Abstract
SUMOylation is a reversible post-translational modification essential for genome stability. Using high-resolution mass spectrometry, we have studied global SUMOylation in human cells and in a site-specific manner, identifying a total of over 4,300 SUMOylation sites in over 1,600 proteins. Moreover, for the first time in excess of 1,000 SUMOylation sites were identified under standard growth conditions. SUMOylation dynamics were quantitatively studied in response to SUMO protease inhibition, proteasome inhibition and heat shock. A considerable amount of SUMOylated lysines have previously been reported to be ubiquitylated, acetylated or methylated, indicating crosstalk between SUMO and other post-translational modifications. We identified 70 phosphorylation and 4 acetylation events in close proximity to SUMOylation sites, and provide evidence for acetylation-dependent SUMOylation of endogenous histone H3. SUMOylation regulates target proteins involved in all nuclear processes including transcription, DNA repair, chromatin remodeling, pre-mRNA splicing and ribosome assembly.
Introduction
Reversible post-translational modification (PTM) of lysine residues in proteins by Small Ubiquitin-like Modifiers (SUMOs) plays a key role in genome stability and transcription 1-3. Important SUMO target proteins in the DNA damage response include PCNA 4, 5, BRCA1 6 and 53BP1 7. SUMOs are conjugated to target proteins via an enzymatic cascade involving a dimeric E1 enzyme (SAE1-2), a single E2 enzyme, Ubc9, and a limited number of E3 enzymes 8. Mice deficient for Ubc9 die at the early post-implantation stage as a result of chromosome condensation and segregation defects, underlining the essential role of SUMOylation to maintain genome stability 9.
Frequently, SUMOylation regulates the function of target proteins by enabling or stabilizing non-covalent protein-protein interactions via SUMO Interaction Motifs (SIMs) 8. Classical examples of this type of interaction include the binding of SUMOylated RanGAP1 to the nucleoporin RanBP2 10 and the binding of the SRS2 helicase to SUMOylated PCNA 11, 12.
Despite great interest in SUMOylation in the fields of genome stability, transcriptional regulation, nuclear organization, signal transduction and from a clinical point of view, global insight in protein SUMOylation is limited. Mass-spectrometry (MS)-based proteomics has enabled global insight in different PTMs in a site-specific manner 13, 14, including phosphorylation 15, 16, acetylation 17, methylation 18, glycosylation 19 and ubiquitylation 20-24. Affinity purification strategies include immunoprecipitation and TiO2 and Fe3+ immobilized metal affinity chromatography. Highly potent SUMO proteases 25, inconvenient C-terminal tryptic SUMO tags and low stoichiometry combined with suboptimal purification methods have hampered the identification of SUMO acceptor lysines in target proteins 14, 26-29.
We set out to develop an efficient purification strategy to study global protein SUMOylation in a site-specific and dynamic manner.
Results
A Strategy for Mapping SUMO Sites in Endogenous Proteins
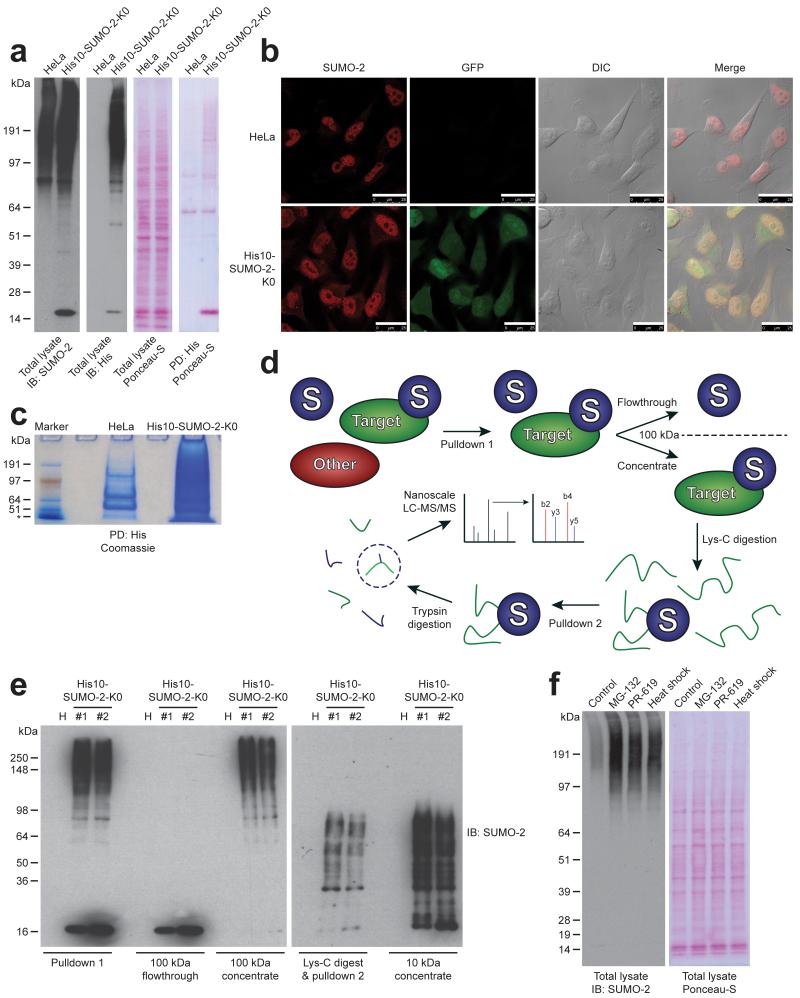
To facilitate the study of SUMOylated proteins, a common method is the employment of epitope-tagged SUMO, in order to allow efficient purification after highly denaturing lysis of cells to inactivate SUMO proteases. We enriched SUMOylated peptides from a HeLa cell line stably expressing His10-tagged SUMO-2. This tag is small and compatible with denaturing buffer conditions. We established these stable cells using a bicistronic lentivirus encoding His10-SUMO-2 and GFP separated by an Internal Ribosome Entry Site (IRES). After infection, a population expressing this construct at low levels was obtained using flow cytometry. We confirmed low expression levels by immunoblotting (Fig. 1A). As expected, the protein located predominantly in the nucleus (Fig. 1B) 30, 31. The His10 tag enabled single round purification with a high yield and purity in contrast to the His6 tag commonly used in the field (Fig. 1C).
Fig. 1.
A strategy for mapping SUMO-2 acceptor lysines in endogenous proteins.
(a) Immunoblot confirming the low level expression of H10-S2-K0 in HeLa cells. Ponceau-S staining is shown as a loading control. Additionally, His10-pulldown was performed to enrich SUMOylated proteins, and Ponceau-S is shown to indicate high-specificity enrichment. The experiment shown was replicated in biological duplicate.
(b) Confocal fluorescence microscopy image confirming the predominantly nuclear localization of H10-S2-K0. GFP; Green Fluorescent Protein. DIC; Differential Interference Contrast. Scale bars represent 25 μm. The experiment shown was replicated in biological duplicate.
(c) Coomassie stain displaying the efficacy of a single-step His10-pulldown performed on approximately 50 million HeLa and H10-S2-K0 cells. The experiment shown was replicated in biological duplicate.
(d) Schematic overview of the H10-S2-K0 SUMOylation site purification strategy. A direct purification step is followed by concentration of SUMOylated proteins, which are subsequently digested with endopeptidase Lys-C. The H10-S2-K0 bearing the SUMOylated peptide is re-purified, concentrated, digested with trypsin, and finally analyzed by high-resolution nanoscale LC-MS/MS.
(e) Immunoblotting analysis was used to confirm the efficiency of the purification steps described in (d). The experiment shown was replicated in biological duplicate.
(f) Immunoblotting analysis of total lysates from cells stably expressing H10-S2-K0 which were mock treated, or treated with MG-132, PR-619 or heat shock. Ponceau-S staining is shown as a loading control. The experiment shown was replicated in biological duplicate.
In order to enrich for SUMOylated peptides, we used a SUMO-2 form that is resistant to cleavage by endopeptidase Lys-C. Lysine-deficient (K0) SUMO-2 behaves very similar to wild-type SUMO-2, except for SUMO polymerization 29. Our purification strategy consisted of a first round of His10-SUMO-2 purification, a filter step to concentrate SUMO-2 conjugates while simultaneously separating SUMO-2 conjugates from free SUMO-2, digestion by Lys-C and a second round of purification followed by a trypsin digest (Fig. 1D and 1E). The second round of purification enabled enrichment at the site-level, greatly reducing the complexity of the final sample.
The C-terminal tryptic fragment of wild-type human SUMO-2 is 32 amino acids and, due to its size, is not compatible with efficient mapping of SUMO-2 acceptor lysines. In contrast, yeast SUMO, Smt3, contains a conveniently located arginine that results in a five amino acid C-terminal tryptic fragment. We have generated a SUMO-2 Q87R mutant mimicking yeast SUMO to enable the identification of SUMO-2 acceptor lysines by mass spectrometry.
SUMOylation is a dynamic process, regulated via crosstalk with the ubiquitin-proteasome system 32, 33, sensitive to heat shock 30 and sensitive to the broad range SUMO – and ubiquitin protease inhibitor, PR-619. Treatments with the proteasome inhibitor MG-132, heat shock and PR-619 resulted in a considerable accumulation of SUMO-2 conjugates (Fig. 1F).
Identification of 4,361 SUMO-2 Sites in 1,606 Proteins
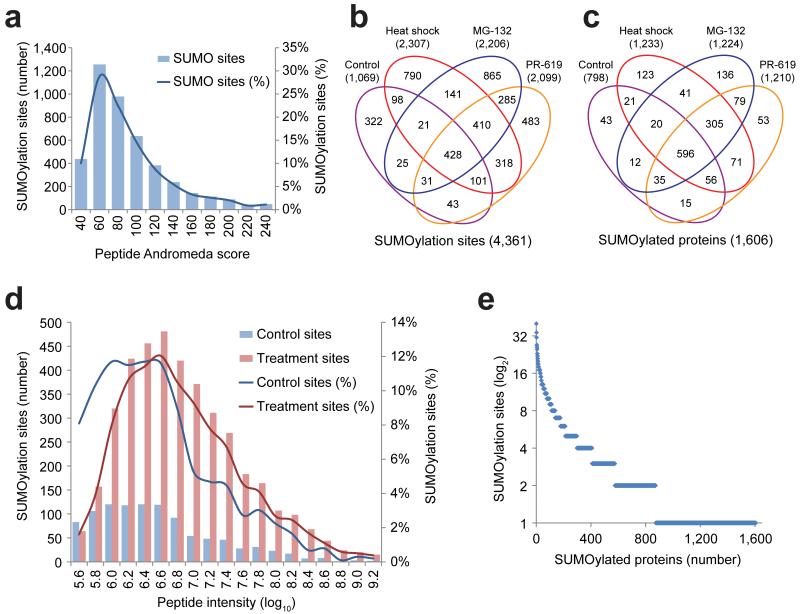
We analyzed tryptic digests of in-solution digested re-purified SUMOylated peptides by nanoscale LC-MS/MS, without further fractionation, and using 2-hour LC gradients. 5,339 SUMOylated peptides corresponding to 4,361 unique SUMOylation sites (Supplementary Table 1) were identified in 1,606 proteins (Supplementary Table 2) at a false discovery rate (FDR) below 1%. Mass accuracy was within 3 ppm for 98.0% of all identified sites, and within 6 ppm for all sites, with an average absolute mass error of 0.77 ppm. The majority of identified SUMOylation sites had an Andromeda peptide score in the range of 60-100 (Fig. 2A). We pinpointed the precise SUMO-2 acceptor lysines in over 98.8% of the SUMOylated peptides (Supplementary Table 1). The overall purity of our method was demonstrated by the average presence of a SUMO-2 acceptor lysine in 25.0% of the peptides identified in the final purified fractions. As negative controls, we attempted identification of SUMOylated lysines after performing site enrichment on the parental HeLa cell line. In addition, we endeavored identification of SUMO sites from HeLa total lysates. In both cases, we did not find a single site.
Fig. 2.
Overview of mass spectrometry results.
(a) Overview of the amount of SUMOylated peptides identified with their respective Andromeda peptide scores. In the range of 400 to 1200 SUMOylation sites were identified from single runs depending on treatment conditions. The control experiments were performed in biological hexuplicate, PR-619 experiments were carried out in biological quintuplicate, and MG-132 and heat shock experiments were performed in biological triplicate. Experiments were measured in technical duplicate, with one 15-cm plate (20 million cells) serving as input for a single run.
(b) Schematic representation of the amount of SUMOylation sites discovered in relation to the cellular treatments used. Over 4,300 SUMOylation sites were identified in total.
(c) Schematic representation of the amount of SUMOylated proteins discovered in relation to the cellular treatments used. Over 1,600 SUMOylated proteins were identified in total.
(d) Overview of the SUMOylated peptides identified under control conditions, as compared to the SUMOylated peptides exclusively discovered after cellular treatments. The absolute and relative amount of SUMOylation sites are plotted against the peptide intensity.
(e) Overview of the amount of SUMOylation sites identified per protein.
This is a major step forward for the field and enables the efficient analysis of SUMOylation at a proteome-wide level in a site-specific manner, only requiring a relatively small amount of cells (~20 million), and for the first time under standard growth conditions. Our dataset includes an extensive number of well-known SUMO target proteins, e.g. RanGAP1, PML, Topoisomerase-1, -2α, -2β, PCNA, BLM, BRCA1, RanBP2, RNF168, SAFB2 (Supplementary Table 2), further confirming the validity of the approach.
In total, we identified 1,069 sites from cells grown under regular cell culture conditions (Fig. 2B and 2C). The dynamic nature of SUMOylation was underlined by the identification of 3,292 additional SUMOylation sites in response to heat shock, proteasome inhibitor treatment and - or SUMO protease inhibitor treatment, representing three quarters (75.5%) of the total identified sites (Fig. 2B). The peak intensities of the SUMOylated peptides identified in response to MG-132, PR-619 or heat shock were higher compared to the control sample, indicating that not only the number of acceptor sites increased upon treatment, but also the stoichiometry of SUMOylation (Fig. 2D).
We used Label-Free Quantification (LFQ) to chart the changes in overall protein SUMOylation after the cellular treatments, and found significant changes in the SUMOylation state of hundreds of proteins (Supplementary Fig. 1A-C and Supplementary Table 3). In order to assess the reproducibility of the methodology, we performed Principle Component Analysis (PCA) to visualize unbiased correlation between all biological replicates (Supplementary Fig. 1D). We performed the control experiment in biological hexuplicate, the PR-619 experiment in biological quintuplicate, and the MG-132 and heat shock experiments in biological triplicate. Using PCA, all same-condition replicates were found to cluster closely together. Additionally, scatter plot analysis was performed to assess Pearson correlation between all experiments (Supplementary Fig. 1E). We found high values of correlation between same-condition samples at the protein level (R: 0.85-0.92) and at the site level (R: 0.66-0.75), as compared to correlation between different-condition (but nonetheless SUMO-enriched) samples. Finally, we performed hierarchical clustering of all biological replicates, highlighting a significant reproducibility between same-condition replicates, and visualizing clusters of proteins that are upregulated or downregulated in SUMOylation dependent on the various cellular stresses (Supplementary Fig. 1F).
In order to confirm the biological validity of the lysine-deficient SUMO mutant for the purpose of mapping SUMO sites in the context of cellular stress conditions, we compared total SUMO pools in HeLa cells expressing either wild-type or the K0-mutant (Supplementary Fig. 2A). No notable differences were observed between the accumulations of SUMOylated protein pools in response to all employed stress conditions, regardless of whether wild-type or lysine-deficient SUMO was employed. Furthermore, we found the amount of co-purifying ubiquitin to be identical in both cell lines, showing that crosstalk between ubiquitin and SUMOylation is not visibly affected.
Further, we validated a set of novel-identified SUMO target proteins, RNF216, SNW1, TCF12, and ZND280D, through pulldown and immunoblot analysis using a cell line stably expressing His10-tagged SUMO-2 wild-type (Supplementary Fig. 2B). Additionally, because we employed the SUMO protease inhibitor PR-619 in SUMO proteomics for the first time, we validated the quantified change in SUMOylation of various target proteins using a wild-type SUMO cell line. This set includes 4 known SUMO targets, FOXM1, HNRNPM, RAD18, and SART1, as well as 2 novel SUMO target proteins, WDR70 and MCM10 (Supplementary Fig. 2C).
SUMOylation sites per protein ranged from a single site in nearly half of all SUMO target proteins, with 10 or more sites in 96 proteins, and 20 or more sites in only 13 proteins – PARP1 (20 sites), ZMYM2 (20), ZNF281 (21), MECOM (22), NSUN2 (22), MKI67 (23), MIS18BP1 (25), TPX2 (26), BLM (27), NKTR (27), FBN1 (31), GTF2I (34), ZNF451 (40)(Fig. 2E). We found most proteins (64.2%) to be conjugated to only one or two SUMO moieties.
We compared all SUMOylated proteins and sites identified in this study to previous studies on SUMOylation. On average, we identified 51% of previously reported MS/MS-identified SUMO target proteins (Supplementary Fig. 3A and Supplementary Table 4), and expanded the known amount of SUMOylated proteins by nearly one thousand. We found 52% of all previously reported MS/MS-identified SUMOylation sites (Supplementary Fig. 3B), as reported in the PhosphoSitePlus database (PSP; PhosphoSitePlus®, www.phosphosite.org, 34) and more recently by Schimmel et al. 35 and Tammsalu et al. 36. Our study expands by over 3,000 the number of known MS/MS-identified SUMOylation sites, and for the first time we identified over 1,000 SUMOylation sites under standard growth conditions.
SUMOylation is a key post-translational modification in all eukaryotes, but is absent in prokaryotes. We studied phylogenetic conservation of SUMOylation with respect to conservation of entire proteomes (Supplementary Fig. 4A). SUMOylated proteins are significantly more conserved than total proteomes. Within orthologues, SUMOylation is the most conserved post-translational modification together with acetylation. When considering conservation of proteins with no orthologues, SUMO is more conserved than phosphorylation, but less than ubiquitylation, acetylation and methylation (Supplementary Fig. 4B). This difference is indicative of an increased frequency of SUMOylation occurring on proteins that are absent in lower eukaryotes.
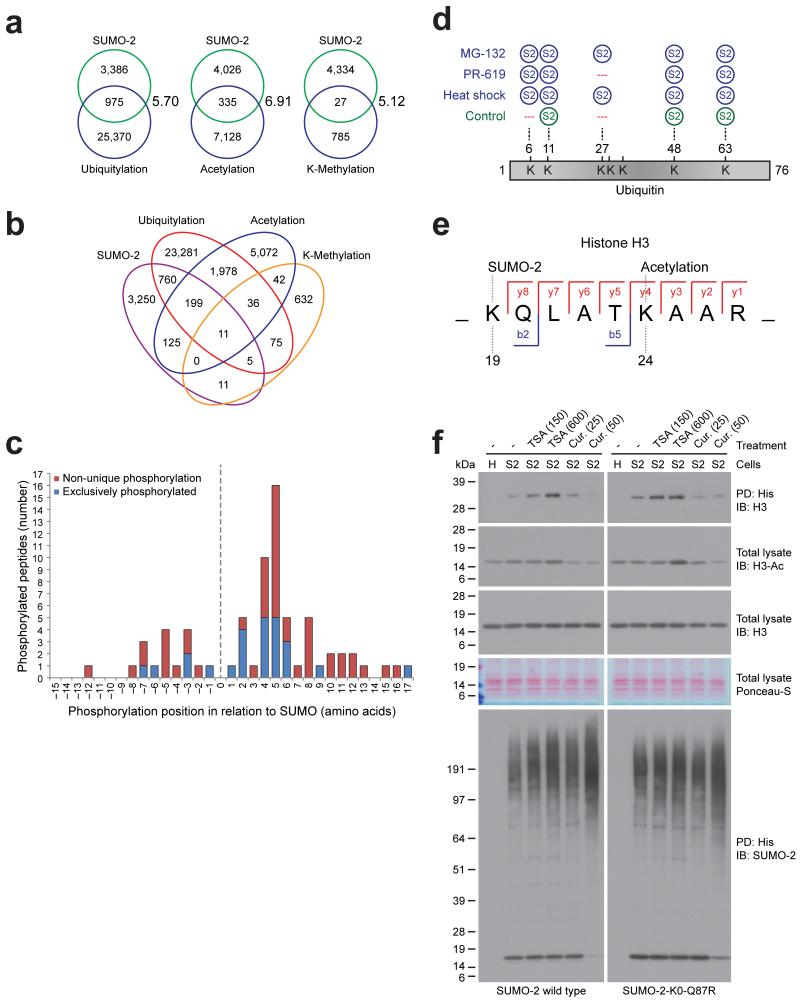
We investigated the potential overlap between the identified SUMO-acceptor lysines with other post-translational lysine modifications. For this purpose, we extracted all known human MS/MS-identified ubiquitylation, acetylation and lysine-methylation sites from PSP, and cross-compared modification sites (Fig. 3A, 3B and Supplementary Table 5). SUMOylation is known to compete with ubiquitin for acceptor lysines in target proteins 37. However, the extent of this crosstalk is currently unclear. We compared the identified SUMO acceptor lysines to acceptor lysines for ubiquitin derived from PSP and found that nearly one in four (22.4%) SUMOylation sites are also known to be ubiquitylated, indicating extensive crosstalk between SUMOylation and ubiquitylation (Fig. 3A and 3B). Overlap between SUMOylation and acetylation and between SUMOylation and lysine-methylation occurs less frequently, although this could be related to the smaller number of acetylation and methylation sites currently identified. From the perspective of all known ubiquitylation, acetylation and methylation sites, SUMOylation occurs on roughly 4% of all these sites, and does not favor one PTM over the other. Considering the observed amount of overlap between SUMO and other PTMs on the same lysines, versus the total amount of lysines in the human proteome, the observed overlap is significant.
Fig. 3.
SUMO is extensively involved in PTM crosstalk.
(a) Schematic representation of the overlap of the identified SUMOylated lysines as compared to the other lysine post-translational modifications ubiquitylation, acetylation and lysine-methylation. Enrichment ratio between observed overlaps and expected overlaps are indicated, and were significant by Fisher Exact Testing with P < 1E-10.
(b) Similar to (a), overlap between SUMOylation, ubiquitylation, acetylation and lysine-methylation.
(c) Schematic overview of phosphorylation sites adjacent to SUMOylated lysines, both identified by mass spectrometry in our screen, and their amino acid spacing in relation to the SUMOylated lysine. Some peptides were exclusively found to be SUMOylated in combination with phosphorylation (blue). Non-unique phosphorylation sites on SUMOylated peptides were also discovered (red).
(d) Schematic representation of the identified SUMOylation sites on ubiquitin.
(e) Schematic representation of the Histone H3 peptide as identified by MS/MS, simultaneously modified by SUMOylation on lysine 19 (H3K18) and acetylation on lysine 24 (H3K23). Identified fragment ions are indicated. A fully annotated high resolution MS/MS spectrum is available as part of Supplementary Data Set 1.
(f) Immunoblot analysis of total lysates and His10-pulldown samples from HeLa cells stably expressing His10-SUMO-2 wild-type or K0-mutant, which were either mock treated, treated with the histone de-acetylation inhibitor Trichostatin A (TSA) at the indicated dose in nM, or treated with the histone acetylation inhibitor curcumin (Cur) at the indicated dose in μM. Ponceau-S staining is shown as a loading control. The experiment shown was replicated in biological duplicate.
Interestingly, crosstalk between SUMOylation and other post-translational modifications includes regulation of enzymatic components including 46 kinases, 33 proteins with intrinsic phosphatase activity, 29 ubiquitin-protein ligase family members, seven ubiquitin proteases, seven acetyltransferases, ten deacetylases, 22 methyltransferases and 13 demethylases (Supplementary Table 6).
We found 23 SUMOylated peptides exclusively in combination with phosphorylation, and an additional 47 SUMOylated peptides together with phosphorylation in a non-unique fashion (Supplementary Table 7). Phosphorylation occurred relatively close to the lysine, and was found both upstream and downstream of the SUMOylation lysine. We found phosphorylation to occur predominantly at or nearby the +5 position (Fig. 3C), in agreement with the earlier described PDSM 29, 38. We observed 5 phosphorylation sites at +2 relative to the SUMOylated lysine, which could serve as the negative charge required for efficient SUMOylation instead of glutamic or aspartic acid.
Furthermore, we found direct modification of endogenous ubiquitin by SUMO-2 on lysines 11, 48, and 63 under control conditions (Fig. 3D). After cellular treatments, we additionally observed SUMO-2 modification of ubiquitin on lysines 6 and 27 (Fig. 3D). Thus, mixed chain formation between ubiquitin and ubiquitin-like family members is more extensive than previously thought. Furthermore, we detected mixed chain formation between all SUMO family members (Supplementary Table 1).
We found three SUMOylated peptides exclusively in combination with acetylation, with two of these events occurring on histones H3 (Fig. 3E) and H4 (Supplementary Table 7). We detected an additional SUMOylated peptide from PML together with acetylation in a non-unique fashion. These sites could indicate acetylation-dependent SUMOylation, suggesting a novel type of crosstalk between these two major modifications. In order to further investigate such a dependency, we treated HeLa cells expressing either wild-type or lysine-deficient SUMO with the histone deacetylases inhibitor Trichostatin A (TSA), known for increasing acetylation of histones, or the histone acetyltransferases inhibitor curcumin, known for decreasing acetylation of histones 39. Subsequently, we performed SUMO enrichment and investigated the SUMOylation state of histone H3 (Fig. 3F). For the first time, we visualized the modification of endogenous histone H3 by SUMO. Furthermore, we found the SUMOylation of histone H3 to be increased upon TSA treatment, correlating with an increase in histone H3 acetylation. Conversely, after treatment with curcumin, we found the SUMOylation of histone H3 to be decreased, correlating with a decrease in histone H3 acetylation. The total pool of conjugated SUMO was found to mildly increase regardless of which inhibitors were used, ruling out a non-specific change in the SUMOylation state of histone H3 (Fig. 3F). Our results demonstrate interplay between the acetylation and SUMOylation of endogenous histone H3.
Insight in the SUMOylation consensus motif
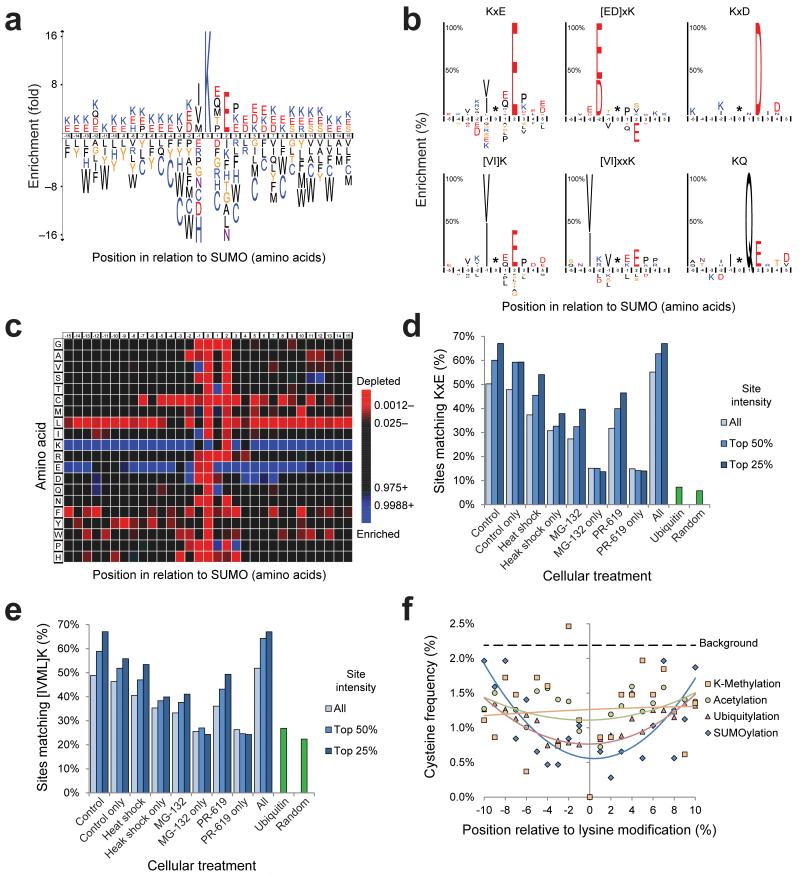
SUMOylation is known to occur on the classical consensus motif ΨK×E 40 or ΨK×[ED] 41, where Ψ is a large hydrophobic amino acid. Previously, we have found that other residues are also used at the Ψ position 29. Our dataset provides an important opportunity to obtain more insight in the SUMOylation consensus motif. More than half of the identified sites from untreated cells matched the consensus motif K×E, whereas K×D type sites were not found to be enriched over background frequency (Fig. 4A). We studied the K×E type SUMOylation motif in RanGAP1, a highly SUMOylated protein 10, 42. Replacing E526 for D resulted in a notable drop in SUMOylation (Supplementary Fig. 5A and 5B). As a negative control, we included our previously published ΔGL RanGAP1 mutant 29.
Fig. 4.
Novel insight in the SUMOylation consensus motif.
(a) IceLogo of all SUMOylation sites identified under control conditions. The height of the amino acid letters corresponds to fold-change. All amino acid changes were significant with P < 0.05 by two-tailed Student’s t test.
(b) SubLogos of various consensus motifs. The height of amino acid letters represents the %-change enrichment or depletion of the motif set as compared to the reference set. All amino acid changes were significant with P < 0.05 by two-tailed Student’s t test.
(c) As (a), but in heatmap format. Green is indicative of a statistical enrichment as compared to randomly expected, and red is indicative of a depletion. All amino acid changes were significant with P < 0.05 by two-tailed Student’s t test.
(d) Overview of the amount of SUMOylation sites matching the short consensus motif K×E in different subsets of sites corresponding to different cellular treatments. Additionally, per subset, the top 25% intense, the top 50% intense, or all sites are shown. Matching of ubiquitin sites to the motif and the randomly expected frequency are also shown.
(e) As (d), but for the short consensus motif [IVML]K.
(f) Schematic representation of the cysteine frequency close to all identified control SUMOylation sites, as well as other PTMs, ranging from −10 to +10 amino acids of the modified lysine. For all PTMs, a 2nd order polynomial trend line was calculated. The background cysteine frequency is indicated.
The acidic residue is not necessarily located two positions downstream of the SUMOylated lysine but can also be found two positions upstream with a higher frequency for aspartic acid compared to the regular SUMOylation motif (Fig. 4B), in agreement with the inverted consensus motif [ED]×K that we proposed based on a very small number of identified SUMOylation sites 29. We confirmed the relevance of the E300 residue in the inverted motif covering Tel SUMOylated lysine 302 (Supplementary Fig. 5C and 5D). Replacing this residue for alanine eliminated SUMOylation, whereas replacing it for aspartic acid did not completely abolish SUMOylation, in agreement with the inverted consensus motif [ED]×K.
We superimposed the 1,069 SUMOylation sites identified under control conditions, including a sequence window ranging from −15 to +15 amino acids, and compensated the amino acid frequency against the randomly expected frequency across the human proteome (Fig. 4A and 4C). We observed the highest degree of enrichment for valine and isoleucine at −1, and glutamic acid at +2, with over half of all control SUMOylation sites adhering to this consensus. When only considering the top 50% and top 25% of most abundant SUMOylation sites for glutamic acid at +2, this frequency increased to 60% and 67% respectively, indicating that the lysines situated in SUMOylation consensus motifs are efficiently SUMOylated (Fig. 4D). We observed a similar trend for the hydrophobic amino acids at −1 (Fig. 4E). A further expanded consensus motif, taking statistical local enrichments into account, amounted to [IVML]-K-[EQMTP]-E-P. Interestingly, the adherence to the consensus motif dropped moderately for sites exclusively mapped after heat shock treatment, and decreased drastically after MG-132 and PR-619 treatment, where sites matching K×E reached as low as 13%, barely higher than the ubiquitin or random lysine frequencies (Fig. 4D).
Considering the 30 amino acid region flanking SUMOylated lysines, it is evident that this region is enriched for lysine and glutamic acid (Fig. 4A and 4C). Thus, SUMOylated lysines are frequently located in regions enriched for charged residues, indicative of solvent exposure. Interestingly, SUMOylated regions are depleted for phenylalanine, tryptophan, tyrosine, leucine, and most notably for cysteine (Fig. 4A and 4C). Since SUMOs are transferred along an enzymatic cascade via thioester formation, the reduced frequency of cysteines near SUMOylated lysines under standard conditions could help to avoid the formation of thioesters between SUMOs and target proteins. Interestingly, a reduced frequency of cysteine can also be observed for ubiquitylated regions, probably for the same reason as proposed for SUMOylation (Fig. 4F). Reduced frequencies of cysteine were less pronounced for regions flanking methylated or acetylated lysines (Fig. 4F).
Insight in SUMOylated protein groups
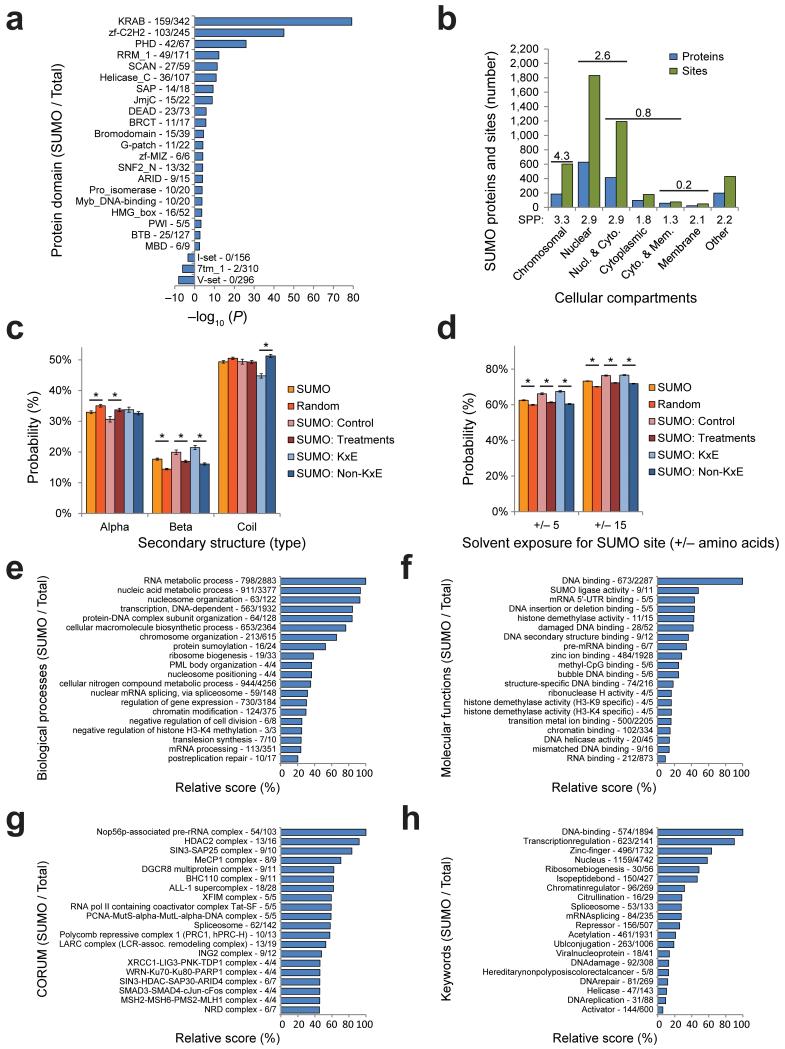
Protein domains that are frequently SUMOylated include the Krüppel associated box (KRAB) domain, which is a repressor domain found in many zinc finger protein-based transcription factors (Fig. 5A). Other domains included zinc fingers, PHD fingers and RRM1, which are domains that serve important roles in binding of DNA, RNA or other proteins, and are often found in nuclear or chromatin-associated proteins.
Fig. 5.
SUMOylation is a predominantly nuclear event, and is involved in many biological processes and protein complexes.
(a) Overview of protein domain families overrepresented or underrepresented in all identified SUMOylated proteins, ranked by −log10 P-value. SUMOylated proteins as compared to total proteins are indicated.
(b) Overview of the amount of SUMOylated proteins and sites in relation to their subcellular localization. Gene Ontology Cellular Compartments (GOCC) enrichment ratios are indicated above categories, and average SUMO sites per protein are indicated below.
(c) Overview of the predicted secondary structure of different subsets of SUMOylated lysines. The random set corresponds to lysines randomly selected from SUMOylated proteins. Differences indicated with an asterisk (*) were significant with P < 0.001. Error bars represent s.e.m. and are based on structural predictions on a per-site basis. For “SUMO” n=4,361. For “Random” n=5,725. For “Control” n=1,070. For “Treatments” n=3,291. For “K×E” n=1,300. For “Non-K×E” n=3,061.
(d) Overview of the predicted solvent exposure of different subsets of SUMOylated lysines, including either −5 to +5 amino acids, or −15 to +15 amino acids. Significance and error bars are synonymous to (c).
(e) All identified SUMOylated proteins were annotated with Gene Ontology Biological Processes terms, and compared against the annotated human proteome. Categories were scored by a combination of enrichment ratio and P-value. The amount of hits as compared to the category size is indicated.
(f) As (e), but for Gene Ontology Molecular Functions.
(g) As (e), but for CORUM complexes.
(h) As (e), but for Keywords.
We investigated the subcellular localization of SUMOylated proteins by Gene Ontology Cellular Compartments classes, and plotted all identified proteins and sites (Fig. 5B). We found SUMOylation to be an almost exclusively nuclear modification, with cytoplasmic modification occurring primarily on proteins that are also annotated as nuclear. Enrichment analysis showed the highest ratio for chromatin-associated proteins, closely followed by all nuclear proteins (Fig. 5B). We found cytoplasmic and membrane proteins to be depleted. Correspondingly, the amount of SUMO sites per protein was also observed to be higher in chromatin-associated and nuclear proteins.
The first identified SUMOylation site, K524 in the nuclear-pore component RanGAP1, is located in an unstructured region of the protein 43. SUMOylation is thought to occur predominantly in unstructured regions 41. To investigate the localized structural properties of proteins around sites of SUMOylation, we in silico folded all 4,361 sites including the 30 amino acid sequence window, as well as over 5,000 lysines randomly chosen from SUMOylated proteins as a reference set. We performed secondary structure prediction of the modified lysine, and classed the structure as α-helix, β-sheet, or otherwise coiled (Fig. 5C). Our results indicate a modest reduction in SUMOylation of α-helices and a significant increase in SUMOylation of β-sheets compared to background frequencies. This trend was most striking for K×E type SUMOylation sites, where a significant decrease in unstructured regions was observed, as a trade-off for an increase in β-sheets. We additionally observed an increased tendency for SUMOylated regions to be solvent exposed (Fig. 5D).
SUMO modifies highly interconnected networks of proteins
Genome stability, transcription and translation are three important biological processes as evidenced by term enrichment analysis for Gene Ontology Biological Processes involving the identified SUMO targets (Fig. 5E). Furthermore, nucleic acid metabolism, chromosome organization, DNA repair, cell cycle regulation, RNA splicing, histone modification, and nuclear body organization are amongst the most enriched processes. For Gene Ontology Molecular Functions, in absolute numbers, we identified 673 DNA binding proteins and 484 Zinc binding proteins as the largest functional groups of SUMO target proteins (Fig. 5F). SUMO also modified significant amounts of subunits from known CORUM protein complexes, including Nop56p pre-rRNA, SIN2-SAP25, BRAF53-BRCA2, LARC, BHC, MeCP1 and HDAC1 and -2 protein complexes (Fig. 5G). Additional analyses by keywords and KEGG terms also highlighted SUMO’s regulation of many pivotal cellular processes (Fig. 5H), including an enrichment for proteins known to be involved in cancer pathways (Supplementary Table 8), such as TP53, MITF, VHL, BRCA2, STAT1, FOS, JUN and SMAD4. The complete term enrichment analysis (Supplementary Table 8), and a fully annotated list of all SUMOylated proteins (Supplementary Table 9), are available as online supporting information.
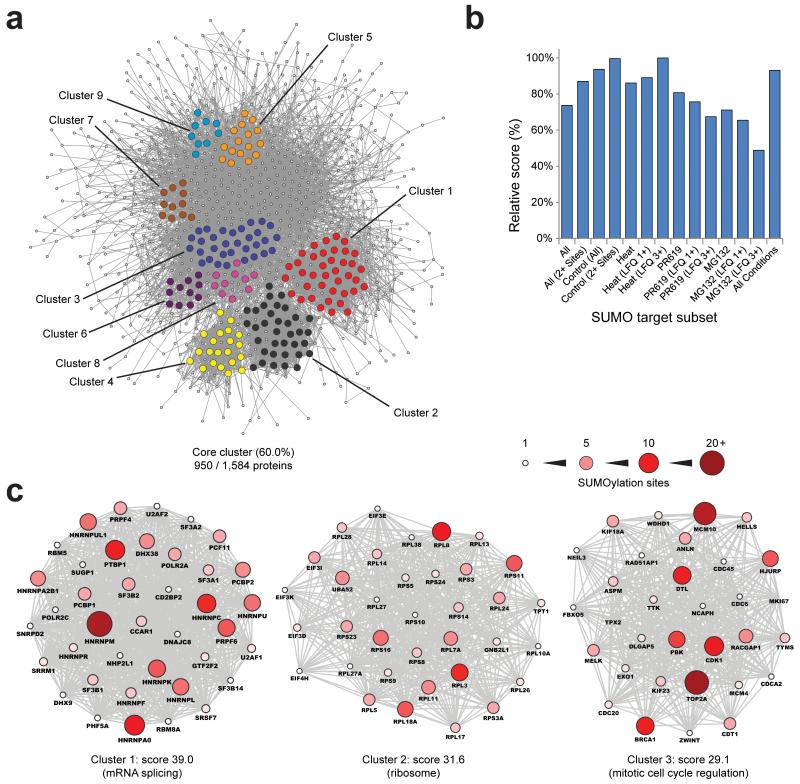
SUMOylated proteins form a very complex, highly organized network of interacting proteins as visualized using a Search Tool for the Retrieval of Interacting Genes and - or Proteins (STRING) network analysis (Fig. 6A). 60% of all identified proteins are part of one main functional cluster, at high STRING confidence. We performed STRING analyses on a per-treatment basis and at high STRING confidence to assess protein-protein interaction enrichment ratios and network participation (Table 1). Overall, we observed ten times more interactions as compared to expected, with the SUMO target proteins from untreated cells and with multiple sites showing the highest degree of enrichment. The strength of the STRING networks, derived from interaction enrichment as well as participation of all proteins, was observed to be greatest for proteins identified under control conditions (Fig. 6B). Strikingly, we observed proteins identified after heat shock, and even more so proteins found to be significantly upregulated in SUMOylation after heat shock, to form a highly coherent STRING network. This stands in contrast to proteins upregulated in SUMOylation after MG-132 or PR-619 treatments, which we found to be less related in their interactions.
Fig. 6.
SUMO modifies highly interconnected functional networks of proteins.
(a) STRING network analysis of all identified SUMOylated proteins, with a STRING interaction confidence of 0.7 or greater. MCODE was used to extract the most highly interconnected functional clusters from the network, which are indicated in different colors.
(b) Overview of relative STRING network score corresponding to Table 1. This score was computed through multiplication of the interaction enrichment ratio, protein network connectivity, and the average STRING confidence of all interactions.
(c) Schematic overview of the three highest scoring MCODE sub-clusters from (a). The size and color of the individual proteins corresponds to the amount of SUMOylation sites identified in the protein. The six additional MCODE clusters are available as Supplementary Fig. 7.
Table 1.
Subsets of SUMO target proteins are differentially interconnected.
Overview of STRING analyses using different subsets of SUMOylated proteins. “Enrichment” is a ratio derived from the observed amount of interactions divided by the expected amount of interactions. “Connected” refers to the percentage of input proteins connected to the core cluster. P values for all individual analyses are < 1E-15. The network displayed in Fig. 6A corresponds to “All”.
| Selection | Proteins | Interactions | Enrichment | Connected |
|---|---|---|---|---|
| All | 1584 | 6801 | 9.4 | 60.0% |
| All (2+ Sites) | 860 | 2765 | 11.6 | 57.6% |
| Control | 790 | 2035 | 12.1 | 58.7% |
| Control (2+ Sites) | 539 | 1202 | 13.4 | 56.4% |
| Heat | 1222 | 5240 | 10.2 | 65.1% |
| Heat (LFQ 1+) | 424 | 987 | 11.9 | 56.8% |
| Heat (LFQ 3+) | 188 | 242 | 14.6 | 52.1% |
| PR-619 | 1197 | 4349 | 10.5 | 59.0% |
| PR-619 (LFQ 1+) | 423 | 793 | 11.7 | 49.4% |
| PR-619 (LFQ 3+) | 151 | 133 | 12.3 | 41.7% |
| MG-132 | 1208 | 4263 | 9.7 | 56.3% |
| MG-132 (LFQ 1+) | 431 | 651 | 11.2 | 45.0% |
| MG-132 (LFQ 3+) | 202 | 145 | 11.6 | 32.2% |
| All conditions | 596 | 1347 | 12.2 | 57.9% |
Subsequently, we performed MCODE analysis, revealing highly interconnected sub-clusters within the core network, including nine sub-clusters with interconnectivity scores ranging from 9 up to 39 (Fig. 6A). Three dominant clusters involve many functionally related proteins from the spliceosome, the ribosome and cell cycle related factors (Fig. 6C). Other clusters contain proteins involved in chromatin remodeling, histone deacetylases, histone methyltransferases, regulation of mitotic prometaphase, regulation of ubiquitin-protein ligases, and proteins involved in ribonucleoprotein complex formation (Supplementary Fig. 7).
We found SUMOylation, ubiquitylation and acetylation sites to significantly overlap (Fig. 3A and 3B). To further investigate this, we performed STRING analysis on the subset of proteins containing these lysines. We found over 70% of these proteins to be situated in a single functional network (Supplementary Fig. 6A, 6B and 6C). Beyond the observed overlap between modification sites, we found these clusters of proteins to be highly modified by SUMO, averaging over 5 SUMOylation sites per protein (Supplementary Fig. 6D). Additionally, we observed a high degree of enrichment for protein-protein interactions as compared to expected (Supplementary Fig. 6E), a higher degree of interaction enrichment as compared to the full SUMO network, and a much higher network participation of all proteins. This resulted in some of the most highest scoring networks when observing proteins containing lysines modified by SUMO and either ubiquitin or acetylation, and by far the strongest network when only considering proteins that are modified on the same lysines by all three of these major PTMs (Supplementary Fig. 6F). Thus, SUMOylation appears to function in concert with other major PTMs, and co-regulates a tight functional cluster of heavily modified and dynamic proteins.
Discussion
We have developed novel methodology for global identification of SUMOylation sites, enabling us to map over 4,300 SUMO acceptor lysines in over 1,600 proteins. This provides detailed insight in the function of this post-translational modification. All nuclear processes are orchestrated by SUMOylation including transcription, DNA repair, chromatin remodeling, pre-mRNA splicing and ribosome assembly.
This is an important step forward compared to the relatively small number of SUMOylation sites that were found previously in cells cultured in standard growth conditions 29, 35 and provides a rich dataset for the scientific community to enable follow-up studies. Our dataset expands the amount of known SUMOylation sites by over 3,000, re-confirms many of the one thousand sites that were recently mapped in response to heat shock 36, and for the first time provides over one thousand sites identified under standard growth conditions. This is particularly relevant for half of the identified sites that are not located in a SUMOylation consensus motif and therefore elude in silico prediction. Furthermore, we have obtained novel insight in the consensus motif for SUMOylation and found that SUMOylation sites are frequently located in domains enriched for charged residues. Moreover, protein regions harboring SUMOylated lysines are depleted for cysteines, possibly to limit thioester formation between SUMOs and target proteins.
We also identified an unprecedented amount of phosphorylation events which occur in close proximity to SUMOylation, with 70 of these events almost matching the total amount of SUMO sites mapped in our screen 4 years ago 29. Moreover, the identification of 4 SUMO and acetyl co-modified peptides, and the acetylation-dependent modification of endogenous histone H3, provides an interesting and novel prospect for crosstalk between these two major PTMs in their regulation of nucleosomes. Previously, it was demonstrated that SUMOylation of histones is linked to transcriptional repression 44, 45. In contrast, acetylation is associated with transcriptional activation. Acetylation-dependent SUMOylation of histone H3 is thus a rather surprising type of crosstalk with unclear significance. Hypothetically, one way to reconcile these differences would be that histone H3 is first acetylated to activate transcription and later SUMOylated to generate a transient activation pattern. It would be interesting to study where acetylated and SUMOylated histone H3 is located on the genome.
SUMO acceptor lysines were for the first time properly compared to identified ubiquitin acceptor lysines and acetylation and methylation sites 17, 24. A significant overlap was found between SUMO acceptor lysines and ubiquitylated, acetylated and methylated lysines. It is known that SUMOylation and ubiquitylation can occur consecutively 46. Furthermore, we found that proteins containing lysines modified by SUMO and other PTMs contain on average twice as many SUMOylation sites, and are much more likely to be functionally interacting. Additionally, our findings provide novel insight in crosstalk between SUMOylation and ubiquitylation since we found that five lysines in ubiquitin can be used as SUMO acceptor lysines, indicating complex heterogeneous SUMO-ubiquitin chains that open up exciting new avenues to investigate mechanisms and biological relevance of this novel type of signal transduction.
Our methodology, in addition to the method recently employed by Tammsalu et al. 36, still relies on the exogenous expression of a SUMO with a mutation inserted close to the C-terminus of SUMO. As such, they cannot identify lysines modified by endogenous and wild-type SUMO, such as those from animal tissues or patient material. Ultimately, the efficient and system-wide identification of SUMO acceptor lysines in an entirely endogenous fashion remains a great challenge to be tackled. However, we have demonstrated that our methodology can accurately and reliably pin-point SUMO acceptor lysines. Furthermore, many other SUMOylation sites remain to be discovered as a result of cell-type specific SUMOylation or stimulus-dependent SUMOylation, which could reliably be investigated with the developed methodology.
Online Methods
Plasmids
The His10-SUMO-2-K0-Q87R we described and used in this manuscript has the following amino acid sequence: MAHHHHHHHHHHGGSMSEERPREGVRTENDHINLRVAGQDGSVVQFRIRRHTPLSRLMRAY CERQGLSMRQIRFRFDGQPINETDTPAQLEMEDEDTIDVFRQQTGG
The His10-SUMO-2 wild-type we described and used in this manuscript has the following amino acid sequence: MAHHHHHHHHHHGGSMSEEKPKEGVKTENDHINLKVAGQDGSVVQFKIKRHTPLSKLMKAYC ERQGLSMRQIRFRFDGQPINETDTPAQLEMEDEDTIDVFQQQTGG
The corresponding nucleotide sequences were cloned in between the PstI and XhoI sites of the plasmid pLV-CMV-IRES-GFP 48.
Cell culture & cell line generation
HeLa and U2-OS cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% FBS and 100 U per mL penicillin and streptomycin (Invitrogen). HeLa cells stably expressing His10-SUMO-2 or His10-SUMO-2-K0-Q87R were generated through lentiviral infection with a virus encoding His10-SUMO-2-IRES-GFP or His10-SUMO-2-K0-Q87R-IRES-GFP. Two weeks after infection, cells were fluorescence-sorted for a low expression level of GFP using a FACSAria II (BD Biosciences). Cells were passed through a 100 μm capillary at a pressure of 138 kPa, and selected for 7.5*102-3*103 GFP BF 530-30-A intensity, which was 2.5-10 times higher than the background cellular auto-fluorescence of 3*102.
Treatments, transfection & lentiviral infection
In order to accumulate SUMOylated proteins, cells were treated with 10 μM MG-132 (Sigma) dissolved in DMSO for 7 hours, treated with 20 μM PR-619 (Millipore) dissolved in DMSO for 7 hours, or incubated at 43°C for 1 hour (heat shock). For increasing acetylation of histones, Trichostatin A (TSA, Sigma) was used at a concentration of 150 nM or 600 nM for 18 hours 39. In order to decrease acetylation of histones, curcumin (Sigma) was used at a concentration of 25 μM and 50 μM for 18 hours 39. For transfection, cells were cultured in DMEM lacking penicillin and streptomycin. Transfections were performed using 2.5 μg of polyethylenimine (PEI) per 1 μg of plasmid DNA, using 1 μg of DNA per 1 million cells. Transfection reagents were mixed in 150 mM NaCl and incubated for 15 minutes before adding it directly to the cells. Cells were split after 24 hours and investigated after 48 hours. Lentiviruses were generated essentially as described previously 49. Infections were performed with a multiplicity of infection of 2 and using a concentration of 8 μg per mL polybrene in the medium. 24 hours after infection the medium was replaced.
Purification of His10-SUMO-2 and His10-SUMO-2-K0-Q87R – Stage 1
Per single MS/MS run to identify SUMO-2 sites, one single fully confluent 15-cm dish of His10-SUMO-2-K0-Q87R (~20 million cells) was prepared. Cells were washed three times on the plate with ice-cold PBS, prior to scraping cells and collecting them in a 15 mL tube. Cells were centrifuged at 250 RCF and re-suspended in ice-cold PBS. Subsequently, the cell pellets were lysed in 10 pellet volumes of guanidine Lysis Buffer (6 M guanidine-HCl, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 8.0). Lysates were subjected to sonication using a microtip sonicator at a power of 30 Watts. Sonication bursts of 5 seconds per 5 mL lysate were used, up to a total sonication time of 15 seconds. Subsequently, lysates were supplemented by addition of imidazole to 50 mM and β-mercaptoethanol to 5 mM. 20 μL (dry volume) Ni-NTA agarose beads (Qiagen) were prepared per 1 mL lysate, by washing them four times with guanidine Lysis Buffer supplemented with imidazole to 50 mM and β-mercaptoethanol to 5 mM. The equilibrated beads were added to the lysates and allowed to tumble overnight at 4°C. Following overnight incubation, beads were pelleted by centrifugation at 500 RCF, and washed for at least 15 minutes using at least 5 bead volumes of the following Wash Buffers in order. Wash Buffer 1: 6 M guanidine-HCl, 0.1% Triton X-100, 10 mM imidazole, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 8.0. Wash Buffer 2: 8 M urea, 0.1% Triton X-100, 10 mM imidazole, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 8.0. Wash Buffer 3: 8 M urea, 10 mM imidazole, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 6.3. Wash Buffer 4: 8 M urea, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 6.3. Wash Buffer 5 (same as Wash Buffer 4): 8 M urea, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 6.3. Subsequently, all Wash Buffer was removed from the beads, and proteins were eluted for 30 minutes using one bead volume of Elution Buffer (7 M urea, 500 mM imidazole, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 7.0). The elution procedure was repeated another two times, and all elutions were pooled and passed through 0.45 μM filters (Ultrafree, Millipore). For mass spectrometric analysis, samples were concentrated over 100 kDa cut-off filters (Vivacon 500, Sartorius Stedim), at 20°C and at 8,000 RCF. Concentration was performed until <10-50 μL of sample remained. After concentration, the proteins remaining on the filters were washed once using 250 μL of Elution Buffer minus imidazole, and re-concentrated. Final concentrated SUMOylated proteins were removed from the filters and were snap frozen and stored at −80°C. The single-stage purified SUMOylated proteins are compatible with both in-gel and in-solution digestion protocols and subsequent mass spectrometric analysis, or may be target-specifically investigated by SDS-PAGE and immunoblotting analysis. Alternatively, re-purification of SUMOylated peptides is performed by following Stage 2 of the protocol.
Purification of His10-SUMO-2-K0-Q87R – Stage 2
Sequencing Grade Endoproteinase Lys-C (Promega) was added to the samples in a 1:25 enzyme-to-protein ratio, and incubated for 4 hours at room temperature, still, and in the dark. Subsequently, another 10 mM of fresh β-mercaptoethanol was added to the samples, followed by an additional amount of Lys-C equal to the first amount. The second incubation was performed overnight, at room temperature, still and in the dark. Next, digests were transferred to 15 mL tubes and diluted with an amount of guanidine Lysis Buffer equal to half the amount used to lyse the initial cell pellet. The samples were then supplemented with addition of imidazole to 50 mM, and β-mercaptoethanol to 5 mM. Next, 40 μL (dry volume) Ni-NTA agarose beads (Qiagen) were prepared per 1 mL sample. The equilibrated beads were added to the lysates and allowed to tumble at 4°C for 5 hours. Following incubation, beads were pelleted by centrifugation at 500 RCF, and washed for 15 minutes using at least 5 bead volumes of the following Wash Buffers in order. Wash Buffer 1: 6 M guanidine-HCl, 0.1% Triton X-100, 10 mM imidazole, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 8.0. Wash Buffer 2: 8 M urea, 0.1% Triton X-100, 10 mM imidazole, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 8.0. Wash Buffer 3: 8 M urea, 10 mM imidazole, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 6.3. Wash Buffer 4: 8 M urea, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 6.3. Wash Buffer 5 (same as Wash Buffer 4): 8 M urea, 5 mM β-mercaptoethanol, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 6.3. Subsequently, all Wash Buffer was removed from the beads, and proteins were eluted for 20 minutes using one bead volume of Elution Buffer (7 M urea, 500 mM imidazole, 100 mM sodium phosphate, 10 mM TRIS, buffered at pH 7.0). The elution procedure was repeated twice, and all elutions were pooled and passed through 0.45 μM filters (Ultrafree, Millipore). Next, samples were concentrated on 10 kDa cut-off spin filters (Vivacon 500, Sartorius Stedim), at 20°C and at 14,000 RCF. Concentration was performed until <10-25 μL of sample remained. After concentration, the proteins remaining on the filters were washed twice using 250 μL of Elution Buffer minus imidazole, and re-concentrated. Final concentrated SUMOylated peptides were removed from the filters and were snap frozen and stored at −80°C. The double-purified SUMOylated peptides are compatible with in-solution digestion protocols and subsequent mass spectrometric analysis aimed at determination of specific sites of protein SUMOylation.
In-solution digestion
SUMOylated peptides were supplemented with ammonium bicarbonate (ABC) to 50 mM. Subsequently, dithiothreitol (DTT) was added to a concentration of 1 mM, and samples were left to incubate at room temperature for 30 minutes. Next, chloroacetamide was added to a concentration of 5 mM, and samples were incubated at room temperature for 30 minutes. After alkylation, another 5 mM of DTT was added, and samples were left to incubate at room temperature for 30 minutes. At this point, samples were gently diluted 4-fold using 50 mM ABC. Subsequently, an amount of Sequencing Grade Modified Trypsin (Promega) was added equal to 25% of the Lys-C initially used in a single digestion step. Digestion with trypsin was performed overnight, at room temperature, still and in the dark.
LC-MS/MS analysis
In-solution digested peptides were cleaned, desalted and concentrated on triple-disc C18 reverse phase StageTips 50, before being eluted twice with 25 μL 80% acetonitrile in 0.1% formic acid. Desalted peptides were vacuum centrifuged at room temperature until 10% of the original volume remained, prior to online nanoflow liquid chromatography-tandem mass spectrometry. The analysis of in-solution digested peptides was performed using an EASY-nLC system (Proxeon) connected to a Q-Exactive (Thermo) using Higher-Collisional Dissociation (HCD) fragmentation. Separation of peptides was performed using 20 cm long analytical columns (ID 75 μm, Polymicro Avantes) packed in-house with 1.8 μm C18 beads (Reprospher 100), using a 120 minute gradient from 5% to 75% acetonitrile in 0.1% formic acid and a flow rate of 250 nL per minute. The mass spectrometer was operated in data-dependent acquisition mode using a top 10 method. Full-scan MS spectra were acquired with a target value of 3E6 and a resolution of 70,000, with a scan range from 300 to 1,750 m/z. HCD tandem MS/MS spectra were acquired using a target value of 1E5, a resolution of 17,500, and a normalized collision energy of 25%. All charges lower than 2 and higher than 6 were rejected, and all unknown charges were rejected. The underfill ratio was set to 0.1%, and a dynamic exclusion of 20 seconds was used. Alternatively, the underfill ratio was set to 1.0% with the dynamic exclusion time set to 10 seconds.
Data processing
MaxQuant version 1.4.1.2 was used to analyze all RAW data 51, 52. The control condition experiment was performed in biological hexuplicate, and measured as 11 technical replicates. The heat shock experiment was performed in biological triplicate, and measured as 6 technical replicates. The MG-132 experiment was performed in biological triplicate, and measured as 6 technical replicates. The PR-619 experiment was performed in biological quintuplicate, and measured as 9 technical replicates. MS/MS spectra were filtered and deisotoped and the 15 most abundant fragments for each 100 m/z were retained. MS/MS spectra were filtered for a mass tolerance of 6 ppm for precursor masses, and a mass tolerance of 20 ppm was used for fragment ions. Peptide and protein identification was performed through matching the identified MS/MS spectra versus a target-decoy version of the complete human Uniprot database, in addition to a database of commonly observed mass spectrometry contaminants. Up to 3 missed tryptic cleavages were allowed. Cysteine carbamidomethylation was set as a fixed peptide modification. Protein N-terminal acetylation, peptide N-terminal carbamylation, methionine oxidation, QQTGG and pyro-QQTGG were set as variable peptide modifications. QQTGG was set as a lysine-specific modification, with a monoisotopic mass of 471.20776, and not allowed to occur at the C-terminal end of peptides. Pyro-QQTGG may spontaneously form out of the tryptic QQTGG remnant as a result of cyclization of the N-terminal glutamine. Pyro-QQTGG (pQQTGG) was set as s lysine-specific modification, with a monoisotopic mass of 454.18121, and not allowed to occur at the C-terminal end of peptides. In order to increase identification confidence, diagnostic peaks were searched within MS/MS spectra corresponding to SUMOylated peptides. To this end, candidate MS/MS spectra were searched for peaks corresponding in m/z to fragmentation of the tryptic QQTGG or pQQTGG remnant present on all SUMOylated peptides. For QQTGG; b5-QQTGG, b4-QQTG, b3-QQT and b2-QQ were accepted as diagnostic peaks. For pQQTGG; b5-pQQTGG, b4-pQQTG, b3-pQQT and b2-pQQ were accepted as diagnostic peaks. In addition to the above variable modifications, lysine acetylation and serine-threonine-tyrosine phosphorylation were individually added as further variable modifications in separate searches. For protein identification, peptides with all above variable modifications were accepted, and protein identification by at least one single SUMO-modified unique peptide was performed. Peptides were accepted with a minimum length of 6 amino acids, a maximum size of 5 kDa, and a maximum charge of 6. The processed data was filtered by posterior error probability (PEP) to achieve a protein false discovery rate (FDR) of below 1%, a peptide-spectrum match FDR of below 1%, and in addition a site decoy fraction of 1% was set. SUMO-site peptides were additionally filtered to have an Andromeda score of at least 40, a localization score of at least 40, a localization probability of at least 90%, and the presence of at least one diagnostic QQTGG or pyro-QQTGG fragment in the MS/MS spectrum.
The mass spectrometry proteomics RAW data have been deposited to the ProteomeXchange Consortium 47, via the PRIDE partner repository with the dataset identifier PXD001061.
Label-Free Quantification (LFQ)
Label-Free Quantification was performed using MaxQuant LFQ. Quantification was performed over 17 biological replicates, including 6 control, 3 heat shock, 3 MG-132 and 5 PR-619 replicates. The standard (not fast) LFQ algorithm was used with otherwise default settings (min. ratio count of 2, min. neighbors of 3, average neighbors of 6). Protein LFQ values were Log2 transformed for further processing. Proteins were filtered for the presence of at least two identifying peptides, and at least one SUMOylated peptide. To ensure biological reproducibility, only proteins detected in at least three biological replicates in any of the four treatment conditions were retained. Subsequently, missing values were imputed using Perseus software, with a down shift of 1.8 (Log2) and a variance of 0.3 (Log2) below the lowest detectable value within each separate replicate. LFQ values were median-averaged within all treatment conditions, and subsequently control condition LFQ values were subtracted from heat shock, MG-132 and PR-619 values to yield the final Log2 ratio.
Statistics
Statistical methods used in this manuscript are detailed in the Online Supplementary Information.
Primary antibodies
Primary antibodies used in this study were Mouse α SUMO-2 (ab81371, Abcam, 1:2,000), Rabbit α SUMO-2 (raised against the C-terminal part of SUMO-2, 1:5,000)31, Mouse α His (HIS-1, H-1029, Sigma, 1:2,500), Mouse α HA (HA.11, MMS-101R, Sanbio,1:1,000), Rabbit α SART-1 (raised against SART-1 peptides, 1:1,000)53, Rabbit α Histone H3 (4499S, Cell Signaling Technology, 1:500), Rabbit α H3-Ac (06-599, Upstate, 1:2,500), Rabbit α RNF216 (A304-111A, Bethyl, 1:2,500), Rabbit α ZNF280D (A303-232A, Bethyl, 1:2,500), Rabbit α SNW1 (A300-784A, Bethyl, 1:2,500), Rabbit α TCF12 (11825S, Cell Signaling Technology, 1:1,000), Rabbit α WDR70 (A301-871A, Bethyl, 1:2,500), Rabbit α FOXM1 (C-20, sc-502, Santa Cruz, 1:1,000), Mouse α HNRNPM (HL374, R3902, Sigma, 1:5,000), Rabbit α RAD18 (A301-340A, Bethyl, 1:2,500), Rabbit α MCM10 (A300-131A, Bethyl, 1:2,500). Validation of antibodies is provided on the manufacturers’ websites, in the cited references and in Antibodypedia.
Electrophoresis and immunoblotting
Protein samples were size-fractionated on Novex 4-12% Bis-Tris gradient gels using MOPS running buffer (Invitrogen). Size-separated proteins were transferred to Hybond-C membranes (Amersham Biosciences) using a submarine system (Invitrogen). Gels were Coomassie stained according to manufacturer’s instructions (Invitrogen). Membranes were stained for total protein loading using 0.1% Ponceau-S in 5% acetic acid (Sigma). Membranes were blocked using PBS containing 0.1% Tween-20 (PBST) and 5% milk powder for one hour. Subsequently, membranes were incubated with primary antibodies as indicated, in blocking solution. Incubation with primary antibody was performed overnight at 4°C. Afterwards, membranes were washed three times with PBST and briefly blocked again with blocking solution. Next, membranes were incubated with secondary antibodies (donkey-anti-mouse-HRP or rabbit-anti-goat-HRP, 1:2,500) for one hour, before washing three times with PBST and two times with PBS. Membranes were then treated with ECL2 (Pierce) as per manufacturer’s instructions, and chemiluminescence was captured using Biomax XAR film (Kodak). A compilation of all uncropped images corresponding to all scans of gels, membranes and films displayed throughout this manuscript is available as Supplementary Data Set 1.
Microscopy
Cells were seeded on glass coverslips, and fixed 24 hours later for 10 minutes in 3.7% paraformaldehyde in PHEM buffer (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2 pH 6.9) at 37°C. After washing with PBS, cells were permeabilized with 0.1% Triton-X100 for 10 minutes, washed with PBST, and blocked using TNB (100 mM TRIS pH 7.5, 150 mM NaCl, 0.5% Blocking Reagent (Roche)) for 30 minutes. Cells were incubated with primary antibody as indicated, in TNB for one hour. Subsequently cells were washed five times with PBST, and incubated with secondary antibodies (Goat α Mouse Alexa 594 (Invitrogen)) in TNB for one hour. Next, cells were washed five times with PBST and dehydrated using alcohol, prior to embedding them in Citifluor (Agar Scientific) containing 400 ng per μL DAPI (Sigma) and sealing the slides with nail varnish. Images were recorded on a Leica SP5 confocal microscope system using 488 nm and 561 nm lasers for excitation, a 63× lens for magnification, and were analyzed with Leica confocal software.
Supplementary Material
Acknowledgements
The authors are grateful for support from the European Research Council, grant number 310913 (A.C.O.V.), the Netherlands Organization for Scientific Research (NWO), grant numbers 70058425, 93511037 and 70059006 (A.C.O.V.) and the Max-Planck Society for the Advancement of Science (M.M.). We would like to acknowledge Dr. Jürgen Cox for his help with MaxQuant and Dr. Kirti Sharma for helpful discussions.
Footnotes
Accession code
The data reported in this paper are provided as Supporting Online Material. The mass spectrometry proteomics RAW data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository 47, with the dataset identifier PXD001061.
The authors declare no conflict of interest.
Reference List
- 1.Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat. Rev. Mol. Cell Biol. 2010;11:479–489. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 3.Gill G. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 5.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 6.Morris JR, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 7.Galanty Y, et al. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–939. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 9.Nacerddine K, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev. Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 10.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 11.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 12.Papouli E, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell Proteomics. 2013 doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vertegaal AC. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem. Rev. 2011;111:7923–7940. doi: 10.1021/cr200187e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 17.Choudhary C, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 18.Guo A, et al. Immunoaffinity Enrichment and Mass Spectrometry Analysis of Protein Methylation. Mol. Cell Proteomics. 2013 doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141:897–907. doi: 10.1016/j.cell.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Kim DY, Scalf M, Smith LM, Vierstra RD. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell. 2013;25:1523–1540. doi: 10.1105/tpc.112.108613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emanuele MJ, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–474. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Povlsen LK, et al. Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat. Cell Biol. 2012;14:1089–1098. doi: 10.1038/ncb2579. [DOI] [PubMed] [Google Scholar]
- 23.Wagner SA, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim W, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hickey CM, Wilson NR, Hochstrasser M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012;13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golebiowski F, et al. System-wide changes to SUMO modifications in response to heat shock. Sci. Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 27.Galisson F, et al. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell Proteomics. 2011;10:M110. doi: 10.1074/mcp.M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamoliatte F, et al. Targeted Identification of SUMOylation Sites in Human Proteins Using Affinity Enrichment and Paralog-specific Reporter Ions. Mol. Cell Proteomics. 2013;12:2536–2550. doi: 10.1074/mcp.M112.025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matic I, et al. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell. 2010;39:641–652. doi: 10.1016/j.molcel.2010.07.026. [DOI] [PubMed] [Google Scholar]
- 30.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 31.Vertegaal AC, et al. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 32.Schimmel J, et al. The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell Proteomics. 2008;7:2107–2122. doi: 10.1074/mcp.M800025-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Tatham MH, Matic I, Mann M, Hay RT. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci. Signal. 2011;4:rs4. doi: 10.1126/scisignal.2001484. [DOI] [PubMed] [Google Scholar]
- 34.Hornbeck PV, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. doi: 10.1093/nar/gkr1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schimmel J, et al. Uncovering SUMOylation Dynamics during Cell-Cycle Progression Reveals FoxM1 as a Key Mitotic SUMO Target Protein. Mol. Cell. 2014 doi: 10.1016/j.molcel.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 36.Tammsalu T, et al. Proteome-Wide Identification of SUMO2 Modification Sites. Sci. Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desterro JM, Rodriguez MS, Hay RT. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- 38.Hietakangas V, et al. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U. S. A. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Balasubramanyam K, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 40.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 41.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- 44.Nathan D, et al. Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev. 2006;20:966–976. doi: 10.1101/gad.1404206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U. S. A. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115:565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 47.Vizcaino JA, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Online Methods
- 48.Vellinga J, et al. A system for efficient generation of adenovirus protein IX-producing helper cell lines. J. Gene Med. 2006;8:147–154. doi: 10.1002/jgm.844. [DOI] [PubMed] [Google Scholar]
- 49.Tiscornia G, Singer O, Verma IM. Production and purification of lentiviral vectors. Nat. Protoc. 2006;1:241–245. doi: 10.1038/nprot.2006.37. [DOI] [PubMed] [Google Scholar]
- 50.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 51.Cox J, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome. Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 52.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 53.Vertegaal AC, et al. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.