Abstract
Breast cancer heterogeneity demands that prognostic models must be biologically driven and recent clinical evidence indicates that future prognostic signatures need evaluation in the context of early compared with late metastatic risk prediction. In pre-clinical studies, we and others have shown that various protein–protein interactions, pertaining to the actin microfilament-associated proteins, ezrin and cofilin, mediate breast cancer cell migration, a prerequisite for cancer metastasis. Moreover, as a direct substrate for protein kinase Cα, ezrin has been shown to be a determinant of cancer metastasis for a variety of tumour types, besides breast cancer; and has been described as a pivotal regulator of metastasis by linking the plasma membrane to the actin cytoskeleton. In the present article, we demonstrate that our tissue imaging-derived parameters that pertain to or are a consequence of the PKC–ezrin interaction can be used for breast cancer prognostication, with inter-cohort reproducibility. The application of fluorescence lifetime imaging microscopy (FLIM) in formalin-fixed paraffin-embedded patient samples to probe protein proximity within the typically <10 nm range to address the oncological challenge of tumour heterogeneity, is discussed.
Keywords: breast cancer, cofilin, ezrin, Förster resonance energy transfer (FRET)/fluorescence-lifetime imaging microscopy (FLIM), protein kinase Cα (PKCα)
Introduction
Breast cancer is the most common malignancy in women. Despite improving survival rates, the global burden of breast cancer remains high with approximately half a million breast cancer related deaths reported worldwide annually. A number of prognostic tools predicting the risk of metastatic relapse are used by oncologists to guide clinical decision-making [1,2]. The accuracy of these prognostic models is far from perfect, and recent clinical evidence indicates that the traditional clinicopathological parameters (e.g. tumour size, lymph node status) used in prognostic models such as Adjuvant Online [1] or the St. Gallen’s Consensus [2] may not correlate well with clinical outcome in some breast cancer subtypes [3,4]. Similarly, although prognostic models using multigene signatures such as Mammaprint [5] or Oncotype Dx [6] have been shown to outperform clinicopathological parameter-based tools in predicting distant metastases [7], a number of studies have also highlighted their shortcomings. Despite molecular estimation of high-risk disease in node-negative breast cancer patients by Oncotype Dx and Mammaprint, 69 % and 44 % of these patients, respectively, experienced long-term disease-free survival.
In recent years, next-generation sequencing approaches have demonstrated the cellular heterogeneity of tumours, comprising distinct subpopulations of cancer cells characterized by specific genomic profiles, and thereby representing the clonal evolution of that tumour [8–10]. In the present article, we focus on protein expression, post-translational modification and protein–protein interaction, and their functional consequences, which offer complementary information to the transcriptome, copy number variation (CNV) and mutational profiles in human breast cancer [11,12]. Currently, the presence/absence of tissue protein markers such as oestrogen receptor (ER), human epidermal receptor 2 [HER2 (ErbB2)] and progesterone receptor (PgR); plus epidermal growth factor receptor (EGFR) status and at least one basal marker [cytokeratin (CK) 5/6], are used to predict cancer progression and guide treatment strategy [13]. Despite the use of these markers to guide stratification of treatment (e.g. anti-ER, HER2 or EGFR targeting inhibitors), the need for improved prognostic and predictive biomarkers remain. For instance, Santagata et al. [14] have recently suggested a new classification based on tissue quantification by multiplex immunofluorescence imaging-based detection of ER, vitamin D receptor (VDR), androgen receptor (AR), CK5 and the proliferation marker Ki67. They showed that this new classification, which is based on defining tumour subtypes according to their similarities with specific normal cell origin subtypes, can be used for disease prognostication.
In the present article, we describe a set of key optical proteomic parameters [15] pertaining to a protein subnetwork which is involved in regulating cancer cell motility, for predicting the time to cancer metastasis among heterogeneous breast cancer patient populations.
Protein kinase Cα (PKCα) in cancer development and metastasis
PKCα (a conventional PKC isoform) belongs to the family of protein kinases initially identified as phospholipid and calcium-dependent kinases [16], which are involved in tumour promotion and progression as a response to stimulation with phorbol ester PMA [17]. More recently, this PKC isoform has been found to be important for maintaining the breast cancer stem cell population [18]. Downstream targets include Raf1 [19] which in turn activates extracellular-signal-regulated kinase 1/2 (ERK1/2), c-Jun N-terminal kinase (JNK) and nuclear factor κB (NF-κB) leading to increased transcription of metalloproteinase-9 and tumour cell migration [20–23]. Among other targets for active PKCα, we have identified β1 integrin, fascin and ezrin [24–26], which form signal complexes on the cell membrane and propagate the signal to the cytoskeleton, triggering a migratory response. Many other PKC targets exist within the motility pathway [27] but are outside the scope of the present article due to space constraints.
Ezrin and cofilin in cancer cell migration
Ezrin [belonging to the ezrin/radixin/moesin (ERM) family of proteins] and cofilin are actin-remodelling proteins playing different roles in reorganization of actin cytoskeleton which results in directional motility of the cell. ERM proteins and cofilin are linked in one gene/signalling network [28,29] and their function depends on the presence of each other [30]. Ezrin expression was found to be necessary for metastasis [31] and its cytoplasmic or nuclear localization correlated with aggressiveness and lymph node positivity in human breast cancer [32,33]. In addition to being a substrate for PKC [25], it can also be activated by ER signalling via the c-Src pathway [34]. The phosphorylation/dephosphorylation cycle of cofilin also plays an important role in actin remodelling which is required for tumour cell protrusion [35], and therefore cell invasive potential [36–38]. In addition to the ERM–cofilin association at a transcriptional level, ERM and the sodium/hydrogen exchanger 1 (NHE-1) have been shown to localize to cofilin-positive invadopodia in a talin-dependent manner to promote invadopodium maturation [39]. This physical association via talin therefore links these two important actin-remodelling proteins in a pathway that can trigger cancer invasiveness. A combined assessment of the activation of these two classes of proteins should provide synergistic information for clinical assessment of the risk of metastasis.
Development and application of imaging assays for prediction of clinical outcome
In pre-clinical studies, we and others have shown that various protein–protein interactions, pertaining to the actin microfilament-associated proteins, ezrin and cofilin, mediate breast cancer cell migration, a prerequisite for cancer metastasis [25, 35, 36, 38, 40, 41]. There is no robust platform to measure these interactions in large-scale clinical sample sets. Our automated imaging platform [42] measures FRET, via the decrease in donor lifetime (reviewed in [43]), by fluorescence lifetime imaging microscopy (FLIM), to directly monitor validated protein–protein interactions [24,26,44,45] and post-translational modifications that include conformational changes, in cultured cells [24,46–50]. A two antibody FRET/FLIM assay to measure endogenous protein–protein interactions (PKC–ezrin) in archived pathological material was developed together with new fluorescence-based assays for measuring the phosphorylation and subcellular localization of ezrin and cofilin (Figures 1 and 2). We hypothesized that these protein interaction/localization-based assays can generate useful information for predicting the likelihood of metastasis due to the biological function pertaining to these cytoskeleton-remodelling molecules.
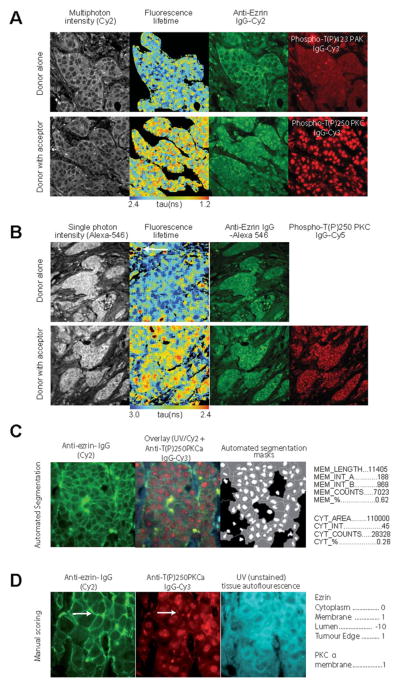
Figure 1. Imaging PKCα–ezrin interaction in FFPE samples.
Representative images of breast cancer tissue stained with anti-ezrin IgG [labelled with Cy2 (A, C and D) or Alexa Fluor® 546 (B)] and anti-PKCα IgG [labelled with Cy3 (A, C and D) or Cy5 (B)]. (A and B) FRET/FLIM images show interaction between proteins (decrease in lifetime, indicated by red pixels in the pseudocolour tumour map). (C and D) Utilization of images for AIS algorithm to generate imaging parameters shown to the right of the images.
Figure 2. Imaging activation status of ezrin and cofilin in FFPE samples.
Representative images of breast cancer tissue stained with anti-ezrin IgG–Cy2 and anti-phospho-ERM IgG–Cy3 (A); and with anti-cofilin IgG–Cy2 and anti-phospho-cofilin IgG–Cy3 (B). Pseudocolour maps show higher co-localization intensities in one sample (upper panel) and lower in another sample (low panel).
Specific FLIM-based ezrin–PKCα–protein interaction detected in formalin-fixed paraffin-embedded (FFPE) tissues
Two- (Figure 1A) and single- (Figure 1B) photon excitation-based acquisition of intermolecular FRET efficiency detected specific protein–protein interactions between ezrin and PKC (see the FLIM/FRET images of invasive breast carcinoma samples that were labelled with fluorescently conjugated anti-ezrin IgG and anti-activated PKCα IgG). Comparison of the corresponding FRET efficiencies measured with either two- or single-photon excitation found no significant difference between the two lifetime acquisition methods and therefore single-photon excitation was chosen to acquire the subsequent FLIM data. Although the immune/inflammatory cell infiltrate (see white arrow in Figure 1B) was autofluorescent, this contributed little (since the number of pixels/area was small proportionally) to the overall mean fluorescence lifetime per tumour. Similarly, the non-specific nuclear staining of the acceptor fluorophore-labelled antibody (anti-activated PKCα IgG) did not interfere with the determination of FRET by FLIM [51,52], which is based on the shortening of donor fluorescence lifetime of the donor fluorophore used to label the anti-ezrin IgG.
Subcellular protein localization and/or phosphorylation quantification
Ezrin–PKCα protein complex formation should result in downstream molecular events such as ezrin phosphorylation, redistribution and stabilization at the membrane [25]. Figures 1 and 2 show ezrin stabilization at the membrane (Figures 1C and 1D), with concomitant ERM phosphorylation (Figure 2A) and activation of PKCα (as shown by Thr250 phosphorylation [53], Figure 1D), preferentially at the membrane of invasive breast carcinoma cells (see white arrow). The subcellular localization of proteins in tissue microarray cores was further quantified by automated image segmentation (AIS) and a manual scoring system. Nine image parameters for the subcellular distribution of ezrin (Figure 1C) across heterogeneous breast tumours were generated in less than 5 s by AIS. The parallel ‘manual’ scoring system (Figure 1D) generated five parameters, describing the subcellular compartment expression levels of both ezrin and PKCα in each tissue core. Further automated co-localization analyses demonstrated an increase in the total ezrin/phospho-ERM and cofilin/phospho-cofilin co-localization intensity at the cell–cell borders and/or edges of invasive tumour cells (Figures 2A and 2B).
Selection of a consensus set of imaging-based covariates for a metastatic predictive model
We next established that the non-FRET-based image parameters (AIS or manual score) pertaining to ezrin and PKC were associated with FRET positivity, which is a measure of ezrin–PKCα interaction. Ezrin–PKCα protein complex formation influences downstream molecular events such as ezrin phosphorylation, redistribution and stabilization at the membrane, which are measured by the non-FRET-based image parameters [25]. Using single-photon FLIM-derived FRET positivity (>0 %) to define a binary outcome, a support vector machine (SVM) [54] predicted FRET positivity using a combination of the ezrin distribution (AIS and manual score) and phosphorylation parameters to an accuracy of ~65 % (Figure 3A). A degree of overfitting was apparent when > four covariates were used to build the SVM.
Figure 3. Utilization of imaging parameters for clinical outcome prediction model.
(A) SVM classification of ezrin–PKCα FRET positivity (1990s cohort, n = 134 patient samples). Prediction accuracy (FRET > 0 or < 0) is shown for a decreasing number of input covariates (AIS and manual scores). Empty points represent the prediction accuracies achieved for training sets; solid points for validation sets. Error bars represent the S.D. across 100 cross-validation iterations. Cross-validation (CV; training:validation ratio, 2:1; 100 iterations) was performed with balanced outcome classes (FRET > 0 or < 0) by randomly selecting an equal number of samples from each class. Ranking of variables was performed by sequentially removing the input variable with the lowest weight when averaged over CV iterations. (B) Kaplan–Meier curve for cofilin/phospho-cofilin co-localization for the 1980s/1990s samples with reported distant metastasis (‘relapse subgroup’). (C–E) Kaplan–Meier curves for ezrin–phospho-ERM co-localization. (C) 1980s/1990s samples with reported distant metastasis (‘relapse subgroup’) showing the upper quartile of values compared with all other samples; (D) all 1980s samples with available co-localization data (shown as upper quartile compared with lower quartile); (E) all 1990s samples with available co-localization data (shown as upper quartile compared with lower quartile). (F) Complexity-optimized Bayesian proportional hazards regression model showing the predictive accuracy for 7-year distant metastasis-free survival for the 1980s cohort. Predictive accuracy is shown for training (empty points) and validation sets (filled points) (CV; training:validation ratio, 1:1; 400 iterations). Models with more than six covariates show a decline in predictive accuracy for validation sets, indicating over-fitting. Models with up to six covariates show a predictive accuracy among validation sets of up to ~70 %. (Horizontal broken line indicates the predictive accuracy expected if all samples are assigned to one class.) (G) Predictive accuracy for 3-year distant metastasis-free survival for 1990s samples with reported metastatic relapse, displayed as for (F). (H and I) Predictive accuracy (training and validation sets, displayed as in F) using up to six covariates from the consensus set derived from analyses in (A and F) and shown for (H) 7-year distant metastasis-free survival in the 1980s group, and (I) 3-year distant metastasis-free survival among the 1990s relapse subgroup.
Next, two independent breast cancer cohorts were imaged for cofilin/phospho-cofilin and ezrin/phospho-ezrin co-localization, along with ezrin localization analysis using AIS (88 patient samples from the 1980s series and a total of 134 patient samples from the 1990s series were available for evaluation). There is a high degree of heterogeneity between the two cohorts (the adjuvant treatments for the two cohorts differed significantly: 1980s compared with 1990s; chemotherapy 0 % compared with 30 %; endocrine 36 % compared with 76 %, respectively). In exploratory univariate analysis, the upper quartile of cofilin/phospho-cofilin co-localization intensities was associated with early distant metastasis among patients with reported relapse [Figure 3B; upper quartile (n = 17) compared with lower values (n = 52); P = 0.022, log-rank test]. This was not the case for ezrin/phospho-ezrin co-localization (Figure 3C); furthermore, exploratory analysis of lower and upper quartiles indicated that lower quartile ezrin/phospho-ERM co-localization may be associated with poorer distant metastasis-free survival in the 1980s series (Figure 3D; P = 0.098, not significant, log-rank test) but not the 1990s series (Figure 3E; P = 0.73, not significant, log-rank test).
The predictive accuracy of all 18 covariates (16 imaging parameters, Figures 1C and 1D, and whether or not the patient had received treatments: tamoxifen or chemotherapy) was assessed for distant metastasis-free survival using a Bayesian proportional hazards regression model with cross-validation for the two temporal cohorts (1980s and 1990s). A step-down procedure was used to iteratively reduce the number of covariates in the model, and over-fitting was observed when greater than six covariates were used to build the model. The top six covariates were identified for the 1980s and 1990s (relapse) cohorts. The predictive accuracy (both training and validation) for 7-year distant metastasis-free survival in the 1980s cohort is shown in Figures 3(F) and 3(G).
There was an overlap between the top four covariates in the SVM analysis for FRET positivity (Figure 3A), and the two separate lists of the top six covariates for predicting metastatic relapse. On the basis of this overlap with SVM analysis, a final consensus set of six covariates (average membrane intensity, relative and absolute; cytoplasm average intensity and number of pixels inside cytoplasm, manual ezrin cytoplasm score and membrane length) was selected and confirmed by re-running the Bayesian proportional hazards regression against each of the two patient cohorts. For the 1980s cohort, the model predicted 7-year distant metastasis-free survival to an accuracy of up to ~70 % (Figure 3H). The same consensus set of covariates was found to be predictive for early relapse (within 3 years) among patients with reported distant metastases in the 1990s cohort (Figure 3I).
Conclusions
The remarkable diversity in breast cancer dictates that prognostic models must be biologically driven. We describe the first optical imaging-based tumour metastatic signature, measuring underlying biological variables which are pertinent in tumour metastases. Our semi-automated tissue imaging platform is capable of performing an integrated analysis of protein phosphorylation, protein–protein interaction and subcellular protein expression/distribution, using FFPE tissue microarrays. Incorporation of protein interaction data was shown to also improve the predictive performance of prognostic gene expression signatures [55,56]. Despite the importance of adjunct information supplied by the protein interactome configuration to improve the existing prognostic signatures for predicting patient outcome [56], this protein interaction information has rarely been incorporated in diagnostic/prognostic assays.
We report that our imaging parameters could predict the metastatic risk for early breast cancer patients with a high level of accuracy (~70 %; Figure 3). Moreover, reproducibility across different temporal cohorts, a prerequisite for any prognostication model to use in ‘real life’ patients, is achieved with our new multivariate, imaging-based metastatic signature. Although prospective validations of prognostic tools are imperative, our study is among the first to compare how well a predictive/prognostic signature performs, between patient cohorts that belong to two time periods (i.e. 1980s compared with 1990s). The clinical implications of this protein network/function-based design, alongside our key points of parameter reduction and time-dependent risk assessment, are likely to provide a novel tool that may be of generic utility not only for future prognostic models but also for studying the effects of different signalling pathways on clinical outcome with the eventual goal of individualization of cancer care. We aim to prospectively investigate the use of our new integrated biomarker platform to delineate and quantify relevant oncogenic protein complexes in clinical specimens within a clinical trial setting.
This new paradigm is likely to be key to improving our understanding of tumour biology and factors relating to recurrence and metastasis as well as characterizing patients for the eventual goal of treatment individualization.
Acknowledgments
We thank Dr Robert Eddy for supplying the anti-cofilin and anti-phospho-cofilin antibodies.
Funding
This work was supported by a European Union Framework Programme 7 HEALTH-2010 grant entitled ‘Imagint’ [grant number 259881 (to G.W. and J.E.B.)], KCL–UCL Comprehensive Cancer Imaging Centre (CCIC) funding (from Cancer Research UK and the Engineering and Physical Sciences Research Council) [grant numbers C1519/A10331 and C1519/A16463 (to K.L., J.E.B., G.O.F. and N.W., as well as the FLIM system)], KCL Breakthrough Breast Cancer Research Unit funding (including the Sarah Greene Fellowship to S.I.), Cancer Research UK [grant number C133/A/1812 to B.V.], U.S. National Institutes of Health grant support [grant numbers CA100324 and CA164468 (to J.C.)], and an endowment fund from Dimbleby Cancer Care to King’s College London [to S.M.A.-B. and T.N.].
Abbreviations
- AIS
automated image segmentation
- CK
cytokeratin
- EGFR
epidermal growth factor receptor
- ER
oestrogen receptor
- ERM
ezrin/radixin/moesin
- FFPE
formalin-fixed paraffin-embedded
- FLIM
fluorescence-lifetime imaging microscopy
- HER2
human epidermal receptor 2
- PKC
protein kinase C
- SVM
support vector machine
References
- 1.Ravdin PM, Siminoff LA, Davis GJ, Mercer MB, Hewlett J, Gerson N, Parker HL. Computer program to assist in making decisions about adjuvant therapy for women with early breast cancer. J Clin Oncol. 2001;19:980–991. doi: 10.1200/JCO.2001.19.4.980. [DOI] [PubMed] [Google Scholar]
- 2.Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009;20:1319–1329. doi: 10.1093/annonc/mdp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wo JY, Chen K, Neville BA, Lin NU, Punglia RS. Effect of very small tumor size on cancer-specific mortality in node-positive breast cancer. J Clin Oncol. 2011;29:2619–2627. doi: 10.1200/JCO.2010.29.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernandez-Aya LF, Chavez-Macgregor M, Lei X, Meric-Bernstam F, Buchholz TA, Hsu L, Sahin AA, Do KA, Valero V, Hortobagyi GN, Gonzalez-Angulo AM. Nodal status and clinical outcomes in a large cohort of patients with triple-negative breast cancer. J Clin Oncol. 2011;29:2628–2634. doi: 10.1200/JCO.2010.32.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 6.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 7.Buyse M, Loi S, van’t Veer L, Viale G, Delorenzi M, Glas AM, d’Assignies MS, Bergh J, Lidereau R, Ellis P, et al. Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst. 2006;98:1183–1192. doi: 10.1093/jnci/djj329. [DOI] [PubMed] [Google Scholar]
- 8.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 9.de Bruin EC, Taylor TB, Swanton C. Intra-tumor heterogeneity: lessons from microbial evolution and clinical implications. Genome Med. 2013;5:101. doi: 10.1186/gm505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jamal-Hanjani M, Thanopoulou E, Peggs KS, Quezada SA, Swanton C. Tumour heterogeneity and immune-modulation. Curr Opin Pharmacol. 2013;13:497–503. doi: 10.1016/j.coph.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 13.Blows FM, Driver KE, Schmidt MK, Broeks A, van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO, Blomqvist C, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: a collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santagata S, Thakkar A, Ergonul A, Wang B, Woo T, Hu R, Harrell JC, McNamara G, Schwede M, Culhane AC, et al. Taxonomy of breast cancer based on normal cell phenotype predicts outcome. J Clin Invest. 2014;124:859–870. doi: 10.1172/JCI70941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelleher MT, Fruhwirth G, Patel G, Ofo E, Festy F, Barber PR, Ameer-Beg SM, Vojnovic B, Gillett C, Coolen A, et al. The potential of optical proteomic technologies to individualize prognosis and guide rational treatment for cancer patients. Targeted Oncol. 2009;4:235–252. doi: 10.1007/s11523-009-0116-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takai Y, Kishimoto A, Iwasa Y, Kawahara Y, Mori T, Nishizuka Y. Calcium-dependent activation of a multifunctional protein kinase by membrane phospholipids. J Biol Chem. 1979;254:3692–3695. [PubMed] [Google Scholar]
- 17.Castagna M, Takai Y, Kaibuchi K, Sano K, Kikkawa U, Nishizuka Y. Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J Biol Chem. 1982;257:7847–7851. [PubMed] [Google Scholar]
- 18.Tam WL, Lu H, Buikhuisen J, Soh BS, Lim E, Reinhardt F, Wu ZJ, Krall JA, Bierie B, Guo W, et al. Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolch W, Heidecker G, Kochs G, Hummel R, Vahidi H, Mischak H, Finkenzeller G, Marme D, Rapp UR. Protein kinase Cα activates RAF-1 by direct phosphorylation. Nature. 1993;364:249–252. doi: 10.1038/364249a0. [DOI] [PubMed] [Google Scholar]
- 20.Shin Y, Yoon SH, Choe EY, Cho SH, Woo CH, Rho JY, Kim JH. PMA-induced up-regulation of MMP-9 is regulated by a PKCα–NF-κB cascade in human lung epithelial cells. Exp Mol Med. 2007;39:97–105. doi: 10.1038/emm.2007.11. [DOI] [PubMed] [Google Scholar]
- 21.Kim JM, Noh EM, Kwon KB, Kim JS, You YO, Hwang JK, Hwang BM, Kim BS, Lee SH, Lee SJ, et al. Curcumin suppresses the TPA-induced invasion through inhibition of PKCα-dependent MMP-expression in MCF-7 human breast cancer cells. Phytomedicine. 2012;19:1085–1092. doi: 10.1016/j.phymed.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Villasana V, Gutierrez-Puente Y, Tari AM. Cyclooxygenase-2 utilizes Jun N-terminal kinases to induce invasion, but not tamoxifen resistance, in MCF-7 breast cancer cells. Oncol Rep. 2013;30:1506–1510. doi: 10.3892/or.2013.2549. [DOI] [PubMed] [Google Scholar]
- 23.Kim J, Thorne SH, Sun L, Huang B, Mochly-Rosen D. Sustained inhibition of PKCα reduces intravasation and lung seeding during mammary tumor metastasis in an in vivo mouse model. Oncogene. 2011;30:323–333. doi: 10.1038/onc.2010.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng T, Shima D, Squire A, Bastiaens PI, Gschmeissner S, Humphries MJ, Parker PJ. PKCα regulates β1 integrin-dependent cell motility through association and control of integrin traffic. EMBO J. 1999;18:3909–3923. doi: 10.1093/emboj/18.14.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng T, Parsons M, Hughes WE, Monypenny J, Zicha D, Gautreau A, Arpin M, Gschmeissner S, Verveer PJ, Bastiaens PI, Parker PJ. Ezrin is a downstream effector of trafficking PKC–integrin complexes involved in the control of cell motility. EMBO J. 2001;20:2723–2741. doi: 10.1093/emboj/20.11.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anilkumar N, Parsons M, Monk R, Ng T, Adams JC. Interaction of fascin and protein kinase Cα: a novel intersection in cell adhesion and motility. EMBO J. 2003;22:5390–5402. doi: 10.1093/emboj/cdg521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Neill AK, Gallegos LL, Justilien V, Garcia EL, Leitges M, Fields AP, Hall RA, Newton AC. Protein kinase Cα promotes cell migration through a PDZ-dependent interaction with its novel substrate discs large homolog 1 (DLG1) J Biol Chem. 2011;286:43559–43568. doi: 10.1074/jbc.M111.294603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patsialou A, Wang Y, Lin J, Whitney K, Goswami S, Kenny PA, Condeelis JS. Selective gene-expression profiling of migratory tumor cells in vivo predicts clinical outcome in breast cancer patients. Breast Cancer Res. 2012;14:R139. doi: 10.1186/bcr3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patsialou A, Bravo-Cordero J, Wang Y, Entenberg D, Liu H, Clarke M, Condeelis JC. Intravital multiphoton imaging reveals multicellular streaming as a crucial component of in vivo cell migration in human breast tumors. Intravital. 2013;2:e25294. doi: 10.4161/intv.25294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marsick BM, San Miguel-Ruiz JE, Letourneau PC. Activation of ezrin/radixin/moesin mediates attractive growth cone guidance through regulation of growth cone actin and adhesion receptors. J Neurosci. 2012;32:282–296. doi: 10.1523/JNEUROSCI.4794-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, Yeung C, Gorlick R, Hewitt SM, Helman LJ. The membrane–cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 32.Arslan AA, Silvera D, Arju R, Giashuddin S, Belitskaya-Levy I, Formenti SC, Schneider RJ. Atypical ezrin localization as a marker of locally advanced breast cancer. Breast Cancer Res Treat. 2012;134:981–988. doi: 10.1007/s10549-012-2017-5. [DOI] [PubMed] [Google Scholar]
- 33.Sarrio D, Rodriguez-Pinilla SM, Dotor A, Calero F, Hardisson D, Palacios J. Abnormal ezrin localization is associated with clinicopathological features in invasive breast carcinomas. Breast Cancer Res Treat. 2006;98:71–79. doi: 10.1007/s10549-005-9133-4. [DOI] [PubMed] [Google Scholar]
- 34.Zheng S, Huang J, Zhou K, Zhang C, Xiang Q, Tan Z, Wang T, Fu X. 17β-Estradiol enhances breast cancer cell motility and invasion via extra-nuclear activation of actin-binding protein ezrin. PLoS ONE. 2011;6:e22439. doi: 10.1371/journal.pone.0022439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tania N, Prosk E, Condeelis J, Edelstein-Keshet L. A temporal model of cofilin regulation and the early peak of actin barbed ends in invasive tumor cells. Biophys J. 2011;100:1883–1892. doi: 10.1016/j.bpj.2011.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nat Rev. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Rheenen J, Condeelis J, Glogauer M. A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J Cell Sci. 2009;122:305–311. doi: 10.1242/jcs.031146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh M, Song X, Mouneimne G, Sidani M, Lawrence DS, Condeelis JS. Cofilin promotes actin polymerization and defines the direction of cell motility. Science. 2004;304:743–746. doi: 10.1126/science.1094561. [DOI] [PubMed] [Google Scholar]
- 39.Beaty BT, Wang Y, Bravo-Cordero JJ, Sharma VP, Miskolci V, Hodgson L, Condeelis J. Talin regulates moesin–NHE-1 recruitment to invadopodia and promotes mammary tumor metastasis. J Cell Biol. 2014;205:737–751. doi: 10.1083/jcb.201312046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Legg JW, Lewis CA, Parsons M, Ng T, Isacke CM. A novel PKC-regulated mechanism controls CD44 ezrin association and directional cell motility. Nat Cell Biol. 2002;4:399–407. doi: 10.1038/ncb797. [DOI] [PubMed] [Google Scholar]
- 41.Prag S, Parsons M, Keppler MD, Ameer-Beg SM, Barber P, Hunt J, Beavil AJ, Calvert R, Arpin M, Vojnovic B, Ng T. Activated ezrin promotes cell migration through recruitment of the GEF Dbl to lipid rafts and preferential downstream activation of Cdc42. Mol Biol Cell. 2007;18:2935–2948. doi: 10.1091/mbc.E06-11-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barber PR, Tullis ID, Pierce GP, Newman RG, Prentice J, Rowley MI, Matthews DR, Ameer-Beg SM, Vojnovic B. The Gray Institute ‘open’ high-content, fluorescence lifetime microscopes. J Microsc. 2013;251:154–167. doi: 10.1111/jmi.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fruhwirth GO, Fernandes LP, Weitsman G, Patel G, Kelleher M, Lawler K, Brock A, Poland SP, Matthews DR, Keri G, et al. How Forster resonance energy transfer imaging improves the understanding of protein interaction networks in cancer biology. ChemPhysChem. 2011;12:442–461. doi: 10.1002/cphc.201000866. [DOI] [PubMed] [Google Scholar]
- 44.Parsons M, Keppler MD, Kline A, Messent A, Humphries MJ, Gilchrist R, Hart IR, Quittau-Prevostel C, Hughes WE, Parker PJ, Ng T. Site-directed perturbation of protein kinase C–integrin interaction blocks carcinoma cell chemotaxis. Mol Cell Biol. 2002;22:5897–5911. doi: 10.1128/MCB.22.16.5897-5911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsons M, Monypenny J, Ameer-Beg SM, Millard TH, Machesky LM, Peter M, Keppler MD, Schiavo G, Watson R, Chernoff J, et al. Spatially distinct binding of Cdc42 to PAK1 and N-WASP in breast carcinoma cells. Mol Cell Biol. 2005;25:1680–1695. doi: 10.1128/MCB.25.5.1680-1695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganesan S, Ameer-Beg SM, Ng TT, Vojnovic B, Wouters FS. A dark yellow fluorescent protein (YFP)-based resonance energy-accepting chromoprotein (REACh) for Forster resonance energy transfer with GFP. Proc Natl Acad Sci USA. 2006;103:4089–4094. doi: 10.1073/pnas.0509922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dadke S, Cotteret S, Yip SC, Jaffer ZM, Haj F, Ivanov A, Rauscher F, III, Shuai K, Ng T, Neel BG, Chernoff J. Regulation of protein tyrosine phosphatase 1B by sumoylation. Nat Cell Biol. 2007;9:80–85. doi: 10.1038/ncb1522. [DOI] [PubMed] [Google Scholar]
- 48.Makrogianneli K, Carlin LM, Keppler MD, Matthews DR, Ofo E, Coolen A, Ameer-Beg SM, Barber PR, Vojnovic B, Ng T. Integrating receptor signal inputs that influence small Rho GTPase activation dynamics at the immunological synapse. Mol Cell Biol. 2009;29:2997–3006. doi: 10.1128/MCB.01008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 50.Carlin LM, Evans R, Milewicz H, Fernandes L, Matthews DR, Perani M, Levitt J, Keppler MD, Monypenny J, Coolen T, et al. A targeted siRNA screen identifies regulators of Cdc42 activity at the natural killer cell immunological synapse. Sci Signal. 2011;4:ra81. doi: 10.1126/scisignal.2001729. [DOI] [PubMed] [Google Scholar]
- 51.Festy F, Ameer-Beg SM, Ng T, Suhling K. Imaging proteins in vivo using fluorescence lifetime microscopy. Mol Biosyst. 2007;3:381–391. doi: 10.1039/b617204k. [DOI] [PubMed] [Google Scholar]
- 52.Wouters FS, Verveer PJ, Bastiaens PI. Imaging biochemistry inside cells. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- 53.Ng T, Squire A, Hansra G, Bornancin F, Prevostel C, Hanby A, Harris W, Barnes D, Schmidt S, Mellor H, et al. Imaging protein kinase Cα activation in cells. Science. 1999;283:2085–2089. doi: 10.1126/science.283.5410.2085. [DOI] [PubMed] [Google Scholar]
- 54.Chang CC, Lin CJ. Training nu-support vector classifiers: theory and algorithms. Neural Comput. 2001;13:2119–2147. doi: 10.1162/089976601750399335. [DOI] [PubMed] [Google Scholar]
- 55.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol. 2009;27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]