Abstract
Purpose.
To comprehensively characterize human corneal endothelial cell (HCEnC) gene expression and age-dependent differential gene expression and to identify expressed genes mapped to chromosomal loci associated with the corneal endothelial dystrophies posterior polymorphous corneal dystrophy (PPCD)1, Fuchs endothelial corneal dystrophy (FECD)4, and X-linked endothelial dystrophy (XECD).
Methods.
Total RNA was isolated from ex vivo corneal endothelium obtained from six pediatric and five adult donor corneas. Complementary DNA was hybridized to the Affymetrix GeneChip 1.1ST array. Data analysis was performed using Partek Genomics Suite software, and differentially expressed genes were validated by digital molecular barcoding technology.
Results.
Transcripts corresponding to 12,596 genes were identified in HCEnC. Nine genes displayed the most significant differential expression between pediatric and adult HCEnC: CAPN6, HIST1H3A, HIST1H4E, and HSPA2 were expressed at higher levels in pediatric HCEnC, while ITGBL1, NALCN, PREX2, TAC1, and TMOD1 were expressed at higher levels in adult HCEnC. Analysis of the PPCD1, FECD4 and XECD loci demonstrated transcription of 53/95 protein-coding genes in the PPCD1 locus, 27/40 in the FECD4 locus, and 35/68 in the XECD locus.
Conclusions.
An analysis of the HCEnC transcriptome reveals the expression of almost 13,000 genes, with less than 1% mapped to chromosomal loci associated with PPCD1, FECD4, and XECD. At least nine genes demonstrated significant differential expression between pediatric and adult HCEnC, defining specific functional properties distinct to each age group. These data will serve as a resource for vision scientists investigating HCEnC gene expression and can be used to focus the search for the genetic basis of the corneal endothelial dystrophies for which the genetic basis remains unknown.
Keywords: corneal endothelium, HCEnC, transcriptome, gene expression, corneal endothelial dystrophy
Transcriptomic analysis of human corneal endothelial cell gene expression identified age-dependent differential expression and candidate genes for the corneal endothelial dystrophies PPCD1, FECD4 and XECD.
Introduction
Corneal endothelial cells maintain corneal clarity by transcribing genes that are important in sustaining homeostatic levels of hydration and solute concentration in the corneal stroma. Human corneal endothelial cells (HCEnC) also express genes that encode extracellular structural proteins that give rise to the Descemet membrane and cell-adhesion molecules that support the semipermeable simple squamous epithelial morphology of the endothelium. In addition, HCEnC express genes that code for proteins involved in solute transport across the semipermeable endothelium, integral to maintaining corneal stromal solute concentrations.
Mutations in genes that encode extracellular matrix proteins, enzymes, and transcription factors have been associated with a number of inherited disorders of the corneal endothelium, collectively known as the corneal endothelial dystrophies. Posterior polymorphous corneal dystrophy (PPCD) mapped to chromosome 10 (PPCD3) has been associated with nonsense and frameshift mutations in the transcription factor ZEB1.1–8 Congenital hereditary endothelial dystrophy (CHED) has been associated with mutations in the sodium-borate cotransporter SLC4A11,9,10 while sequence variants in ZEB1,11,12 SLC4A11,13,14 COL8A2,15 TCF4,16 LOXHD1,17 and AGBL118 have been associated with Fuchs endothelial corneal dystrophy (FECD). However, the genetic basis of at least three corneal endothelial dystrophies remains to be determined: PPCD mapped to chromosome 20 (PPCD1),19–22 FECD mapped to chromosome 9 (FECD4),12 and X-linked endothelial dystrophy (XECD).23 Therefore, identification of the genes expressed in the human corneal endothelium that are mapped to these chromosomal loci would provide a list of positional gene candidates that would narrow the search for the genetic basis of these corneal endothelial dystrophies.
A complete characterization of the HCEnC transcriptome has been a goal of vision science researchers for the last decade, who have used serial analysis of gene expression (SAGE),24,25 gene arrays,25–27 and next-generation sequencing (RNA-seq) technologies.28–30 We present a new approach and build on the previously published HCEnC transcriptome studies. Defining the normal and age-dependent HCEnC transcriptome will be useful for elucidation of the genetic basis of inherited disorders of the corneal endothelium and for facilitating the development of novel strategies for managing endothelial dysfunction, such as validating the transcriptome of cultured corneal endothelial cells prior to transplantation.
Methods
Ribonucleic Acid Isolation and Purification
Eleven corneas from six pediatric (4, 6, 10, 11, 17, and 18 years old) and five adult (53, 56, 57, 64, and 70 years old) donors were obtained from various eye banks affiliated with the Vision Share consortium of eye banks (Vision Share, Apex, NC, USA) with the following characteristics: mean central corneal endothelial cell density of 2973 cells/mm2 (range, 2114–3937 cells/mm2); mean death to preservation time of 6 hours 57 minutes (range, 3 hours 5 minutes–11 hours 49 minutes); and mean death to removal of Descemet membrane and corneal endothelium time of 2.9 days (range, 2–4 days) (Table 1). Total RNA was isolated from the corneal endothelium using TriReagent (Life Technologies, Grand Island, NY, USA) and subsequently purified with the RNeasy Clean-Up Kit (Qiagen, Valencia, CA, USA). The integrity of the isolated RNA was analyzed using the Agilent 2100 Electrophoresis Bioanalyzer System (Agilent Technologies, Inc., Santa Clara, CA, USA) and found to be of sufficient quantity (approximately 1 μg) and quality (RNA integrity number in the range of 8.3–9.0) for analysis using the Affymetrix gene chip arrays.
Table 1.
Donor Tissue Information
|
Age |
Age Group |
Cause of Death |
Endothelial Cells/mm2 |
Storage Medium |
Reason Not Suitable for Transplantation? |
Death to Cooling |
Death to Preservation |
Death to Harvest |
| 4 | Preschooler | Brain death | 3937 | Life4C | PA in respiratory cx | 1 h 49 min | 8 h 34 min | 4 d |
| 6 | Preschooler | Drowning | 3067, 3390 | Life4C | PA in respiratory cx | NA | 3 h 5 min | 4 d |
| 10 | Preadolescent | Seizure | 2924, 2725 | Life4C | PA in respiratory cx | NA | 3 h 59 min | 2 d |
| 11 | Preadolescent | Probable OD | 2923, 3115 | Optisol GS | SA-positive sputum | 3 h 35 min | 8 h 12 min | 3 d |
| 15 | Adolescent | Cardiac arrhythmia | 3096 | Optisol GS | Contact lens attached | 2 h 48 min | 11 h 13 min | 2 d |
| 17 | Adolescent | Severe head injury | 3184, 3184 | Optisol GS | Submersion under water | 3 h 27 min | 2 h 21 min | 3 d |
| 53 | Adult | CHF | 2959, 2793 | Life4C | Possible IV drug use | NA | 5 h 15 min | 3 d |
| 56 | Adult | SDH | 2923 | Optisol GS | Cocci (staph?) in blood | 1 h 11 min | 4 h 26 min | 2 d |
| 57 | Adult | Liver cancer | 2114, 2183 | Optisol GS | Paracentral tear in DM | 2 h 20 min | 8 h 10 min | 2 d |
| 64 | Adult | Probable MI | 2793, 2762 | Optisol GS | Possible HRB* | 3 h 57 min | 11 h 49 min | 3 d |
| 70 | Adult | MOSF | 3215, 3205 | Optisol GS | Possible hepatitis A† | 6 h 28 min | 9 h 29 min | 4 d |
OD, overdose; CHF, congestive heart failure; SDH, subdural hematoma; MI, myocardial infarction; MOSF, multiple organ system failure; PA, Pseudomonas aeruginosa; cx, microbiological culture; SA, Staphlococcus aureus; HRB, high-risk behavior.
Next of kin were unable to complete high-risk behavior questionnaire relating to the deceased.
Next of kin indicated resolved hepatitis A infection, but were unable to confirm infection status.
Transcriptome Analysis
Samples of total RNA from each donor cornea were processed and analyzed separately (i.e., samples were not pooled). Total RNA from pediatric and adult normal endothelium was hybridized to the Affymetrix GeneChip 1.1ST array (Affymetrix, Inc., Santa Clara, CA, USA), and raw intensity values stored in CEL files were imported into Partek Genomics Suite software (PGS) (Partek, Inc., St. Louis, MO, USA). Raw values were normalized using the Robust Multi-array Average (RMA) method with background correction, and the adjusted intensity values were output as log2 transformed values.31,32 An additional background threshold was calculated by averaging the log2 values for genes predicted to have no transcription in HCEnC. Subsequently, transcripts corresponding to normalized probe intensity values at or below the background value of 3.8 were considered as not expressed. Principal component analysis (PCA) and hierarchical clustering (HC) were performed to identify variation and relationships among samples. Principal component analysis and HC analysis revealed that the pediatric samples clustered into three distinct groups that are hereafter referred to as preschooler (4 and 6 years old), preadolescent (10 and 11 years old), and adolescent (17 and 18 years old). The CEL files containing the raw signal intensity values and the RMA normalized and log2 transformed signal intensity values are available from the GEO DataSets database (accession number GSE58315; National Center for Biotechnology Information [NCBI], Bethesda, MD, USA).
Differential Gene Expression Analysis
Differential gene expression was determined for all genes using PGS. Three separate statistical (P value) and fold-change (FC) settings were used to obtain lists of significant differentially expressed genes between pediatric and adult endothelium. In order to correct for multiple comparisons and reduce the number of false positives, a false discovery rate (FDR) adjusted P value of 0.05 was used in combination with a FC > 3. To obtain a larger number of genes for validation by digital molecular barcoding technology (NanoString Technologies, Seattle, WA, USA), a non-FDR P value of 0.005 and FC > 2.9 were applied. The third settings were the least stringent, with a non-FDR P value of 0.05 and FC > 2, and were used to generate a list of differentially expressed genes for gene ontology (GO) analysis.
Digital Molecular Barcoding Analysis
A custom multiplexed NanoString nCounter Elements expression assay (NanoString Technologies) comprising the nine differentially expressed genes was assembled. All reagents and software were from NanoString Technologies, unless stated otherwise. The procedure was performed according to the NanoString nCounter Single Cell Gene Expression and Elements Gene Expression Assay Protocols. Briefly, 1 ng total RNA was used for the amplification (10 cycles) of nine separate transcripts using Multiplexed Target Enrichment oligonucleotides (Integrated DNA Technologies, San Diego, CA, USA). Subsequently, 5 μL amplification reaction was mixed with 20 and 5 μL nCounter reporter probes and biotinylated nCounter capture probes (Integrated DNA Technologies), respectively. Hybridization was performed at 65°C for approximately 16 to 20 hours. Excess probe was washed away using a two-step magnetic bead–based purification technique on the nCounter Prep Station. Biotinylated capture probe–bound samples were immobilized and recovered on a streptavidin-coated cartridge using the nCounter PrepStation. To determine transcript species (i.e., barcode pattern) and abundance (i.e., fluorescence intensity), a max-density scan (encompassing 555 fields of view) was performed using the nCounter Digital Analyzer. Data analysis was performed using the nSolver and GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA, USA).
Gene Ontology Analysis
Gene ontology analysis was performed for the identification of significantly enriched functional groups using the GO Enrichment option available in the PGS software (Partek, Inc.). A list of differentially expressed genes between pediatric and adult corneal endothelium was generated utilizing a non-FDR P value of 0.05 and FC cutoff of 2. Significantly enriched GO functional groups were defined as having an enrichment score equal to or greater than 3 (P value > 0.05) and were required to be composed of at least three genes.
Corneal Endothelial Dystrophy Disease Locus Analysis
The genes located within the PPCD1 locus (bordered by the D20S182 and D20S106 genetic markers), the FECD4 locus (bordered by the D9S1681 and D9S1684 genetic markers), and the XECD locus (bordered by the DXS8057 and DXS1047 genetic markers) were identified using the most recent build of the human genome (Annotation 105) and the Map Viewer resource available through the NCBI website (http://www.ncbi.nlm.nih.gov/mapview/ [in the public domain]). These lists of genes were then compared to the list of expressed genes corresponding to the PPCD1, FECD4, and XECD loci obtained using the PGS software to identify positional candidate genes for each of these corneal endothelial dystrophies.
Statistical Analysis
The two-tailed unpaired t-test was utilized for identifying a significant difference in the means of the expression levels (barcode counts) of transcripts in pediatric versus adult corneal endothelium as determined by digital molecular barcoding technology. Differences in the means of gene expression in the different age groups of ex vivo endothelium were considered significant (P ≤ 0.05) by rejection of the null hypothesis. One-way analysis of variance (ANOVA) was used to identify significantly differentially expressed genes in the microarray data of the pediatric versus adult samples.
Results
Multidimensional Transcriptome Analysis
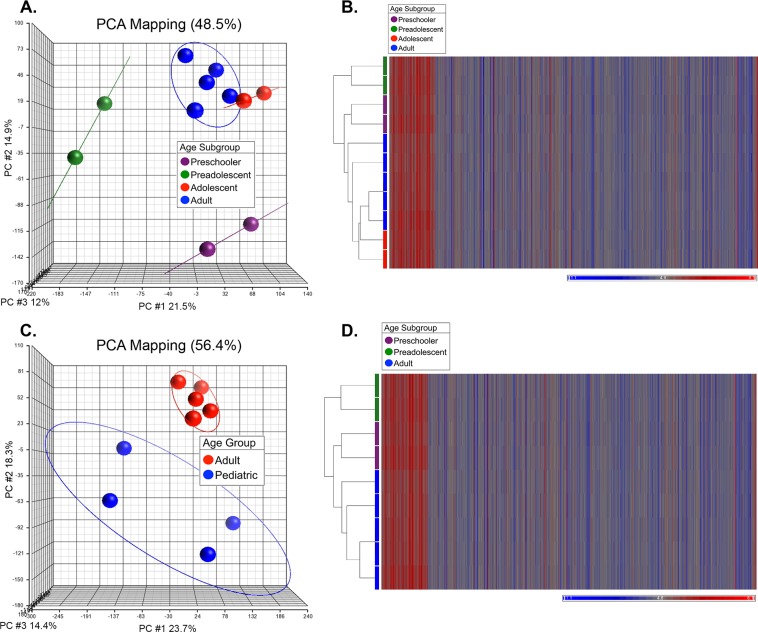
Principal component analysis and HC showed distinct relationships between the pediatric and adult samples (Fig. 1). An initial analysis of all of the samples demonstrated that the six pediatric samples segregated into three distinct groups. We observed that the groupings were not random and that they depended on the age of the donor. As such, the six pediatric samples were categorized into three distinct age groups, which were defined as preschooler (4 and 6 years old), preadolescent (10 and 11 years old), and adolescent (17 and 18 years old). The two adolescent samples clustered near the adult samples, suggesting that the transcriptomes of the two adolescent samples were more similar to the adult than to the other two pediatric groups. Therefore, they were subsequently excluded from the pediatric group for the additional analyses (Figs. 1A, 1B). An analysis of the remaining samples demonstrated that the first three principal components (PC1: 23.7%; PC2: 18.3%; PC3: 14.4%) accounted for 56.4% of the variance observed between the gene expression data sets (Fig. 1C). Principal component analysis demonstrated three distinct groups that correlated with age. Hierarchical clustering recapitulated the results obtained with PCA, with gene expression correlated to age (Fig. 1D). Differential gene expression analysis was performed on two HCEnC groups: pediatric (preschooler and preadolescent) and adult.
Figure 1.
Principal component analysis and hierarchical clustering of ex vivo HCEnC gene expression data sets. (A) PCA demonstrated three distinct groupings, with the adolescent (17 and 18 years, red spheres) HCEnC group having a closer relationship to adult samples (53–70 years, blue spheres) compared to preadolescents (10 and 11 years, green spheres) and preschoolers (4 and 6 years, violet spheres). (B) Hierarchical clustering confirmed the relationships observed with PCA, with the adolescent HCEnC samples (red bars) demonstrating a closer relationship to adult HCEnC samples (blue bars). (C) After removal of the HCEnC samples from the adolescent group, PCA demonstrated three distinct groupings, two of which were analyzed together as the pediatric group (preschooler and preadolescent, 4–11 years, blue spheres) and the adult group (53–57 years, red spheres). Subsequent data analysis was performed on two groups defined by the colored ovals (pediatric and adult). (D) Hierarchical clustering confirmed the relationships observed in the PCA analysis, with the preschooler (violet bar) and the preadolescent (green bar) HCEnC samples having clustered separately from the adult (blue bar) HCEnC.
Whole-Genome Gene Expression
The pediatric and adult samples were combined and analyzed for whole-genome gene transcription. Annotation with the RefSeq Transcripts database (NCBI) demonstrated 20,253 unique gene symbols, of which 12,596 were considered expressed in the corneal endothelium with an expression value above the background threshold of 3.8 (Table 2, top 100 unique probeset IDs comprising 83 unique genes). Four of the six genes that have been associated with the endothelial corneal dystrophies (COL8A2, SLC4A11, ZEB1, and TCF4) were expressed in the corneal endothelium, while two (LOXHD1 and AGBL1) were not (Table 3).
Table 2.
Most Highly Expressed Genes in Normal Human Corneal Endothelial Cells*
|
Probeset ID |
Gene Symbol |
Gene Type |
Mean Intensity |
| 7917645 | RN28S1 | rRNA | 12.35 |
| 8165665 | COX2 | Protein coding | 12.34 |
| 8165690 | CYTB | Protein coding | 12.23 |
| 7978905 | SDCCAG1 | Protein coding | 12.22 |
| 8165661 | COX1 | Protein coding | 12.21 |
| 8165686 | ND5 | Protein coding | 12.05 |
| 8170360 | FTH1 | Protein coding | 11.94 |
| 8165653 | ND1 | Protein coding | 11.89 |
| 8165669 | COX3 | Protein coding | 11.87 |
| 8165648 | C7orf11 | Protein coding | 11.84 |
| 7953385 | GAPDH | Protein coding | 11.81 |
| 7938777 | LDHA | Protein coding | 11.78 |
| 7912198 | ENO1 | Protein coding | 11.70 |
| 8165676 | ND4 | Protein coding | 11.69 |
| 7941272 | MALAT1 | ncRNA | 11.68 |
| 7942791 | RN28S1 | rRNA | 11.59 |
| 7986765 | RPL5 | Protein coding | 11.58 |
| 8165674 | SH3KBP1 | Protein coding | 11.54 |
| 7954310 | RN18S1 | rRNA | 11.50 |
| 8158952 | EEF1A1 | Protein coding | 11.37 |
| 7967563 | UBC | Protein coding | 11.35 |
| 8050548 | LAPTM4A | Protein coding | 11.35 |
| 8138531 | EEF1A1 | Protein coding | 11.34 |
| 7981978 | SNORD116-15 | snoRNA | 11.28 |
| 8109821 | RPL10 | Protein coding | 11.28 |
| 7952325 | HSPA8 | Protein coding | 11.20 |
| 7965467 | RPL41 | Protein coding | 11.11 |
| 7982129 | RPL41 | Protein coding | 11.11 |
| 7961514 | MGP | Protein coding | 11.09 |
| 8061136 | PTMA | Protein coding | 11.08 |
| 7922301 | MYOC | Protein coding | 11.08 |
| 8137008 | C7orf11 | Protein coding | 11.08 |
| 8076511 | RPL5 | Protein coding | 10.99 |
| 8165672 | RFC1 | Protein coding | 10.92 |
| 8116520 | GNB2L1 | Protein coding | 10.91 |
| 8102587 | C4orf31 | Protein coding | 10.90 |
| 8154305 | SELT | Protein coding | 10.88 |
| 7915472 | SLC2A1 | Protein coding | 10.87 |
| 8075691 | RPL41 | Protein coding | 10.80 |
| 8107470 | PTMA | Protein coding | 10.78 |
| 8069644 | APP | Protein coding | 10.77 |
| 8173941 | TSPAN6 | Protein coding | 10.77 |
| 8025395 | RPS28 | Protein coding | 10.76 |
| 7904254 | ATP1A1 | Protein coding | 10.74 |
| 8165658 | ND2 | Protein coding | 10.74 |
| 7942824 | RPS28 | Protein coding | 10.73 |
| 8092970 | APOD | Protein coding | 10.73 |
| 8005471 | RPS28 | Protein coding | 10.73 |
| 8066262 | SNORA71D | snoRNA | 10.71 |
| 7942586 | RPS3 | Protein coding | 10.70 |
| 7921916 | RGS5 | Protein coding | 10.67 |
| 8178435 | IER3 | Protein coding | 10.65 |
| 7989501 | CA12 | Protein coding | 10.64 |
| 7981976 | SNORD116-14 | snoRNA | 10.63 |
| 8113120 | TOB2 | Protein coding | 10.62 |
| 8165707 | TOB2 | Protein coding | 10.62 |
| 8109750 | RPLP0 | Protein coding | 10.62 |
| 8064613 | SLC4A11 | Protein coding | 10.60 |
| 8120249 | RN7SK | snRNA | 10.57 |
| 8102831 | C4orf49 | Protein coding | 10.56 |
| 7977775 | DAD1 | Protein coding | 10.52 |
| 8159642 | TUBB2C | Protein coding | 10.51 |
| 8081431 | ALCAM | Protein coding | 10.51 |
| 8164165 | HSPA5 | Protein coding | 10.50 |
| 8117377 | HIST1H1E | Protein coding | 10.49 |
| 8034512 | SNORD41 | snoRNA | 10.48 |
| 8061364 | RPL41 | Protein coding | 10.48 |
| 8105432 | RPL41 | Protein coding | 10.48 |
| 8042335 | VDAC2 | Protein coding | 10.47 |
| 7981988 | SNORD116-20 | snoRNA | 10.46 |
| 8067955 | CXADR | Protein coding | 10.44 |
| 8110522 | CANX | Protein coding | 10.41 |
| 8086752 | SNORD13 | snoRNA | 10.39 |
| 8038086 | RPL18 | Protein coding | 10.36 |
| 8142468 | TPM3 | Protein coding | 10.35 |
| 7902398 | SNORD45A | snoRNA | 10.35 |
| 7981982 | SNORD116-17 | snoRNA | 10.34 |
| 7981986 | SNORD116-17 | snoRNA | 10.34 |
| 7929816 | SCD | Protein coding | 10.33 |
| 8000217 | SMG1 | Protein coding | 10.32 |
| 7978644 | NFKBIA | Protein coding | 10.31 |
| 7958130 | HSP90B1 | Protein coding | 10.31 |
| 7919193 | NUDT4P1 | Pseudogene | 10.30 |
| 8035829 | RPL34 | Protein coding | 10.30 |
| 8179704 | IER3 | Protein coding | 10.30 |
| 8124848 | IER3 | Protein coding | 10.30 |
| 8117995 | TUBB | Protein coding | 10.30 |
| 8177858 | TUBB | Protein coding | 10.30 |
| 8179174 | TUBB | Protein coding | 10.30 |
| 8146216 | VDAC3 | Protein coding | 10.30 |
| 8022814 | HNRNPA1 | Protein coding | 10.26 |
| 7938329 | SNORA23 | snoRNA | 10.26 |
| 7899480 | SNORA73A | snoRNA | 10.26 |
| 8109222 | RPL7 | Protein coding | 10.25 |
| 8119993 | HSP90AB1 | Protein coding | 10.22 |
| 8116929 | RPL15 | Protein coding | 10.22 |
| 8030980 | ZNF525 | Protein coding | 10.22 |
| 8012110 | GABARAP | Protein coding | 10.21 |
| 7982597 | THBS1 | Protein coding | 10.21 |
| 8005547 | SNORD3A | snoRNA | 10.21 |
rRNA, ribosomal RNA; ncRNA, non-coding RNA; snoRNA, small nucleolar RNA; snRNA, small nuclear RNA.
Top 100 transcript IDs by level of expression comprising 83 genes.
Table 3.
Expression in Normal Corneal Endothelium of Genes Associated With Corneal Endothelial Dystrophies
|
Probeset ID |
Gene Symbol |
Cytoband |
Corneal Dystrophy |
Mean Intensity* |
| 8064613 | SLC4A11 | 20p13 | CHED, FECD | 10.60 |
| 8023415 | TCF4 | 18q21.2 | FECD | 9.44 |
| 7914880 | COL8A2 | 1p34.3 | FECD | 7.28 |
| 7926916 | ZEB1 | 10p11.22 | PPCD3, FECD | 6.13 |
| 8023080 | LOXHD1 | 18q21.1 | FECD | 3.41† |
| 7985741 | AGBL1 | 15q25.3 | FECD | 3.13† |
Mean intensity is given in log2 values.
Intensity values are below the background threshold of 3.8.
Identification of Differentially Expressed Genes
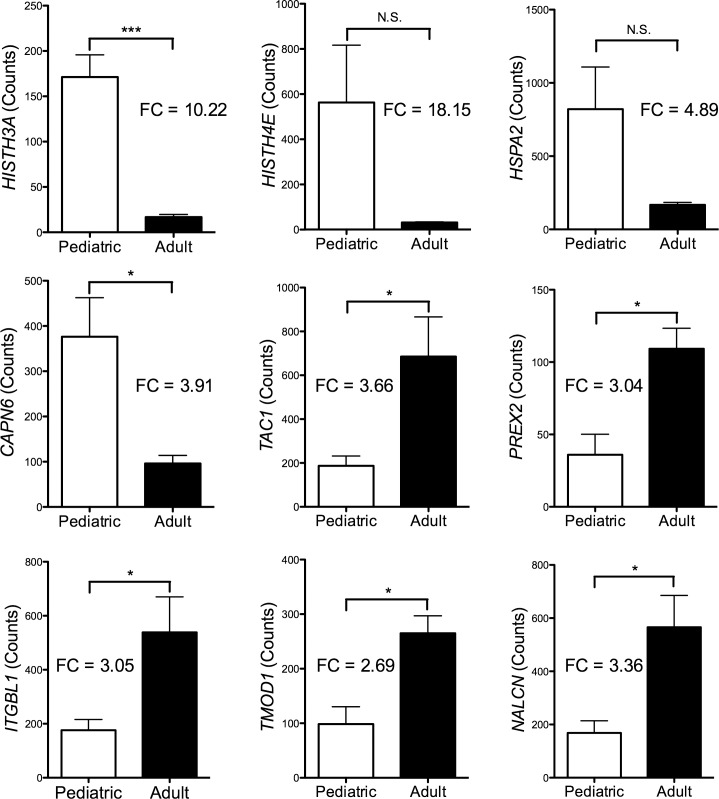
Of the 12,596 genes that were identified as expressed by HCEnC, a total of 60 genes (Fig. 2) demonstrated significant differential expression (non-FDR P value < 0.05, >2 fold change) between pediatric and adult corneal endothelium. A total of 30 genes had higher transcript abundance in pediatric endothelium, and 30 genes had higher transcript abundance in adult endothelium (Supplementary Table S1). After more stringent filtering criteria were applied, 9 genes demonstrated highly significant differential expression (non-FDR P value < 0.005 and >2.9 fold change) between pediatric and adult HCEnC (Table 4). Only HIST1H3A, which was expressed at higher levels in the pediatric HCEnC, survived the most stringent criteria (FDR P value < 0.05 and >3 fold change). All 9 genes demonstrated a fold change greater than 2.5 by digital molecular barcoding analysis, while statistical analysis confirmed the significant differential expression of 7 of the 9 genes (Fig. 3). HIST1H4E and HSPA2 did not show statistical significance, but did demonstrate a trend toward significance with P values of 0.08 and 0.06, respectively.
Figure 2.
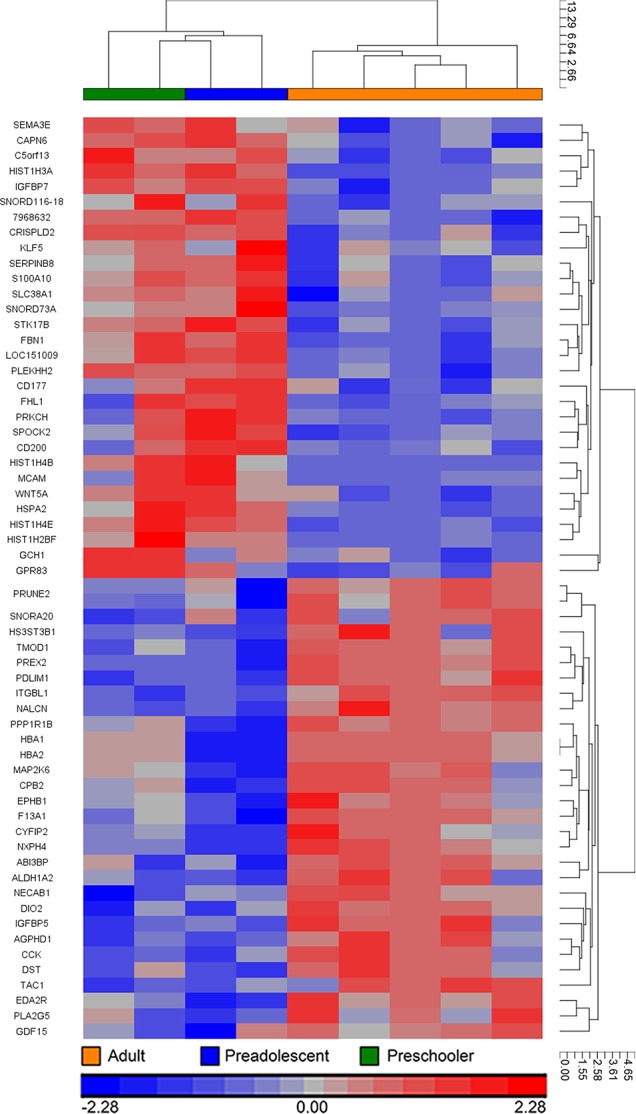
Hierarchical clustering analysis was performed for genes that demonstrated significant (non-FDR P value = 0.05, FC > 2) differential expression between pediatric (preschooler and preadolescent) and adult endothelium.
Table 4.
Highly Differentially Expressed Genes Between Pediatric and Adult HCEnC
|
Probeset ID |
Gene Symbol |
Fold Change* |
P
Value* |
Cytoband |
| Upregulated in pediatric HCEnC | ||||
| 8117330 | HIST1H3A | 5.19082 | 4.0E-06 | 6p22.1 |
| 8117402 | HIST1H4E | 3.90932 | 2.1E-04 | 6p22.1 |
| 7975076 | HSPA2 | 3.11007 | 2.5E-03 | 14q24.1 |
| 8174527 | CAPN6 | 2.92365 | 3.4E-03 | Xq23 |
| Upregulated in adult HCEnC | ||||
| 8134420 | TAC1 | 7.10805 | 4.3E-03 | 7q21-q22 |
| 8146794 | PREX2 | 3.89636 | 2.9E-04 | 8q13.2 |
| 7969861 | ITGBL1 | 3.61241 | 7.3E-05 | 13q33 |
| 8156706 | TMOD1 | 2.98584 | 2.5E-03 | 9q22.3 |
| 7972601 | NALCN | 2.95067 | 4.9E-04 | 13q32.3 |
Differential gene expression between pediatric and adult HCEnC that showed a high probability of rejecting the null hypothesis (P ≤ 0.005), a fold change greater than or equal to 2.9, and an absolute expression intensity value greater than background intensity (3.8) was considered significant.
Figure 3.
Differentially expressed genes were validated by digital molecular barcoding technology. The fold change (FC) for each gene was greater than 2. Transcript levels for the TAC1, PREX2, ITGBL1, TMOD1, and NALCN genes were higher in adult samples (black bars), while transcript levels for the HIST1H3A, HISTH4E, HSPA2, and CAPN6 genes were higher in pediatric samples (white bars). (*P < 0.05, ***P < 0.001).
Gene Ontology Analysis
Gene transcripts that showed significant differential expression between the pediatric and adult group were analyzed for functional group enrichment. The 30 genes with significantly higher transcript levels in pediatric samples showed significant enrichment for six functional groups, of which four (extracellular matrix, proteinaceous extracellular matrix, calcium ion binding, and blood coagulation) were unique to pediatric endothelium (Table 5). The 30 genes that showed significantly higher expression in adult samples demonstrated significant enrichment for seven functional groups, of which five (heparin binding, axon, positive regulation of apoptosis, intracellular signal transduction, and signal transduction) were unique to adult endothelium (Table 5).
Table 5.
Gene Ontology in Pediatric and Adult Corneal Endothelium
|
Function |
GO ID |
Type |
Enrichment Score |
Genes |
| GO functional groups enriched in pediatric HCEnC | ||||
| Extracellular matrix | 31012 | Cellular component | 6.80 | CRISPLD2,FBN1,IGFBP7 |
| Proteinaceous extracellular matrix | 5578 | Cellular component | 5.85 | FBN1,SPOCK2,WNT5A |
| Extracellular region | 5576 | Cellular component | 4.51 | CRISPLD2, FBN1, HIST1H3A, IGFBP7, SEMA3E, SPOCK2, WNT5A |
| Calcium ion binding | 5509 | Molecular function | 4.44 | FBN1,GCH1,S100A10,SPOCK2 |
| Extracellular space | 5615 | Cellular component | 3.92 | FBN1, IGFBP7, SEMA3E, WNT5A |
| Blood coagulation | 7596 | Biological process | 3.72 | CD177,HIST1H3A,PRKCH |
| GO functional groups enriched in adult HCEnC | ||||
| Extracellular region | 5576 | Cellular component | 9.80 | CCK, F13A1, GDF15, HBA1, IGFBP5, ITGBL1, MAP2K6, NXPH4, PLA2G5, SERPINA5, TAC1 |
| Heparin binding | 8201 | Molecular function | 7.01 | ABI3BP,PLA2G5,SERPINA5 |
| Axon | 30424 | Cellular component | 6.32 | DST,EPHB1,TAC1 |
| Positive regulation of apoptosis | 43065 | Biological process | 6.29 | ALDH1A2,CCK,MAP2K6 |
| Extracellular space | 5615 | Cellular component | 5.13 | CCK, CPB2, GDF15, SERPINA5, TAC1 |
| Intracellular signal transduction | 35556 | Biological process | 4.53 | IGFBP5,PPP1R1B,PREX2 |
| Signal transduction | 7165 | Biological process | 3.45 | CCK,GDF15,IGFBP5,MAP2K6,PP1R1B |
Bold text represents functional groups that were unique to the respective age group.
Identification of Genes Expressed in Corneal Endothelial Dystrophy Loci
Posterior Polymorphous Corneal Dystrophy-1.
The PPCD1 locus as described by Yellore et al.21 in a large North American Caucasian pedigree is composed of 204 genes, 94 of which are protein coding and 110 of which are noncoding (pseudogenes, micro RNAs, lincRNAs, and so on). All of the protein-coding genes and 26% (29/110) of the pseudogenes and noncoding RNAs are represented by oligonucleotide probes on the Affymetrix GeneChip 1. array. Fifty-six percent (53/94) of the protein-coding genes and 34% (10/29) of the represented pseudogenes and noncoding RNAs demonstrated signal intensity greater than 3.8 (Supplementary Table S2).
Fuchs Endothelial Corneal Dystrophy-4.
The FECD4 locus is composed of 99 genes, 41 of which are protein coding and 58 of which are noncoding. Of the protein-coding genes, 98% (40/41) were represented on the array, while 19% (11/58) of the noncoding genes had probes on the 1. array. Forty percent (16/40) of the represented protein-coding genes and 55% (6/11) of the represented pseudogenes and noncoding RNAs demonstrated signal intensity greater than background (Supplementary Table S2).
X-Linked Endothelial Corneal Dystrophy.
The XECD locus is composed of 181 genes, of which 68 are protein coding and 113 are putative or noncoding. All of the protein-coding genes and 20% (23/113) of the putative or noncoding genes are represented by oligonucleotide probes on the 1. array. Fifty-one percent (35/68) of the protein-coding genes and 35% (8/23) of the represented putative or noncoding genes demonstrated signal intensity greater than background (Supplementary Table S2).
Discussion
Transcriptome analysis of the corneal endothelium has been previously reported in in vitro27 and ex vivo24–26 HCEnC. Recent reports demonstrate the utility of next-generation sequencing (NGS) technology for transcriptome analysis of human corneal cells.28–30 However, many of these studies were performed using a low number of samples or samples pooled from multiple donor corneas, which limited the ability to perform robust statistical analyses. In addition, obtaining tissue from different sources (academic pathology department and eye bank in one study30) may introduce a batch effect from the different tissue handling and storage conditions.
To address the technical variables that may have negatively impacted the results of prior studies and to build upon their findings, we utilized an experimental design that controlled for several technical factors, including tissue handling and processing, and used a larger sample size to enable robust statistical analysis. First, we obtained both pediatric and adult corneas from commercial eye banks, which use a standardized protocol for tissue recovery and storage. Second, we obtained high-quality RNA from HCEnC of one or both corneas from a single donor by using a recently developed technique for removing the corneal endothelium and Descemet membrane (DM) from the donor cornea, thus eliminating the need to create pooled samples from corneas obtained from multiple individuals.33 Third, we used the Affymetrix GeneChip Human Gene 1.1 ST array, which provides comprehensive whole-transcript coverage of well-annotated protein-coding genes. We do, however, acknowledge the limitations of array-based transcriptome analysis, including that the detection of genes is dependent on the design of the array and that only genes known at the time of the array design are detectable, preventing the identification of novel genes. However, as gene annotation is essentially complete, the inability to identify novel genes is not expected to significantly impact the ability of microarray analysis to accurately characterize the corneal endothelial transcriptome.34,35
After accounting for several technical variables, we performed a whole-genome gene transcription analysis and identified a total of 12,596 expressed genes. This represents approximately 62% (12,596/20,253) of the primarily protein-coding genes in the human genome, which is consistent with a recent report showing that 60% to 70% of protein-coding genes are expressed in human and mouse tissues.36 These data were then analyzed for age-associated differential gene expression between pediatric and adult HCEnC to reveal genes that play temporal but central roles in HCEnC biology.
The eye, and in particular the cornea, undergoes substantial changes during a person's lifetime. The initial postnatal reduction in corneal endothelial cell density, a result of the increase in the size of the cornea coupled with low or no HCEnC mitotic activity, subsequently slows to approximately 0.6% per year.37,38 However, a recent report has proposed a role for UV-induced oxidative stress in the loss of HCEnC.39 Thus, activation of cellular processes in response to such extrinsic/intrinsic stimuli will invariably lead to de novo gene transcriptional activity. Results from our differential gene expression analysis have demonstrated that many genes are uniquely expressed in pediatric and adult HCEnC, while GO analysis identified functional groups associated with those genes. The functional groups enriched in pediatric HCEnC demonstrate an important role for cell–matrix and cell–cell interactions in these cells distinct from adult HCEnC. Thus, pediatric HCEnC may be in the process of establishing an intact and integral endothelial monolayer mediated by cell–cell adhesion molecules such as cadherins.40 In contrast, the adult HCEnC data set was enriched for functional groups indicating that these cells have developed characteristics of senescent cells and have become sensitized to apoptotic stimuli. The observation that adult HCEnC have reduced proliferative capacity may be attributed to the role that heparins, which have been shown to reduce corneal endothelial growth factor availability, play in the extracellular concentration of mitogenic factors.41
A recent study that used NGS technology for transcriptome analysis showed a large number of gene expression changes between fetal and adult human corneal endothelium.30 The findings identify key biological functions for two anatomically and functionally distinct tissues. Although the fetal corneal endothelium can be identified as cuboidal epithelium lining the undeveloped posterior cornea, given the absence of DM, which at this stage of development is simply a discontinuous homogenous acellular layer, it is not surprising that the fetal corneal endothelial transcriptome differs significantly from that of the adult. Herein we build upon previous reports by comparing gene expression of pediatric (preschooler and preadolescent) and adult corneal endothelium, which are anatomically and morphologically similar to each other and presumably functionally indistinct. We identified nine highly significantly differentially expressed genes between pediatric and adult corneal endothelium. NALCN, TAC1, and TMOD1 (upregulated in adult corneal endothelium) have been associated with neuronal function.42–44 As neurons are generally regarded as a terminally differentiated nonproliferating cell type, the increased expression of these genes in adult HCEnC may be representative of the terminally differentiated state of adult HCEnC. PREX2 (upregulated in adult corneal endothelium) has been associated with increased cell proliferation by inhibiting PTEN,45 yet its role in G-protein–coupled receptor signaling suggests a potential role in a wider array of functions not related to cell proliferation. Knockdown of CAPN6 message has been demonstrated to promote cell migration and spreading,46 while the core histone genes HIST1H3A and HIST1H4E are highly expressed during the S1 phase of the cell cycle and play a positive role in cell division.47,48 Thus, downregulation of these genes in the adult corneal endothelium could in part explain the increase in HCEnC polymegethism and pleomorphism seen with increasing age. In addition, the downregulation of heat shock gene HSPA2 may remove a critical protein involved in protection against various stressors.49–51 Taken together, the upregulation of neuron-associated genes (NALCN, TAC1, and TMOD1) and adhesion-associated genes (ITGBL1), in combination with the downregulation of genes associated with cell migration (CAPN6), cell division (HISTH1H3A, HIST1H4E), and cell survival (HSPA2) in the adult corneal endothelium, may prove to be a characteristic gene expression profile of HCEnC that are in functional decline and sensitized to mediators of cell death.
Determining the gene expression profile of cells that show a prominent disease phenotype is crucial in the process of identifying the genetic basis for the disease and elucidating the mechanisms that result in the disease phenotype. With regard to the PPCD3 locus on chromosome 10, only 20 of the 69 positional candidate genes are expressed in the HCEnC. Not only is ZEB1 one of these 20 genes, it is one of the most highly expressed. Thus, had the expression data that we report been available prior to the identification of ZEB1 mutations in PPCD3, ZEB1 would have been among only a handful of candidate genes selected for screening in families with PPCD3. It is therefore reasonable to expect that the genes involved in the pathogenesis of PPCD1, FECD4, and XECD are also expressed in HCEnC and that corneal endothelial expression should be used as a filtering strategy when evaluating coding region variants identified in positional candidate genes with NGS.
In conclusion, we have identified a number of differentially expressed genes in HCEnC, each of which is associated with one of two HCEnC age groups (pediatric and adult). These results have provided, for the first time, insight into the different functional characteristics of anatomically and morphologically indistinct HCEnC from two donor age groups. We have presented transcriptomic evidence that pediatric and adult HCEnC possess a set of distinct functional properties, but follow-up investigations are needed to determine the full functional roles of the genes identified in this study. On the basis of these results, we recommend that investigators studying gene expression in diseased tissue consider using age-matched controls for the most accurate comparison. In addition, we encourage investigators who have reported families with FECD4, XECD, and PPCD1 linked to the aforementioned loci to use the expression data that we present to prioritize screening of the positional candidate genes for each of these corneal endothelial dystrophies.
Acknowledgments
Supported by National Eye Institute Grants R01 EY022082 (AJA) and P30 EY000331 (Core Grant), an unrestricted grant from Research to Prevent Blindness, and a grant from the Gerald Oppenheimer Family Foundation Center for the Prevention of Eye Disease (AJA).
Disclosure: R.F. Frausto, None; C. Wang, None; A.J. Aldave, None
References
- 1. Krafchak CM, Pawar H, Moroi SE, et al. Mutations in TCF8 cause posterior polymorphous corneal dystrophy and ectopic expression of COL4A3 by corneal endothelial cells. Am J Hum Genet. 2005; 77: 694–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aldave AJ, Yellore VS, Yu F, et al. Posterior polymorphous corneal dystrophy is associated with TCF8 gene mutations and abdominal hernia. Am J Med Genet A. 2007; 143A: 2549–2556. [DOI] [PubMed] [Google Scholar]
- 3. Liskova P, Tuft SJ, Gwilliam R, et al. Novel mutations in the ZEB1 gene identified in Czech and British patients with posterior polymorphous corneal dystrophy. Hum Mutat. 2007; 28: 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vincent AL, Niederer RL, Richards A, Karolyi B, Patel DV, McGhee CN. Phenotypic characterisation and ZEB1 mutational analysis in posterior polymorphous corneal dystrophy in a New Zealand population. Mol Vis. 2009; 15: 2544–2553. [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen DQ, Hosseini M, Billingsley G, Heon E, Churchill AJ. Clinical phenotype of posterior polymorphous corneal dystrophy in a family with a novel ZEB1 mutation. Acta Ophthalmol. 2010; 88: 695–699. [DOI] [PubMed] [Google Scholar]
- 6. Bakhtiari P, Frausto RF, Roldan AN, Wang C, Yu F, Aldave AJ. Exclusion of pathogenic promoter region variants and identification of novel nonsense mutations in the zinc finger E-box binding homeobox 1 gene in posterior polymorphous corneal dystrophy. Mol Vis. 2013; 19: 575–580. [PMC free article] [PubMed] [Google Scholar]
- 7. Liskova P, Palos M, Hardcastle AJ, Vincent AL. Further genetic and clinical insights of posterior polymorphous corneal dystrophy 3. JAMA Ophthalmol. 2013; 131: 1296–1303. [DOI] [PubMed] [Google Scholar]
- 8. Lechner J, Dash DP, Muszynska D, et al. Mutational spectrum of the ZEB1 gene in corneal dystrophies supports a genotype-phenotype correlation. Invest Ophthalmol Vis Sci. 2013; 54: 3215–3223. [DOI] [PubMed] [Google Scholar]
- 9. Vithana EN, Morgan P, Sundaresan P, et al. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2). Nat Genet. 2006; 38: 755–757. [DOI] [PubMed] [Google Scholar]
- 10. Desir J, Moya G, Reish O, et al. Borate transporter SLC4A11 mutations cause both Harboyan syndrome and non-syndromic corneal endothelial dystrophy. J Med Genet. 2007; 44: 322–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mehta JS, Vithana EN, Tan DT, et al. Analysis of the posterior polymorphous corneal dystrophy 3 gene, TCF8, in late-onset Fuchs endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2008; 49: 184–188. [DOI] [PubMed] [Google Scholar]
- 12. Riazuddin SA, Zaghloul NA, Al-Saif A, et al. Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet. 2010; 86: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vithana EN, Morgan PE, Ramprasad V, et al. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008; 17: 656–666. [DOI] [PubMed] [Google Scholar]
- 14. Riazuddin SA, Vithana EN, Seet LF, et al. Missense mutations in the sodium borate cotransporter SLC4A11 cause late-onset Fuchs corneal dystrophy. Hum Mutat. 2010; 31: 1261–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gottsch JD, Zhang C, Sundin OH, Bell WR, Stark WJ, Green WR. Fuchs corneal dystrophy: aberrant collagen distribution in an L450W mutant of the COL8A2 gene. Invest Ophthalmol Vis Sci. 2005; 46: 4504–4511. [DOI] [PubMed] [Google Scholar]
- 16. Wieben ED, Aleff RA, Tosakulwong N, et al. A common trinucleotide repeat expansion within the transcription factor 4 (TCF4, E2-2) gene predicts Fuchs corneal dystrophy. PLoS One. 2012; 7: e49083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Riazuddin SA, Parker DS, McGlumphy EJ, et al. Mutations in LOXHD1, a recessive-deafness locus, cause dominant late-onset Fuchs corneal dystrophy. Am J Hum Genet. 2012; 90: 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riazuddin SA, Vasanth S, Katsanis N, Gottsch JD. Mutations in AGBL1 cause dominant late-onset Fuchs corneal dystrophy and alter protein-protein interaction with TCF4. Am J Hum Genet. 2013; 93: 758–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heon E, Mathers WD, Alward WL, et al. Linkage of posterior polymorphous corneal dystrophy to 20q11. Hum Mol Genet. 1995; 4: 485–488. [DOI] [PubMed] [Google Scholar]
- 20. Gwilliam R, Liskova P, Filipec M, et al. Posterior polymorphous corneal dystrophy in Czech families maps to chromosome 20 and excludes the VSX1 gene. Invest Ophthalmol Vis Sci. 2005; 46: 4480–4484. [DOI] [PubMed] [Google Scholar]
- 21. Yellore VS, Papp JC, Sobel E, et al. Replication and refinement of linkage of posterior polymorphous corneal dystrophy to the posterior polymorphous corneal dystrophy 1 locus on chromosome 20. Genet Med. 2007; 9: 228–234. [DOI] [PubMed] [Google Scholar]
- 22. Hosseini SM, Herd S, Vincent AL, Heon E. Genetic analysis of chromosome 20-related posterior polymorphous corneal dystrophy: genetic heterogeneity and exclusion of three candidate genes. Mol Vis. 2008; 14: 71–80. [PMC free article] [PubMed] [Google Scholar]
- 23. Schmid E, Lisch W, Philipp W, et al. A new, X-linked endothelial corneal dystrophy. Am J Ophthalmol. 2006; 141: 478–487. [DOI] [PubMed] [Google Scholar]
- 24. Gottsch JD, Bowers AL, Margulies EH, et al. Serial analysis of gene expression in the corneal endothelium of Fuchs' dystrophy. Invest Ophthalmol Vis Sci. 2003; 44: 594–599. [DOI] [PubMed] [Google Scholar]
- 25. Gottsch JD, Seitzman GD, Margulies EH, et al. Gene expression in donor corneal endothelium. Arch Ophthalmol. 2003; 121: 252–258. [DOI] [PubMed] [Google Scholar]
- 26. Jun AS, Liu SH, Koo EH, Do DV, Stark WJ, Gottsch JD. Microarray analysis of gene expression in human donor corneas. Arch Ophthalmol. 2001; 119: 1629–1634. [DOI] [PubMed] [Google Scholar]
- 27. Serbecic N, Lahdou I, Scheuerle A, Hoftberger R, Aboul-Enein F. Function of the tryptophan metabolite, L-kynurenine, in human corneal endothelial cells. Mol Vis. 2009; 15: 1312–1324. [PMC free article] [PubMed] [Google Scholar]
- 28. Cheong YK, Ngoh ZX, Peh GS, et al. Identification of cell surface markers glypican-4 and CD200 that differentiate human corneal endothelium from stromal fibroblasts. Invest Ophthalmol Vis Sci. 2013; 54: 4538–4547. [DOI] [PubMed] [Google Scholar]
- 29. Chng Z, Peh GS, Herath WB, et al. High throughput gene expression analysis identifies reliable expression markers of human corneal endothelial cells. PLoS One. 2013; 8: e67546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen Y, Huang K, Nakatsu MN, Xue Z, Deng SX, Fan G. Identification of novel molecular markers through transcriptomic analysis in human fetal and adult corneal endothelial cells. Hum Mol Genet. 2013; 22: 1271–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003; 4: 249–264. [DOI] [PubMed] [Google Scholar]
- 32. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003; 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Price MO, Giebel AW, Fairchild KM, Price FW Jr. Descemet's membrane endothelial keratoplasty: prospective multicenter study of visual and refractive outcomes and endothelial survival. Ophthalmology. 2009; 116: 2361–2368. [DOI] [PubMed] [Google Scholar]
- 34. Ezkurdia I, Juan D, Rodriguez JM, et al. The shrinking human protein coding complement: are there fewer than 20,000 genes? [published online ahead of print January 17, 2014]. bioRxiv. 10.1101/001909. [Google Scholar]
- 35. Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ramskold D, Wang ET, Burge CB, Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009; 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bahn CF, Glassman RM, MacCallum DK, et al. Postnatal development of corneal endothelium. Invest Ophthalmol Vis Sci. 1986; 27: 44–51. [PubMed] [Google Scholar]
- 38. Bourne WM, Nelson LR, Hodge DO. Central corneal endothelial cell changes over a ten-year period. Invest Ophthalmol Vis Sci. 1997; 38: 779–782. [PubMed] [Google Scholar]
- 39. Joyce NC, Harris DL, Zhu CC. Age-related gene response of human corneal endothelium to oxidative stress and DNA damage. Invest Ophthalmol Vis Sci. 2011; 52: 1641–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wheelock MJ, Johnson KR. Cadherins as modulators of cellular phenotype. Annu Rev Cell Dev Biol. 2003; 19: 207–235. [DOI] [PubMed] [Google Scholar]
- 41. Fannon M, Forsten-Williams K, Dowd CJ, Freedman DA, Folkman J, Nugent MA. Binding inhibition of angiogenic factors by heparan sulfate proteoglycans in aqueous humor: potential mechanism for maintenance of an avascular environment. FASEB J. 2003; 17: 902–904. [DOI] [PubMed] [Google Scholar]
- 42. Ren D. Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron. 2011; 72: 899–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shanley L, Lear M, Davidson S, Ross R, MacKenzie A. Evidence for regulatory diversity and auto-regulation at the TAC1 locus in sensory neurones. J Neuroinflammation. 2011; 8: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fath T, Fischer RS, Dehmelt L, Halpain S, Fowler VM. Tropomodulins are negative regulators of neurite outgrowth. Eur J Cell Biol. 2011; 90: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leslie NR. P-REX2a driving tumorigenesis by PTEN inhibition. Sci Signal. 2009; 2: . [DOI] [PubMed] [Google Scholar]
- 46. Tonami K, Kurihara Y, Arima S, et al. Calpain-6, a microtubule-stabilizing protein, regulates Rac1 activity and cell motility through interaction with GEF-H1. J Cell Sci. 2011; 124: 1214–1223. [DOI] [PubMed] [Google Scholar]
- 47. Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991; 60: 827–861. [DOI] [PubMed] [Google Scholar]
- 48. Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002; 14: 692–699. [DOI] [PubMed] [Google Scholar]
- 49. Gyrd-Hansen M, Nylandsted J, Jaattela M. Heat shock protein 70 promotes cancer cell viability by safeguarding lysosomal integrity. Cell Cycle. 2004; 3: 1484–1485. [DOI] [PubMed] [Google Scholar]
- 50. Hut HM, Kampinga HH, Sibon OC. Hsp70 protects mitotic cells against heat-induced centrosome damage and division abnormalities. Mol Biol Cell. 2005; 16: 3776–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scieglinska D, Piglowski W, Mazurek A, et al. The HspA2 protein localizes in nucleoli and centrosomes of heat shocked cancer cells. J Cell Biochem. 2008; 104: 2193–2206. [DOI] [PubMed] [Google Scholar]