.1 Cover Slip Aminosilanization
Cover slips (18 mm × 18 mm × 0.170 mm) (No. 1.5, Zeiss Cat. No. 474030-9000-000; see Note 2).
Porcelain cover glass holders (Thomas, Part No. 8542E40-TS; see Note 3).
Clean blunt-nosed forceps (clean in ethanol and flame prior to use).
Double-deionized water (ddH20) purification system (see Note 4).
Mucaso™ detergent solution, 2 % (v/v): 98 % double-deionized water, 2 % Mucasol fast alkaline cleaning agent (Merz Hygiene GmbH), by volume.
APTES (3-aminopropyl triethoxysilane, also called APS; Sigma Cat. No. 440140 or A3648) or AEAPTES (N-(2-aminoethyl)-3-aminopropyl triethoxysilane, Gelest Cat. No. SIA0590.5) (see Note 5).
Acetone (Sigma, cat. no. 270725; see Note 6).
Spectrophotometric grade ethanol, 200 proof (Sigma, Cat. No. 459828; see Note 6).
Five plastic jars, 250 mL each, made of polymethylpentene (PMP) (Nalgene). Label each of the jars and lids with the following: ddH20, Mucasol, Ethanol, Acetone, Silane (see Note 7).
Bath sonicator with temperature control (Fisher Scientific, FS30D) and holder for jars (see Note 8).
Plasma cleaner (PDC-001, Harrick Plasma).
Oven or hot plate (see Note 9).
Vacuum desiccator.
Personal protective equipment: nitrile gloves, eye protection, lab coat, etc.
2.2 Microscope Slide Chamber Construction
Microscope slides (3″ × 1″ × 1.0 mm).
Parafilm® “M” (Bemis, see Note 10).
Forceps (tweezers): one sharp-nosed and one blunt-nosed.
Heat gun (e.g., Fit Gun-3, Alpha Wire) or heating block.
2.3 Slide Chamber Glutaraldehyde Treatment
2.4 Labeling of Tubulin with Cy3 Dye
See Note 13:
Bovine brain tubulin (1 mg/vial, Cytoskeleton, Inc.; see Note 14).
Cy3™ monofunctional N-hydroxysuccinimide (NHS) ester reactive dye (10 mg/mL): dissolve one vial of Cy3 monofunctional NHS ester dye (0.2 mg, GE Healthcare) in 20 μL anhydrous DMSO. Store at −20°C in the dark (see Note 15).
PM buffer: 100 mM PIPES and 1 mM MgCb, pH ~7.
Beckman Coulter Optima™ TLX Ultracentrifuge (120,000 maximum rpm).
Beckman TLA-100 fixed angle rotor (7 × 20 mm, 0.2 mL tube).
2.5 Cy3-Labeled MT Polymerization
BRB80 buffer: 80 mM PIPES (Sigma P6757), 2 mM MgCb, and 1 mM EGTA, pH ~7.
Unlabeled bovine brain tubulin (10 μL aliquots of 10 mg/mL): dissolve 1 vial of 1 mg lyophilized powder (Cytoskeleton, Inc.) in 100 μL BRB80 buffer (final concentration 10 mg/mL). Aliquot and flash freeze. Store aliquots at −80°C (see Note 16).
Cy3-labeled tubulin (1.6 μL aliquots of 30 μg tubulin, labeled/unlabeled ~1:2; each preparation will be slightly different - adjust accordingly).
Guanosine-5′-triphosphate (GTP; 25 mM, with equimolar MgSO4).
Anhydrous DMSO.
Paclitaxel (Taxol) (2 mM): dissolve 1 vial of lyophilized powder (0.2 μmol/vial; Cytoskeleton, Inc.) in 100 μL anhydrous DMSO. Store at −20°C. The working concentration is 10–20 μM.
Dithiothreitol (DTT) (1 M): dissolve DTT in ddH20 and store at −20°C. The working concentration is 1 mM.
Glycerol cushion (BRB80 with 60 % glycerol): 80 mM PIPES, 2 mM MgCb, 1 mM EGTA, and 60 % glycerol (v/v).
Beckman Coulter Optima™ TLX Ultracentrifuge (120,000 maximum rpm).
Beckman TLA-100 fixed angle rotor (7 ×20 mm, 0.2 mL tube).
2.6 β-Casein Preparation
Bovine β-casein, 1 g (Sigma, Cat. No. C6905), store at −20°C upon arrival.
50 mL of 20 mM Tris-HCl buffer, pH ~7, chilled.
0.5 M NaOH for pH adjustment.
pH test strips (Fisher Scientific).
Ultracentrifuge with Beckman Ti 70.1 rotor and 10.4 mL centrifuge tubes (Beckman, Cat No. 355603).
50 mL syringe with 0.22 11m filter.
Glass Pasteur pipette with long tip.
2.7 Sample Preparation: MT Attachment and Surface Passivation
BRB80 buffer (see Subheading 2.5).
25 mg/mL bovine β-casein (see Note 17 and Subheading 3.6).
10 mM paclitaxel (Sigma) in DMSO.
1 M DTT (see Subheading 2.5).
Glutaraldehyde-functionalized slide chamber.
Cy3-labeled MTs (see Subheading 3.5).
Pieces of filter paper cut into strips ~ 2 in. long and ~0.5 in. wide.
Vacuum grease (see Note 18) and cotton-tipped applicator.
3 Methods
3.1 Cover Slip Aminosilanization
This protocol describes how to amino-functionalize glass cover slips for subsequent treatment with glutaraldehyde. These cover slips are used as substrates for MT immobilization. The glass is first cleaned and activated (Fig. 2) and then treated with an aminopropyltriethoxysilane. The relevant chemistry [21, 26, 29, 32-36] is presented in Fig. 3 and the aminosilane surface coating strategy is outlined in Fig. 4 (see Note 19):
Fill bath sonicator with water and set temperature to 45°C.
Set oven to 110°C.
Rinse out the Mucasol jar with ddH2O and fill it nearly to the top with 2 % Mucasol.
Rinse off the cover slip holders and put in the cover slips. Wear gloves to avoid getting finger oils on the glass (handle only the edges) and use forceps to place the cover slips in the holders. From this point forward, avoid any direct contact with the cover slips, and handle only the holders. We typically prepare two racks (24 cover slips) at a time.
Put cover slip holders in the Mucasol jar (one holder on top of the other), fill to the top with 2 % Mucasol, and cap the jar. Optional: For stability, insert a plastic “wedge” (a piece of a 15-mL conical tube works well) in the jar to prevent the cover slip holders from moving.
Submerge the body of the Mucasol jar in the bath sonicator (just up to the cap; see Note 8).
Run sonicator on degas setting for 5 min. Then sonicate for 25 min.
Rinse the ddH2O jar well with ddH2O and then fill it. Using forceps, transfer cover slip racks directly from the Mucasol jar to the ddH2O jar. Rinse under running ddH2O for about 3–5 min, gently swirling and periodically dumping the water from the jar. Minimize exposure to the air (see Note 20).
After rinsing, fill the ddH2O jar and sonicate for 5 min. Rinse again briefly. This is an acceptable place to stop and resume at a later time (leave the cover slips in water until proceeding).
Set a paper towel on a flat, stable surface next to a clean piece of aluminum foil that is big enough to wrap around the cover slip holder. Remove one cover slip holder at a time from the jar and tap it on the paper towel a few times to remove water (firmly, but without knocking the cover slips out of their slots). There should be very little water left. Immediately place the holder on the aluminum foil and wrap the foil around it. The holder should be protected from dust, while still allowing some air to circulate. Keep the holder in the foil until step 17.
Put all racks in the oven until all cover slips are completely dry (~30 min). They should look completely clean by eye.
Remove racks and let them cool for a few minutes. Then place them in the plasma cleaner (still in the aluminum foil covers). Evacuate the chamber to ~800 μtorr and turn on the RF coil on “high” for 5 min (see Note 21). Then turn off the cleaner, return the chamber to atmospheric pressure, and remove the cover slips.
Optional: test one cover slip by placing a drop of approximately 20 μL of ddH2O on the center of the glass surface. It should spread evenly over the glass without beading at all, indicating the highly hydrophilic nature of the plasma-treated surface (Fig. 2c).
Leave the racks (still covered) on the bench for 15 min (see Note 22).
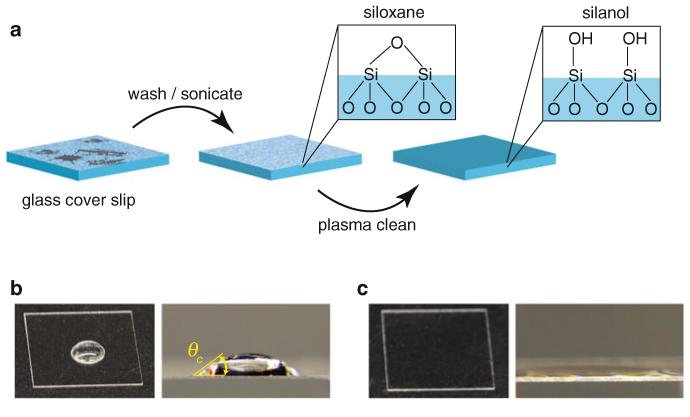
Fig. 2.
Glass preparation for aminosilanization. (a) Borosilicate glass cover slips are sonicated in an alkaline (pH ~11.5) phosphate detergent to remove gross surface contaminants (and possibly a very thin surface layer of the silicon dioxide network that forms the glass structure). Next, plasma cleaning removes any residual organic contaminants and converts surface siloxanes to silanol groups that are more reactive with aminopropyltriethoxysilanes. (b) Prior to plasma cleaning, 20 μL of ddH2O deposited on the cover slip surface forms a bead (left), which, when viewed from the side (right), forms a dome shape with a non-negligible contact angle, θc (even if one attempts to spread the drop over the surface). (c) Following plasma cleaning, 20 μL of ddH2O flows evenly over the highly hydrophilic glass surface and does not form a bead (left). The contact angle is greatly reduced (and difficult to observe; right)
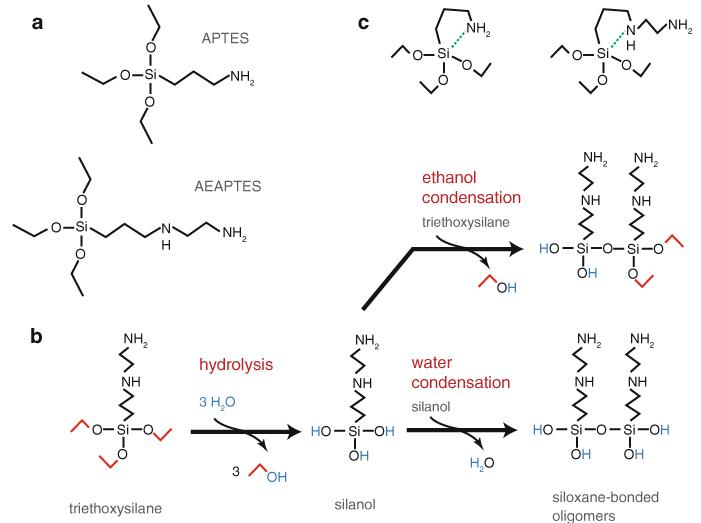
Fig.
3 Aminopropyltriethoxysilane structures and reaction scheme. (a) Chemical structures of 3-aminopropyltriethoxysilane (APTES) and N-(2-aminoethyl)-3-aminopropyltriethoxysilane (AEAPTES). (b) General reaction scheme for aminopropyltriethoxysilanes, shown for the specific case of AEAPTES. Exposure to water leads to hydrolysis of the ethoxy groups, yielding two alcohols, a silanol and an ethanol leaving group. Silanols can undergo condensation reactions either with each other (water condensation) or with unhydrolyzed silanes (ethanol condensation) to form siloxane bonds and yield oligomers. The water released by the condensation reaction can participate in hydrolysis of additional ethoxy groups. Thus, even small amounts of water can catalyze silane polymerization. Uncontrolled polymerization yields complex, disordered networks that form a viscous sol–gel [36]. Note that the reaction scheme shown here is a simplified, conceptual summary and that hydrolysis and condensation can potentially occur on different parts of each molecule simultaneously. Silane polymerization is catalyzed by the addition of ammonia (not shown). (c) Aminopropyltriethoxysilanes for five-membered ring structures (left APTES, right AEAPTES), allowing the amino group to intramolecularly catalyze silane polymerization, even in the absence of water
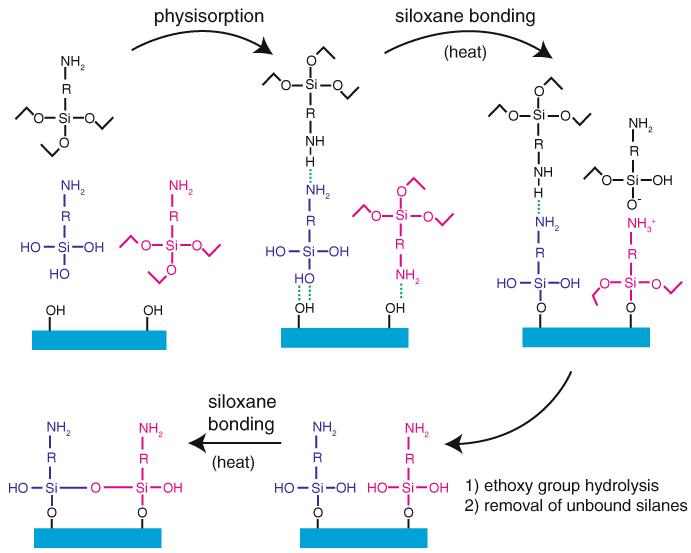
Fig. 4.
Aminopropyltriethoxysilane functionalization of glass using APTES (R=–(CH2)3–) or AEAPTES (R=–(CH2)3–NH(CH2)2–). An appropriately cleaned glass surface containing a high density of silanol groups is exposed to the aminosilane in nearly anhydrous acetone, yielding a diverse mixture of free silanes and silanols in solution, which then physisorb onto the surface via hydrogen bonding and/or ionic interactions (several configurations in addition to the ones shown are possible). The addition of heat drives the formation of siloxane bonds between the physisorbed silanes/silanols and the glass surface by supplying energy and removing condensation products (water and ethanol) by evaporation. This reaction is catalyzed by the terminal amine group. Rinsing in ethanol and water removes any remaining physisorbed aminosilane deposits and leads to hydrolysis of remaining ethoxy groups on the bound silanes, converting them to silanols. A final heating/drying helps these silanol groups form intramolecular siloxane linkages (siloxane bonds may also form via reaction of adjacent ethoxy and silanol groups, as during the initial bonding to the surface). This conceptual scheme does not illustrate several concurrent pathways that also lead to stable binding of aminosilanes to the glass surface (e.g., aminosilane oligomerization in solution, followed by physisorption and binding to the surface). The final product of this treatment is a glass surface densely covered in covalently attached amines
Perform steps 15–22 in the fume hood:
-
15
Fill the Acetone jar with 125 mL acetone and the Silane jar with 100 mL acetone. Measure an additional 125 mL in a graduated cylinder for use in step 18.
-
16
Add 2 mL aminosilane (APTES or AEAPTES) to make a 2 % v/v solution (μ80 mM). Gently swirl the liquid in the jar to mix well (see Note 23 regarding safety!).
-
17
Using a metal forceps, dip each cover slip rack in the Acetone jar several times. Immediately dip each rack in the Silane jar for 10 s, gently agitating the jar by hand (see Note 24). After treating the first rack, immediately proceed to step 18 before repeating steps 17 and 18 for the second rack.
-
18
Remove the cover slip rack and again dip repeatedly in the Acetone jar to remove excess, weakly physisorbed aminosilane.
-
19
Dump the contents of the Silane jar into an appropriate waste container. Transfer the contents of the Acetone jar to the Silane jar, and fill the Acetone jar with the remaining 125 mL of acetone in the graduated cylinder. Dip the cover slip racks repeatedly in the Acetone jar.
-
20
Tap each cover slip rack dry on a paper towel, cover in aluminum foil, and bake for 1 h at 110°C.
-
21
Dispose of the contents of the Silane jar. Swirl the acetone in the Acetone jar to rinse the walls thoroughly, and then transfer it to the Silane jar. Rinse the walls well. Rinse forceps in the acetone briefly to remove aminosilane, and dispose of the acetone (see Note 25).
-
22
Remove cover slip racks from the oven and let them cool for a few minutes. Then place them in the Ethanol jar and fill it with spectrophotometric ethanol (ethanol can be reused over several preparations). Sonicate for 10 min (see Note 26).
-
23
Transfer the cover slip racks to the ddH2O jar and rinse well in ddH2O. Sonicate 5 min in ddH2O and rinse again briefly.
-
24
Repeat steps 10 and 11.
-
25
Store the aminosilanized cover slips in the cover slip holders in a vacuum desiccator containing Drierite (anhydrous CaSO4) (see Note 27).
3.2 Microscope Slide Chamber Construction
This protocol describes constructing a microscope slide chamber for use in single-molecule microscopy. The chamber consists of an aminosilanized cover slip fixed to a standard microscope slide by two strips of Parafilm. The chamber is subsequently treated with glutaraldehyde to facilitate the covalent binding of MTs to the cover slip. After flowing reagents into the chamber, it is sealed using vacuum grease:
Optional: clean microscope slides in 2 % Mucasol, as done for cover slips prior to silanization.
Cut a sheet of Parafilm into strips of ~5 mm in width (see Note 28).
Cut two pieces from the Parafilm strips of ~1 inch (2.5 cm) in length.
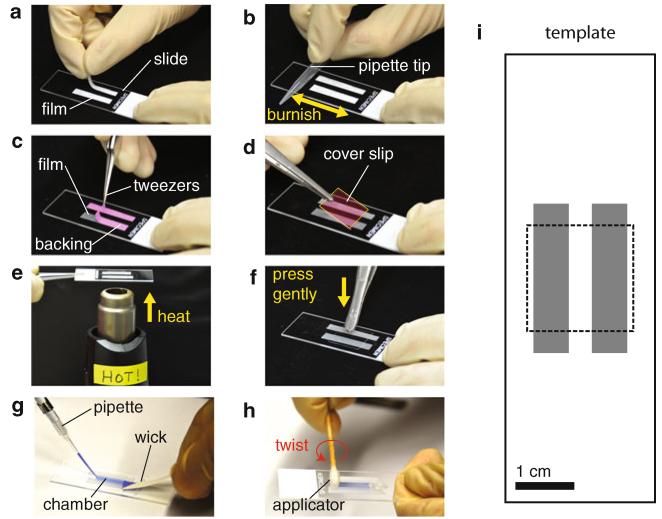
Wear gloves. Place a slide on top of a photocopy of the template provided in Fig. 5i. Hold it firmly in position with one hand. With the other hand, put each piece of Parafilm (film side down, so it sticks) carefully along the edge of the gray rectangle that demarcates the chamber (Fig. 5a). Once the pieces are in the proper position, push down lightly in a few spots with a finger to help them stick.
Use a small cylinder (e.g., a 1 mL plastic pipette tip) to burnish the film onto the glass (Fig. 5b). Rub with even pressure across both strips several times to make good adhesion (see Note 29).
Use the pincers on a pair of sharp-nosed forceps to “pick” at the paper backing on one of the strips of Parafilm until it begins to separate. Once the backing is separated from the Parafilm, use the tweezers to pull it off completely (Fig. 5c; see Note 30). Remove the backing from both pieces of Parafilm.
Position the slide over the template again. Use a forceps to pick up an aminosilanized cover slip by the corner (so as not to contaminate the “useful” area that will form the surface of the chamber). Carefully place it in position on the template (Fig. 5d, see Note 31).
Lightly tap the cover slip with the back of the tweezers over the part touching the Parafilm (this is just to make it stick weakly so it does not fall off).
Pick up the whole slide chamber with the tweezers, cover-slip-side-up. With the heat gun on the table pointing upward, turn it on “high.” Hold the center of the slide over the heat gun, about 1/2in. away (Fig. 5e). As the Parafilm heats, it becomes transparent. Wait ~1 s after this happens and remove the slide quickly.
Hold the slide chamber firmly with forceps (or set on a clean surface). Then quickly use the back of the tweezers to lightly press or tap on the cover slip regions above the Parafilm (Fig. 5f; this forms a tight seal between the two pieces of glass). Set the slide chamber aside to cool (Parafilm will turn opaque).
Store the chamber in the vacuum desiccator for later use, or proceed immediately to glutaraldehyde treatment (see Note 32).
Fig. 5.
Slide chamber preparation. (a) Parafilm is applied to a glass slide and (b) burnished with a clean pipette tip so it adheres to the glass. (c) The paper backing is removed with a pair of sharp-nosed tweezers (paper backing is pseudo-colored pink in this image to enhance visibility). (d) An aminosilanized glass cover slip is carefully placed on top of the exposed film strips (the cover slip is pseudo-colored pink and outlined in yellow in this image to enhance visibility) and lightly pressed down (not shown), and (e) the slide chamber is heated briefly (cover-slip-side-up) until the Parafilm becomes transparent. (f) The cover slip is then gently pressed with a tweezers in order to form a tight bond with the parafilm. After treating the chamber with glutaraldehyde (not shown), (g) microtubule solution and then trapping assay solution are introduced from one end of the chamber while simultaneously using a filter paper “wick” on the opposite end to help draw the solution through (in the photograph, a blue dye is used instead of trapping solution in order to enhance visibility). (h) The chamber is then sealed using a cotton-tipped applicator saturated in vacuum grease. By twisting the applicator at the entrance of the chamber, grease is swept into the mouth of the chamber, forming a perfect seal. (i) Using a template during steps (a-d) helps ensure consistent chamber volumes and cover slip placement. The template provided has a chamber width of 4 mm, yielding a volume of ~10 μL (Color figure online)
3.3 Slide Chamber Glutaraldehyde Treatment
Here we provide a protocol for activating the aminosilanized surface of the slide chamber with glutaraldehyde (Fig. 6), to enable subsequent covalent attachment of MTs (see Note 33). This is a widely used method for protein immobilization [30], and similar methods have been used by others for optical trapping assays [31]. We generally prepare several slides at once and store them dry for later use (similar to other reports [37, 38]), but this procedure may also be done immediately prior to an experiment:
Prepare a “humidity chamber” by placing a damp Kimwipe in a tight-sealing box large enough to hold several slide chambers. An empty pipette tip box works well (see Note 34).
Remove the 25 % glutaraldehyde from the freezer (one 10 μL aliquot per 3 slide chambers to be treated). Centrifuge briefly and then dilute 1:2 in chilled, ultrapure ddH2O (e.g., 10 μL glutaraldehyde + 20 μL ddH2O) to yield ~8 % glutaraldehyde (see Note 35).
Set each slide chamber to be treated cover-slip-side-up in the humidity chamber.
Fill each slide chamber with glutaraldehyde solution using a pipette. Pipette the solution into the mouth of the chamber, and it will be drawn in via capillary action (see Fig. 5g; no filter paper is needed for this step). Use approximately 10 μL per chamber, adding solution just to the point that ~1 μL starts to pool at the outlet of the chamber.
Cover the humidity chamber and incubate at room temperature for 30 min.
After the incubation, use a folded Kimwipe to wick the solution out of each chamber (it helps to hold the slide upright, drawing the solution out of the bottom end). Remove all of the solution (see Note 36).
To remove residual unreacted glutaraldehyde, rinse each chamber three times with 200 μL of ultrapure ddH2O, wicking it through with Kimwipes (see Note 37).
Dry each chamber with filtered, compressed air. Use a pipette tip on the end of the air line to direct air through the chamber and hold a Kimwipe on the opposite end to collect the water. Blow air only in the direction of solution flow during steps 6 and 7. When completely dry, store the chambers in the vacuum desiccator.
Fig. 6.
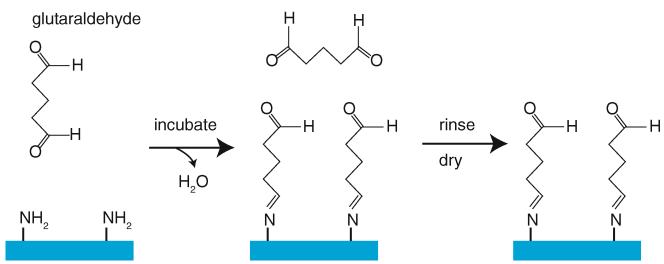
Glutaraldehyde treatment of amino-functionalized glass. The bifunctional glutaraldehyde binds surface amine groups via a hydrolysis reaction, thus functionalizing the surface with formyl groups that will bind surface-exposed Iysines on proteins. Extensive rinsing removes any free glutaraldehyde that could yield unwanted reactions in the final assay. Finally, the glass is thoroughly dried and stored under vacuum
3.4 Labeling of Tubulin with Cy3 Dye
This protocol explains how to label purified tubulin with the cyanine dye Cy3 (Fig. 7). This enables visualization of fluorescent MTs in the microscope using 532 nm laser excitation. The protocol is modified from the one given by Peloquin et al. [39]:
Add 20 μL cold PM buffer to 1 mg bovine brain tubulin (1 vial of Cytoskeleton tubulin). Dissolve and mix well; keep the tube on ice.
Add 1 μL of 25 mM Mg-GTP and 2.5 μL DMSO to the tubulin; incubate at 37°C for 15 min.
Mix 2 μL Cy3 monofunctional NHS ester with the polymerized tubulin and incubate at 37°C for 40 min in the dark (see Note 38).
Transfer the mixture to a clean 0.2 mL TLA-100 tube. Remove the excess dye by centrifugation for 5 min at 37°C at 37,000 rpm (53,000 rcf, average; k-factor48.1).
Carefully remove the supernatant, then add 40 μL cold PM buffer to resuspend the pellet. Incubate on ice for 15 min to depolymerize the MTs. Cool rotor to 4 °c.
Spin the solution for 5 min at 4 °C at 37,000 rpm (53,000 rcf, average; k-factor 48.1) to remove insoluble fraction.
Move supernatant to a clean new tube, add 2 μL of 25 mM Mg-GTP and 5 μL DMSO, and then incubate at 37°C for 15 min in the dark. Warm rotor to 37 °c.
Spin the solution for 5 min at 37°C at 37,000 rpm (53,000 rcf, average; k-factor 48.1).
Discard supernatant, add 25 μL cold PM buffer, then incubate on ice for 15 min. Cool rotor to 4°C.
Spin the solution at 37,000 rpm (53,000 rcf, average; k-factor 48.1) for 5 min at 4°C.
Move supernatant to a clean 1.5 mL microcentrifuge tube. Use Nanodrop or other microliter UV spectrometer to determine the concentration of both tubulin and Cy3 dye, and calculate the labeling ratio (equivalent to labeled/unlabeled tubulin). Aliquot to appropriate amount, flash freeze, and store at −80°C.
Fig. 7.
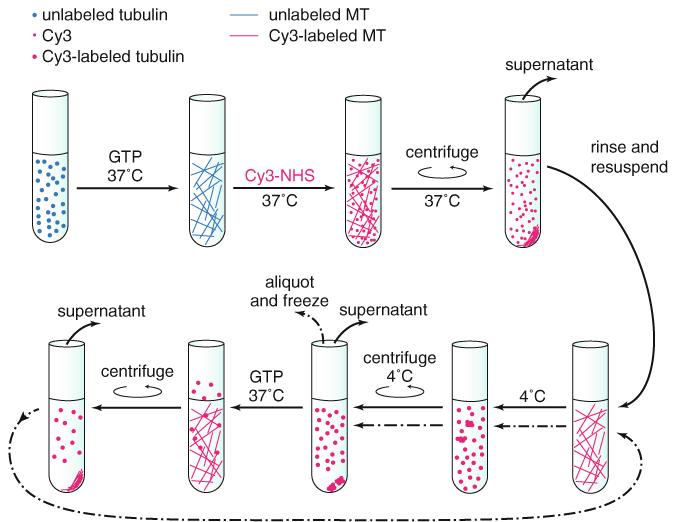
Labeling of tubulin with Cy3 dye. Tubulin is first polymerized into MTs. Cy3 conjugated to the reactive N-hydroxysuccinimide (NHS) is then added, which facilitates attachment of the dye to amine groups on the surface of the MT. After pelleting the MTs and removing excess dye, the MTs are resuspended and depolymerized in the cold. The insoluble fraction is then pelleted and the supernatant transferred to a new tube in which the tubulin is repolymerized, pelleted, and depolymerized again, followed by aliquotting and snap freezing. This strategy ensures that the tubulin is labeled in regions other than the key interfaces required for polymerization and that the final product contains only tubulin capable of cyclic polymerization and depolymerization (and thus unperturbed by the attached dye)
3.5 Cy3-Labeled MT Polymerization
This procedure yields fluorescently labeled MTs suitable for optical trapping and other single-molecule assays (Fig. 8).
Fig. 8.
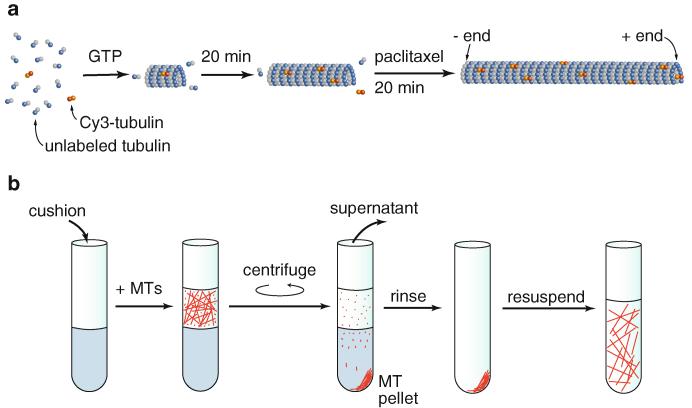
MT preparation. (a) Addition of GTP to free tubulin (αβ-tubulin dimers) induces MT polymerization (promoted by addition of glycerol [40, 41] and 37°C temperature). When preparing fluorescent MTs, the small amount <5 %) of tubulin labeled with the organic fluorophore Cy3 incorporates randomly into the MT lattice and is distributed sparsely enough that the dye molecules do not affect motor interaction with the MTs in the optical trapping assay. Initially, MTs exhibit dynamic instability [42, 43]. Paclitaxel greatly enhances polymerization and stabilizes the MTs [44-49] (the DMSO in which paclitaxel is dissolved also enhances MT polymerization [50]). (b) Removal of residual-free tubulin and very short MT fragments is accomplished by sedimentation through a 60 %-glycerol “cushion.” First, MTs are layered carefully on top of the cushion. Following centrifugation, the free tubulin and very short MTs remaining in the supernatant are removed. The MT pellet is then resuspended in buffer. These MTs are stable for days to weeks at room temperature
Skip steps 1–4 if 20 μL aliquots of 10 mg/mL unlabeled tubulin is already prepared.
Set ultracentrifuge to 4 °C with TLA-100 rotor inside.
Spin a vial of lyophilized, unlabeled tubulin (1 mg) briefly in tabletop centrifuge at maximum speed. Put the vial on ice, and add 100 μL BRB80 (to make μ10 mg/mL tubulin solution). Pipette gently to dissolve, avoiding formation of air bubbles.
Transfer the tubulin solution to a Beckman centrifuge tube (343775 or 342303), and centrifuge at 80,000 rpm (250,000 rcf, average; k-factor 10.3) for 10 min. Transfer the supernatant to a 0.5 mL microcentrifuge tube on ice, leaving any pellet (precipitated and/or polymerized tubulin) behind.
Make 20 μL aliquots, flash freeze in liquid nitrogen, and store at −80°C.
Skip step 5 if 5 μL aliquots of 10 mg/mL (labeled/unlabeled ≈ 1:22) tubulin is available.
-
5
Remove 1 aliquot each of Cy3-labeled tubulin and unlabeled tubulin from the freezer. Thaw quickly in the hand and immediately place on ice. Combine the labeled and unlabeled tubulin (this gives approximately 10 mg/mL total tubulin, labeled/unlabled≈ 1:22; see Note 39). Make 5 μL aliquots, flash freeze, and store at −80°C.
-
6
Thaw a 5 μL tubulin aliquot from step 5.
-
7
Dilute 1 μL of 1 M DTT in 49 μL ddH2O, to yield 20 mM DTT.
-
8
Prepare polymerization buffer: 2 μL glycerol cushion (see Note 40), 1 μL of 25 mM Mg-GTP, 6.5 μL BRB80, 0.5 μL of 20 mM DTT (from step 7). Add 5 μL polymerization buffer to the 5 μL tubulin aliquot. Mix gently with the pipette (avoid bubbles).
-
9
Incubate the tubulin solution for 20 min in a 37°C water bath to start polymerizing the tubulin into MTs. Protect from light to avoid photobleaching of the fluorescent dye.
-
10
Dilute 1 μL of 2 mM paclitaxel in 9 μL DMSO (200 μM paclitaxel). Add 1.1 μL of this to the MTs (20 μM paclitaxel final). Mix gently.
-
11
Incubate MTs at 37°C for an additional 20 min (see Note 41). During the incubation, set the ultracentrifuge to 25°C (see Note 42) with the TLA-100 rotor inside.
-
12
In a Beckman centrifuge tube (343775 or 342303), add 60 μL of glycerol cushion and 0.7 μL of 2 mM paclitaxel and mix (for a balance tube, use 60 μL glycerol cushion and 10 μL H2O).
-
13
Carefully layer the 10 μL of MTs onto the glycerol cushion (do not disturb the cushion with the pipette tip). Mark the outside edge of the tube with a permanent marker to help find the pellet position after the centrifugation.
-
14
Centrifuge at 80,000 rpm (250,000 ref, average; k-factor 10.3) for 10 min. During the centrifugation, prepare the wash buffer: 150 μL BRB80 plus 1.52 μL of 2 mM paclitaxel.
-
15
Following the centrifugation, the pellet, containing the MTs, will be toward the outside wall of the tube (the side marked in step 13) and should have a slightly pink color. Remove ~50 μL of supernatant/cushion, keeping the pipette at the top of the liquid and moving down as it is withdrawn (see Note 43).
-
16
Rinse the walls of the tube above the remaining cushion with 50 μL of wash buffer, and carefully remove it. Then remove the remaining cushion, pipetting from the side of the tube opposite the pellet.
-
17
Gently rinse the pellet with an additional 50 μL of wash buffer, and carefully remove it without disturbing the pellet.
-
18
Resuspend the pellet in 10 μL of wash buffer (~5 mg/mL tubulin; see Note 44). The final mixture will be fairly viscous. Transfer it to a small microcentrifuge tube shielded from the light (use a black tube or cover it in aluminum foil). These MTs are stored at room temperature and can be used in optical trapping assays for a week or more.
3.6 β-Casein Preparation
This procedure yields 25 mg/mL β-casein for use in passivating the aminosilanized cover slip surface:
Remove the β-casein bottle from the freezer and place it on ice.
Add 25 mL of Tris-HCl buffer to bottle. Stir gently with a serologic pipette to dissolve as much as possible. Avoid malcing bubbles.
Use ~15 μL of solution to check the pH on a pH strip. Add NaOH dropwise (~18 drops) until the pH reaches ~8 (see Note 45).
Add another 5 mL of Tris buffer. As solution is added, rinse off the pipette from step 1 to prevent loss of protein. Swirl to dissolve and readjust pH to ~8 with NaOH (~12 drops).
Place the bottle on a rocker in a 4 °C cold room for 1 h. Readjust pH to 8 with NaOH (~4 drops).
Return bottle to rocker in cold room for 2 h more. Readjust pH to 8.
Gently swirl bottle and rotate to get any residue off the walls and into solution. Let the bottle sit upright for 5 min and wash the walls with 5 mL Tris buffer.
Using a serologic pipette, transfer the entire protein solution from the bottle to a 50 mL conical tube. Wash the walls of the bottle with 3 mL of Tris buffer and add it to the conical tube. Then add Tris buffer to bring total volume in tube to 40 mL.
Centrifuge the solution in the ultracentrifuge at 65,000 rpm (388,000 rcf, average; k-factor 42.3) for 20 min at 4°C.
Carefully remove the tubes from the centrifuge. There will be a faint cloudy layer at the top and a very small gray pellet at the bottom. Use the Pasteur pipette to withdraw the middle, clear layer to a clean 50 mL conical tube on ice. This solution should look completely clear.
Sterile filter the solution to a clean conical tube on ice. Aliquot, snap freeze, and store at −80°C (large aliquots can be stored for subsequent thawing and realiquotting).
3.7 Sample Preparation: MT Attachment and Surface Passivation
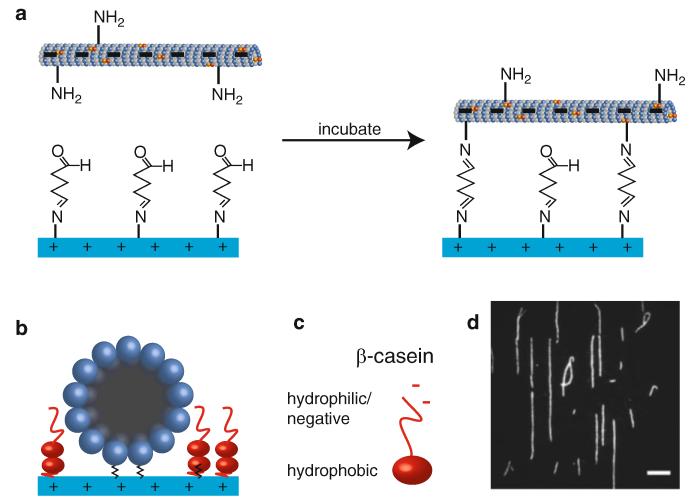
The procedure below describes how to attach MTs to the glass cover slip and block the remaining surface to prevent unwanted adsorption of microspheres during subsequent optical trapping experiments (Fig. 9). At the near-neutral pH of most assay buffers, MTs have negative surface charge (especially at the C-terminal, glutamate-rich tubulin “E-hook” with which kinesin interacts) [51-54], whereas aminosilanized surfaces (pKa = 7-10 [55, 56]) are protonated and thus positively charged. The MTs are thus rapidly adsorbed to the glass, whereupon they covalently bind the formyl groups left from previous glutaraldehyde treatment. During the MT incubation, reagents for the desired assay should be prepared (e.g., molecular motors bound to optical trapping beads in the presence of ATP; see Chapter 10). Following blocking with β-casein, the reagents are introduced to the flow chamber, the chamber ends are sealed, and the sample is taken to the microscope for the experiment.
Fig. 9.
MT immobilization in aminosilane-/glutaraldehyde-treated slide chambers. (a) The negatively charged MT is attracted to the positively charged glass surface. Following incubation, reactive formyl groups attached to the glass bind to amines on surface-exposed Iysines on the MT, thus covalently linking the MT to the cover slip. (b) End-on view of MT attached to glass, with remaining surface passivated with β-casein (introduced to the chamber at 2 mg/mL). The β-casein, an amphiphile capable of adsorption on a variety of surfaces, forms a bilayer on the glass surface that prevents unwanted interactions with trapping microspheres, and its lysine residues react with any remaining formyl groups. (c) The β-casein N-terminal hydrophilic region is negatively charged, allowing favorable interactions with the positively charged glass surface. The top layer extends this hydrophilic region into the solution, forming a “brush” layer on the surface that prevents trapping beads from sticking. (d) Fluorescence image of Cy3-labeled MTs covalently bound to the cover slip in a microscope flow chamber. They are very well aligned with the long axis of the chamber due to combination of the laminar flow induced when filling the chamber and the highly favorable initial adhesion to the positively charged glass. Scale bar: 10 11m
Prepare the following freshly at the beginning of a set of experiments:
“BRB/Tx”: 350 μL of BRB80, 0.5 μL of 10 mM paclitaxel. Keep at room temperature.
Blocker: 156 μL of BRB/Tx, 14 μL of 25 mg/mL β-casein. Keep at room temperature.
“MT30”: 0.5 μL MT stock (10 mg/mL), 14.5 μL BRB/Tx (i.e., 30× dilution). Keep at room temperature and protect from the light (can be used for several days).
Do the following for each experiment:
-
4
Mix the MT30 suspension gently to evenly distribute the MTs. Add 0.5–0.8 μL MT30 (depending on the desired MT density on the cover slip) to 10 μL BRB/Tx. Flow this into a glutaraldehyde-treated flow chamber, using a piece of filter paper (see Fig. 5g) waiting on the opposite end in order to get very good flow through the chamber (this aligns the MTs with the chamber’s long axis) (see Note 36 regarding the direction of flow).
-
5
Incubate the MTs in the chamber for 20–30 s and flush the chamber with 40 μL of BRB/Tx. Incubate 20 min or longer (see Notes 46 and 47).
-
6
During the MT incubation, prepare the reagents for the main assay (see, e.g., Chapter 10 for a protocol to measure kinesin force generation), timed so that all incubations end at approximately the same time.
-
7
About 1 min before the end of the incubations, flush the flow chamber with 40 μL of Blocker and leave it to incubate.
-
8
Flow the assay solution into the flow chamber. Dry the ends with a Kimwipe, wiping away from the center of the chamber and taking care not to suck solution out of the chamber itself. Seal the chamber with vacuum grease as shown in Fig. 5h (see Note 48). Avoid getting any grease on the surface of the cover slip that will contact the objective. When finished, wipe away any excess grease (see Note 49).
Acknowledgements
We thank Johan Andreasson, Robert Coleman, and Wei Chen for helpful discussions regarding aminosilanization and Laura E.K. Nicholas for assistance with photography and figure illustration. The authors are supported by the National Institutes of Health grant R01GM098469. M.P.N. received support from the NIH-funded Medical Scientist Training and Molecular Biophysics Training programs (NIH grants T32GM007288 and T32GM008572, respectively) at the Albert Einstein College of Medicine.
Footnotes
An additional technique [57], reported to be very effective at attaching MTs while preventing nonspecific absorption, is coating the glass surface with a highly hydrophobic silane. Anti-tubulin antibodies are then adsorbed to the glass, and the remaining surface blocked with Pluronic® F-127 (Sigma), a poloxamer (triblock copolymer) of polypropylene glycol (which is hydrophobic and sticks to the hydrophobic surface) flanked on either side by polyethylene glycol (which is hydrophilic and forms a protective “brush” layer in the solvent above the glass) [57]. F-127 has been used to minimize bead-surface interactions in optical trapping experiments in the absence of protein [58, 59], but we have not tested this method.
Zeiss cover slips are manufactured by Schott/Marienfeld Superior (Lauda-Konigshofen, Germany). While other cover slips will probably work acceptably, we use these because of the high quality of the glass and more importantly because of their highly precise thickness of 170 ± 5 μm. Individual No. 1.5 cover slips within typical lots from most manufacturers can vary in thickness from 160 to 190 μm, potentially leading to a significant deviation from the 170 μm cover slip thickness for which most oil-immersion microscope objectives are designed. This can induce unwanted spherical aberration that broadens the width of the focused laser beams (e.g., optical trapping beams) in all dimensions, as well as degrading imaging quality in fluorescence microscopy. This problem can be avoided by manually measuring each cover slip with a micrometer before use (and discarding those that are too thin or thick) or by purchasing cover slips like the ones recommended, for which there is very little variability in thickness.
We find porcelain cover slip holders the most convenient because they fit nicely in our jars and tend to sit very stably on the bench. However, they are expensive. Cheaper racks made of other materials may be substituted as long as they are resistant to 100 % acetone and ethanol and can withstand temperatures up to 110°C. Polytetrafluoroethylene (Teflon®) and polypropylene (e.g., Wash-N-Dry™ polypropylene racks) are acceptable alternatives.
Many water purifiers have a low flow rate, making the rinsing steps in the cover slip preparation protocol more time consuming and less effective. If the flow rate is less than ~10 mL/s, we advise filling a clean polypropylene carboy with ddH2O and using the spigot on the carboy to dispense the water.
We have had excellent results with both APTES and AEAPTES, but we prefer AEAPTES due to its increased stability against self-catalyzed hydrolysis by its terminal amine [26]. It is critical to avoid contact with humidity during storage, to prevent polymerization of the aminosilane (even small amounts of water can catalyze extensive polymerization over time). Preferably, the reagents should be stored under nitrogen or argon in a bottle sealed with a rubber septum (e.g., Sigma Cat. No. 440140), so that no direct contact between the stock solution and room air occurs. Alternatively, we aliquot the aminosilanes into 2 mL volumes, flash freeze in liquid nitrogen, and store in an airtight plastic bag with Drierite desiccant (anhydrous CaSO4) at −80°C until use. If this method is used, the aliquot should be brought to room temperature in a desiccator, to prevent water condensation. Some authors recommend distilling APTES regularly to remove polymers [60]. However, we have obtained satisfactory results with aminosilanes properly stored for several months. See also Note 24.
We use very high (>99 %) purity reagents for cover slip cleaning and surface preparation, in order to avoid contaminating the glass surface with undesired chemicals (especially organic molecules, which may interfere with surface chemistry by binding the substrate and/or lead to background fluorescence during microscopy).
Any non-glass jars that are resistant to the chemicals used can be substituted (e.g., other jars composed of PMP). We recommend using a separate jar for each reagent (water, Mucasol solution, ethanol, acetone, and silane solution) to minimize cross-contamination and any potential unwanted reactions that might result. Keep jars tightly closed when not in use.
To suspend the jars in the sonicator bath, we use a simple homemade holder fashioned from a polystyrene box lid. Use a scalpel to cut a recess in the lid and a hole through which the jar body (but not the lid) can pass. Set this holder on top of the sonicator, and insert the jar, such that as much of the jar as possible is submerged in the bath without making physical contact with any of the walls of the sonicator.
The cover slips are baked at approximately 110°C a few different times during preparation. If an oven is unavailable, we have found that a hot plate or other heater can be used as an “oven” by simply placing an aluminum foil “tent” overtop of it. Simply adjust the heating element such that the air temperature inside the tent is approximately 110°C. For whatever baking method is used, it is important that the inside of the oven be clean. Baldng cover slips in an oven with dust, or other contaminants that get vaporized during heating, can foul the glass surface and thereby ruin the effects of the prior cleaning steps.
We prefer to use Parafilm in place of the double-sided tape often employed in constructing microscope slide chambers because it attracts fewer contaminants (e.g., dust), it is easier to place on the slide, and it allows slides to be prepared in large numbers for later use (versus having to apply the tape immediately before placing the cover slip). In addition, we have observed glue dissolving along the edges of the chamber over time in flow chambers made with tape (visible as whitish streaks along the chamber edges). This is in agreement with work by Schäffer et al. [58], who reported direct evidence of solution contamination in flow chambers constructed with tape.
Glutaraldehyde can polymerize extensively in aqueous solution (see [27, 28] for a comprehensive review). We store it at −20°C to limit this behavior.
The compressed air typically available at the lab bench is generally not suitable for drying chambers, as it may contain abundant contaminants (e.g., particulate matter, water vapor, and oil droplets from air pumps). Instead, highly pure, compressed nitrogen gas is generally used, with an oil-free filter system at the output of the air line. However, in practice, we have found that the bench air in our lab can be used by employing the following system: A ~1 m hose filled with Drierite is attached to the air line, with two 0.2 μm syringe filters in series at the output. This provides air of sufficient dryness and absence of contaminants for preparing chambers for routine optical trapping studies.
Labeling tubulin with Cy3 is not absolutely necessary, and Cy3-labeled tubulin may be substituted with another fluorescently labeled tubulin, e.g., carboxytetramethylrhodamine (TAMRA)-labeled tubulin (Cytoskeleton, Inc.). However, Cy3 is brighter and more photostable than rhodamine derivatives, allowing a lower labeled/unlabeled tubulin ratio to be used while still easily visualizing MTs. Finally, whereas rhodamine MTs suffer light-induced structural damage resulting from the formation of reactive oxygen species (ROS) [61, 62], we have found Cy3 MTs to be less affected (we also use an oxygen scavenging system to eliminate ROS in either case).
Tubulin may be purified in the laboratory relatively easily, which is generally far more economical. Traditionally, the approach is to purify tubulin by cycles of polymerization, sedimentation, and depolymerization [63, 64] (followed by removal of MAPs with a phosphocellulose column), and abundant protocols are available both via laboratory websites and the literature [65-67]. Recently, a one-step, high-efficiency purification method was developed [68] that may also prove useful (and may permit the isolation of tubulin from a wider variety of sources).
DMSO is preferred over ddH2O as solvent: NHS ester has a fast hydrolysis rate in water, especially at high pH (after hydrolysis, it can no longer react with NH2 groups of amino acids); moreover, the dye is more soluble in DMSO.
The vial should always be kept on ice, since the tubulin will polymerize at higher temperature.
Caseins (chemical properties reviewed in refs. [69, 70]), the major proteins in milk [71], are highly amphipathic, giving them surfactant-like properties [72, 73]. As reported previously [20], casein is very effective in passivating an aminosilanized surface to prevent protein binding. In our hands, 1 mg/mL β-casein was superior to BSA (even up to 10 mg/mL) and Pluronic F127 in blocking the aminosilanated cover slip surface and preventing trapping microspheres from binding. Free microspheres do not bind to the cover slip surface, and there are no measureable binding forces when a trapped bead is dragged along the surface. We use β-casein rather than whole casein because it is available in higher purity and we found it dissolves much better. Moreover, it is resistant to denaturation/precipitation induced by DTT (β-casein contains no cysteine residues [71]) or elevated temperatures [74], and forms stable micelles even at low pH [75]. Although it is a natively unstructured protein [76, 77], β-casein nevertheless has a domain structure, with a highly negatively charged N-terminal region and a hydrophobic C-terminus. This allows it to interact favorably with both hydrophilic and hydrophobic surfaces [72, 76, 78, 79], forming a “brush” layer on the surface with the hydrophilic N-termini pointing up into the solvent (Fig. 9a, b). These properties make it an excellent choice for blocking APTES surfaces, since different parts of the protein can interact favorably with both the positively charged amine and the aliphatic chain of APTES.
Slide chamber ends should be sealed to prevent solution evaporation (which can markedly change concentrations), limit exposure to oxygen (which can produce ROS, especially in the presence of microsphere and fluorescent dye interactions with laser light), and provide physical closure of the chamber. Vacuum grease is highly preferred to nail polish for this purpose, since it can be applied more precisely (thus limiting the possibility for damage to valuable microscope optics by accidental contact) and because it is immiscible in aqueous solution (as opposed to the acetone solvent used in nail polish). When using nail polish as sealant, Schäffer et al. [58] reported contamination of the chamber solution, and we observe regions of casein precipitation near the chamber edges that grow toward the center over minutes (presumably with similar effects on other proteins). This is accompanied by nonspecific binding of trapping microspheres to the cover slip and MTs. These problems are not encountered when using vacuum grease.
The optimal conditions (e.g., temperature, solvent vs. vapor deposition, solvent type, silane concentration, etc.) for silane deposition are still a topic of debate and ongoing research [25, 26]. Based on our review of the literature, and our desire to keep our protocol as simple as possible, we use a solution-phase deposition in acetone solvent. Although aqueous deposition protocols are available, we chose to avoid this method because silanes can polymerize extensively when exposed to water. In addition, given the pKa of surface silanol groups (pKa ~4 or ~9, depending on the silanol configuration) on glass [80-82] and amino groups on aminosilanes (pKa,~10) [55, 83], in aqueous solution near neutral pH, the surface silanols carry a negative charge, while the aminosilane carries a positive charge [24, 32, 60, 83]. This causes the silane to physisorb with its amino group toward the glass, which prevents the associated silanol groups on the glass from participating in condensation reactions with APTES silanol groups (though possibly limiting unwanted silane polymerization on the glass surface [24]).
The purpose of the cleaning steps is to remove gross surface contaminants (e.g., dust, oil, and glass particles remaining from the manufacturing process), leaving the glass surface as clean as possible prior to the final plasma cleaning. During the rinsing step, the main goal is to remove all traces of Mucasol, while preventing exposure to any dust that may be in the air.
Plasma cleaning (see refs. [84, 85] for very informative reviews and ref. [86] for discussion of the effects on glass) removes surface contaminants (particularly hydrocarbons) from the glass and induces silanol formation [86] on the surface, thereby rendering it very hydrophilic [87] and amenable to silanization. Plasma cleaners work by exciting the molecules in a low-pressure gas with a radio-frequency electromagnetic wave. The energized gas emits ultraviolet radiation and eventually forms a plasma consisting of ions, free electrons, and reactive radical species that bombard contaminated surfaces. As contaminants are broken down, they rapidly vaporize, leaving the surface extremely clean. Various mixtures of gas can be used, including room air (i.e., ~78 % nitrogen, ~21 % oxygen, ~1 % argon), which is what we use. Historically, similar cleaning processes have been accomplished by chemical means (reviewed by Kern [88]), in particular “piranha” solution (a 3:1 mixture of sulfuric acid and hydrogen peroxide). This solution is corrosive, violently reactive with organic matter, and potentially explosive [26, 56, 57]. Although piranha is still widely used and can replace plasma cleaning, we highly recommend the latter based on its efficacy, ease, safety, and environmental friendliness.
Following plasma cleaning, the cover slips are left in room air briefly in order to allow a layer of water to adsorb on the surface. During the subsequent treatment with aminosilane, this promotes hydrolysis of amino silane ethoxy groups to silanols specifically near the surface, rather than throughout the solution, thereby favoring surface binding rather than polymerization of unbound aminosilane.
Aminosilanes are corrosive to the skin, eyes, and mucous membranes. Be familiar with the material safety data sheet (MSDS). Handle only in the fume hood and wear appropriate personal protective equipment (nitrile gloves, eye/face protection, and lab coat). If storing under nitrogen or argon (e.g., using a container with a rubber septum seal), use extreme care to prevent spraying bottle contents or overpressurizing the bottle with compressed gas. Any spills or droplets of aminosilane solutions should be cleaned promptly with a Kimwipe with acetone or ethanol. If allowed to dry, spills form white films that are difficult to remove. We perform all aminosilane work on absorbent pads with plastic backing.
Deposition times for aminosilanes vary considerably in the literature and in the technical documentation provided by manufacturers, from minutes to many hours. Prolonged reaction times may lead to formation of complex, branched silane networks and agglomerates on the treated surface [22, 24, 60, 89] that expand upon exposure to water [60]. These networks constitute a dynamic gel that can restructure continuously [36]. In our hands, this short 10 s incubation yields excellent results. For longer incubations (e.g., 30 min, or even 1 min, if using improperly stored aminosilanes, which may have partially polymerized), we have observed behavior consistent with the formation of an aminosilane gel on the surface that hinders interactions between subsequently attached MTs and motors during both optical trapping and single-molecule fluorescence experiments. Motors are unable to bind to and move along particular MTs when cover slips are treated with aminosilane for these longer incubation times. Interestingly, in the absence of initial glutaraldehyde surface treatment, it is precisely the MTs that remain stably bound after introducing β-casein that are unable to support movement, possibly due to the MTs embedding within the silane gel network (see also Note 46). We therefore recommend as short an incubation time as needed to support reliable MT attachment and molecular motor motility on all MTs tested (prior to glutaraldehyde treatment, β-casein should release virtually all MTs from the glass, consistent with the absence of any significant gel network on the surface).
This protocol creates a significant amount of acetone waste. This could be decreased using vapor-phase silanization (rather than the solution-phase silanization employed here), which has also been reported to be less sensitive to ambient conditions (e.g., humidity) [26]. However, this requires additional equipment (e.g., a vacuum chamber compatible with organic vapors). Since the waste acetone should be free of significant contaminants except for aminosilane, and since aminosilanes boil at a much higher temperature than acetone (>200 °C), the acetone could be recycled, e.g., via purification using a combination of Drierite (anhydrous CaSO4, to remove water) and fractional distillation [90, 91].
Washing the silanized cover slips in ethanol and water has three purposes: (1) removal of loosely bound aminosilane (sometimes visible as small white deposits after the initial baldng), (2) hydrolysis of any remaining ethoxy groups in order to form silanols and promote intramolecular siloxane bond formation, and (3) final cleaning to remove any surface contaminants.
Aminosilanized surfaces are easily contaminated and thereby inactivated by organic impurities adsorbed from the air [92]. In addition, carbon dioxide has a well-studied capacity to adsorb onto and react with these surfaces (e.g., refs. [93, 94]), forming carbamates and bicarbonates that will not facilitate glutaraldehyde attachment. In a protocol similar to ours, Jeney et al. [19] recommend storing aminosilanized cover slips for no more than 2 days in order to ensure strong MT attachment. Although storing the treated glass in water or other solution may limit adsorption of airborne contaminants and CO2, the amino silane amine groups can catalyze siloxane bond hydrolysis and release from the surface (i.e., the reverse of the condensation reaction they catalyze during binding) [23, 26, 95], which is more facile in solution (the use of AEAPTES rather than APTES somewhat limits this problem [26]). Storage in vacuum significantly prolongs the activity of aminosilanized surfaces [92, 96]. We store aminosilanized cover slips in a vacuum desiccator with Drierite to remove any residual humidity and leave the vacuum line on in order to ensure the lowest pressure possible over extended periods. Avoid opening the desiccator unnecessarily. Cover slips stored in this manner retain activity for days to weeks. In fact, we have used cover slips stored for over one month. If cover slips are stored for more than 1–2 weeks, we usually clean them by repeating steps 22-25 (sonication in ethanol and water) before using them.
A paper cutter works well to make long, straight, clean cuts. Whatever cutting tool is used, it should be very sharp. Dull cuts will crush the film into the paper backing along the edge and make it difficult to separate them. Furthermore, ragged edges can disrupt smooth fluid flow through the chamber, thereby interfering with good MT alignment on the glass. When cutting, keep the film side up, with the paper backing on the table, so the Parafilm stays clean. These strips can be stored in a clean plastic bag until ready for use.
When burnishing the Parafilm, do not press too hard, as this can either deform the film or make it difficult to remove the backing later. Many slides with two pieces of Parafilm attached can be prepared at once and stored indefinitely in a dust-free box or bag for later use.
Pull parallel to the glass surface more than upward, to avoid pulling the film itself off the glass. If the film does begin to separate from the slide, this indicates it was not burnished sufficiently. Simply press it back down and burnish it again.
Place one edge down over the template and then carefully remove the tweezers, letting the cover slip drop into place. Make an effort to position it perfectly on the first attempt, to avoid sliding the cover slip around on the Parafilm and contaminating the aminosilanized surface. To avoid contamination, it is also advisable to use separate forceps for removing the Parafilm backing and for handling the cover slips.
We prefer to immediately treat the chambers with glutaraldehyde, since the density of amine groups on the surface with which the glutaraldehyde can react will be greatest for newer slide chambers. In addition, treatment with glutaraldehyde may prolong the activity of the chamber by preventing loss of active amines, as happens slowly for an untreated aminosilanized surface. If the slide chambers will be stored for more than a few days, we prefer to seal the ends with small pieces of plastic cling wrap (carefully burnished with a cotton-tipped applicator to form a tight seal). This limits exposure to any residual gases or contaminants in the desiccator and is easily removed prior to use.
The use of cross-linkers such as glutaraldehyde is often a matter of concern. We wish to emphasize that the procedure employed here immobilizes glutaraldehyde on the glass surface, with all free glutaraldehyde subsequently removed. During MT immobilization, only the bottom surface of the MT (which the motor would not make contact with anyhow) is chemically altered, leaving the upper surface in its native form. In addition, it is worth noting that previous reports have demonstrated kinesin motility on glutaraldehyde-fixed MTs at rates similar to those untreated for MTs [97]. Finally, all remaining glutaraldehyde on the glass surface is blocked with β-casein (which contains several lysine residues [71] capable of reacting with the glutaraldehyde) before any motors are introduced.
The purpose of the humidity chamber is to limit evaporation of the slide chamber contents during incubation.
We measure the pH of the 8 % glutaraldehyde solution to be approximately 4.5. This pH is favorable for preventing glutaraldehyde polymerization [27, 28, 30], but is more acidic than that commonly employed for glutaraldehyde reactions, and somewhat unfavorable for reaction with aminosilane surface amine groups (pKa≈7–10 [55, 56]), which must be deprotonated in order to react. On the other hand, it is beneficial if a large portion of the surface amines do not react, so that the aminosilanized surface retains its positive charge. It also is likely unnecessary to create a very high density of surface aldehydes for MT binding, and possibly even undesirable, given the effects of glutaraldehyde on MT morphology in tissue fixation [98].
The most effective method for using a Kimwipe to withdraw the solution is to fold the wipe several times in a rectangular shape and then press the corner of this rectangle firmly into the corner of one of the mouths of the slide chamber. Do not remove the wipe until all solution has been withdrawn. From this point forward) always flow solutions in the same direction through each chamber. This avoids drawing any contaminants from the widing paper into the chamber.
Use a 200 μL pipette so that flow remains constant during each rinse. Try to flow the water through the chamber as rapidly as possible. If persistent bubbles form (thereby preventing adequate rinsing of parts of the cover slip surface), use the filtered air or a nitrogen stream to blow all the solution out of the chamber, and rinse it again. Make sure the entire surface of the cover slip is washed.
Increasing the concentration of functionalized dye in solution does not necessarily lead to better labeling yields. Tubulin with multiple dyes attached may not polymerize well or may irreversibly aggregate [39].
If using rhodamine-labeled tubulin rather than Cy3-labeled tubulin, a ratio of 1:10 labeled/unlabeled is recommended.
Paclitaxel, which is a potent stimulator of MT polymerization, is added after initially polymerizing with only glycerol and GTP, because adding paclitaxel initially leads to the formation of many short (<1 μm) MTs [44], rather than fewer longer filaments. The latter is more desirable for the optical trapping assay. MT length can be controlled to some extent by adjusting the incubation period prior to adding paclitaxel.
MTs polymerize best at 37°C but are stable in the presence of paclitaxel at room temperature.
The purpose of centrifuging the MT solution through the glycerol cushion is to remove any unpolymerized tubulin (which will remain in the supernatant) and very short MT fragments (which will remain in the cushion). These “contaminants” can interfere with MT binding to the cover slip and clutter the surface of the cover slip with unwanted fluorescence. Following the centrifugation, the objective is to remove all protein except that in the pellet before resuspending the MTs. Take care not to (a) disturb the pellet or (b) mix the upper and lower levels of the supernatant.
Resuspension of the MT pellet can take several minutes. Pipette solution up and down above the pellet, and avoid sticking the pipette tip directly into the pellet, especially in the beginning, as this can cause shearing of the MTs. Avoid maldng bubbles.
Casein is acidic and will lower the pH as it dissolves. It becomes less soluble as the pH approaches the casein isoelectric point of ~4.6.
Although the MTs immediately and rigidly attach to the charged glass surface, the covalent linkage via glutaraldehyde is slower (minutes to hours [28, 100]). This reaction must be allowed to complete before adding the Blocker solution (2 mg/mL β-casein). Otherwise, β-casein tends to compete with the MTs for attachment to the cover slip (and likely also shields the positive charge of the aminosilane). This results in floppy attachment of the MTs to the glass surface, which makes finding a well-aligned and stationary MT very difficult. On the other hand, if the β-casein is added after the MTs have attained a covalent linkage, they remain immobilized upon and rigidly attached to the glass. With planning, the MT incubation on the cover slip can be done during other tasks (e.g., microscope alignment) so that it does not introduce a delay. When performing consecutive experiments, we usually take a 2-min break from the microscope to start the MT incubation for the next slide. Alternatively, several slide chambers can be prepared with MTs at once and kept in a humidity chamber until ready for use. We have stored slide chambers with bound MTs for several hours, or even overnight, with no loss of capacity to support motility.
If surface-bound microspheres are needed for calibration or as fiducial markers, a very low concentration of protein-coated microspheres (e.g., the same ones used for optical trapping) can be added directly after the MTs. These will also form covalent linkages with the glass and remain very stably attached.
The best seal is made when a small amount of vacuum grease enters the mouth of the chamber. This seal can last for days or even weeks without significantly affecting the volume of liquid in the chamber. Perhaps nonintuitively, forcing more grease into the chamber mouth actually yields a worse seal: the accompanying increase in pressure in the chamber eventually forces buffer through the weakest part of the seal, thus brealdng it and allowing buffer evaporation and oxygen entry.
If any grease is accidentally left of the cover slip surface, it can be cleaned using a very small amount of spectrophotometric grade ethanol with a Kimwipe or cotton-tipped applicator. Wipe in a single motion away from the center of the chamber.
References
- 1.Yildiz A, Tomishige M, Vale RD, et al. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 2.Reck-Peterson SL, Yildiz A, Carter AP, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gennerich A, Carter AP, Reck-Peterson SL, et al. Force-induced bidirectional stepping of cytoplasmic dynein. Cell. 2007;131:952–965. doi: 10.1016/j.cell.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho C, Reck-Peterson S, Vale R. Cytoplasmic dynein’s regulatory ATPase sites affect processivity and force generation. J Biol Chem. 2008;283:25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Nad Acad Sci USA. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guydosh NR, Block SM. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher K, Schnitzer MJ, Block SM. Single kinesin molecules studied with a molecular force clamp. Nature. 1999;400:184–189. doi: 10.1038/22146. [DOI] [PubMed] [Google Scholar]
- 8.Baralc P, Rai A, Rai P, et al. Quantitative optical trapping on single organelles in cell extract. Nat Methods. 2013;10:68–70. doi: 10.1038/nmeth.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vershinin M, Carter BC, Razafsky DS, et al. Multiple-motor based transport and its regulation by Tau. Proc N ad Acad Sci USA. 2006;104:87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helenius J, Brouhard G, Kalaidzidis Y, et al. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 11.Rice S, Lin AW, Safer D, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 12.Gestaut DR, Graczyk B, Cooper J, et al. Phosphoregulation, lattice diffusion, and depolymerization-driven movement of the Daml complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nalcata T, Hirokawa N. Point mutation of adenosine triphosphate-binding motif generated rigor kinesin that selectively blocks anterograde lysosome membrane transport. J Cell Biol. 1995;131:1039–1053. doi: 10.1083/jcb.131.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crevel IM-TC, Nyitrai M, Alonso MC, et al. What kinesin does at roadblocks: the coordination mechanism for molecular walking. EMBO J. 2003;23:23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adio S, Jaud J, Ebbing B, et al. Dissection ofkinesin’s processivity. PLoS One. 2009;4:e4612. doi: 10.1371/journal.pone.0004612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schroeder HW, III, Mitchell C, Shuman H, et al. Motor number controls cargo switching at actin-microtubule intersections in vitro. Curr Biol. 2010;20:687–696. doi: 10.1016/j.cub.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor AP, Webb RI, Barry JC, et al. Adhesion of microbes using 3-aminopropyl triethoxy silane and specimen stabilisation techniques for analytical transmission electron microscopy. J Microsc. 2000;199:56–67. doi: 10.1046/j.1365-2818.2000.00692.x. [DOI] [PubMed] [Google Scholar]
- 18.Turner DC, Chang C, Fang K, et al. Selective adhesion of functional microtubules to patterned silane surfaces. Biophys J. 1995;69:2782–2789. doi: 10.1016/S0006-3495(95)80151-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeney S, Florin EL, Harber JK. Use of photonic force microscopy to study single-motor-molecule mechanics. Methods Mol Biol. 2001;164:91–108. doi: 10.1385/1-59259-069-1:91. [DOI] [PubMed] [Google Scholar]
- 20.Brown TB, Hancock WO. A polarized microtubule array for kinesin-powered nanoscale assembly and force generation. Nano Lett. 2002;2:1131–1135. [Google Scholar]
- 21.Kanan SM, Tze WTY, Tripp CP. Method to double the surface concentration and control the orientation of adsorbed (3-aminopropyl)dimethylethoxysilane on silica powders and glass slides. Langmuir. 2002;18:6623–6627. [Google Scholar]
- 22.Howarter JA, Youngblood JP. Optimization of silica silanization by 3-aminopropyltriethoxysilane. Langmuir. 2006;22:11142–11147. doi: 10.1021/la061240g. [DOI] [PubMed] [Google Scholar]
- 23.Asenath SE, Chen W. How to prevent the loss of surface functionality derived from aminosilanes. Langmuir. 2008;24:12405–12409. doi: 10.1021/la802234x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim J, Seidler P, Wan LS, et al. Formation, structure, and reactivity of aminoterminated organic films on silicon substrates. J Colloid Interface Sci. 2009;329:114–119. doi: 10.1016/j.jcis.2008.09.031. [DOI] [PubMed] [Google Scholar]
- 25.Aissaoui N, Bergaoui L, Landoulsi J, et al. Silane layers on silicon surfaces: mechanism of interaction, stability, and influence on protein adsorption. Langmuir. 2012;28:656–665. doi: 10.1021/la2036778. [DOI] [PubMed] [Google Scholar]
- 26.Zhu M, Lerum MZ, Chen W. How to prepare reproducible, homogeneous, and hydrolytic ally stable aminosilane-derived layers on silica. Langmuir. 2012;28:416–423. doi: 10.1021/la203638g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walt DR, Agayn VI. The chemistry of enzyme and protein immobilization with glutaraldehyde. Trends Anal Chem. 1994;13:425–430. [Google Scholar]
- 28.Migneault I, Dartiguenave C, Bertrand MJ, et al. Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques. 2004;37:790–802. doi: 10.2144/04375RV01. [DOI] [PubMed] [Google Scholar]
- 29.Hermanson GT. Bioconjugate techniques. 2nd edn Academic; New York: 2008. [Google Scholar]
- 30.Betancor L, López-Gallego F, Alonso-Morales N, et al. Glutaraldehyde in protein immobilization. Methods Biotechnol. 2006;22:57–64. [Google Scholar]
- 31.Clancy BE, Behnke-Parks WM, Andreasson JOL, et al. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iler RK. The chemistry of silica: solubility, polymerization, colloid and surface properties and biochemistry of silica. Wiley-Interscience; Hoboken, NJ: 1979. [Google Scholar]
- 33.Rother D, Sen T, East D, et al. Silicon, silica and its surface patterning/activation with alkoxy- and amino-silanes for nanomedical applications. N anomedicine. 2011;6:281–300. doi: 10.2217/nnm.10.159. [DOI] [PubMed] [Google Scholar]
- 34.Der Voort PV, Vansant EF. Silylation of the silica surface: a review. J Liq Chrom Relat Tech. 1996;19:2723–2752. [Google Scholar]
- 35.Witucki GL. A silane primer: chemistry and applications of alkoxysilanes. J Coating Tech Res. 1993;65:57–60. [Google Scholar]
- 36.Brinker C. Hydrolysis and condensation of silicates: effects on structure. J Non-Cryst Solids. 1988;100:31–50. [Google Scholar]
- 37.Tan W, Desai TA. Layer-by-layer microfluidics for biomimetic three-dimensional structures. Biomaterials. 2004;25:1355–1364. doi: 10.1016/j.biomaterials.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 38.Diao J, Ren D, Engstrom JR, et al. A surface modification strategy on silicon nitride for developing biosensors. Anal Biochem. 2005;343:322–328. doi: 10.1016/j.ab.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 39.Peloquin J, Komarova Y, Borisy G. Conjugation of fluorophores to tubulin. Nat Methods. 2005;2:299–303. doi: 10.1038/nmeth0405-299. [DOI] [PubMed] [Google Scholar]
- 40.Keates RAB. Effects of glycerol on microtubule polymerization kinetics. Biochem Biophys Res Commun. 1980;97:1163–1169. doi: 10.1016/0006-291x(80)91497-7. [DOI] [PubMed] [Google Scholar]
- 41.O’Brien ET, Erickson HP. Assembly of pure tubulin in the absence of free GTP: effect of magnesium, glycerol, ATP, and the nonhydrolyzable GTP analogues. Biochemistry. 1989;28:1413–1422. doi: 10.1021/bi00429a070. [DOI] [PubMed] [Google Scholar]
- 42.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 43.Gardner MK, Zanic M, Howard J. Microtubule catastrophe and rescue. Curr Opin Cell Biol. 2013;25:14–22. doi: 10.1016/j.ceb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carlier MF, Pantaloni D. Taxol effect on tubulin polymerization and associated guanosine 5′-triphosphatehydrolysis. Biochemistry. 1983;22:4814–4822. doi: 10.1021/bi00289a031. [DOI] [PubMed] [Google Scholar]
- 45.Orr GA, Verdier-Pinard P, McDaid H, et al. Mechanisms of taxol resistance related to microtubules. Oncogene. 2003;22:7280–7295. doi: 10.1038/sj.onc.1206934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schiff PB, Fant J, Horwitz SB. Promotion of microtubule assembly in vitro by taxol. Nature. 1979;277:665–667. doi: 10.1038/277665a0. [DOI] [PubMed] [Google Scholar]
- 47.Schiff PB, Horwitz SB. Taxol stabilizes microtubules in mouse fibroblast cells. Proc Natl Acad Sci USA. 1980;77:1561–1565. doi: 10.1073/pnas.77.3.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson WC, Wilson L, Purich DL. Taxol induces microtubule assembly at low temperature. Cell Motil. 1981;1:445–454. doi: 10.1002/cm.970010405. [DOI] [PubMed] [Google Scholar]
- 49.Kumar N. Taxol-induced polymerization of purified tu bulin. Mechanism of action. J Biol Chem. 1981;256:10435–10441. [PubMed] [Google Scholar]
- 50.Robinson J, Engelborghs Y. Tubulin polymerization in dimethyl sulfoxide. J Biol Chem. 1982;257:5367–5371. [PubMed] [Google Scholar]
- 51.Balcer NA, Sept D, Joseph S, et al. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Minoura I, Muto E. Dielectric measurement of individual microtubules using the electro orientation method. Biophys J. 2006;90:3739–3748. doi: 10.1529/biophysj.105.071324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sackett DL, Bhattacharyya B, Wolff J. Promotion of tubulin assembly by carboxyterminal charge reduction. Ann NY Acad Sci. 1986;466:460–466. doi: 10.1111/j.1749-6632.1986.tb38424.x. [DOI] [PubMed] [Google Scholar]
- 54.Redeker V, Melki R, Promé D, et al. Structure of tubulin C-terminal domain obtained bv subtilisin treatment. The major ex and β tubuiin isotypes from pig brain are glutamylated. FEBS Lett. 1992;313:185–192. doi: 10.1016/0014-5793(92)81441-n. [DOI] [PubMed] [Google Scholar]
- 55.Bezanilla M, Manne S, Laney DE, et al. Adsorption of DNA to mica, silylated mica, and minerals: characterization by atomic force microscopy. Langmuir. 1995;11:655–659. [Google Scholar]
- 56.Van der Maaden K, Sliedregt K, Kros A, et al. Fluorescent nanoparticle adhesion assav: a novel method for surface pKa determination of self-assembled monolayers on silicon surfaces. Langmuir. 2012;28:3403–3411. doi: 10.1021/la203560k. [DOI] [PubMed] [Google Scholar]
- 57.Gell C, Bormuth V, Brouhard GJ, et al. Microtubule dvnamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods Cell Biol. 2010;95:221–245. doi: 10.1016/S0091-679X(10)95013-9. [DOI] [PubMed] [Google Scholar]
- 58.Schäffer E, Nørrelyldee SF, Howard J. Surface forces and drag coefficients of microspheres near a plane surface measured with optical tweezers. Langmuir. 2007;23:3654–3665. doi: 10.1021/la0622368. [DOI] [PubMed] [Google Scholar]
- 59.Tolić-Nørrelvldee SF, Schäffer E, Howard J, et al. Calibration of optical tweezers with positional detection in the back focal plane. Rev Sci Instrum. 2006;77:103101–103111. [Google Scholar]
- 60.Vandenberg ET, Bertilsson L, Liedberg B, et al. Structure of 3-aminopropyl triethoxv silane on silicon oxide. J Colloid Interface Sci. 1991;147:103–118. [Google Scholar]
- 61.Vigers GP, Coue M, McIntosh JR. Fluorescent microtubules break up under illumination. J Cell Biol. 1988;107:1011–1024. doi: 10.1083/jcb.107.3.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo H, Xu C, Liu C, et al. Mechanism and dynamics of brealeage of fluorescent microtubules. Biophys J. 2006;90:2093–2098. doi: 10.1529/biophysj.105.071209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weisenberg RC. Microtubule formation in vitro in solutions containing low calcium concentrations. Science. 1972;177:1104–1105. doi: 10.1126/science.177.4054.1104. [DOI] [PubMed] [Google Scholar]
- 64.Borisy GG, Olmsted JB. Nucleated assembly of microtubules in porcine brain extracts. Science. 1972;177:1196–1197. doi: 10.1126/science.177.4055.1196. [DOI] [PubMed] [Google Scholar]
- 65.Miller HP, Wilson L. Preparation of microtubule protein and purified tubulin from bovine brain by cycles of assembly and disassembly and phospho cellulose chromatography. Methods Cell Biol. 2010;95:3–15. doi: 10.1016/S0091-679X(10)95001-2. [DOI] [PubMed] [Google Scholar]
- 66.Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization–depolymerization in a high-molarity buffer. Protein Expr Purif. 2003;32:83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 67.Gell C, Friel CT, Borgonovo B, et al. Purification of tubulin from porcine brain. Methods Mol Biol. 2011;777:15–28. doi: 10.1007/978-1-61779-252-6_2. [DOI] [PubMed] [Google Scholar]
- 68.Widlund PO, Podolski M, Reber S, et al. One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol Biol Cell. 2012;23:4393–4401. doi: 10.1091/mbc.E12-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fox PF, McSweeney PLH. Advanced dairy chemistry: protein. Springer; New York: 2003. [Google Scholar]
- 70.Fox PF, McSweeney PLH. Dairy chemistry and biochemistry. Springer; New York: 1998. [Google Scholar]
- 71.Eigel WN, Butler JE, Ernstrom CA, et al. Nomenclature of proteins of cow’s mille fifth revision. J Dairy Sci. 1984;67:1599–1631. [Google Scholar]
- 72.Nylander T, Wahlgren NM. Forces between adsorbed layers of β-casein. Langmuir. 1997;13:6219–6225. [Google Scholar]
- 73.Mikheeva LM, Grinberg NV, Grinberg VY, et al. Thermodynamics of micellization of bovine β-casein studied by high-sensitivity differential scanning calorimetry. Langmuir. 2003;19:2913–2921. [Google Scholar]
- 74.Thorn DC, Meehan S, Sunde M, et al. Amvloid fibril formation bv bovine mille κ-casein and its inhibition by the molecular chaperones αS- and β-casein. Biochemistry. 2005;44:17027–17036. doi: 10.1021/bi051352r. [DOI] [PubMed] [Google Scholar]
- 75.Portnaya I, Ben-Shoshan E, Cogan U, et al. Self-assembly of bovine β-casein below the isoelectric pH. J Agric Food Chem. 2008;56:2192–2198. doi: 10.1021/jf072630r. [DOI] [PubMed] [Google Scholar]
- 76.Evers CHJ, Andersson T, Lund M, et al. Adsorption of unstructured protein β-casein to hydrophobic and charged surfaces. Langmuir. 2012;28:11843–11849. doi: 10.1021/la300892p. [DOI] [PubMed] [Google Scholar]
- 77.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 78.Kull T, Nylander T, Tiberg F, et al. Effect of surface properties and added electrolyte on the structure of β-casein layers adsorbed at the solid/aqueous interface. Langmuir. 1997;13:5141–5147. [Google Scholar]
- 79.Verma V, Hancock WO, Catchmark JM. The role of casein in supporting the operation of surface bound kinesin. J Biol Eng. 2008;2:14. doi: 10.1186/1754-1611-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ong S, Zhao X, Eisenthal KB. Polarization of water molecules at a charged interface: second harmonic studies of the silica/water interface. Chem Phys Lett. 1992;191:327–335. [Google Scholar]
- 81.Fan H-F, Li F, Zare RN, et al. Characterization of two types of silanol groups on fused-silica surfaces using evanescent-wave cavity ring-down spectroscopy. Anal Chem. 2007;79:3654–3661. doi: 10.1021/ac062386n. [DOI] [PubMed] [Google Scholar]
- 82.Hair ML, Herd W. Acidity of surface hydroxyl groups. J Phys Chem. 1970;74:91–94. [Google Scholar]
- 83.Balladur V, Theretz A, Mandrand B. Determination of the main forces driving DNA oligonucleotide adsorption onto aminated silica wafers. J Colloid Interface Sci. 1997;194:408–418. doi: 10.1006/jcis.1997.5123. [DOI] [PubMed] [Google Scholar]
- 84.Belkind A, Gershman S. Plasma cleaning of surfaces. Vacuum Coating and Technology November. 2008:46–57. [Google Scholar]
- 85.Isabell TC, Fischione PE, O’Keefe C, et al. Plasma cleaning and its applications for electron microscopy. Microsc Microanal. 1999;5:126–135. doi: 10.1017/S1431927699000094. [DOI] [PubMed] [Google Scholar]
- 86.DeRosa RL, Schader PA, Shelby JE. Hydrophilic nature of silicate glass surfaces as a function of exposure condition. J Non-Cryst Solids. 2003;331:32–40. [Google Scholar]
- 87.Bhattacharya S, Datta A, Berg JM, et al. Studies on surface wettability of poly(dimethyl) siloxane (PDMS) and glass under oxygen-plasma treatment and correlation with bond strength. J Microelectromech Syst. 2005;14:590–597. [Google Scholar]
- 88.Kern W. The evolution of silicon wafer cleaning technology. J Electrochem Soc. 1990;137:1887–1892. [Google Scholar]
- 89.Williams EH, Davydov AV, Motayed A, et al. Immobilization of streptavidin on 4H-SiC for biosensor development. Appl Surf Sci. 2012;258:6056–6063. [Google Scholar]
- 90.Armarego WLF, Chai C. Purification of laboratory chemicals. Butterworth-Heinemann; Oxford: 2012. [Google Scholar]
- 91.Weires NA, Johnston A, Warner DL, et al. Recycling of waste acetone by fractional distillation. J Chem Educ. 2011;88:1724–1726. [Google Scholar]
- 92.Siqueira Petri DF, Wenz G, Schunk P, et al. An improved method for the assembly of amino-terminated monolayers on SiO2 and the vapor deposition of gold layers. Langmuir. 1999;15:4520–4523. [Google Scholar]
- 93.Leal O, Bolivar C, Ovalles C, et al. Reversible adsorption of carbon dioxide on amine surface-bonded silica gel. Inorg Chim Acta. 1995;240:183–189. [Google Scholar]
- 94.Serna-Guerrero R, Da’na E, Sayari A. New insights into the interactions of CO2 with amine-functionalized silica. Ind Eng Chem Res. 2008;47:9406–9412. [Google Scholar]
- 95.Etienne M, Walcarius A. Analytical investigation of the chemical reactivity and stability of aminopropyl-grafted silica in aqueous medium. Talanta. 2003;59:1173–1188. doi: 10.1016/S0039-9140(03)00024-9. [DOI] [PubMed] [Google Scholar]
- 96.Möller R, Csaki A, Kohler JM, et al. DNA probes on chip surfaces studied by scanning force microscopy using specific binding of colloidal gold. Nucleic Acids Res. 2000;28:e91. doi: 10.1093/nar/28.20.e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Turner D, Chang C, Fang K, et al. Kinesin movement on glutaraldehyde-fixed microtubules. Anal Biochem. 1996;242:20–25. doi: 10.1006/abio.1996.0422. [DOI] [PubMed] [Google Scholar]
- 98.Cross AR, Williams RC., Jr Kinky microtubules: bending and brealdng induced by fixation in vitro with glutaraldehyde and formaldehyde. Cell Motil Cytoskeleton. 1991;20:272–278. doi: 10.1002/cm.970200403. [DOI] [PubMed] [Google Scholar]
- 99.Shelansld ML, Gaskin F, Cantor CR. Microtubule assembly in the absence of added nucleotides. Proc Natl Acad Sci USA. 1973;70:765–768. doi: 10.1073/pnas.70.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Habeeb AFSA, Hiramoto R. Reaction of proteins with glutaraldehyde. Arch Biochem Biophys. 1968;126:16–26. doi: 10.1016/0003-9861(68)90554-7. [DOI] [PubMed] [Google Scholar]