Abstract
The evolutionarily conserved Mediator complex is a critical coactivator for RNA polymerase II (Pol II)-mediated transcription. Here, we report the reconstitution of a functional 15-subunit human core Mediator complex and its characterization by functional assays and chemical cross-linking coupled to mass spectrometry (CX-MS). Whereas the reconstituted head and middle modules can stably associate, only with incorporation of MED14 into the bi-modular complex does it acquire basal and coactivator functions. This results from a dramatically enhanced ability of MED14-containing complexes to associate with Pol II. Altogether, our analyses identify MED14 as both an architectural and a functional backbone of the Mediator complex. We further establish a conditional requirement for metazoan-specific MED26 that becomes evident in the presence of heterologous nuclear factors. This general approach paves the way for systematically dissecting the multiple layers of functionalities associated with the Mediator complex.
INTRODUCTION
Activation of genes transcribed by eukaryotic RNA polymerase II (Pol II) entails a complex functional interplay between general transcription factors (GTFs), gene- and cell-type specific activators and an array of coactivators1. Whereas Pol II and GTFs can form a preinitiation complex (PIC) on core promoter elements that exhibits low-level (basal) activity in vitro, activators can greatly stimulate PIC function through coactivator recruitment. Among the diverse types of coactivators described, the multi-subunit Mediator complex has emerged as perhaps the most critical coactivator that facilitates PIC establishment and function2. Although initially identified and characterized as a cofactor that bridges activators and the Pol II machinery1, the metazoan Mediator has also been shown to stimulate basal (activator-independent)3-5 and negative (co-repressor)2,6 functions under certain conditions. More recently, given the multi-step nature of the transcription process, Mediator has been further implicated in coordinating mechanistic transitions from the chromatin opening to the PIC establishment phase7-9 and, potentially, from the initiation to elongation phase10-12. Additionally, evidence exists to suggest Mediator involvement in other transcriptionally relevant processes such as facilitation of enhancer-promoter communication by stabilization of chromatin loops through interactions with lncRNA13 or cohesin14, and transcription-coupled DNA repair15. Mediator’s critical role in the cell is also underscored by reports that tie mutations in its various subunits to human disease16,17.
These diverse Mediator-associated functions are reflected in its complex subunit architecture. The 2 MDa metazoan Mediator consists of 30 subunits, many of which are evolutionarily conserved from yeast to human18. However, consistent with the increased complexity of metazoan transcriptional programs relative to those in yeast, the extent of homology ranges from about 50% for a handful of the most conserved subunits (e.g., MED7 and MED31) to much weaker relationships for the remainder18. Further, the metazoan complex contains additional, metazoan-specific subunits (e.g., MED26 and MED30). The overall structure of the complex, both in yeast and human, is modular, with the subunits organized into head, middle, tail and kinase subcomplexes2. The subunits comprising the head and middle modules are tightly associated with each other and constitute a stable core; they have been implicated in interactions with the Pol II machinery. By contrast, the individual subunits of the tail module are relatively loosely associated with each other; and specific promoter- or enhancer-bound activators mainly, but not exclusively, target individual tail subunits19. The kinase module reversibly associates with the core complex and broadly tends to confer repressive properties to the Mediator.
Substantial progress has been made in our understanding of structure-function relationships for the Mediator, especially in yeast. Thus, previous studies of yeast Mediator provided crystal structures for both the head and partial middle modules20-24 and a model for protein interactions within the middle module based on cross-linking25. Yeast two-hybrid screens also led to predictions for the protein interaction networks within the head and middle modules26. Most recently, EM analyses of the yeast Mediator have suggested a model for how individual subunits are organized within the complex27,28. However, without any demonstration of the minimal set of subunits required for the assembly of transcriptionally active Mediator or the identification and pin-pointing of the critical roles of individual essential subunits, these studies have not led to an understanding of the identity and mechanism of action of the active core Mediator components. Furthermore, understanding of the metazoan complex has also been hampered in part due to technical difficulties in manipulating this complex. These include its large size, heterogeneity, the presence of many essential subunits and limited yields upon purification from cell extracts.
Thus far, the metazoan Mediator complex has been functionally characterized mainly in in vitro biochemical assays using preparations obtained from nuclear extract of HeLa cell lines that stably express wild-type or mutant versions of selected subunits. However, in order to obtain a detailed structure-function understanding of the metazoan Mediator complex, it is necessary to dissect it at the level of individual subunits, modules and multi-module assemblies, and to make correlations with their roles in the transcriptional processes. The inherent modularity of the Mediator and the ability to isolate an active form (the PC2 complex) that lacks the kinase module and several tail subunits, but is enriched with respect to the metazoan-specific MED264,29, makes it feasible to undertake a reconstitution-based approach to establish structure-function relationships for the Mediator.
Here, toward the generation of a minimal active core Mediator complex and the isolation of homogeneous preparations in desirable yields, we used the efficient Multibac baculovirus expression system30 to jointly express Mediator subunits that are found in the active PC2 form of the Mediator. We first reconstituted separately the head and middle modules. We found that although these modules can stably associate with each other, the resulting bi-modular complex is inactive in transcriptional assays unless MED14 is also incorporated. Mechanistically, we show that MED14 addition to the complex markedly enhances its interaction with Pol II. However, this complex is unable to support activity in extract-based assay systems unless complemented with MED26 -- suggesting that this subunit allows the Mediator to operate in the context of additional factors present in the extract. We also report an in-depth cross-linking coupled mass spectrometric analysis (CX-MS) of the reconstituted core complex that, while also revealing other interactions, further highlights the key structural role of MED14 in bridging all the main modules of the Mediator complex. Our results are discussed in the context of a recent study focused solely on the architecture of yeast and human Mediator 27.
RESULTS
Reconstitution of the head-middle bi-modular complex
We initiated the reconstitution by first generating the middle module through co-expression of Flag-tagged-MED7 (f-MED7), MED19, MED4, Myc-MED21, MED31, MED9 and His-MED10 in insect cells. Because of the conditional requirement for MED131 we did not include this subunit in our initial analysis. Sequential affinity chromatography (Supplementary Fig. 1a,b) yielded a MED4--MED7--MED10--MED21--MED31 complex containing all essential subunits of the middle module (Fig. 1a,b). The MED9 subunit (non-essential in yeast32,33) failed to express and was not required for Mediator function in our transcription assays (below), and thus was omitted in further reconstitutions. MED19, while expressed, showed no association with the middle module. We similarly reconstituted the head module of the human Mediator by co-expressing f-MED17, MED6, MED8, MED11, MED18, MED19, MED20, MED22, and the metazoan-specific MED30, which previously was not assigned to any module. Following purification, we obtained a head module complex (MED6--MED8--MED11--MED17--MED18--MED20--MED22--MED30) (Fig. 1c,d) that contains all of the input subunits except MED19, whose association with the complex is likely dependent on a metazoan-specific subunit(s) not included in our reconstitutions.
Fig. 1. Reconstitution of human Mediator sub-complexes.
(a) SDS-PAGE analysis (Coomassie-blue staining) of baculovirus-expressed reconstituted middle module. (b) Western blot analysis of middle module subunits. (c) SDS-PAGE analysis (Coomassie-blue staining) of the reconstituted head module. (d) Western blot analysis of head module subunits. (e) SDS-PAGE analysis (Coomassie-blue staining) of the bi-modular head+middle (H+M) complex following purification on M2-agarose (via f-MED17) and HA-agarose (via HA-MED7). (f) SDS-PAGE analysis (Silver staining) of a MED14-containing head+middle (H+M+14) complex purified as in panel e, except that the MED14 was Flag-tagged. (g) SDS-PAGE analysis (Coomassie-blue staining) of the H+M+14+26 complex purified as in panel f. Asterisks in panels f and g point to contaminating polypeptides. See also Supplementary Data Set 1.
To reconstitute a complex containing both the head and middle modules (“H+M”), we co-expressed the subunits of the two modules. Sequential selection through f-MED17 (head module) and HA-MED7 (middle module) subunits followed by Superose 6 gel filtration revealed a stable interaction between the head and middle modules (Fig. 1e and Supplementary Fig. 1d). The resulting H+M preparation (Supplementary Fig. 1d) contained stoichiometric amounts of all the subunits except MED18 and MED20, which in yeast are non-essential32,33 and form a labile heterodimer23 and thus tend to dissociate upon gel filtration (Supplementary Fig. 1e; see further below). Interestingly, we observed that separately purified head and middle modules do not associate to form a bi-modular complex when mixed together (data not shown), possibly indicating the strict requirement for co-expression of subunits constituting the modules.
MED14 is critical for basal and activated transcription
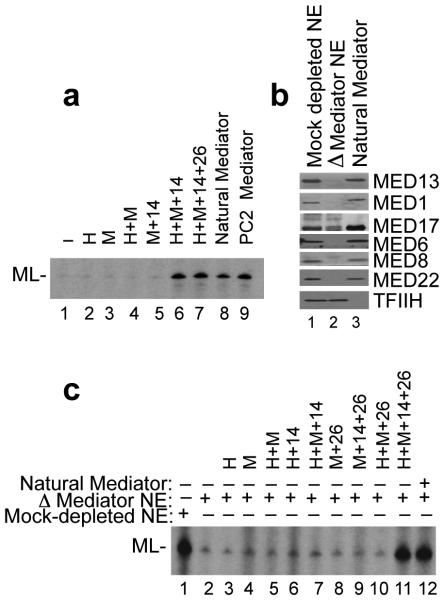
Natural Mediator purified from human cells stimulates both basal and activator-dependent transcription in nuclear extract.3-5. We therefore tested if the H+M preparation stimulated basal transcription in our two standard in vitro transcription assays34 containing either (i) purified general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH), coactivator PC4 and RNA Pol II (Fig. 2a) or (ii) unfractionated HeLa cell nuclear extract (NE) immuno-depleted for the Mediator complex (Fig. 2b, lane 1 vs. lane 2). Since the H+M preparation, as well as the independent head and middle modules, failed to show any activity in either assay (Fig. 2a, lanes 1-4; Fig. 2c, lane 5 vs. lane 2), we sought to include additional subunits in our reconstitution. We started with MED14, which despite its previous assignment to the tail module35, is present in stoichiometric amounts in our PC2 preparations that otherwise tend to be deficient in tail components2.
Fig. 2. Critical roles of MED14 and MED26 in Mediator-stimulated basal transcription.
(a) Autoradiogram of in vitro transcription reactions from a template (ML) containing the adenovirus major late core promoter. Reactions were performed using purified GTFs (IIA, IIB, IID, IIE, IIF, and IIH), Pol II, and PC4 and the indicated Mediator sub-complexes (H, head; M, middle; 14, MED14; 26, MED26). (b) Western blot analysis of HeLa nuclear extract (NE) immuno-depleted of Mediator by anti-MED30 antibody (ΔMediator NE). (c) Autoradiogram of in vitro transcription reactions from the ML template using control (mock-depleted) or ΔMediator NE. Mediator sub-complexes were added to the transcription reactions as indicated. See also Supplementary Data Set 1.
An H+M+14 complex was reconstituted and affinity-selected via MED14 to ensure that the resulting homogeneous preparation contains stoichiometric MED14 (Fig. 1f). When tested in the in vitro transcription assay with purified factors (GTFs, Pol II and PC4), this 14-subunit complex effected a considerable stimulation of basal transcription (Fig. 2a, lane 6 vs. lane 1). Importantly, the fold-stimulation was equivalent to that elicited both by a natural Mediator preparation that contains a complete set of subunits (lane 6 vs. lane 8) and by its PC2 form (lane 6 vs. lane 9). We therefore conclude that the subunits contained in the H+M+14 preparation define the active human core Mediator complex.
MED26 requirement for Mediator function in nuclear extract
While active in the defined assay system, the H+M+14 preparation was unable to restore basal transcription when added back to Mediator-depleted nuclear extract (Fig. 2c, lane 7 vs. lane 1). Because the extract contains a more natural complement of various nuclear factors, this result indicated a requirement for another Mediator subunit(s) to overcome an apparent constraint by a negative cofactor(s) present in the nuclear extract36. Although MED26-containing PC2 is a small fraction of the total cellular Mediator population in HeLa cells, extracts from which this sub-population is depleted fail to support in vitro transcription4. Further, MED26-containing Mediator preparations have a higher Pol II content4,37 and MED26 can recruit the super elongation complex to promoters12,38. We therefore generated variant complexes containing MED26. We found that MED26 associates with the middle, but not the head, module (Supplementary Fig. 2a and 2b), consistent with the recent report from the Asturias group27, and that it could be stably incorporated into H+M+26 (Supplementary Fig. 2c) and H+M+14+26 (Fig. 1g and Supplementary Fig. 3a) complexes. Importantly, in the Mediator-depleted extract (Fig. 2c, lane 11 vs. lane 12), as in the pure system (Fig. 2a, lane 7 vs. lanes 8 and 9), the H+M+14+26 complex restored basal transcription to the same level as a natural Mediator preparation. Inclusion of MED26 into other partial complexes failed to restore transcription in the extract-based assay (Fig. 2c, lanes 8-10), suggesting that the additional requirement for MED26 in this context is superimposed upon a more fundamental structural dependency on MED14. Importantly, in the extract-based assay, the H+M+14+26 (but not the H+M+14) complex exhibited a clear coactivator function for the transcriptional activator p53, which is known to interact with the MED17 subunit39 (Fig. 3a, lane 8 vs. lanes 6 and 4). In control experiments, no coactivator function was seen for the thyroid hormone receptor, which functions as a heterodimer with RXR and targets the missing MED1 subunit40 (Fig. 3b, lane 12 vs. lane 6).
Fig. 3. Critical role of MED14 and MED26 in Mediator coactivator function.
(a) Autoradiogram of in vitro transcription reactions performed as in Fig. 2C. Extract-based reactions contained the p53-responsive template 5×p53REML and a control template (ML). The activator (p53) and Mediator sub-complexes (H+M+14 and H+M+14+26) were added as indicated. (b) Autoradiogram of in vitro transcription reactions performed as in panel a, except that the TR--RXR heterodimer was used as the activator together with the TR-responsive template 5xTREML. An irrelevant activator (AML1-ETO) was included as control. See also Supplementary Data Set 2.
MED14 is crucial for Mediator-Pol II interaction
To understand the mechanism whereby MED14-containing Mediator complexes are rendered active, we used co-immunoprecipitation to investigate the interaction of the head with TFIID and the interaction of the head, H+M, and H+M+14 complexes with Pol II (Fig 4). Consistent with previous studies41, the head module alone interacted with TFIID (Fig. 4b, lane 7). However, in contrast to the case in yeast42, neither the head module (whether in the presence (Fig. 4c, lane 10) or absence (Fig. 4b, lane 5 andFig. 4c, lane 8 ) of TFIIF) nor the H+M complex (Fig. 4d, lane 5) was able to bind to Pol II. By contrast, and importantly, the H+M+14 complex bound up to 75% of input Pol II (Fig. 4d, lane 6). Hence, a critical function of MED14 is to render H+M capable of efficiently interacting with Pol II and thereby stimulating transcription.
Fig. 4. MED14-dependent Mediator-Pol II interaction and MED26-dependent Pol II recruitment in nuclear extract.
(a) SDS-PAGE analysis (silver staining) of purified preparations of Pol II, TFIID and natural Mediator used in the binding assays. (b) Western blot analysis of Mediator interaction assays. Binding reactions included the Mediator head module and Pol II or TFIID, as indicated. Anti-MED30 immunoprecipitates were probed for TFIID (TBP), Pol II (RBP1), and selected Mediator subunits. (c) Western blot analysis of Mediator head interaction assays in the presence of TFIIF. Binding reactions were as in panel b, except that they also included TFIIF as indicated. (d) Western blot analysis of Mediator-Pol II interaction assays. Binding reactions included Pol II and the indicated recombinant Mediator sub-complexes (H, H+M, or H+M+14). Anti-MED30 immunoprecipitates were probed for Pol II (RPB1 and RBP6) and Mediator (MED7, MED14, MED30) subunits. For lanes 1-3 (inputs), longer Western blot exposures are also included. (e) Western blot analysis of an immobilized template recruitment assay to assess Pol II recruitment. Reactions were done with control or Mediator-depleted Hela NE (ΔMediator NE) and were supplemented with various recombinant Mediator sub-complexes. Recruitment of Pol II (RPB1) and TFIID (TBP) was monitored. See also Supplementary Data Set 2.
MED26 overcomes a Pol II recruitment restriction
To understand the basis for the conditional requirement of MED26 for Mediator function in nuclear extract, we performed an immobilized template assay in which we monitored Pol II recruitment to a promoter (Fig. 4e). For this purpose, DNA-bound beads were incubated with control or Mediator-depleted nuclear extract. In the latter case, the reactions were further supplemented with our various Mediator preparations. Consistent with our previous results43, Pol II recruitment was abolished in Mediator-depleted extracts (Fig. 4e, lane 2 vs. lane 1). Interestingly, neither the H+M (lane 3) nor the H+M+14 (lane 4) complex, which interacted strongly with purified Pol II (Fig. 4d), was able to induce Pol II recruitment. By contrast, and paralleling the results of the in vitro transcription experiment (Fig. 2c and Fig. 3), the H+M+14+26 complex was able to induce Pol II recruitment (Fig. 4e, lane 6 vs. lane 1). Therefore, we conclude that the conditional requirement of MED26 in nuclear extract reflects a restriction at the level of Pol II recruitment to the promoter that MED26 allows the Mediator to overcome.
Molecular architecture of the core Mediator complex
To dissect the molecular architecture of the Mediator complex, we chemically conjugated the reconstituted H+M+14+26 complex by amine-specific, isotopically labeled disuccinimidyl suberate (DSS) (Supplementary Fig. 5a) and applied high-resolution mass spectrometry (CX-MS)44 to identify cross-linked peptides. We identified 277 unique cross-links (Supplementary Table 1, 2), which were used to build a spatial connectivity map of the complex (Fig. 5). Remarkably, the cross-linked lysines represent 60% of the total lysines of the reconstituted H+M+14+26 complex (Supplementary Fig. 5b, c). The data reveal an extensive network of contacts between subunits within each of the head and middle modules, as well as between subunits of the two modules (Fig. 5). The intra-modular cross-linking data are in good agreement with the published studies of yeast Mediator modules22-25. Especially for the head module, a region (amino acids ~150-300) toward the N-terminus of human MED17 is also a structural hub within the module, cross-linking with MED6, MED8 (Supplementary Table 2), MED11, MED22, and MED30. Consistent with their previously proposed hinge function within the middle module20,25, MED7 and MED21 contact each of the other constituent subunits. MED18, MED20, and MED26, being sub-stoichiometric, were not scored by CX-MS.
Fig. 5. Molecular architecture of the reconstituted Mediator complex revealed by chemical cross-linking and mass spectrometry (CX-MS).
Residue-specific cross-linking map of the Mediator complex obtained by cross-linking and mass spectrometry. Except for MED14, for which intra-subunit cross-links (> 200 residues apart) are shown, only inter-subunit cross-links are depicted. Middle module subunits are depicted in light blue; head module subunits are in gold; MED14 in purple. See Supplementary Tables 1 and 2 for the complete cross-linking dataset.
Importantly, the CX-MS data reveal inter-modular contacts between MED17 in the head module and MED10 and MED21 in the middle module. Furthermore, relevant to the critical role of MED14 in Mediator function, this subunit cross-linked to both head (MED6, MED17 [Supplementary Table 2]) and middle (MED7) components, thus serving to further bridge the two modules. We also identified several intra-subunit cross-links between N- and C- terminal residues of MED14 in the active core Mediator complex, indicating that this large (170 kDa) subunit may potentially fold back upon itself and facilitate its interaction with the head and middle modules and perhaps also Pol II (Supplementary Table 2). Alternatively, this cross-linking pattern might arise from a tendency of MED14 to form (transient) dimers.
In complementary experiments to validate the CX-MS data, we generated a series of partial derivatives of the head and middle modules by selective omission of subunits and performed immunoprecipitations with selected subunit combinations (Supplementary Fig. 6). We confirmed CX-MS-identified interactions of MED7 and MED21 with various middle subunits (Supplementary Fig. 6a-c) and detected an additional interaction between MED21 and MED31 (Supplementary Fig. 6a, lane 8). Similarly, for the head module (Supplementary Fig. 6d), we identified complex formation between MED11 and MED22 (Supplementary Fig. 6d, lanes 6, 7), between MED11, MED22 and MED17 (Supplementary Fig. 6d, lane 6), between MED6 and MED17 (Supplementary Fig. 6d, lane 4), between MED8 and MED17 (Supplementary Fig. 6d, lane 3 vs. lanes 4 and 5) and between MED8 and MED18 (Supplementary Fig. 6d, lane 3 vs. lane 5). Importantly, as in yeast, MED18 and MED20 were found to form a heterodimer that is anchored to the head via MED8 (Supplementary Fig. 6d, lane 8 and lane 3 vs. lane 4).
Consistent with the cross-linking data for MED17, H+M formation was dependent on the presence of MED17 (Supplementary Fig. 2b). Moreover, MED17 also co-purified with the middle module (Supplementary Fig. 6f), identifying it as a major link between the head and middle modules. Related, this series of analyses also identified an additional interaction (between MED17 and MED7) contributing to the head-middle interaction (Supplementary Fig. 6g).
Notably, through co-expression of MED14 with either head or middle module subunits, and consistent with the crosslinking data, we established that MED14 could independently associate with the head and middle modules (Supplementary Fig. 4a-b). Also of note, MED14 also interacted with the MED24 and MED16 subunits of the tail module, which, however, were not included in our reconstitutions (Supplementary Fig. 4c). These results further implicate MED14 as the essential backbone of the Mediator that bridges its three main modules. A composite subunit interaction network for the human core Mediator complex deduced from various approaches is shown in Fig. 6. At a gross level, the deduced interactions among the subunits and the general architecture of the human core Mediator complex these data are in good agreement with the data of Tsai et al27.
Fig. 6. Schematic representation of subunit interactions in the human core Mediator complex based on composite data from CX-MS and biochemical approaches.

Intra-module cross-links in the middle module are in light blue; intra-module cross-links in the head are in brown; MED14 cross-links are in purple; inter-module cross-links between head and middle modules are in dark blue.
DISCUSSION
In this paper, we describe a reconstitution-based approach aimed at the generation of Mediator complexes that display various functionalities previously ascribed to the Mediator. We also generated a detailed spatial connectivity map of the active core Mediator complex through CX-MS and pair-wise interaction analyses of selected subunits. These structural and functional studies converge to highlight a critical role for MED14 in Mediator architecture and activity. Although head-middle interactions yield a stable complex, MED14 association with these two modules is necessary to reconstitute a functionally active 14-subunit core Mediator complex. In this complex the MED14 subunit is the one most critical for facilitating a very strong Pol II interaction that correlates with the acquisition of both basal and selective activator (p53)-dependent transcription activity. Our results also show that MED26, while not required for core Mediator function in an assay with purified factors, is essential (along with MED14) for core Mediator function in a nuclear extract. Thus, our approach has allowed us, uniquely, to identify the minimal components of the active core-Mediator complex and to understand the underlying mechanism and dissect the roles of Mediator subunits in a minimal purified system versus a nuclear extract containing a more natural complement of nuclear factors.
MED14 has been viewed as a tail component, albeit one that bridges the tail to the bulk complex35. Our protein-protein interaction and CX-MS data establish that MED14 interacts with tail subunits (MED16, MED24), as well as head (MED6, MED8, MED17) and middle (MED7, MED10) module subunits. Thus, MED14 appears to furnish the architectural features necessary for integrating three separate modules of the Mediator into a single functional entity. This model of MED14 as an architectural backbone of the Mediator complex is in good conformity with the recent cryo-EM analysis of Tsai et al27, which revealed that density attributable to MED14 spans the length of the natural yeast Mediator complex and makes multiple contacts with subunits of the tail, middle and head modules. Neither our present study nor the Tsai et al study27 addressed how MED14 relates to the dissociable kinase module.
In a major extension of the solely architectural focus of the Tsai et al study27, we show further that MED14 is critically required for the function of the core Mediator. Previously, the isolated head module of the yeast Mediator was reported to interact with Pol II (via MED17) and to stimulate basal activity42,45. It therefore was initially somewhat surprising that our reconstituted head-middle bi-modular complex was unable to support even the most rudimentary Mediator activity of stimulating basal transcription. Indeed, the metazoan head-middle complex (as well as the head complex alone) failed to interact with Pol II in our hands. Only when MED14 was incorporated into this assembly through coexpression did the complex interact with Pol II and acquire basal transcription activity. Beyond its stimulation of basal transcription, the resulting complex also exhibits a selective coactivator function for p53, which interacts with the head subunit MED17. Thus, MED14 is not simply an architectural backbone of the Mediator complex, but plays a critical role in facilitating transduction of the necessary signals within the Mediator-PIC assembly. Reciprocally, the results suggest that the tail module may serve principally as an activator target site for Mediator recruitment, with no additional role in core Mediator-enhanced transcription.
How might MED14 contribute to Pol II interaction and ultimately to stimulation of transcription? Most simply, this could be due to a direct physical interaction between Pol II and the large surface furnished by the MED14 backbone. However, and although the most recent study cryo-EM study from the Asturias group does not shed additional light on which features of the yeast Mediator complex are responsible for holoenzyme formation, they previously proposed46 a multistep model in which the Pol II CTD first interacts with the head module and then comes to rest within a cavity formed by the head, middle and tail modules of the remodeled Mediator complex. While the EM analyses did not allow precise delineation of contacts, the density now identified as the MED14 backbone does not seem to be in direct contact with Pol II in the published images46. Furthermore, to our knowledge, neither prior yeast genetic nor other studies have implicated MED14 in Pol II interactions. Therefore, the alternative possibility remains that MED14 effects on Pol II binding are indirect and are related to the documented inter-module movements that occur upon Pol II binding46,47. It is likely that in the absence of MED14, the otherwise stable head-middle complex, which may yet be responsible for a majority of the Pol II contacts, is incapable of acquiring the necessary conformation on its own.
Consistent with its structural complexity, and superimposed on its overlapping core functions of stimulating basal transcription and mediating activation signals, Mediator can coordinate the action of numerous cofactors that impinge upon the transcriptional machinery2. Thus, in contrast to its activity in transcription assays reconstituted with pure factors, the H+M+14 complex failed to function in HeLa cell nuclear extract. Previous studies have shown that whereas Mediator acts mainly to stimulate transcription in the purified systems, its requirement in extracts is absolute3,10. This suggests that in the cellular milieu, an important role of the Mediator is to overcome the effects of negatively acting cofactors that include DSIF10, Gdown148, and potentially NC249. Our finding that a MED26-containing complex can function in nuclear extract to stimulate both basal and activator-dependent transcription suggests a role for this subunit in counteracting negative cofactors. This is consistent with our prior observation that even though the MED26-containing subpopulation of the Mediator (PC2) constitutes a very small fraction of the total Mediator, its depletion from HeLa cell nuclear extract leads to abrogation of transcription activity50.
MED26 has been implicated in interactions with TFIID and the P-TEFb- and ELL-containing Super Elongation Complex, leading to a model in which this subunit functions in a hand-off from the initiation to the elongation machinery12. However, our mechanistic dissection reveals that, collectively, the cofactors in the extract impose an even earlier restriction at the level of Pol II recruitment to the promoter and that MED26-containing Mediator overcomes the restriction. This observation suggests a function for MED26 at the earliest stages of the transcription process that precede involvement of the elongation machinery. It does not, however, preclude a subsequent additional role for MED26 at the initiation-to-elongation transition or elongation stages. The precise mechanism whereby MED26-containing Mediator overcomes the effect of negative factors is unclear. However, its localization in the middle module relatively distant from the Pol II binding cavity27 argues against direct interactions with Pol II. Possibilities include MED26-dependent recruitment of activities that neutralize the negative cofactors or freezing of Mediator in conformations that favor Pol II interactions and disallow negative cofactor interference. Note that even in this context, the MED14 requirement persists, consistent with its mechanistically distinct and essential role.
A recent study in Drosophila has suggested that the MED26 requirement is stage-specific51. Thus, it remains unclear whether our results reflect a general MED26 requirement or cell type-specific (HeLa) regulation. Nonetheless, our ability to generate compositionally defined Mediator complexes that carry out functions over and above Mediator’s core functions nicely illustrates how it is feasible to recapitulate increasingly complex metazoan-specific regulatory functions by building ever-larger Mediator derivatives. As we expand the scope of these studies and reconstitute larger derivatives of the core Mediator complex, we hope to obtain a better understanding of the full range of Mediator functions, including those that go awry in diseased states.
ONLINE METHODS
cDNA cloning of Mediator subunits
For sub-cloning into baculovirus expression vectors, we used existing cDNAs for MED4, MED6, MED7, MED10, MED14, MED16, MED18, MED20, MED21, and MED241-4. For the remaining, we isolated new cDNA clones from HeLa cells. Total RNA from HeLa cells was purified and cDNA prepared by reverse transcription using oligo-dT primers. The resulting cDNA was amplified with appropriate PCR primers to generate individual clones for MED8, MED9, MED11, MED19, MED22, MED26, MED30, and MED31. Interestingly, at least two variants were seen for MED8 and MED22. We selected the shortest variant cDNAs of each for expression.
Reconstitution of human Mediator complexes
In order to obtain near-stoichiometric complexes, Mediator subunit cDNAs were cloned into pFBDM and pUCDM transfer vectors5. Various tags (histidine, myc, HA or Flag) were inserted into different subunits of the Mediator to facilitate downstream purification. The transfer vectors were integrated into a single bacmid (through both transposition and cre-lox recombination) for single virus generation. The resulting viruses were amplified in Sf9 cells. For protein production, Hi5 cells were infected with the amplified viruses. Infected cells were homogenized in BC500 (500 mM KCl, 10 mM Tris-Cl pH7.9, 20% glycerol, 0.1 mM EDTA, 3.5 mM β-mercaptoethanol and 0.1 mM PMSF supplemented protease inhibitors pepstatin (0.5 μg/ml) and leupeptin (0.5 μg/ml). After ultracentrifugation (20,000 rpm in a Type 45 Ti rotor for 30 minutes), the lysate was diluted to 300 mM KCl. The extract was then purified through various combinations of affinity (anti-Flag M2 agarose and anti-HA agarose for Flag-tagged and HA-tagged subunits, respectively), ion exchange (typically SP-Sepharose) and gel filtration (Superose 6) chromatography. For both M2 and HA beads, elution was with 0.5 mg/ml of the corresponding peptide.
Optimization of the reconstitution protocol entailed countless viral titrations, as well as identifying, by trial-and-error, which subunit to tag. The following summarizes our reconstitution protocol for one of the largest Mediator variants reported here. The individual cDNAs for subunits of the Mediator head module (MED6, MED8, MED11, MED18, MED19, MED20, MED22, and MED30) were inserted into the pFBDM and pUCDM transfer vectors and the resulting transfer vectors were integrated into a single bacmid for virus generation. Individual cDNAs for subunits of the middle module were also inserted into the pFBDM and pUCDM transfer vectors (HA-MED7, MED4, MED21, His:MED10, MED31, MED9 and MED26) and integrated into another bacmid for production of the second virus. pFBDM MED17 and pFBDM Flag MED14 were integrated into two different bacmids to form the third and the fourth viruses, which were amplified in Sf9 cells. For protein production in Hi5 cells, scaled up cultures were infected with the virus cocktail. A typical yield of pure core Mediator complex from 500 ml of infected cells was 100 μg.
Purification of transcription factors, activators and coactivators
Purification of the general transcription factors was essentially as described6. Recombinant TFIIB, TFIIE and TFIIF were expressed in bacteria and purified as described before6. Baculovirus-expressed TFIIA was purified from insect cells, as were the various transcriptional activators (p53, TRα and RXRα). Pol II, TFIIH, and TFIID were purified from corresponding stable HeLa cell lines that expressed epitope-tagged subunits. For routine use, Mediator was also similarly affinity purified from a HeLa cell line that stably expressed the core subunit MED106. The PC2 form of Mediator was affinity-purified from the phosphocellulose P11 0.85 M fraction of nuclear extract from a cell line that expresses Flag-tagged MED261.
In vitro transcription assays
In vitro transcription assays using purified factors or nuclear extract were performed essentially as described previously6. Transcription reactions typically contained 50 ng of test templates. All templates contained G-less cassettes downstream of the adenovirus major late (ML) core promoter. The template 5Xp53REML further contained 5 copies of a p53 response element and the 5XTREML template 5 copies of a thyroid response element. Reactions were initiated by adding protein factors to the reaction mixes, which contained α32P-UTP or α32P-CTP as the labeled nucleotide triphosphate. Reactions took place for 50 minutes at 30°C and then were processed and analyzed by electrophoresis on 5% polycrylamide-50% urea gels and autoradiography. For reactions with Mediator-depleted nuclear extract, HeLa cell nuclear extract7 was immunodepleted using antigen-purified anti-MED30 antibody (below), as described6.
Immunoprecipitation assays
Antigen-purified MED30 antibody was coupled to protein A-Sepharose beads. The beads were washed with BC200, and added to binding reactions containing various Mediator derivatives and either Pol II or TFIID. After incubation for 2 hours, the beads were washed again in BC200 plus 0.1% NP-40 and eluted. The immunoprecipitates were analyzed by Western blotting.
Immobilized template recruitment assays
A PCR-generated biotinylated adenovirus major late (Ad ML) promoter-containing DNA fragment was bound to Dynabeads M-280 Streptavidin (Invitrogen 11205-D), as recommended by the manufacturer. The beads were incubated with either mock-depleted or Mediator-depleted nuclear extracts in the presence or absence of variant Mediator preparations. The reaction mixes were set up as for in vitro transcription but were scaled up 10-fold, as described8. Following incubation and washing, the bound material was eluted by boiling in SDS-PAGE sample buffer and characterized by immunoblotting.
Antibodies
Antibodies against most of the Mediator subunits were from our Laboratory’s previously published collection1. MED18 (sc-161835), MED8 (sc-103619), MED22 (sc-107739), and MED14 (sc-9419) were purchased from Santa Cruz. MED30 antibody used for immunodepletion and co-immunoprecipitation was affinity purified by chromatography against bacterially expressed antigen6.
Chemical cross-linking and mass spectrometry
The purified complex was chemically cross-linked by 1 mM isotopically labeled disuccinimidyl suberate (d0:d12 with 1:1 ratio, Creative Molecules) for 45 minutes at 4°C with constant agitation. The reaction was then quenched in 50 mM ammonia bicarbonate. After disulfide reduction and cysteine alkylation, the cross-linked complex was digested both in solution and in gel with trypsin to identify cross-linked peptides10,11. For in-solution digestion, ~50-100 μg of purified complex was digested with 2 μg of trypsin (Promega) in 1M urea with ~2% acetonitrile (ACN) and 0.1% Rapigest (Waters) at 37°C. After 12-16 hours of incubation an additional 1-2 μg trypsin was added to the digest and incubated for a further 4 hours. The resulting proteolytic peptide mixture was purified using a C18 cartridge (Sep-Pak, Waters), lyophilized and fractionated by peptide size exclusion chromatography9. For in-gel digestion, ~50 ug purified complex was resuspended and heated in 2X LDS loading buffer. The sample was cooled at room temperature for cysteine alkylation and separated by electrophoresis in a 4-12% SDS PAGE gel. The gel region above ~220 kDa was sliced, crushed into small pieces and digested in-gel by trypsin. After extraction and purification, the resulting proteolytic peptide mixture was dissolved in 20 μl of a solution containing 30% ACN and 0.2% formic acid (FA) and fractionated by peptide SEC (Superdex Peptide PC 3.2/30, GE Healthcare) using off-line HPLC separation with an auto sampler (Agilent Technologies). Three SEC fractions in the molecular mass range of ~2.5 kDa to 8 kDa were collected and analyzed by LC/MS.
Purified peptides were dissolved in the sample loading buffer (5 % MeOH, 0.2% FA) and loaded onto a self-packed PicoFrit® column with integrated electrospray ionization emitter tip (360 O.D, 75 I.D with 15 μm tip, New Objective). The column was packed with 8 cm of reverse-phase C18 material (3 µm porous silica, 200 angstrom pore size, Dr. Maisch GmbH). Mobile phase A consisted of 0.5% acetic acid and mobile phase B of 70% ACN with 0.5% acetic acid. The peptides were eluted in a 150-min LC gradient (8 % B to 46% B, 0- 118 min, followed by 46% B -100% B, 118-139 min and equilibrated with 100% A until 150 min) using a HPLC system (Agilent), and analyzed with a LTQ Velos Orbitrap Pro mass spectrometer (Thermo Fisher). The flow rate was ~200 nl/min. Spray voltage was set at 1.9-2.2 kV. Capillary temperature was 275°C and ion transmission on Velos S lenses was set at 35%. The instrument was operated in the data-dependent mode, where the top eight-most abundant ions were fragmented by higher energy collisional dissociation/HCD (HCD energy 27-33, 0.1 ms activation time) and analyzed in the Orbitrap mass analyzer. The target resolution for MS1 was 60,000 and 7,500 for MS2. Ions (370- 1700 m/z) with charge state of >3 were selected for fragmentation. A dynamic exclusion of (15”/ 2 / 55”) was used. Other instrumental parameters include: “lock mass” at 371.1012 Da, the minimal threshold of 5,000 to trigger an MS/MS event, Ion trap accumulation limits were 105 and 106 respectively for the linear ion trap and orbitrap. The max ion injection time for the LTQ was set at 200 ms. The max ion injection time for orbitrap was 500 ms for full scan, and 500-700 ms for MS2.
The raw data were transformed to MGF (mascot generic format) and searched by pLink software12 with a database containing sequences of the protein subunits of human Mediator complex and BSA. Other search parameters included: mass accuracy of MS1 ≤ 10 ppm (parts per million) and MS2 ≤ 20 ppm for the initial database search, cysteine carboxymethylation as a fixed modification, methionine oxidation as a variable modification, and a maximum of two trypsin miscleavages was allowed. The results were filtered at 5% false discovery rate (FDR) and were subjected to manual verification of the resulting MS/MS spectra based on the following criteria: for positive identifications, both peptide chains must contain at least five amino acids and for both peptide chains, the major MS/MS fragmentation peaks must be assigned and followed a pattern that contains a continuous stretch of fragmentations. The appearance of dominant fragment ions N-terminal to proline and C-terminal to aspartic acid and glutamic acid for arginine-containing peptides was generally expected13,14. A total of 277 unique cross-linked peptides were identified as a result.
Supplementary Material
ACKNOWLEDGEMENTS
We thank T. Richmond, Institute of Molecular Biology & Biophysics ETH Zurich, for the Multibac baculovirus system, J. Fernandez-Martinez (M. P. Rout lab) for assistance with the off-line Agilent HPLC system, and M. Guermah for discussion. Funding for this work was provided by US Department of Defense grant W81XWH-13-1-0172 (R.G.R.) and by US National Institute of Health grants CA129325 (R.G.R.), 1RC1GM090929 (R.G.R. and S.M.) and P41 GM103314 (B.T.C.). M.A.C. was supported by an American Cancer Society Eastern Division - New York Cancer Research Fund Postdoctoral Fellowship.
Footnotes
AUTHOR CONTRIBUTIONS
M.A.C., S.M., R.G.R., Y.S., and B.T.C designed the experiments and wrote the manuscript. M.A.C. carried out biochemical experiments including cDNA preparations, reconstitutions, in vitro transcriptions and co-IP experiments. D.L. helped M.A.C. in the generation of partial head module complexes in the Fig. 6d. Y.S. carried out the CX-MS experiments. The authors declare no competing financial interests.
References
- 1.Roeder RG. Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 2005;579:909–915. doi: 10.1016/j.febslet.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nature reviews. Genetics. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek HJ, Malik S, Qin J, Roeder RG. Requirement of TRAP/mediator for both activator-independent and activator-dependent transcription in conjunction with TFIID-associated TAF(II)s. Mol Cell Biol. 2002;22:2842–2852. doi: 10.1128/MCB.22.8.2842-2852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malik S, Baek HJ, Wu W, Roeder RG. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittler G, Kremmer E, Timmers HT, Meisterernst M. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2001;2:808–813. doi: 10.1093/embo-reports/kve186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Critical reviews in biochemistry and molecular biology. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black JC, Choi JE, Lombardo SR, Carey M. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell. 2006;23:809–818. doi: 10.1016/j.molcel.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 8.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 9.Lin JJ, Lehmann LW, Bonora G, Sridharan R, Vashisht AA, Tran N, Plath K, Wohlschlegel JA, Carey M. Mediator coordinates PIC assembly with recruitment of CHD1. Genes & Dev. 2011;25:2198–2209. doi: 10.1101/gad.17554711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Proc Natl Acad Sci. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nock A, Ascano JM, Barrero MJ, Malik S. Mediator-regulated transcription through the +1 nucleosome. Mol Cell. 2012;48:837–848. doi: 10.1016/j.molcel.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, Florens L, Seidel CW, Lin C, Smith ER, Shilatifard A, Conaway RC, Conaway JW. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, Taatjes DJ, Dekker J, Young RA. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010 doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eyboulet F, Cibot C, Eychenne T, Neil H, Alibert O, Werner M, Soutourina J. Mediator links transcription and DNA repair by facilitating Rad2/XPG recruitment. Genes & Dev. 2013;27:2549–2562. doi: 10.1101/gad.225813.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiano C, Casamassimi A, Rienzo M, de Nigris F, Sommese L, Napoli C. Involvement of Mediator complex in malignancy. Biochimica et biophysica acta. 2014;1845:66–83. doi: 10.1016/j.bbcan.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Spaeth JM, Kim NH, Boyer TG. Mediator and human disease. Seminars in cell & developmental biology. 2011;22:776–787. doi: 10.1016/j.semcdb.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blazek E, Mittler G, Meisterernst M. The mediator of RNA polymerase II. Chromosoma. 2005;113:399–408. doi: 10.1007/s00412-005-0329-5. [DOI] [PubMed] [Google Scholar]
- 20.Baumli S, Hoeppner S, Cramer P. A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J Biol Chem. 2005;280:18171–18178. doi: 10.1074/jbc.M413466200. [DOI] [PubMed] [Google Scholar]
- 21.Koschubs T, Seizl M, Larivière L, Kurth F, Baumli S, Martin DE, Cramer Pl. Identification, structure, and functional requirement of the Mediator submodule Med7N/31. The EMBO J. 2009;28:69–80. doi: 10.1038/emboj.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lariviere L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P. Structure of the Mediator head module. Nature. 2012;492:448–451. doi: 10.1038/nature11670. [DOI] [PubMed] [Google Scholar]
- 23.Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL, Asturias FJ, Kornberg RD, Takagi Y. Architecture of the Mediator head module. Nature. 2011;475:240–243. doi: 10.1038/nature10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson PJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc Natl Acad Sci U S A. 2012;109:17931–17935. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lariviere L, Plaschka C, Seizl M, Petrotchenko EV, Wenzeck L, Borchers CH, Cramer P. Model of the Mediator middle module based on protein cross-linking. Nucleic Acids Res. 2013;41:9266–9273. doi: 10.1093/nar/gkt704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai KL, Tomomori-Sato C, Sato S, Conaway RC, Conaway JW, Asturias FJ. Subunit architecture and functional modular rearrangements of the transcriptional mediator complex. Cell. 2014;157:1430–1444. doi: 10.1016/j.cell.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang X, Sun Q, Ding Z, Ji J, Wang J, Kong X, Yang J, Cai G. Redefining the modular organization of the core Mediator complex. Cell Res. 2014;24:796–808. doi: 10.1038/cr.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik S, Gu W, Wu W, Qin J, Roeder RG. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol Cell. 2000;5:753–760. doi: 10.1016/s1097-2765(00)80254-3. [DOI] [PubMed] [Google Scholar]
- 30.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nature biotechnology. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 31.Ge K, Guermah M, Yuan CX, Ito M, Wallberg AE, Spiegelman BM, Roeder RG. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature. 2002;417:563–567. doi: 10.1038/417563a. [DOI] [PubMed] [Google Scholar]
- 32.Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 34.Malik S, Roeder RG. Isolation and functional characterization of the TRAP/mediator complex. Methods in enzymology. 2003;364:257–284. doi: 10.1016/s0076-6879(03)64015-2. [DOI] [PubMed] [Google Scholar]
- 35.Dotson MR, Yuan CX, Roeder RG, Myers LC, Gustafsson CM, Jiang YW, Li Y, Kornberg RD, Asturias FJ. Structural organization of yeast and mammalian mediator complexes. Proc Natl Acad Sci of the United States of America. 2000;97:14307–14310. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malik S, Barrero MJ, Jones T. Identification of a regulator of transcription elongation as an accessory factor for the human Mediator coactivator. Procs Natl Acad Sci of the United States of America. 2007;104:6182–6187. doi: 10.1073/pnas.0608717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, Conaway JW, Conaway RC. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 38.Conaway RC, Conaway JW. The Mediator complex and transcription elongation. Biochimica et biophysica acta. 2013;1829:69–75. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 40.Yuan CX, Ito M, Fondell JD, Fu ZY, Roeder RG. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lariviere L, Geiger S, Hoeppner S, Röther S, Strässer K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat Struct Mol Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 42.Cai G, Chaban YL, Imasaki T, Kovacs JA, Calero G, Penczek PA, Takagi Y, Asturias F. Interaction of the mediator head module with RNA polymerase II. Structure. 2012;20:899–910. doi: 10.1016/j.str.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baek HJ, Kang YK, Roeder RG. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–15181. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 44.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science. 2011;331:1451–1454. doi: 10.1126/science.1200188. [DOI] [PubMed] [Google Scholar]
- 46.Tsai KL, Sato S, Tomomori-Sato C, Conaway RC, Conaway JW, Asturias FJ. A conserved Mediator-CDK8 kinase module association regulates Mediator-RNA polymerase II interaction. Nat Struct Mol Biol. 2013;20:611–619. doi: 10.1038/nsmb.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes & Dev. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jishage M, Malik S, Wagner U, Uberheide B, Ishihama Y, Hu X, Chait BT, Gnatt A, Ren B, Roeder RG. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol Cell. 2012;45:51–63. doi: 10.1016/j.molcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemaire M, Xie J, Meisterernst M, Collart MA. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Molecular microbiology. 2000;36:163–173. doi: 10.1046/j.1365-2958.2000.01839.x. [DOI] [PubMed] [Google Scholar]
- 50.Malik S, Baek HJ, Wu W, Roeder RG. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marr SK, Lis JT, Treisman JE, Marr MT., 2nd The Metazoan-Specific Mediator Subunit 26 (Med26) Is Essential For Viability And Is Found At Both Active Genes And Pericentric Heterochromatin In Drosophila. Mol Cell Biol. 2014 doi: 10.1128/MCB.01365-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 1.Malik S, Baek HJ, Wu W, Roeder RG. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol Cell Biol. 2005;25:2117–2129. doi: 10.1128/MCB.25.6.2117-2129.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malik S, Gu W, Wu W, Qin J, Roeder RG. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol Cell. 2000;5:753–760. doi: 10.1016/s1097-2765(00)80254-3. [DOI] [PubMed] [Google Scholar]
- 3.Gu W, Malik S, Ito M, Yuan CX, Fondell JD, Zhang X, Martinez E, Qin J, Roeder RG. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol Cell. 1999;3:97–108. doi: 10.1016/s1097-2765(00)80178-1. [DOI] [PubMed] [Google Scholar]
- 4.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 5.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nature biotechnology. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
- 6.Malik S, Roeder RG. Isolation and functional characterization of the TRAP/mediator complex. Methods in enzymology. 2003;364:257–284. doi: 10.1016/s0076-6879(03)64015-2. [DOI] [PubMed] [Google Scholar]
- 7.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malik S, Wallberg AE, Kang YK, Roeder RG. TRAP/SMCC/mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol Cell Biol. 2002;22:5626–5637. doi: 10.1128/MCB.22.15.5626-5637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leitner A, Reischl R, Walzthoeni T, Herzog F, Bohn S, Förster F, Aebersold R. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014126. M111 014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, Fernandez-Martinez J, Tjioe E, Pellarin R, Kim SJ, Williams R, Schneidman D, Sali A, Rout MP, Chait BT. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Algret R, Fernandez-Martinez J, Shi Y, Kim SJ, Pellarin R, Cimermancic P, Cochet E, Sali A, Chait BT, Rout MP, Dokudovskaya S. Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway. Mol Cell Proteomics. 2014 doi: 10.1074/mcp.M114.039388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, Luo SK, Ding YH, Wang LH, Hao Z, Xiu LY, Chen S, Ye K, He SM, Dong MQ. Identification of cross-linked peptides from complex samples. Nature methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 13.Qin J, Chait BT. Matrix-assisted laser desorption ion trap mass spectrometry: efficient isolation and effective fragmentation of peptide ions. Analytical chemistry. 1996;68:2108–2112. doi: 10.1021/ac951163m. [DOI] [PubMed] [Google Scholar]
- 14.Michalski A, Neuhauser N, Cox J, Mann M. A systematic investigation into the nature of tryptic HCD spectra. Journal of proteome research. 2012;11:5479–5491. doi: 10.1021/pr3007045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.