Abstract
The retained N-terminal methionine (Met) residue of a nascent protein is often N-terminally acetylated (Nt-acetylated). Removal of N-terminal Met by Met-aminopeptidases frequently leads to Nt-acetylation of the resulting N-terminal Ala, Val, Ser, Thr and Cys residues. Although a majority of eukaryotic proteins, for example, more than 80% of human proteins, are cotranslationally Nt-acetylated, the function of this extensively studied modification is largely unknown. Here we found, using the yeast Saccharomyces cerevisiae, that the Nt-acetylated Met residue could act as a degradation signal (degron), targeted by the Doa10 ubiquitin ligase. Moreover, Doa10 also recognized the Nt-acetylated Ala, Val, Ser, Thr and Cys residues. Several examined proteins of diverse functions contained these N-terminal degrons, termed AcN-degrons, which comprise a prevalent class of degradation signals in cellular proteins.
Many eukaryotic proteins are acetylated at the α-amino group of their N-terminal residues (fig. S1A) (1). Previous studies of Nα-terminal acetylation (Nt-acetylation) characterized Nt-acetylated proteins and Nα-terminal acetyltransferases (Nt-acetylases) that catalyze this cotranslational modification (2-7). Owing to the design of the genetic code, nascent proteins contain N-terminal Met. A retained N-terminal Met that is followed by “acetylation-permissive” residues is usually Nt-acetylated (fig. S1A) (5-7). Met-aminopeptidases cleave off the N-terminal Met if the residue at position 2 has a small enough side chain, resulting in N-terminal Ala, Val, Ser, Thr, Cys, Gly or Pro (fig. S1B) (8). With the near-exception of Gly and Pro, these N-terminal residues are often Nt-acetylated, similarly to N-terminal Met (5-7). In cell extracts, some Nt-acetylated proteins can be degraded by the ubiquitin (Ub) system (9). However, no cognate Ub ligases have been identified, and it has been assumed that the relevant degradation signals were internal (not N-terminal) (9). Currently, the prevalent view of Nt-acetylation is that this modification protects proteins from degradation. To the contrary, we report here that Nt-acetylation creates specific degradation signals (degrons) that are targeted by a novel branch of the Ub-dependent N-end rule pathway.
Destabilizing N-terminal sequences
The N-end rule relates the in vivo half-life of a protein to the identity of its N-terminal residue (10-20). N-terminal degradation signals of the N-end rule pathway are called N-degrons. Their main determinant is a destabilizing N-terminal residue of a protein (fig. S1C). Recognition components of the N-end rule pathway are called N-recognins. An N-recognin is an E3 Ub ligase that can target for polyubiquitylation at least a subset of N-degrons (13, 15, 18, 19). The N-end rule of the yeast S. cerevisiae comprises 12 destabilizing, unacetylated N-terminal residues (out of the fundamental set of 20 amino acids) (12-15, 18, 19). Among these residues, 8 are primary destabilizing residues, i.e., recognized directly by the Ubr1 N-recognin, whereas the other 4 N-terminal residues, called secondary or tertiary destabilizing residues, must be modified through deamidation and/or arginylation before the corresponding proteins can be targeted by Ubr1 (fig. S1C).
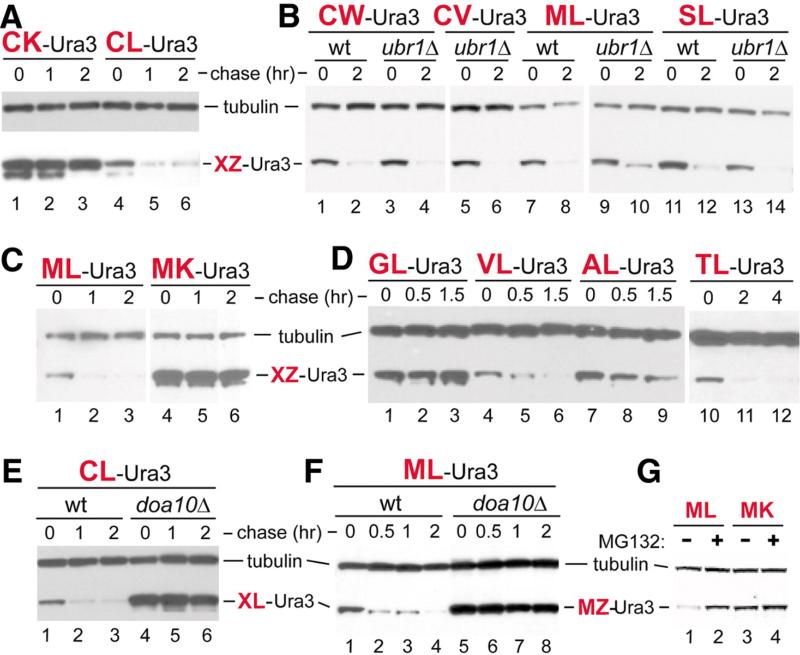
In mammalian cells, N-terminal Cys of N-end rule substrates can be oxidized, by nitric oxide (NO) and oxygen, and thereafter arginylated by an arginyl-transferase. The resulting N-terminal Arg is recognized by Ubr1-type N-recognins (15, 16). In contrast, N-terminal Cys appeared to be a stabilizing residue in S. cerevisiae, which lacks NO synthases (11). That study also classified N-terminal Met, Ala, Val, Ser and Thr as stabilizing residues in S. cerevisiae (11). One caveat in these assignments is the possible influence of sequences downstream of the reporter's N-terminus. To determine whether N-terminal Cys can be destabilizing in yeast, we performed a screen in ura3 S. cerevisiae with Cys-Z-eK-Ura3 reporters, produced by deubiquitylation (10, 21) of Ub-Cys-Z-eK-Ura3. Z denotes a varied residue at position 2, and eK (extension (e) containing lysine (K)) denotes a ~40-residue sequence upstream of Ura3. The eK extension (fig. S1D) has a technically valuable property of lacking internal degrons while containing “ubiquitylatable” Lys residues (10-12). This screen identified Cys-Z-eK-Ura3 fusions (Z=Leu, Val, Pro) with low Ura3 activity. We examined these fusions using a cycloheximide (CHX)-chase assay, in which a protein is analyzed by immunoblotting as a function of time after the inhibition of translation by CHX (18, 19). The above three reporters were short-lived in vivo (t1/2 < 1 hr), in contrast to GL-eK-Ura3 (N-terminal Gly) and CK-eK-Ura3 (Lys at position 2), which were long-lived (Fig. 1A, B, D, fig. S2B, and fig. S3A, D). Other non-basic residues at position 2 also yielded short-lived CZ-eK-Ura3 (Z=Trp, Glu, Gly, Ile) (Fig. 1B and fig. S3A).
Fig. 1.
Destabilizing N-terminal residues. (A) CHX chases, for 0, 1, and 2 hours in wild-typeS. cerevisiaeexpressing CK-eK-Ura3 (lanes 1 to 3) or CL-eK-Ura3 (lanes 4 to 6). Cell extracts were fractionated by SDS–polyacrylamide gel electrophoresis, followed by immunoblotting with anti-Ha and anti-tubulin, the latter a loading control. (B) As in (A) but chases for 0 and 2 hours with XZ-eK-Ura3 (X = Cys, Met, or Ser; Z = Trp, Val, or Leu) in wild-type versus ubr1Δ cells. (C) As in (A) but with MZ-eK-Ura3 (Z = Leu or Lys) in wild-type cells. (D) As in (A) but chases for 0, 0.5, and 1.5 hours with XL-eK-Ura3 (X = Gly, Val, Ala, or Thr) in wild-type cells. (E) As in (A) but with CL-eK-Ura3 in wild-type cells (lanes 1 to 3) versus doa10Δ cells (lanes 4 to 6). (F) As in (E) but chases for 0, 0.5, 1, and 2 hours with ML-eK-Ura3. (G) Lanes 1 and 2, short-lived ML-eK-Ura in the MG132-sensitive pdr5Δ S. cerevisiae, in the absence and presence of MG132, respectively. Lanes 3 and 4, same as lanes 1 and 2 but with long-lived MK-eK-Ura3.
Remarkably, several other XL-eK-Ura3 reporters (X=Met, Ser, Val, Ala, Thr) were also short-lived in vivo, like CL-eK-Ura3 and in contrast to long-lived MK-eK-Ura3 (Lys at position 2), MR-eK-Ura3 (Arg at position 2), GL-eK-Ura3 (N-terminal Gly) and PL-eK-Ura3 (N-terminal Pro) (Fig. 1A-D, fig. S2A, B, and fig. S3B, C, E). We also performed 35S-pulse-chases (18, 19) with CL-eK-Ura3 and ML-eK-Ura3 versus CK-eK-Ura3 and MK-eK-Ura3 (fig. S4A-C). The techniques of CHX-chases and 35S-pulse-chases are complementary, as the former method monitors degradation of all molecules of a specific protein, whereas the latter assay measures degradation of newly formed (pulse-labeled) molecules. 35S-pulse-chases confirmed the instability of XL-eK-Ura3 (X=Cys, Met) and stability of XK-eK-Ura3 (X=Cys, Met) (Fig. 1A, C, fig. S2A, B, and fig. S4A-C). The degradation of ML-eK-Ura3 was proteasome-dependent, as the MG132 proteasome inhibitor significantly increased the level of the normally short-lived ML-eK-Ura3 but not of the long-lived MK-eK-Ura3, whose levels were high both in the presence and absence of MG132 (Fig. 1G).
The Doa10 ubiquitin ligase recognizes acetylated N-terminal residues
To search for a Ub ligase(s) that mediates the degradation of XL-eK-Ura3 (X=Met, Ala, Val, Ser, Thr, Cys), we expressed CL-eK-Ura3 in S. cerevisiae mutants that lacked specific E3 or E2 enzymes (fig. S4D). Strikingly, CL-eK-Ura3 became long-lived in the absence of Doa10 (Fig. 1E and fig. S4D, E). Moreover, other short-lived XL-eK-Ura3 proteins (Z=Met, Ser, Val) were also stable in doa10Δ cells (Fig. 1F and fig. S4F, G). Doa10 is a transmembrane E3 Ub ligase that functions with the Ubc6/Ubc7 E2s and resides in the endoplasmic reticulum (ER) and inner nuclear membrane (INM) (22-24). To address the above results, we focused on MATα2, a physiological substrate of Doa10.
The 24 kDa MATα2 contains more than one degradation signal and has an in vivo half-life of 5-10 min (13, 25, 26). MATα2 represses transcription of a-specific genes in α-cells, whereas in a/α diploids the MATα2-MATa1 complex represses haploid-specific genes (27, 28). The 67-residue N-terminal region of MATα2, termed Deg1, has been shown to harbor a Doa10-dependent degron (22-24). MATα2 is absent from databases of Nt-acetylated proteins (5, 6), possibly because of its short in vivo half-life. We expressed full-length MATα2 in doa10Δ ubc4Δ yeast and analyzed purified MATα2 using mass spectrometry (LC-MS/MS). The results (fig. S5A) indicated virtually complete Nt-acetylation of MATα2 (no MATα2 that lacked Nt-acetylation could be detected), in agreement with Nt-acetylation of other proteins containing the N-terminal Met-Asn (5-7). Similar LC-MS/MS of the Doa10-targeted, purified ML-eK-Ura3 (fig. S5B, C) indicated the Nt-acetylation of this reporter, in agreement with Nt-acetylation of other proteins containing the N-terminal Met-Leu (5-7). We also observed Nt-acetylation of a Deg1-bearing reporter that was purified from E. coli and incubated with S. cerevisiae extracts (fig. S6A).
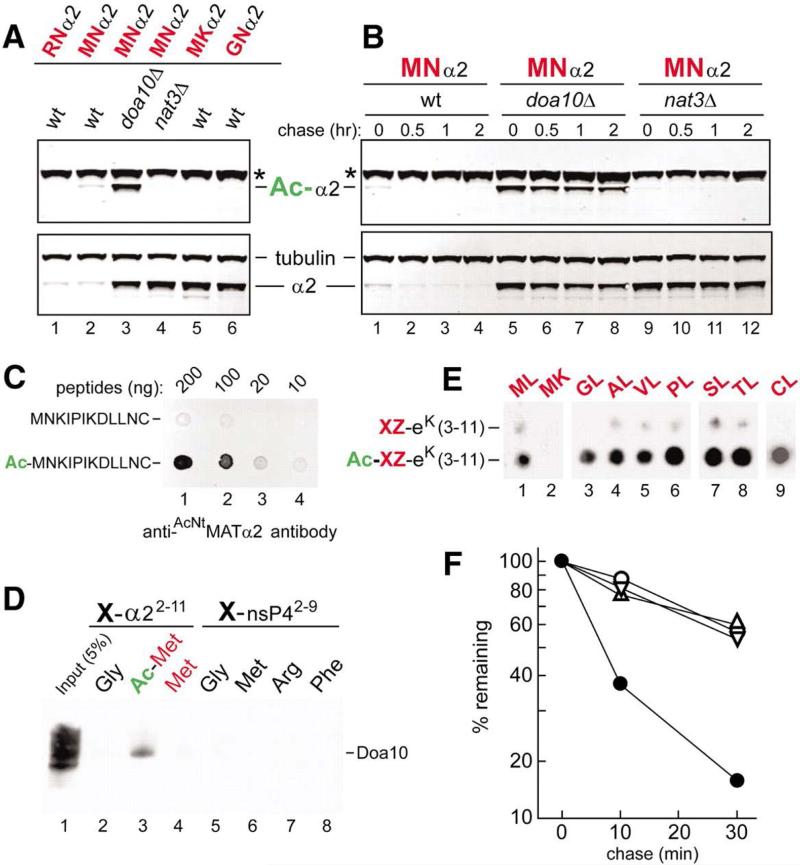
In addition, we produced an antibody, termed anti-AcNtMATα2, that recognized the Nt-acetylated N-terminal sequence of MATα2 (Fig. 2C) and was specific for the Nt-acetylated, ha-tagged, MATα2-derived MN-α23-67-eK-Ura3 reporter, denoted as MNα2 (Fig. 2A, B). Anti-AcNtMATα2 and anti-ha (the latter antibody recognized both Nt-acetylated and unacetylated MNα2) were used to immunoblot extracts of cells that expressed MNα2. Wild-type cells contained barely detectable steady-state levels of either total or Nt-acetylated MNα2 (Fig. 2A), owing to its rapid degradation (see below). By contrast, nat3Δ cells, which lacked the cognate NatB Nt-acetylase, contained high levels of unacetylated MNα2 (detected by anti-ha antibody) and almost no Nt-acetylated MNα2 (Fig. 2A). Similar patterns were observed in wild-type cells that expressed MKα2 (Lys at position 2) or GNα2 (N-terminal Gly) (Fig. 2A). As shown by proteome-scale analyses, S. cerevisiae proteins containing Lys at position 2 are virtually never Nt-acetylated, and few proteins that bear N-terminal Gly are Nt-acetylated (5-7). Most significantly, high levels of Nt-acetylated MNα2 were present in doa10Δ cells (Fig. 2A), owing to metabolic stabilization of Nt-acetylated MNα2 in the absence of Doa10 (see below). These data (Fig. 2A) were in agreement with the LC-MS/MS results that MATα2 was Nt-acetylated (fig. S5A).
Fig. 2.
Doa10 as an N-recognin. (A) Extracts from wild-type, doa10Δ, and nat3Δ S. cerevisiae that expressed XZ-α23-67-eK-Ura3 (XZα2) (X = Met, Arg, or Gly; Z = Asn or Lys) were immunoblotted with anti-AcNtMATα2 (which selectively recognized Nt-acetylated MNα2) or (separately) with anti-Ha, which recognized both Nt-acetylated and unacetylated MNα2, or with anti-tubulin. XZα2 (“α2”), Nt-acetylated XZα2 (“Ac-α2”), and tubulin are indicated. Asterisks denote a protein cross-reacting with anti-AcNtMATα2. (B) As in (A) but CHX chases for 0, 0.5, 1, and 2 hours with MNα2, in wild-type, doa10Δ, and nat3Δ cells. (C) Indicated amounts of the Ntacetylated Ac-MNKIPIKDLLNC peptide versus its unacetylated counterpart were spotted onto membrane and assayed for their binding to anti-AcNtMATα2. (D) X-peptide pulldown with peptides XNKIPIKDLLNC (X = Met, AcMet, or Gly) (lanes 2 to 4) or XIFSTDTGPGGC (X = Gly, Met, Arg, or Phe) (lanes 5 to 8) and extract of S. cerevisiae that expressed Doa10myc13. Lane 1, input extract (5%). (E) SPOT assay with purified, flag-tagged Doa10f and spot-arrayed synthetic peptides XZ-eK(3-11) (X = Gly, Ala, Val, Pro, Ser, Thr, or Cys; Z = Leu or Lys) and their Nt-acetylated XZ-eK(3-11)counterparts. XZ residues are indicated at the top of the membrane. (F) Quantitation, using a PhosphorImager, of 35S-pulse chases with MATα2f and its mutant derivatives (fig. S6, B and C). Solid circles, MNMATα2f; open circles,MKMATα2f; upright triangles, GNMATα2f (initially MGNMATα2f) in ubc4Δ cells; inverted triangles, MNMATα2f in ubc4Δ doa10Δ cells.
To determine whether the Doa10 Ub ligase recognizes the Nt-acetylated Met (AcNtMet), we employed the X-peptide assay (15) with synthetic peptides XNKIPIKDLLNC (X=Met, AcMet, Gly). Except for C-terminal Cys and the varied N-terminal residues, these peptides were identical to the N-terminal region of MATα2. Immobilized peptides were incubated with extract from yeast that expressed myc13-tagged Doa10, followed by elution of the bound proteins and immunoblotting with anti-myc antibody. Doa10myc13 bound to the MATα2 peptide with N-terminal AcNtMet but not to the otherwise identical peptides with unmodified N-terminal Met or with N-terminal Gly (Fig. 2D). Additional controls, which did not bind to Doa10myc13, were peptides XIFSTDTGPGGC (X=Gly, Met, Arg, Phe) derived from the N-terminus of nsP4, a Sindbis viral protein (13) (Fig. 2D). Thus Doa10 recognizes the AcNtMet residue and does not have a significant affinity for downstream sequences of MATα2 or nsP4.
Doa10 specificity was also analyzed using the SPOT technique, in which synthetic XZ-eK(3-11) peptides and their Nt-acetylated AcXZ-eK(3-11) counterparts were C-terminally linked to a membrane as “dots” in equal molar amounts. SPOT peptides were identical to the N-terminal region of eK (fig. S1D), with varied residues at positions 1 and 2. A SPOT assay with C-terminally flag-tagged Doa10f indicated the recognition of AcNtMet by Doa10, in agreement with the results of the X-peptide assay (Fig. 2D, E). SPOT also indicated a highly preferential binding of Doa10f to other Nt-acetylated (versus unacetylated) AcXZ-eK(3-11) peptides (X=Gly, Ala, Val, Pro, Ser, Thr, Cys), including Nt-acetylated Gly and Pro (Fig. 2E). Thus, Gly and Pro are (largely) stabilizing in the N-end rule (Fig. 1D and fig. S3E) because N-terminal Gly and Pro are Nt-acetylated in relatively few proteins (5-7). Interestingly, Doa10 did not bind to N-terminal AcNtMet if it was followed by Lys at position 2 (Fig. 2E). Thus, the metabolic stability of XK-eK-Ura3 (X=Met, Cys) containing Lys at position 2 (e.g., Fig. 1A, C) stems not only from the absence of Nt-acetylation (5-7) but also from the rejection, by Doa10, of Lys at position 2 (Fig. 2E).
The Doa10-dependent AcN-degron of MATα2
Taking advantage of the specificity of anti-AcNtMATα2 for Nt-acetylated MNα2, we performed CHX-chases as well, in addition to steady-state assays (Fig. 2A, B). MNα2 was short-lived in wild-type cells. Even “time-zero” samples, at the time of addition of CHX, contained barely detectable levels of either Nt-acetylated or total MNα2 (Fig. 2B). By contrast, MNα2 was a long-lived protein in doa10Δ and nat3Δ cells, but for different reasons: in doa10Δ cells, which lacked the cognate Ub ligase, MNα2 was long-lived despite its Nt-acetylation, whereas in nat3Δ cells, which lacked the cognate Nt-acetylase, the largely unacetylated MNα2 was long-lived because the targeting by Doa10 required Nt-acetylation (Fig. 2B).
MATα2 contains yet another degradation signal, targeted by an unknown E3 in conjunction with the Ubc4 and (to a minor extent) Ubc5 E2s (25, 26). This degron is nearly inactive in ubc4Δ cells (23, 26). By contrast, the Doa10 Ub ligase functions with the Ubc6/Ubc7 E2s and remains active in ubc4Δ cells (22). In 35S-pulse-chases with C-terminally flag-tagged full-length MATα2f, the rapid degradation of wild-type MNMATα2f in ubc4Δ cells (t1/2 ≈ 9 min) was substantially decreased in doa10Δ ubc4Δ cells (t1/2 ≈ 35 min) (Fig. 2F and fig. S6B, C). A Lys residue at position 2 in a polypeptide chain is known to preclude Nt-acetylation in S. cerevisiae, and few proteins that bear N-terminal Gly are Nt-acetylated (5, 6). The absence of Nt-acetylation in MKMATα2f (Lys at position 2) or GNMATα2f (Gly at position 1) decreased the rate of MATα2 degradation in wild-type cells (Fig. 2F and fig. S6B, C). Most tellingly, the extent of this decrease, in comparison to degradation of Nt-acetylated MNMATα2f in wild-type cells, was indistinguishable from the decrease of MNMATα2f degradation in doa10Δ cells, which lacked the Doa10 Ub ligase (Fig. 2F and fig. S6B-E). In addition to indicating that the sole degron targeted by Doa10 in MATα2 is its AcN-degron, these results were also in agreement with technically independent evidence that utilized the anti-AcNtMATα2 antibody to prove that the Nt-acetylation of MNα2 was required for its targeting by Doa10 (Fig. 2A-C).
AcN-degrons in cellular proteins
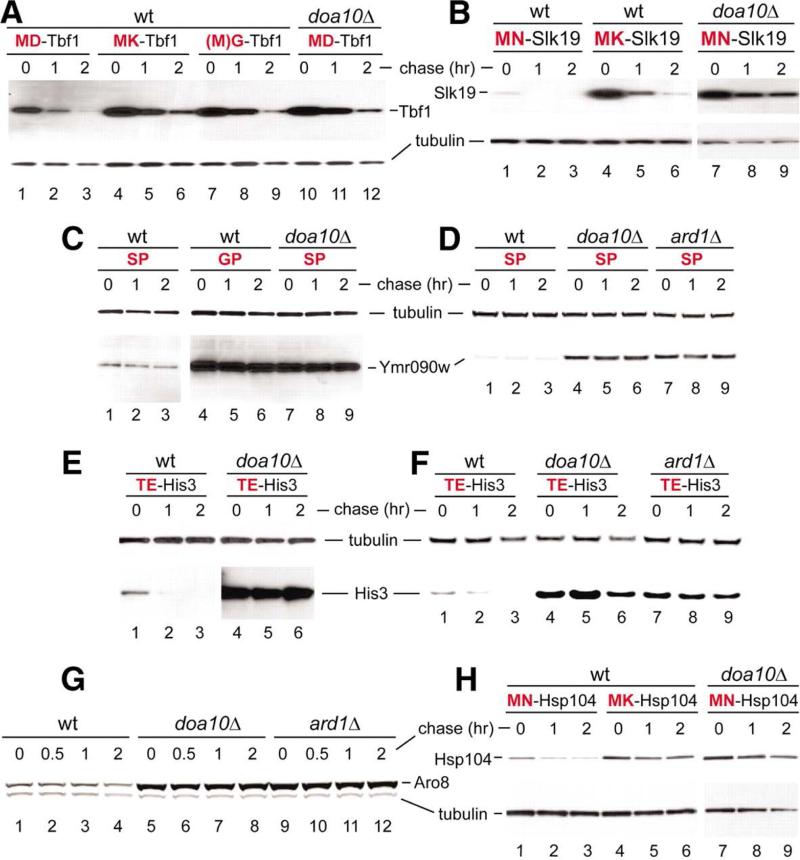
As expected, given the presence of AcN-degron in MATα2, both full-length MATα2f and MNα2 were strongly stabilized in nat3Δ cells, which lacked the cognate NatB Nt-acetylase (Fig. 2A, B and fig. S6D, E). Besides MATα2, our survey of S. cerevisiae proteins has encompassed, thus far, Tbf1, a regulator of telomeres; Slk19, a regulator of chromosome segregation; Ymr090w, a cytosolic protein of unknown function; His3, an enzyme of histidine biosynthesis; Pop2, a subunit of mRNA-deadenylating complexes; Hsp104, a chaperone; Tho1, an RNA-binding regulator; Ubp6, a deubiquitylating enzyme; and Aro8, an aromatic aminotransferase (Fig. 3, fig. S2C, D, and fig. S7) (29).
Fig. 3.
AcN-degrons in yeast proteins. (A) Lanes 1 to 3, CHX chase for 0, 1, and 2 hours in wild-type S. cerevisiaeexpressing Tbf1ha. Lanes 4 to 6, 7 to 9, and 10 to 12, analogous patterns but in doa10Δ cells withMKTbf1ha (Lys at position 2), GTbf1ha(initially MGTbf1ha), and wild-type Tbf1ha, respectively. (B) As in (A) but with wild-type Slk19ha and its mutant derivatives in wild-type versus doa10Δ cells. (C) As in (A) but with wild-type Ymr090wha and its mutant derivatives in wild-type versus doa10Δ cells. (D) As in (C) but with wild-type Ymr090wha in wild-type versus doa10Δ and ard1Δ cells. (E) As in (A) but with wild-type His3ha in wild-type versus doa10Δ cells. (F) As in (E), with wild-type His3ha in wild-type versus doa10Δ and ard1Δ cells. (G) As in (A) but CHX chases for 0, 0.5, 1, and 2 hours with wild-type Aro8hain wild-type versus doa10Δ and ard1Δ cells. (H) As in (A) but with wild-type Hsp104ha and its mutant derivatives in wild-type versus doa10Δ cells.
Wild-type Tbf1, Slk19, Pop2, Hsp104, Tho1, Ubp6 and Aro8 are known to be Nt-acetylated (5, 6). In contrast, the testing of His3 and Ymr090w stemmed from our 2-D electrophoretic analyses, including 35S-pulse-chases. The resulting patterns contained a number of protein spots with significantly higher levels of 35S in samples from doa10Δ versus wild-type cells (fig. S8). We examined three of these spots using MALDI-MS fingerprinting techniques and identified His3, Ymr090w, and Aro8 as putative substrates of Doa10 (fig. S8). The testing for AcN-degrons in Tbf1, Slk19, Ymr090w, His3, Pop2, Hsp104, Tho1, Ubp6 and Aro8 (this analysis included second-residue mutants of some of these proteins) involved CHX-chases in the presence versus absence of a cognate Nt-acetylase or the Doa10 Ub ligase. As shown in Fig. 3, fig. S2C, D and fig. S7, we identified AcN-degrons in all of these proteins (in addition to MATα2), except Pop2 and Tho1 (see also (29)).
Discussion
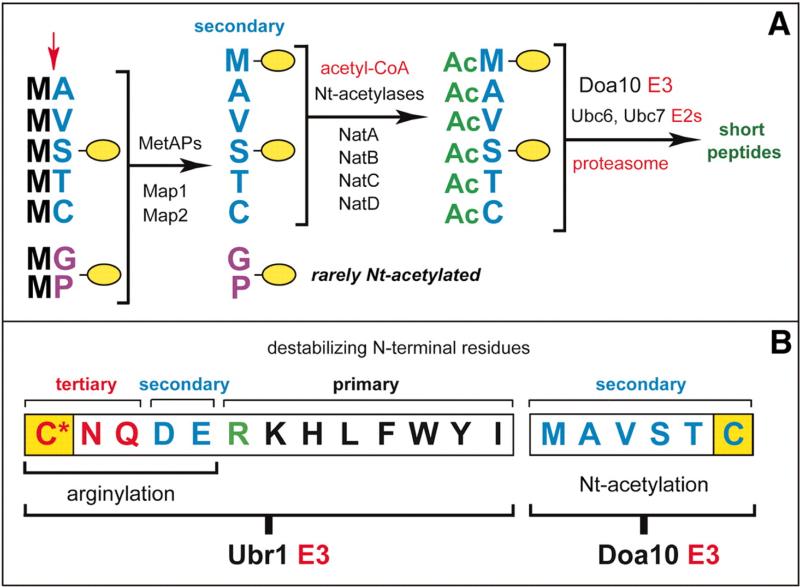
Our results, summarized in Fig. 4, revealed the function of Nt-acetylation, producing the largest increase in the scope of the N-end rule pathway since its discovery more than two decades ago (10-13). At present, only ~10 proteins in all eukaryotes have been identified that require, or are inferred to require, Nt-acetylation for their in vivo roles, which are unrelated to protein degradation (ref. (29) and refs. therein). In contrast, the creation of degradation signals by Nt-acetylation (Fig. 4) is relevant, in principle, to all Nt-acetylated proteins. N-terminal Met, Ala, Val, Ser, Thr, and Cys are shown here to function as secondary destabilizing residues in the N-end rule pathway, in that they must be Nt-acetylated before their recognition by the S. cerevisiae Doa10 Ub ligase as N-degrons, termed AcN-degrons, that require Nt-acetylation (Fig. 4). Out of 20 amino acids in the genetic code, 18 are now known to function as destabilizing N-terminal residues in the N-end rule pathway (Fig. 4 and fig. S1C). More than 50% of proteins in S. cerevisiae and more than 80% of proteins in human cells are Nt-acetylated (5-7). Thus, remarkably, the majority of eukaryotic proteins harbor a specific degradation signal from the moment of their birth. Putative metazoan counterparts of the yeast Doa10 Ub ligase (22-24) include human TEB4 (30), indicating the likely relevance of our results to all eukaryotes.
Fig. 4.
Nα-terminal acetylases, Met-aminopeptidases, and the Doa10 branch of the N-end–rule pathway. (A) The Doa10-mediated branch of the S. cerevisiaeN-end–rule pathway (see fig. S1C for the Ubr1-mediated branch of this pathway). The red arrow on the left indicates the MetAP-mediated removal of N-terminal Met. This Met is retained if a residue at position 2 is nonpermissive (too large) for MetAPs. If the retained N-terminal Met or N-terminal Ala, Val, Ser, Thr, and Cys are followed by acetylation-permissive residues, the above N-terminal residues are usually Ntacetylated (5–7). The resulting N-degrons are termed AcN-degrons. The term “secondary” refers to the necessity of modification (Nt-acetylation) of a destabilizing N-terminal residue before a protein can be recognized by a cognate Ub ligase (fig. S1C). Proteins containing AcN-degrons are targeted for ubiquitylation (and proteasome-mediated degradation) by the Doa10 E3 Ub ligase. Although Gly or Pro can be made N-terminal by MetAPs, and although Doa10 can recognize Ntacetylated Gly and Pro (Fig. 2E), few proteins with N-terminal Gly or Pro are Nt-acetylated (5– 7). (B) The Ubr1 and Doa10 branches of the N-end–rule pathway. Both branches target, through different mechanisms, the N-terminal Cys residue (yellow rectangles), with oxidized Cys marked by an asterisk.
The Nt-acetylation is largely cotranslational, apparently irreversible, and involves a majority of cellular proteins. What functions are subserved by such a massive production of degradation signals (AcN-degrons) in nascent proteins if many of these proteins are destined for long half-lives? We suggest that a major role of these degradation signals involves quality control mechanisms and regulation of protein stoichiometries in a cell. A key feature of such mechanisms would be conditionality of AcN-degrons. If a nascent Nt-acetylated protein can fold its N-terminal domain rapidly enough, or if this protein either interacts with a “protective” chaperone such as Hsp90 or becomes assembled into a cognate multisubunit complex, the cotranslationally created AcN-degron of this protein may become inaccessible to the Doa10 Ub ligase. Consequently, the degradation of this protein would be decreased or precluded. In contrast, delayed or defective folding of a protein's N-terminal domain (because of oxidative, heat or other stresses, or a conformation-perturbing mutation, or non-stoichiometric levels of cognate protein ligands) would keep an AcN-degron exposed (active) and thereby increase the probability of the protein's destruction.
The discovery that Nt-acetylation is a part of the N-end rule pathway (Fig. 4) has also revealed the physiological functions of Nt-acetylases and Met-aminopeptidases. Nt-acetylases produce AcN-degrons, while the upstream Met-aminopeptidases make possible these degradation signals, all of them except the one mediated by Nt-acetylated Met (Fig. 4). Nt-acetylases and Met-aminopeptidases are universally present, extensively characterized and essential enzymes whose physiological roles were largely unknown. These enzymes are now functionally understood components of the N-end rule pathway (Fig. 4 and fig. S1C).
Although the bulk of Nt-acetylation is cotranslational (4), posttranslational Nt-acetylation is likely to be extensive as well. A number of proteases can specifically cleave a variety of intracellular proteins, resulting in C-terminal fragments that often bear destabilizing N-terminal residues of the Ubr1-mediated branch of the N-end rule pathway (fig. S1C). Such fragments are often short-lived in vivo, thereby regulating specific circuits (reviewed in (13)). Given the major expansion of the N-end rule in the present work (Fig. 4), most in vivo-produced C-terminal fragments of intracellular proteins should now be viewed, a priori, as putative targets of the Doa10 or Ubr1 branches of the N-end rule pathway.
The topologically unique location of N-terminal residues, their massive involvement in proteolysis, and their extensive modifications make N-degrons a particularly striking example of the scope and subtlety of regulated protein degradation (Fig. 4 and fig. S1C).
Supplementary Material
Acknowledgments
We thank J. Zhou (Caltech) for MS analyses, and C. Brower for comments on the paper. We are grateful to members of the Varshavsky laboratory for their advice in the course of this study, and particularly thank O. Batygin for her technical assistance. This work was supported by grants from the NIH and the March of Dimes Foundation to A.V.
References and Notes
- 1.Jörnvall H. J. Theor. Biol. 1975;55:1. doi: 10.1016/s0022-5193(75)80105-6. [DOI] [PubMed] [Google Scholar]
- 2.Mullen JR, et al. EMBO J. 1989;8:2067. doi: 10.1002/j.1460-2075.1989.tb03615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park EC, Szostak JW. EMBO J. 1992;11:2087. doi: 10.1002/j.1460-2075.1992.tb05267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gautschi M, et al. Mol. Cell. Biol. 2003;23:7403. doi: 10.1128/MCB.23.20.7403-7414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polevoda B, Sherman F. J. Mol. Biol. 2003;325:595. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 6.Arnesen T, et al. Proc. Natl. Acad. Sci. USA. 2009;106:8157. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goetze S, et al. PLoS Biol. 2009;7:e1000236. doi: 10.1371/journal.pbio.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frottin F, et al. Mol. Cell. Proteomics. 2006;5:2336. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Mayer A, Siegel NR, Schwartz AL, Ciechanover A. Science. 1989;244:1480. doi: 10.1126/science.2544030. [DOI] [PubMed] [Google Scholar]
- 10.Bachmair A, Finley D, Varshavsky A. Science. 1986;234:179. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 11.Bachmair A, Varshavsky A. Cell. 1989;56:1019. doi: 10.1016/0092-8674(89)90635-1. [DOI] [PubMed] [Google Scholar]
- 12.Varshavsky A. Proc. Natl. Acad. Sci. USA. 1996;93:12142. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varshavsky A. J. Biol. Chem. 2008;283:34469. doi: 10.1074/jbc.X800009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogk A, Schmidt R, Bukau B. Trends Cell Biol. 2007;17:165. doi: 10.1016/j.tcb.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Tasaki T, Kwon YT. Trends Biochem. Sci. 2007;32:520. doi: 10.1016/j.tibs.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Hu R-G, et al. Nature. 2005;437:981. doi: 10.1038/nature04027. [DOI] [PubMed] [Google Scholar]
- 17.Hu R-G, Wang H, Xia Z, Varshavsky A. Proc. Natl. Acad. Sci. USA. 2008;105:76. doi: 10.1073/pnas.0710568105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang C-S, Varshavsky A. Proc. Natl. Acad. Sci. USA. 2008;105:19188. doi: 10.1073/pnas.0808891105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang C-S, Shemorry A, Varshavsky A. Proc. Natl. Acad. Sci. USA. 2009;106:2142. doi: 10.1073/pnas.0812316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Piatkov KI, Brower CS, Varshavsky A. Mol. Cell. 2009;34:686. doi: 10.1016/j.molcel.2009.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varshavsky A. Meth. Enzymol. 2005;399:777. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 22.Swanson R, Locher M, Hochstrasser M. Genes Dev. 2001;15:2660. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deng M, Hochstrasser M. Nature. 2006;443:827. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- 24.Ravid T, Kreft SG, Hochstrasser M. EMBO J. 2006;25:533. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hochstrasser M, Varshavsky A. Cell. 1990;61:697. doi: 10.1016/0092-8674(90)90481-s. [DOI] [PubMed] [Google Scholar]
- 26.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Cell. 1993;74:357. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 27.Johnson AD. Curr. Op. Genet. Dev. 1995;5:552. doi: 10.1016/0959-437x(95)80022-0. [DOI] [PubMed] [Google Scholar]
- 28.Zill OA, Rine J. Genes Dev. 2008;22:1704. doi: 10.1101/gad.1640008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Supporting material on Science Online.
- 30.Hassink G, et al. Biochem. J. 2005;388:647. doi: 10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.