Abstract
Cellular mechanisms of stress sensing and signaling represent the initial plant responses to adverse conditions. The development of high-throughput “Omics” techniques has initiated a new era of the study of plant molecular strategies for adapting to environmental changes. However, the elucidation of stress adaptation mechanisms in plants requires the accurate isolation and characterization of stress-responsive proteins. Because the functional part of the genome, namely the proteins and their post-translational modifications, are critical for plant stress responses, proteomic studies provide comprehensive information about the fine-tuning of cellular pathways that primarily involved in stress mitigation. This review summarizes the major proteomic findings related to alterations in the wheat proteomic profile in response to abiotic stresses. Moreover, the strengths and weaknesses of different sample preparation techniques, including subcellular protein extraction protocols, are discussed in detail. The continued development of proteomic approaches in combination with rapidly evolving bioinformatics tools and interactive databases will facilitate understanding of the plant mechanisms underlying stress tolerance.
Keywords: wheat, proteomics, abiotic stress, review
Introduction
Wheat is one of the major food crops worldwide, and the glutens and storage proteins in wheat grain are the single greatest source of protein in the human diet (Gill et al., 2004). In particular, the wheat species Triticum aestivum L. provides one-fifth of the total calories the world's population (Reynolds et al., 2010). To further improve wheat yields, it is necessary to develop varieties of wheat that can be managed using methods that preserve local environments and natural resources (Ribeiro et al., 2013). To meet this challenge, the integration of wheat genomics, transcriptomics, and proteomics with rapidly evolving bioinformatics tools and interactive databases is required.
The genome of common wheat is large (17 Gb) (Safár et al., 2010) and complex due to numerous polyploidy events that occurred between 8000 and 10,000 years ago (Gupta et al., 2008; Brenchley et al., 2012). The wheat genome is essentially comprised of the DNA of three different primitive species, which may explain the great capacity of wheat plants to adapt to various ecological conditions (Brenchley et al., 2012). The sequencing of the wheat genome is enabling a more effective and focused approach to the breeding of high-yielding varieties with increased tolerance to environmental stresses. The International Wheat Genome Sequencing Consortium recently published a chromosome-based draft sequence of the bread wheat genome (Brenchley et al., 2012), an accomplishment that is expected to facilitate the breeding of varieties that are tolerant to the biotic and abiotic stresses that cause yield losses. However, because knowledge of a genomic sequence alone does not indicate how a plant interacts with the environment, and not all open reading frames correspond to a functional gene (Ribeiro et al., 2013), proteomics approaches are critical for understanding plant mechanisms of stress tolerance.
The present review highlights the major proteomic findings in studies examining wheat acclimation responses to abiotic stresses. Moreover, the strengths and weaknesses of different sample preparation techniques, including subcellular protein extraction protocols, are discussed. The continued development and application of these proteomics techniques will provide new insights into the underlying mechanisms of stress tolerance in wheat.
Sample preparation techniques
Total protein extraction
Sample preparation is the most crucial aspect of proteomics analysis. The preparation of total proteins from plants is considerably more challenging compared to other organisms due to the abundance of plant proteases and other compounds such as polyphenol, polysaccharides, starch, lipids, and secondary metabolites, which interfere with protein detection by causing proteolytic breakdown, streaking and charge heterogeneity. Moreover, certain tissues contain highly abundant proteins that hamper the isolation, separation, visualization, and accurate identification of the complete proteome. For example, the presence of the extremely abundant photosynthetic CO2 fixation enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) in leaves not only limits the dynamic resolution of low-abundance target proteins, but also impairs the detection of other proteins and affects the electrophoretic mobilities of neighboring protein species (Herman et al., 2003). Different fractionation techniques based upon the physiological or biochemical properties of RuBisCO have been used to reduce or remove this enzyme by polyethylene glycol and DTT from total leaf protein extracts (Kim et al., 2001; Cho et al., 2008; Widjaja et al., 2009). For example, an affinity column containing anti- RuBisCO large subunit antibody and protein A-Sepharose as a resin effectively eliminated RuBisCO from protein extracts of rice chloroplasts (Hashimoto and Komatsu, 2007). Krishnan and Natarajan (2009) developed a relatively fast and simple fractionation technique using 10 mM calcium and 10 mM phytate to precipitate 85% of the total RuBisCO from soluble protein extract of soybean leaf. These techniques can also be used for the preparation of wheat proteins less contaminated with RuBisCO.
In contrast, accurate quantitation of RuBisCO itself is another challenge for proteomics study of plant stress response. Capillary electrophoresis is often used as an effective method for recovery of RuBisCO from the crude plant extract. In this process RuBisCO extracted from plant leaves is first completely denatured into small and large subunits in presence of SDS. The SDS-RuBisCO complexes are then separated by using an uncoated fused-silica capillary filled with a replaceable polymer solution, with detection at 220 nm (Chen et al., 2000). However, presence of phenolic compounds in plant extract often limits the use of this method. Interestingly, addition of insoluble polyvinylpolypyrrolidone during leaf extraction was found to be effective in effective removal of phenols thus, allowing accurate quantitation of RuBisCO (Warren et al., 2000). Leech and Marrison (1996) exploited cyto-immunofluorescence technique for accurate quantitation of RuBisCO per chloroplast in wheat. In this process, PEG-embeded transverse leaf sections were first hybridized with RuBisCO antisera followed by a secondary antibody conjugated to fluorescein isothiocyanate (FITC). The fluorescence output was measured against the standard curve prepared through rocket immunoelectrophoresis. In this method, RuBisCO levels in the leaf tissue were quantified by measuring rocket peak heights and then comparing these with the known concentrations of purified wheat RuBisCO protein (Laurell, 1966).
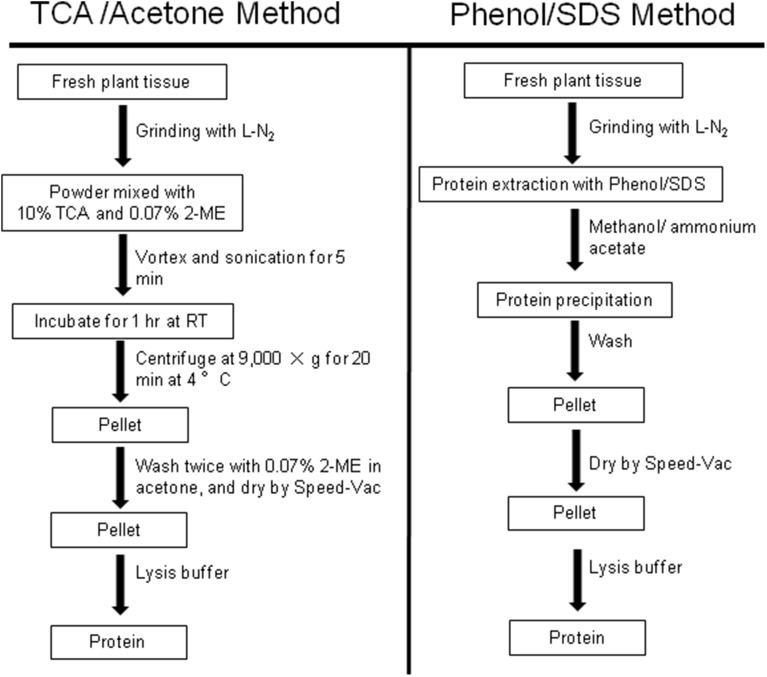
Standardization of sample preparation protocols to optimize protein yields and overcome the physicochemical limitations inherent to most techniques (Table 1). The recently developed trichloroacetic acid/acetone precipitation method (Kim et al., 2010; Shin et al., 2011) has been shown in wheat to be highly reproducible compared to the other methods (Figure 1). Although the final protein pellet is occasionally resistant to solubilization in this method, protein contaminants and plant pigments are effectively removed under acidic and/or hydrophobic conditions, resulting in high-quality gels. Another popular approach for total protein preparation is phenol extraction, which has a strong capacity for the removal of contaminants (Figure 1) (Bancel et al., 2010). In this protocol, it is critical that the sample is kept at very low temperature during the extraction, and the phenolic phase must be carefully recovered after each centrifugation step. The efficient solubilization of extracted proteins is an important step in all proteomic sample preparation methods to achieve sufficient protein concentrations. Moreover, the selection of the most suitable protocol is dependent on protein abundance, molecular weight, charge, hydrophobicity, post-translational processing and modifications, and presence of inhibitory molecules. To date, however, no specific protocol has been developed that is effective for all protein extractions.
Table 1.
Summary of wheat proteome analyses examining plant responses to abiotic stresses and others.
| Stress/Conditions | Treatment time and dose | Cultivar | Organ/Organelle | Proteomic technologies | Stress induced modulation of metabolic pathways | Differentially expressed protein classification | References | |
|---|---|---|---|---|---|---|---|---|
| Functions | Localization | |||||||
| Flooding | 7 d | Bobwhite line SH 9826 | Seminal root | 2-DE, nano LC-MS/MS | Antioxidant defense | StrRes | – | Haque et al., 2011 |
| Flooding | 2 d | Shiroganekomugi | Root | 2-DE, nano LC-MS/MS | Carbohydrate (glycolysis) | EnMet, ProtMet, SigTran, Tranp | Cell wall | Kong et al., 2010 |
| Drought | 100 d | Opata, Nesser | Root | iTRAQ | Energy metabolism, Replication, repair | EnStr, OxiRed, Trans, | Mem,Cyto, Cell wall, Mito, Nucl, Plast, Vacu | Alvarez et al., 2014 |
| Drought | 7 d | Ofanto | Leaf | 2-DE, MALDI-TOF | Carbohydrate (glycolysis, gluconeogenesis) | PTR, StrRes, TCA, ROSsca, AAB, GG | – | Caruso et al., 2009 |
| Drought | 7 d | Katya, Sadovo, Zlatitza, Miziya | Leaf | SDS-PAGE, 2-DE | Energy (photosynthesis) | EnMet, EnvDevS | Chlo | Demirevska et al., 2009 |
| Drought | 9 d | Keumkang | Leaf | 2-DE, MALDI-TOF/TOF | Energy (photosynthesis) | Photo | Chlo | Kamal et al., 2013a |
| Drought | 10, 15, 20, and 25 d | Janz, Kauz | Seed | 2-DE, MALDI-TOF | Carbohydrate metabolism | ROSsca, CarMet, SigTran | – | Jiang et al., 2012 |
| Drought | 14, 24 d | Kukri, Excalibur, RAC87 | Leaf | iTRAQ | Energy (photosynthesis) | Photo, GG, ProtF, Tranp, EnStr | – | Ford et al., 2011 |
| Drought | 20% PEG | Hanxuan 10 and Ningchun 47 | Leaf | nano LC-MS/MS | Antioxidant defense | DRM, SigTran, StrRes, ROSsca | – | Zhang et al., 2014 |
| Heat and Drought | 10 d | Vinjett | Kernel | 2-DE, MALDI-TOF | Carbohydrate (glycolysis) | CarboMet, STP | – | Yang et al., 2011 |
| High temperature | 37°C d, 28°C N/10 d, 20 d | Butte 86 | Endosperm | 2-DE, QSTAR PULSAR-TOF | Carbohydrate metabolism | CarboMet, NitMet, ProtMet, StrRes, STP, SigTran, Tranp, Trans | – | Hurkman et al., 2009 |
| Salt | 150 mM NaCl/1 d, 2 d, 3 d | Keumkang | Leaf | 2-DE, LTQ-FTICR-MS | Energy (photosynthesis) | Photo, StrRes | Chlo | Kamal et al., 2012a |
| Salt | 1.0, 1.5, 2.0, and 2.5% NaCl in HS/2 d | Zhengmai 9023 | Leaf | 2D-DIGE/ Q-TOF-MS | Carbohydrate metabolism | CarMet, ProtF, Tranp, ROS, ATP | – | Gao et al., 2011 |
| Salt | 200 mM | Wyalkatchem, Janz | Shoot | 2-DE, LC-MS/MS | – | – | Mito | Jacoby et al., 2010 |
| Aluminum | 250 μM/2 d, 3 d | Atlas-66, Fredrick | Root | SDS-PGE, Immunoblot | Signaling pathway | Oxi | – | Delisle et al., 2001 |
| Aluminum | 100, 150 μM/5 d | Keumkang | Root | 2-DE, LTQ-FTICR-MS | Energy (glycolysis) | Gly, Tranp, SigTran, StrRes, EnMet | – | Oh et al., 2014 |
| Copper | 100 μM/3 d | Yumai 34 | Root, Leaf | 2-DE, HPLC-Chip/ ESI-Q-TOF/MS/MS | Energy (photosynthesis), antioxidant defense | StrRes, SigTran, ProtMet, CarMet, Photo, EnMet | – | Li et al., 2013 |
| Protein profiling | 20 d | Keumkang | Leaf | SDS-PAGE, LTQ-FTICR | Energy (photosynthesis) | COB, DevPro, DRM, ProtF, ProtMet, StrRes, Tranp, Trans | Chlo | Kamal et al., 2012b |
| Protein profiling | Mature seed | Wild type (AA, BB, DD genome) | Seed | SDS-PAGE, nano LC-MS/MS | Carbohydrate metabolism | StrRes, EnMet, ProtS, CGD, COD, ProtF, SigTran, STP, Tranp | – | Kim et al., 2010 |
| Cadmium | 10, 100, and 200 μM | Yangmai 15 | Leaf | IPG, MALDI-TOF | Energy (photosynthesis) | Oxi, ProtMet, Photo | – | Wang et al., 2011 |
| Cadmium | 0.5 mM/L | Yangmai 13 | Leaf | IPG, MALDI-TOF | Antioxidant defense | ROSsca | – | Ge et al., 2009 |
AAB, amino acid biosynthesis; ATP, ATP synthase; CarMet, carbohydrate metabolism; CGD, cell growth and division; COB, cell organization, COD, cellular organization and development; DevPro, developmental process; DRM, DNA and RNA metabolism; EnMet, energy metabolism; EnStr, environmental stress; EnvDevS, environmental and developmental signals; GG, glycolysis and gluconeogenesis; Gly, glycolysis; NitMet, nitrogen metabolism; Oxi, oxidative stress; OxiRed, oxidation-reduction process; Photo, photosynthesis; ProtF, protein folding; ProtMet, protein metabolism; ProtS, protein synthesis; PTR, post-transcriptional regulation; ROSsca, ROS scavenging; SigTran, signal transduction; STP, storage proteins; StrRes, stress response; TCA, calvin cycle; Tranp, transport; Trans, translation, Chlo, chloroplast; Mem, membrane; Cyto, cytoplasm Nucl, nucleus; Mito, mitochondria; Plast, plastid; Vacu, vacuole; HS, hoagland solution.
Figure 1.
Flowchart showing the steps of plant protein extraction. For trichloroacetic acid (TCA)/acetone, fresh plant tissues are first ground to powder in liquid nitrogen. The powder is transferred to TCA and 2-mercaptoethanol (2-ME) in acetone, vortexed, and the resulting suspension is sonicated. After a 1-h incubation with vortexing, the sample is centrifuged and the obtained pellet is washed twice with 2-ME in acetone. The washed pellet is dried using a Speed-Vac concentrator, and resuspended in lysis buffer for analysis (Kim et al., 2010). For phenol/SDS extraction methods, dried powder of plant tissue is resuspended in phenol/SDS buffer, and the mixture is then vortexed thoroughly. After separating the phenol phase by centrifugation, the upper phenol phase is glycera to fresh tubes. At least 5 volumes of cold methanolic ammonium acetate is added to the phenol phase and the resulting mixture is stored at −20°C. Precipitated proteins are recovered by centrifugation and then washed twice each with cold methanolic ammonium acetate and cold acetone prior to analysis (Bancel et al., 2010).
Subcellular protein extraction
Cell wall purification
The cell wall is an important subcellular organelle for the modulation of stress signals and exhibits changes in protein abundance in response to environmental stress. Cell wall proteins have been extracted and purified from wheat roots under flooding stress (Feiz et al., 2006). The purification of plant cell walls is hampered by a number of technical difficulties such as contamination from other organelles. Thus, characterization of the cell wall proteome remains challenging and requires a combination of various treatment and analytical approaches (Watson et al., 2004). For Arabidopsis, Jamet et al. (2008) described a protocol for purifying soluble and weakly bound cell wall proteins with only low levels of contamination by intracellular proteins, thus providing a more acurate description of protein functions in the apoplast (Jamet et al., 2008). However, extraction methods developed for Arabidopsis may not be useful for other species. A procedure for extracting and purifying cell wall proteins was adopted for wheat seedling roots, and the purity of the cell wall protein extract was confirmed by measuring the activity of glucose-6-phosphate dehydrogenase (Kong et al., 2010). Structural differences in the cell walls between species could have consequences to cell susceptibility to rupture by infiltration techniques, whereas compositional differences in the matrix, such as the content of homogalacturonic acid, might require the use of different extraction buffers for more effective protein release from the matrix. This implies that for the first proteomic studies of a plant species, the level of contamination of the cell wall extract must be monitored carefully and the extraction protocol adjusted accordingly to maximize its content of cell wall proteins (Komatsu and Yanagawa, 2013).
Chloroplast and mitochondria isolation
Photosynthesis in plant, algae, and cyanobacteria uses the energy from sunlight to covert carbon dioxide and water into chemical energy. Chloroplast isolation and purification from various organs or cell types are critical steps for the profiling of the chloroplast proteome. Intact chloroplasts of wheat have been isolated and purified from fully developed leaves on Percoll gradients (D'Amici et al., 2009; Kamal et al., 2012b). On the other hand, mitochondria have been considered the most attractive targets for subcellular proteomics because of the wide range of functions that they perform in cells. According to Jacoby et al. (2010), mitochondria were isolated from shoots in wheat using differential centrifugation followed by polyvinylpyrrolidone-40 gradient. Most of the proteins in the chloroplast envelop membrane and mitochondria membrane is hydrophobic in nature, so that it is technically difficult to analyze them by gel-based proteomic technique. This problem can, however, be resolved by an approach involving protein extraction with organic solvents together with a gel-free proteomic technique.
Plasma membrane purification
In studies involving membrane proteomics, designing suitable method of extraction and identification of entire set of hydrophobic proteins remains a challenge (Komatsu et al., 2009). Isolation of microsomal membrane fraction through “differential centrifugation” is the most common procedure in comprehensive plasma membrane proteomic analysis. The microsomal fraction, collected as the last pellet of ultracentrifuge protocol is the fraction of interest as it usually contains desired membranes such as endoplasmic reticulum, plasma membranes, Golgi apparatus, vacuolar membranes, and different types of endosomal vesicles (Abas and Luschnig, 2010). Using similar ultracentrifuge protocol, Basu et al. (1994) successfully fractionated microsomal membrane proteins from aluminum stressed wheat roots. Ahsan et al. (2012) also effectively isolated root microsomal proteins to examine Cd uptake and translocation in two contrasting Cd-accumulating soybean cultivars. In contrast to the above mentioned conventional ultracentrifugation method, Abas and Luschnig (2010) standardized a method for isolating microsomal-type membranes from Arabidopsis using lower relative centrifugal force of a microcentrifuge. This protocol incorporates specific manipulation of sample density throughout the procedure, with minimal preclearance, minimal volumes of extraction buffer, and minimal sedimentation path length. It also avoids losses during “preclearance” step, thereby ensuring maximal membrane yield.
Flooding-induced changes in wheat proteome composition
Root proteomics of early stage wheat under flooding stress
Cell wall proteins are important both for maintenance of cell structure and for responses to abiotic and biotic stresses; especially flooding stress induced the cell wall loosening in the early stage of plant growth (Komatsu and Yanagawa, 2013). However, neither wheat cell wall proteomics nor any cell wall response to flooding stress identified by proteomics in plants has been previously studied. A procedure for extracting and purifying plant cell wall proteins as adopted for wheat seedling roots, and the purity of the cell wall protein extract was assessed by measuring the activity of glucose-6-phosphate dehydrogenase. To identify flooding-stress responsive proteins in the wheat cell wall, gel-based and mass spectrometry (MS)-based proteomic techniques were applied. A total of 18 and 15 proteins were shown to accumulate in response to flooding by the former and latter proteomic techniques, respectively (Table 1). Among the accumulated proteins detected at lower levels in response to flooding, most were related to the glycolysis pathway and cell wall structure and modification (Kong et al., 2010). In contrast, the cell wall proteins of highest abundance after flooding treatment belonged to the category of defense and disease-response proteins (Kong et al., 2010). In addition to the identified accumulated proteins, a number of root proteins, including methionine synthase, β-1,3-glucanase, β-galactosidase, and β-glucosidase, were decreased in wheat in response to flooding. Among them, methionine synthase plays a pivotal role in methionine synthesis, which is essential for plant cell growth (Huang et al., 2006). The other enzymes, namely β-1,3-glucanases, β-galactosidase, and β-glucosidase belong to the glycosyl hydrolase family of proteins, are involved in the modification of cell wall polysaccharides (Lee et al., 2007). The decrease of these proteins suggests that wheat seedlings respond to flooding stress by restricting cell growth to avoid energy consumption. Thus, by coordinating methionine assimilation and cell wall hydrolysis, cell wall proteins appear to play critical roles in flooding stress acclimation in wheat (Kong et al., 2010).
Proteomics of seminal roots of wheat under flooding stress
Seminal roots of seedlings play an important role in nutrient acquisition. The length and number of seminal roots may be particularly important in the acquisition of immobile nutrients by increasing soil exploration as well as inter-root competition (Zhu et al., 2006). Plant roots are particularly sensitive to oxygen deficit caused by waterlogging because they must absorb oxygen from the soil. In wheat, the seminal root system is the first organ that experiences severe waterlogging (Oyanagi, 2008). Hence, a rapid and well-organized response to the environment is essential in root meristems in low-oxygen soil (Haque et al., 2011). In Table 1, to examine these responses, proteins were extracted from wheat roots under flooding stress, separated by two-dimensional polyacrylamide gel electrophoresis (2-DE), and analyzed by nano-liquid chromatography (LC)-MS/MS. Sixteen proteins in 10 protein spots were significantly changed in the seminal roots of wheat in response to flooding stress (Haque et al., 2011).
Among the wheat root proteins that are increased during flooding stress, ADP-ribosylation factor 1 is known to play a role in vesicular trafficking and induction of phospholipase D activity (Memon, 2004). Vesicular trafficking drives directional root hair tip growth in Arabidopsis (Yoo et al., 2008) and induces the synthesis of matrix and cellulosic polysaccharides during cell wall construction (Lanubile et al., 1997). ADP-ribosylation factor 1 might be associated with alteration of the cell and cell wall structure during aerenchyma formation in roots under hypoxic conditions. The ubiquitin-conjugating enzyme spm2, which targets proteins for degradation in proteasomes via the ubiquitination reaction (Nandi et al., 2006), was also increased in the seminal roots of wheat under flooding conditions. Supermine modulates the expression of genes encoding redox components and is involved in protein folding, secretion, and degradation, and host defense. The increased activity of this enzyme in the seminal roots of wheat exposed to flooding might have an important role in protein degradation during cell degeneration in the process of aerenchyma formation in the root cortex (Haque et al., 2011).
Peroxidases are involved in lignification, suberization, auxin catabolism, wound healing, and defense against pathogen infection. The increase in peroxidaes might be associated with the higher lignin content in the cell wall of root cortex cells under flooding stress during aerenchyma formation (Erdmann et al., 1986), as well as host defense responses (Haque et al., 2011). Pathogenesis-related protein 1.2 has also been implicated as flooding-stress responsive proteins in the roots of wheat. The corresponding gene is specifically expressed in roots, and is up-regulated by rice blast fungus infection and various abiotic stresses, such as drought and salinity (Qu et al., 2006). The accumulation of this protein in the seminal roots of wheat suggests its involvement in adaptive responses to hypoxic conditions (Haque et al., 2011, 2014).
Changes in wheat proteome composition under drought and salinity stresses
Changes in wheat proteome composition under salt stress
Excess soil salinity is an ever-present threat to crop yields, particularly in arid and semi-arid zones. Extreme low temperature in winter wheat at either autumn seedling stage prior to verbalization or early spring crown stage can cause severe crop damage and reduce production (Xu et al., 2013). High temperatures during grain filling might be affected the dough properties and quality characteristics of wheat. Responses to high temperature have been related to changes in protein composition at both quantitative and qualitative levels (Majoul et al., 2003). Na+ and Cl− ions are affected synergistically to salt toxicity in wheat to exert maximum damage to growth. The toxicity is expressed as a reduction in both plant growth and the photosynthetic apparatus (Martin and Koebner, 1995). High levels of soil C1− can interfere with uptake of nitrate, leading to N starvation (Grattan and Grieve, 1992). Salinity, which is primarily due to the presence of excess Na+, is considered to be the single most widespread soil toxicity problem limiting global agricultural production, including wheat Excessive Na+ imparts both ionic and osmotic stresses to plant cells. Acclimation to salinity involves the synthesis of compatible solutes as well as adjustments in ion transport, including ion uptake, extrusion, and sequestration. This kind of proceedings ultimately lead to detoxification and a restitution of cellular homeostasis, allowing for protecting the plant under salinity condition (Chaves et al., 2009). Salinity stress were induced stomatal shutting, response inhibition due to decline descend activity, decreased RuBisCO levels, displacement of critical cations from endo-membranes structures, and swelling and disorganization of the grana lead to reduced photosynthesis (Peng et al., 2009). In addition, high salt concentrations (Na+) may directly affect stomatal conductance by reducing guard cell turgor and intercellular CO2 partial pressure (Kamal et al., 2012a).
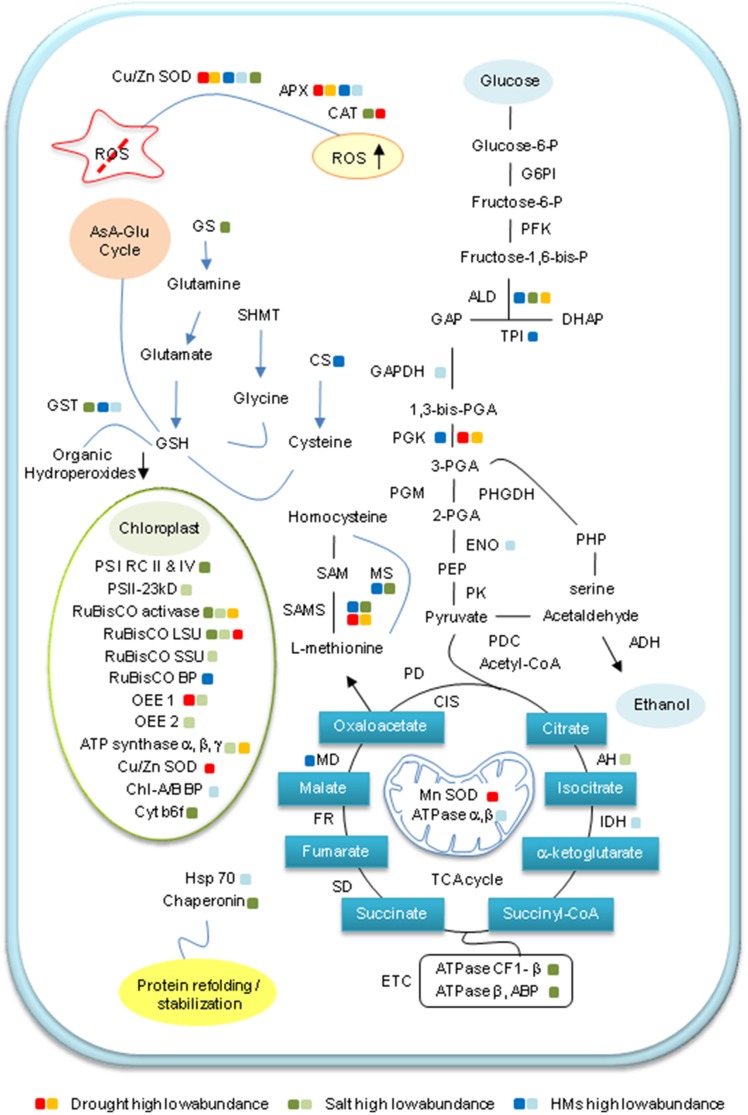
In Table 1, a number of proteomic techniques have been used to identify salinity-induced changes in protein networks and thereby gain insight into wheat salinity stress responses (Wang et al., 2008; Peng et al., 2009; Gao et al., 2011). The impact of salt stress on protein expression patterns in wheat chloroplasts was recently monitored by 2-DE coupled with linear quadruple trap-Fourier transform ion cyclotron resonance (LTQ-FTICR) hybrid MS (Kamal et al., 2012a). Most differentially changed proteins exhibited higher abundances following an initial decrease on exposure to salt stress. This rapid change in protein abundance may represent a counterbalancing effect among the identified proteins. In addition, salinity stress significantly reduced the photosynthesis and transpiration rates, stomatal conductance, relative water content, and levels of Chl a and b, although proline levels were increased. In addition, photosystem I reaction center subunits II and IV, oxygen-evolving proteins (OEEs), and cytochrome b6–f (Cyt b6–f) complex decreased on exposure to salt stress (Figure 2).
Figure 2.
Abiotic stress-induced changes in major metabolic pathways of wheat. The proposed scheme is based on the results of proteomic studies examining changes in the wheat proteome in response to abiotic stress. High- and low-abundance proteins under the three different stress conditions (drought, salt, and heavy metals [HM]) are represented by dark- and light-shaded colored boxes, respectively. ADH, alcohol dehydrogenase; AH, aconitate hydratase; ALD, aldolase; APX, ascorbate peroxidase; AsA-Glu, ascorbate glutathione; CAT, catalase; CS, cysteine synthase; ENO, enolase; GAPDH, glyceraldehydes 3-phosphate dehydrogenase; G6PI, Glucose-6-phosphate isomerase; GS, glutamine synthetase; GSH, reduced glutathione; GST, glutathione-S-transferase; HM, heavy metal; Hsp, heat shock proteins; IDH, isocitrate dehydrogenase; LSU, large subunit; MD, malate dehydrogenase; MS, methionine synthase; OEE, oxygen-evolving enhancer protein; PFK, Phosphofructokinase; PGK, phosphoglycerate kinase; PHP, 3-phosphohydroxypyruvate; PK, pyruvate kinase; PS I, photosystem I; ROS, reactive oxygen species; RuBisCO, ribulose-1,5-bisphosphate carboxylase oxygenase; SAMS, S-adenosylmethionine synthetase; SAM, S-adenosylmethionine; SD, succinate dehydrogenase; SOD, superoxide dismutase; SSU, small subunit; and TPI, triose-phosphate isomerase.
RuBisCO is a stroma-localized protein and constitutes up to 50% of all chloroplast proteins. The expression of RuBisCO in wheat chloroplasts is strongly inhibited by salt treatment. In NaCl-treated rice, 19 different fragments of the RuBisCO large subunit have been detected, suggesting that it might be degraded during stress (Yan et al., 2006). Among differentially expressed chloroplast proteins in salt-stressed wheat seedlings, nucleoprotein might be concerned for chloroplast mRNAs processing (Baginsky and Gruissem, 2002) as well as light-induced activation of translation (Trebitsh and Danon, 2001) was identified. Salt stress treatments have paradoxical impacts on the thylakoid CF1-CF0 complex (ATP synthase). For instance, ATP synthase (α, β, γ, and ε) was modulated by salinity stress in wheat, and maize chloroplasts (Zörb et al., 2009), but changed in unevenly. The α and γ subunits generally increased at initial stage, whereas the ε subunit decreased over time. In contrast, the β subunit was changed dynamically under salt stress in chloroplast of wheat (Kamal et al., 2012a).
Proteome analysis of salt-stressed wheat chloroplasts revealed that five subunits of catalase (CAT), which is well-characterized antioxidant enzyme that protects cells from the toxic effects of hydrogen peroxide, displayed increased levels (Nishikawa et al., 2009). Superoxide dismutase (SOD) and CAT inactivate superoxide radicals (O−2) and H2O2, which are prevented the configuration of the most reactive form of reactive oxygen species (ROS), the hydroxyl radical (-OH). SOD dismutase the superoxide into oxygen and H2O2, while CAT converts H2O2 into water and oxygen. Because these enzymes are responsible for detoxifying ROS, monitoring their activities can be used indirectly to predict ROS production in plant cells (Yang and Poovaiah, 2002). However, the WoLF PSORT sub-cellular predictor indicates that all of the CAT subunits, with the exception of CAT3, are located in chloroplasts (Kamal et al., 2012a).
In addition to CAT, chlorophyll a-b binding proteins (CAB) were unevenly changed under salt stress in wheat chloroplasts (Kamal et al., 2012a). Tsugane et al. (1999) showed in Arabidopsis that expression of the light-harvesting complex, which is normally decreased at the mRNA level, but increased under salinity conditions. The up-regulation might be involved for changing the carbon flux in response resulting decreased the photosynthesis and osmotic adjustment (Li et al., 2011). Consistent with this speculation, the levels of the photosynthetic protein ribulose-bisphosphate carboxylase were decreased by salt stress in chloroplast of wheat, whereas a few proteins increased. Salt concentration and RuBisCo activity were changed reversely. Salt stress increases oxidation and decreases carboxylation activities of RuBisCO, and causes to decrease in severity of CO2 fixation (Sivakumar et al., 2000). RuBisCO activities were decreased by salinity may due to sensitivity of RuBisCO to Cl− (Seemann and Critchley, 1985). The eukaryotic translation initiation factor complexes (eIF 3i, 5A-1/2, and 5A-3) were engaged in protein synthesis, which is stimulated the mRNA binding and methionyl-tRNAi to the 40S ribosome. eIF 5A-1/2, 5A-3, and eIF-3i were dynamically changed under salt stress in chloroplast of wheat (Kamal et al., 2012a). Thus, alteration of the salt concentration in the environment leads to increased or decreased translational activity in a cell or an organism depending on the nature of the stimulus (Burks et al., 2001).
In wheat, aldolases may therefore play a role in acclimating wheat seedlings to anaerobic conditions by reducing oxidative stress (Kamal et al., 2012a). The three isoforms of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) identified to date, GAPA, GAPB, and GAPC, were decreased in wheat seedlings after 3 days of salt stress (Kamal et al., 2012a). In rice, the expression of mRNA for aldolases is enhanced by salt stress (Salekdeh and Komatsu, 2007). For example, fructose-bisphosphate aldolase, which catalyze the cleavage of fructose-1-6-bisphosphate into D-glyceraldehyde-3-phosphate, which is produced from dihydroxyacetone phosphate and ATP. Glyceraldehyde-3-phosphate dehydrogenase (GAPA, GAPB, and GAPC) decreased in chloroplast of wheat seedlings (Kamal et al., 2012a). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) might be involved to abiotic stress by the glycolysis in plant (Hancock et al., 2005). These results are consistent with barley and rice under high-salt stress, whereas GAPDH decreased (Ueda et al., 2006). Sucrose synthase 4 is a membrane-associated form of the cytoplasmic enzymes that synthesize starch and involved in respiration and cell wall protein synthesis under salt stress in chloroplasts of wheat (Pang et al., 2010). Malate dehydrogenase (MDH) also displays a decrease in levels under salt stress in wheat (Kamal et al., 2012a). The chloroplastic NADP-dependent MDH is required for C4 photosynthesis to avoid the less-efficient photorespiration process in plants. NADP-MDH converts oxaloacetate to malate in the chloroplasts of mesophyll cells for its delivery to bundle sheath cells in C4 plants (Cushman, 1993).
ATPases are integral transport proteins that couple the hydrolysis of ATP and movement of protons across membranes to generate electrochemical gradients (Palmgren and Harper, 1999). The action of ATPases can influence salt stress mechanism by changing the membrane potential and proton gradient. The uptake, exclusion, and sequestration of Na+ and other ions are influenced by the above factors. V-type proton ATPase subunits, including VHA-B1, VHA-B2, and VHA-B3, were gradually decreased in chloroplast of wheat (Kamal et al., 2012a). NaCl helps to decrease the plasma membrane ATPases activity in both salt-sensitive and salt-tolerant wheat and tomato (Mansour et al., 2003). Cytoplasmic proteins might have important role for improving the unfavorable effects of high Na+ concentrations by the binding of ions. These changes in pump activities in response to salt exposure are not necessarily adaptive, but may be physiological consequences of salt stress, which causes Na+ ions to interfere with membrane integrity and alter protein activities (Mansour et al., 2003).
The abundance of 3-isopropylmalate dehydrogenase, which is involved in leucine biosynthesis, decreased in wheat chloroplasts after 1 day of salt-stress treatment (Kamal et al., 2012a). In contrast, glutamate dehydrogenase 1 and glutamine synthetase are increased. Glutamine synthetase is associated with the assimilation of enzyme for ammonia, and glutamate dehydrogenases 1 make bridge between carbon and nitrogen metabolism through the amination of 2-oxoglutarate to form glutamate (biosynthetic reaction) or the deamination of glutamate, generating ammonium and 2-oxoglutarate (catabolic reaction). Glutamate dehydrogenase 1 and glutamine synthetase along with other enzymes play vital roles in sustaining the balance between carbon and nitrogen assimilation (Miflin and Habash, 2002). Fumarate hydratase and isocitrate dehydrogenase in the citrate cycle were also unevenly changed in wheat (Kamal et al., 2012a). Isocitrate dehydrogenase supplies NADPH for plant defenses against oxidative stress (Millar et al., 2001) and generates 2-oxoglutarate, which is consumed in the glutamine synthetase–glutamate synthase cycle in carbon-limiting conditions that are experienced by plants under salinity stress (Hodges et al., 2003).
Germin-like protein was dynamically changed in wheat chloroplasts, but had increased at later stage under salt stress (Kamal et al., 2012a). Germin-like proteins are localized in the apoplast that are slackly allied with the cell wall matrix. Germin-like proteins are not only involved in germination, but also in biotic or abiotic stress in plants (Soussi et al., 1998; Schafleitner and Wilhelm, 2002). Germin like proteins has also been associated with SOD activity and shown oxalate oxidase activity, which is often increased by the stress exposure through the production of free-radicals. (Woo et al., 2000). In addition, decreased abundances of photosynthesis-responsive proteins might be the consequence of continuous and highly elevated Na+ ion uptake in the leaf and transpiration stream due to the reduced photosynthetic competence of plants under salt stress (Abbasi and Komatsu, 2004). The 3-oxoacyl-[acyl-carrier-protein] synthase I protein are involved in fatty-acid biosynthesis and was found to be unevenly changed under salt stress in wheat (Kamal et al., 2012a). This protein catalyzes the organic reaction of fatty-acid synthesis by the combining of two carbon acyl acceptor groups from malonyl-acyl carrier protein, resulting the produce the fatty acids from C-10 to C-16 and C-18 (Siggaard-Andersen et al., 1991). Alpha-1, 4-glucan-protein synthase [UDP-forming] is an ancestor of glycogen biosynthesis that is instigated by glycogen initiator synthase to catalyze the transfer of glucose from UDP-glucose to an acceptor protein, which was increased in chloroplast of wheat (Kamal et al., 2013b).
Carbonic anhydrase also was increased in wheat in response to salt treatment. This enzyme has important functions in assisting the transport of carbon dioxide and protons within the intracellular space, across biological membranes, and into the layers of the extracellular space. Besides, carbonic anhydrase is associated with respiration and photosynthesis in eukaryotes by catalyzing the rapid inter-conversion of carbon dioxide and water into carbonic acid, protons, and bicarbonate ions. The switch of bicarbonate to carbon dioxide facilitates its transport into cells, whereas the alteration to bicarbonate assists to trap carbon dioxide inside the cell (Hacisalihoglu et al., 2003).
Several pyridoxal phosphate-binding proteins, including cysteine synthase, PDX1.2, and PDX1.3, were shown to be differentially expressed in wheat chloroplasts in response to salt stress decreased at 2 d while increased at 3 d. These results suggest that pyridoxal phosphate-binding proteins respond to salt stress by inducing cysteine biosynthesis as a protective measure against high ion concentrations (Youssefian et al., 1993). Ferredoxin–NADP reductase and NAD(P)H–quinone oxidoreductase were also induced in salt-stressed wheat chloroplasts, a response that was also reported in tomato leaves (Zhou and Sauve, 2009). S-adenosylmethionine synthase and jasmonate O-methyltransferase generate methyl jasmonate by the methylation of jasmonate that are acted as cellular valve of various developmental courses and defense retorts in Arabidopsis (Seo et al., 2001).
The chloroplast translation elongation factor (EF)-Tu was increased in salt-stressed wheat plants. EF-Tu is a 46-kDa protein that actively binds and transports appropriate codon-specific aminoacyl-tRNAs to the aminoacyl site of the ribosome. EF-Tu from Escherichia coli interacts with unfolded and denatured proteins in a similar manner as molecular chaperones (Caldas et al., 1998). Uroporphyrinogen decarboxylase was increased, as has been observed in rice (Mock and Grimm, 1997). However, this enzyme, which is thought to be a cell death-responsive protein (Zang and Komatsu, 2007), was decreased in the chloroplasts of salt-stressed wheat. Stress-associated protein 7, which contains zinc finger A20 and AN1 domains, was increased in wheat chloroplasts during stress. This pattern was also found in rice and Arabidopsis (Vij and Tyagi, 2006).
The chloroplast protein nudix hydrolase 20, which belongs to a family of proteins that catalyze the hydrolysis of nucleoside diphosphates, exhibits fluctuating levels during stress periods in wheat (Kamal et al., 2012a). In Arabidopsis, levels of nudix hydrolase increase following oxidative stress (Jambunathan and Mahalingam, 2006). Although this protein was originally proposed to have housekeeping-functions such as RNA processing, valve of calcium channel, and directive of ERK signaling (McLennan, 2006). Serine/threonine protein kinase was increased in chloroplast of wheat that has also been connected to biotic stress as well as dehydration (Chinnusamy et al., 2004). This kinase might have important functions in abiotic and biotic stress-signaling pathways. Supporting this speculation, SnRK family (SNF-1 related protein kinases) were also increased or activated under osmotic stress (Veeranagamallaiah et al., 2008).
In Table 1, separation of wheat (cv. Zhengmai 9023) leaf proteins by 2-DE followed by their identification using Q-TOF MS has revealed that salt-responsive proteins are mainly involved in membrane transport, ROS detoxification, ATP synthesis, carbon metabolism, and protein folding (Gao et al., 2011). The higher abundances of H+-ATPases, glutathione S-transferase, ferritin, and triosephosphate isomerase might be responses salt tolerance mechanism at the basis of molecular levels in Zhengmai 9023. Biotic and abiotic stresses often lead to the production of excess amounts of ROS (singlet oxygen, superoxide radical, hydroxyl radical, and hydrogen peroxide) that are protecting the oxidative damage. Hence, ROS (antioxidants and antioxidant enzymes) are associated to suspend the cascades of abandoned oxidation in cell organelles (Shigeoka et al., 2002).
Glutathione S-transferases (GSTs) participated in various cellular metabolisms including scavenging the ROS. GST has been draw attention in stress responses such as pathogen attack, oxidative stress, heavy-metal toxicity, salt stress. Ferritin protein were also increased significantly under salt stress in wheat (Jiang et al., 2007), Arabidopsis (Ndimba et al., 2005), and rice (Parker et al., 2006). It is worked as iron-reserve proteins and plays a defending responsibility against the lethal effects of iron in cells. Ferrous iron and H2O2 produce the hydroxyl radicals through Fenton reactions that are most hazardous type of ROS in plant cells that are potentially neutralized by the ferritin. Triosephosphate isomerase generates the reversible alteration of dihydroxyacetone phosphate to glyceraldehyde-3-phosphate and is involved in many metabolic pathways, including glycolysis, the Calvin cycle, and glycerol metabolism (Kamal et al., 2015). Glucose catabolism might be amplified under salt stress in wheat for supplementary energy for the detoxification and repair of damage caused by oxidative molecules (Gao et al., 2011).
Changes in wheat proteome composition under drought stress
Drought resulting from a shortage of water induces osmotic stress, which is a major limiting factor for plant growth, development and quality crop production (Mohammadi et al., 2012). Osmotic stress causes a reduction in CO2 fixation, thereby decreasing NADP+ regeneration by the Calvin cycle. As a consequence, the photosynthetic electron transport chain becomes over-reduced, leading to the formation of excess ROS, predominantly superoxide radicals and singlet oxygen, which impair the function of chloroplast proteins involved in photosynthesis (Ashraf and Harris, 2013). The mechanisms involved in the drought response have been extensively studied at the protein level using proteomics over the last few decades. However, only a few researcher studies on chloroplast of wheat proteome under drought stress (Kamal et al., 2013a). Redox regulation, oxidative stress response, signal transduction, protein folding, secondary metabolism, and photosynthesis related proteins were respond under drought stress in chloroplast of wheat. The levels of metabolism-related proteins were increased under polyethylene glycol-treated and drought-stressed in leaves of wheat seedlings, whereas the levels of proteins related to energy production and protein synthesis were decreased. Notably, the root was initiated to be the mainly drought-responsive organ, as it displayed the largest changes in protein abundance in response to drought stress (Mohammadi et al., 2012).
Proteins involved in carbon metabolism generally demonstrate marked changes in response to water deficit (Figure 2). Starch biosynthesis and accumulation in endosperm cells primarily occur during the grain filling stage. Among starch-synthesizing enzymes, ADP-glucose pyrophosphorylase is considered to be the rate-limiting enzyme, as it generates the sugar nucleotide ADP-glucose and inorganic pyrophosphate from glucose-1-phosphate and ATP as the first step in the synthetic process (Dai, 2010). In Table 1, wheat grain proteome analysis by linear and non-linear 2-DE and MALDI-TOF MS revealed that both the small and large subunits of ADP-glucose pyrophosphorylase, in addition to ascorbate peroxidase (APX) and G beta-like protein, were down-regulated in drought-sensitive variety Janz, whereas the level of the enzymes in the tolerant variety Kauz did not show any significant changes in response to drought stress (Jiang et al., 2012). In contrast, CAT isozyme 1, WD40 repeat protein, late-embryogenesis subunit (LEA), and alpha-amylase inhibitors displayed higher abundances in Kauz, but were down-regulated or unchanged in Janz under drought conditions. Higher abundance of sucrose synthase together with unaltered levels of ADP-glucose pyrophosphorylase results in higher starch synthesis even under drought conditions. This finding indicates that these enzymes are strongly associated with the higher drought resistance of the Kauz variety of wheat (Jiang et al., 2012).
RuBisCO, the most abundant leaf protein plays a major role in CO2 fixation and photorespiration in C3 plants (Jensen and Bahr, 1977). It constitutes a large pool of stored leaf nitrogen (15–30%) that is rapidly remobilized under stress and senescence (Feller et al., 2008). The activity of RuBisCO is regulated by RuBisCO activase protein, which removes the tightly bound sugar-phosphates from the active centers of RuBisCO, leading to its reactivation (Gutteridge and Gatenby, 1995; Spreitzer and Savucci, 2002). It is considered that the impaired activity of RuBisCO under drought condition may be associated with the reduction in ATP concentration (Tezara et al., 1999). RuBisCO activase is sensitive to the high temperature that is often associated with drought (Crafts-Brandner and Salvucci, 2000).
Drought has been shown to induce metabolic impairment such as decrease in the RuBisCO activity (Tezara et al., 2002; Bota et al., 2004). This declined activity is associated with low stomatal conductance and chloroplast CO2 concentration and is not induced by decreased relative water content under water stress (Flexas et al., 2006). The effects of drought stress on the amount of wheat RuBisCO protein differ among reports, from slightly enhanced to no change (Demirevska et al., 2008). The RuBisCO level also found to be increased during leaf expansion and reached the highest after full leaf expansion (Ishida et al., 1997). However, during leaf senescence, RuBisCO is degraded and its nitrogen is re-mobilized and translocated into growing organs and used for their growth. Studies have indicated that a majority of RuBisCO are degraded within chloroplasts as RuBisCO levels are rapidly decreased in conjunction with the loss of chloroplasts in barley and wheat leaves during senescence (Wardley et al., 1984). Interestingly, magnitude of the decrease in RuBisCO content during senescence was found to be enhanced by exogenous abscisic acid (ABA) in rice leaves (Fukayama et al., 2010). The SDS-dependent proteases induced by ABA might be responsible for senescence related degradation of chloroplast proteins including RuBisCO and RuBisCO activase. Notably, in wheat leaves, both drought and exogenous ABA pretreatment resulted in increased expression of RuBisCO activase (Asghari and Ebrahimzadeh, 2006). A separate study on proteome changes in wild and modern wheat leaves upon exposure to drought also revealed elevated level of RuBisCO both at protein and transcript levels (Budak et al., 2013). When separated on 2D gels, several subunits of RuBisCO are located in different gel areas, and likely represent different splice variants, post-translationally modified isoforms, or cleaved isoforms of the same protein (Weiss and Gorg, 2007). The RuBisCO subunits are unevenly expressed in the chloroplasts of drought-stressed wheat seedlings (Kamal et al., 2013a). Isoforms of RuBisCO activase might have significant roles in alleviating and scheming proteolysis (Schwartz et al., 1995) and in maintaining chloroplast functioning during drought stress (Huo et al., 2004).
H+-transporting two-sector ATPase and membrane-bound ATP synthase subunit b were localized in plasma membranes that are dynamically changed under drought stress in chloroplast of wheat (Kamal et al., 2013a). Non-phosphorylated ATPase is implicated in ion transportation. F-type enzymes in the inner mitochondrial and thylakoid membranes also function in ATP synthesis. Additionally, V- and A-type enzymes contained analogous structure, whereas pump H+ ions are not involved in the synthesis of ATP (Flexas et al., 2006). A number of integral membrane proteins, include members of the chloride carrier/channel (ClC) family, were also increased in wheat exposed to drought stress. The members of the ClC family are response to voltage-regulated in ion channels that provide a range of physiological responsibilities including cell volume regulation, membrane potential stabilization, signal transduction, and trans-epithelial transport (Worden et al., 2009; Ford et al., 2011). ClC family in plants might have function as anion channels for nitrate homeostasis. Analysis using the WolF PSORT sub-cellular location predictor indicates that CLC proteins have 14% similarity with phytoene synthase proteins (Kamal et al., 2013a), which are located in chloroplasts (Römer et al., 1993).
Cytochrome b6-f complex is an enzyme located in the thylakoid membrane that is intervened electron transfer between photo-systems (PS) I and II including cyclic electron flow around PSI and state transitions (Hurt and Hauska, 1981). In wheat plants exposed to drought, the iron-sulfur subunit (petC) of the cytochrome b6-f complex was increased after 3 days of treatment, as has been observed in drought-stressed rice (Ali and Komatsu, 2006). However, petC was decreased after 6 and 9 days of drought stress, which leads to photosynthesis degradation and alters chloroplast structure, as well as promoting leaf senescence (Kamal et al., 2013a). In contrast, the expression of chloroplast OEE1 was increased after 9 days of drought stress. Murota et al. (1994) reviewed the function of OEE1in salt adaptation in photo-autotrophically cultured green tobacco cells, mangrove, and rice (Sugihara et al., 2000).
Adenylate kinase (ADK) is a small ubiquitous enzyme involved in the metabolism of purine nucleotides and is essential for cell maintenance and growth. In drought-stressed wheat, ADK levels were increased after 3 days, but had decreased after 6 and 9 days, possibly due to cell death (Kamal et al., 2013a). As ADK participates in ATP biosynthesis, drought-tolerant genotypes may provide more ATP for maintaining cellular activities under drought stress (Gong et al., 2010). Although plastid stromal ADK plays important roles in the coordination of metabolism and growth, the importance of this isoform is suggested to be tissue-dependent (Carrari et al., 2005). ADK is a key enzyme in energy metabolism, as it catalyzes a reversible transphosphorylation reaction that converts ADP to ATP and AMP, and is critical for many processes in living cells (Pradet and Raymond, 1983).
In Table 1, a shotgun proteomic approach for investigating quantitative changes in protein abundance of two drought-tolerant and one intolerant cultivar of Australian bread wheat revealed a higher abundance of CAT and three isoforms of SOD, namely chloroplastic and cytosolic Cu/Zn-SOD, and mitochondrial Mn-SOD, in response to water deficit stress (Ford et al., 2011). Moreover, a coordinated stress-induced down regulation of proteins involved in photosynthesis and the Calvin cycle was observed, irrespective of cultivar, suggesting this may be a defense strategy to avoid excess ROS generation (Figure 2).
ABA-responsive marker proteins, such as LEA proteins and PP2C family phosphatases, are strongly increased in different plant species in response to ABA and abiotic stresses such as cold and drought and constitute the core proteins of ABA signaling pathways (Vaseva et al., 2010). A total of six LEA proteins, ABA-responsive proteins, and protein phosphatases were identified as ABA-responsive proteins in wheat roots under drought stress (Alvarez et al., 2014). In addition, numerous proteins involved in secondary metabolism, including those related to jasmonic acid, lignin, oxylipin, and phenylpropanoid metabolic processes, are increased in response to ABA treatment in wheat roots under drought stress, (Alvarez et al., 2014).
Drought and high-temperature stress adversely affect wheat seed yields and quality, particularly the composition of seed storage proteins that form during the grain filling stage. Water stress during the grain development stage markedly affects the levels and composition of grain carbohydrates, and storage protein synthesis and accumulation, resulting in poor grain quality (Jiang et al., 2007). However, the content of albumin and gliadin are increased in grains in response to drought, whereas globulin and glutenin do not markedly change (Zhang et al., 2013). Similarly, several α-gliadins, γ-gliadins, and low-molecular-weight glutenins increased in wheat grains exposed to high temperature, whereas α/β-gliadin, ω-gliadin, and globulins decreased (Yang et al., 2011). 1-Cys peroxiredoxin was differentially regulated under drought and high-temperature stress. In wheat exposed to high temperature and drought stress, differentially changed proteins were predominantly involved in stress/defense, signaling pathways, redox regulation, and energy metabolism (Yang et al., 2011). High-temperature stress often increases stomatal conductance, respiration, leaf transpiration, and oxidative stress (Rizhsky et al., 2002), shortens the duration of grain filling, and enhances gluten protein accumulation and starch synthesis (Hurkman et al., 2009). Plants respond to heat by signaling via ABA, ethylene and salicylic acid, scavenging of ROS via the production of antioxidants, and transcriptional activation of stress-related proteins (Wahid et al., 2007). A number of proteins, including heat shock, carbohydrate metabolism, and storage proteins, were changed in the wheat proteome in response to high temperature. Albumin proteins involved in primary metabolism were not changed significantly under high temperature (Yang et al., 2011).
Responses of wheat to heavy metal stresses
Responses of wheat to aluminum stress
Aluminum (Al) toxicity is a major constraint for agricultural crop production on acid soils, which are estimated to comprise over 50% of the world's potentially arable lands, (Yang et al., 2007). Among the various Al toxicity symptoms, the most sensitive response is the inhibition of root elongation. Toxic levels of Al in acid soils hinder root growth and cause a significant reduction in yields of Al-sensitive crops (Dechassa et al., 2011). In addition, Al stress leads to a number of cellular, physiological, and biochemical disorders (Kochian, 1995; Yamamoto et al., 2002; Suping et al., 2009). Based on the assessment off morphological and physiological features, root lengths and weights were shown to be severely reduced by Al stress (150 uM) in wheat, and the Al ion concentration within tissues was significantly increased (Oh et al., 2014). In Table 1, in proteomics-based experiments examining the effects of Al stress on root proteins of wheat, 47 differentially change proteins were identified by LTQ-FTICR MS after separation by 2-DE. Of these proteins, 19 proteins were significantly increased, whereas 28 proteins were decreased (Oh et al., 2014).
β-amylases are exoamylases that release maltose from the non-reducing ends of glucans or dextrins by cleavage of α(1–4) linkages. Although plant β-amylase is considered to be a key enzyme for catalyzing the breakdown of starch, the enzyme was shown to be located outside the plastids, and its role in starch breakdown remains unclear (Bancel et al., 2010). S-adenosylmethionine synthetase is related to the ethylene-mediated inhibition of root growth and may also be involved in the alteration of cell wall structures and polymers in the roots of wheat under Al stress (Fukuda et al., 2007). Oxalate oxidase catalyzes the oxidation of oxalic acid to CO2, Ca2+, and H2O2, and is stored as insoluble calcium salt upon reaction with molecular oxygen (Lane et al., 1993). The Al–induced activation of oxalate oxidase in wheat roots may be involved in detoxifying H2O2 (Delisle et al., 2001). When exposed to Al, tolerant plants secrete organic acid anions to chelate and immobilize Al3+ at the root surface, thereby preventing the excess accumulation of Al3+ in the root system (Kobayashi et al., 2005, 2007). Malate dehydrogenase (MDH) was increased in the roots of wheat under Al stress. MDH catalyzes a reversible reaction that forms malate and NAD from oxaloacetate and NADH, and is therefore a key enzyme in the TCA cycles of prokaryotes and eukaryotes (Ding and Ma, 2004).
Cysteine synthase is a key enzyme for mediating Al tolerance and is increased in the roots of wheat exposed to Al. Cysteine synthase is required for cysteine biosynthesis in plants and is one of the most important enzymes in sulfur assimilation leading to the production of antioxidants and metal chelators, such as glutathione, metallothionein, and phytochelatin (Yang et al., 2007). Triosephosphate isomerase is increased in wheat roots under Al stress, whereas glyceraldehyde-3-phosphate dehydrogenase is decreased in roots (Figure 2). Glyceraldehyde-3-phosphate dehydrogenase catalyzes the conversion of glyceraldehyde-3-phosphate into 1,3-bisphosphoglycerate, which is then converted to 3-phosphoglycerate by the action of phosphoglycerate kinase, which is an important transferase enzyme in glycolysis pathway and is also increased in wheat roots under Al stress (Watson et al., 1982; Zhou et al., 2009). Chloroplast phosphoglycerate kinase is encoded as a polyprotein precursor that is comprised of at least four subunits, which have been separated and shown to be conserved tetrapeptides (Nowitzki et al., 2004).
The activity of APX is increased in response to Al stress and to other abiotic stresses such as salinity, chilling, metal toxicity, drought, and heat. APX is reported to have a possible role in detoxifying H2O2 in the cells of various plant species (Davis and Swanson, 2001; Bueno and Piqueras, 2002). Quinone reductase was differentially changed under AL stress in wheat (Oh et al., 2014). It is a homodimeric FAD-containing enzyme that catalyzes the obligatory NAD(P)H-dependent two-electron reduction of quinones to protect cells against the toxic and neoplastic effects of free radicals and ROS arising from one-electron reductions in tomato under Al stress (Zhou et al., 2009). Methionine synthase was changed under Al stress in leaves of wheat (Oh et al., 2014). It catalyzes the transfer of a methyl group from methyltetrahydrofolate to homocysteine, generating tetrahydrofolate and methionine, which is converted to adenosylmethionine (AdoMet), which serves as a methyl donor in numerous biosynthetic reactions (Matthews et al., 1998). Elongation factor 1-gamma was changed under Al stress in leaves of wheat (Oh et al., 2014) that is involved in translational control by linking the alpha and beta subunits of eukaryotic elongation factor during the GDP to GTP exchange reaction (Thornton et al., 2003). Succinyl CoA ligase, which catalyzes the reversible conversion of succinyl-CoA to succinate, is increased in the roots of wheat under Al stress (Drummond et al., 2001). The formation of a nucleoside triphosphate molecule from an inorganic phosphate molecule and a nucleoside diphosphate molecule were facilitated by this enzyme. Succinyl CoA ligase is also a key catalyst in the citric acid cycle, which occurs in the matrix of mitochondria (Chakrabarty, 1998; Oh et al., 2014).
Plant annexins form a multigene family, are responsive to drought, salt, and cold stresses in Arabidopsis (Cantero et al., 2006). Different isoforms of annexin are reported to possess different enzyme or other protein activities, including phosphodiesterase, peroxidase, F-actin binding, and calcium channel activities. Plant annexins may also participate in the regulation of callose and cellulose synthase activity. Annexins have been implicated in wheat under cold stress (Breton et al., 2002). UDP-d-glucuronate decarboxylase catalyzes the formation of UDP-D-xylose from UDP-D-glucuronate in an irreversible reaction with the help of UDP-D-glucuronate decarboxylase (Zhang et al., 2005). Fructose-bisphosphate aldolase is increased in wheat roots under Al stress (Oh et al., 2014) that are catalyzes the cleavage of fructose 1-6-bisphosphate into the glycolytic intermediates D-glyceraldehyde-3-phosphate and dihydroxyacetone phosphate (Johnson et al., 1989). This enzyme might be facilitated the acclimation to Al through the synthesis of ATP in glycolysis and ethanolic fermentation (Andrews et al., 1994).
Responses of wheat to Cadmium and copper stresses
Cadmium (Cd) is a non-essential element that is highly toxic to plants, even at very low concentrations. Plants readily absorb Cd from the soil through membrane-bound cation transporters, which are primarily responsible for the uptake of essential trace elements (Welch and Norvell, 1999). Within cells, Cd2+ disturbs normal cellular functions by displacing Zn2+, Ca2+, and Fe2+ from proteins, leading to enzyme inactivation and the disruption of vital biochemical pathways related to sugar metabolism, nitrate absorption, and photosynthesis. Plants suffering from Cd toxicity exhibit leaf chlorosis, wilting, growth inhibition, and cell death (Vassilev et al., 1995; Sanità di Toppi and Gabbrielli, 1999; Clemens, 2006). Morphological and physiological analyses of wheat exposed to high Cd concentrations showed that leaf elongation was decreased, and that H2O2 and malondialdehyde (substrate of lipid peroxidation) levels significantly increased (Wang et al., 2011). Further, 2-DE analysis led to the identification of proteins that were differentially changed in response to Cd stress. A number of oxidative stress-related proteins such as APX, glutathione-S-transferase (GST), and SOD are increased in response to Cd accumulation (Figure 2, Table 1). GST plays a role in the oxidative stress response by activating GSH-dependent peroxidases, which is able to detoxify metabolites arising from oxidative damage (Edwards et al., 2000). Additionally, GST is thought to be directly involved in heavy metal detoxification in plant cells by forming complexes with metal ions, such as the GSH-Cd complex (Adamis et al., 2004; Mishra et al., 2009). The increases in APX, SOD, and GST in response to Cd stress might play an important role antioxidant defense.
H+-transporting two-sector ATPase alpha and beta chain was increased by Cd and 1,2,4- Trichlorobenzene (TCB) stresses in leaves of wheat, which is essential to maintain the ATP high level required by the stressed cells. The vacuolar proton-ATPase (V-ATPase) subunit A was increased by Cd and TCB in wheat (Ge et al., 2009) that are plays an important role in plant responses to environmental stresses (Fukuda et al., 2004). V-ATPase uses the energy derived from the hydrolysis ATP to establish the electrochemical gradient of H+ across vacuolar membrane (Sun-Wada et al., 2003), which is also the driving force for the accumulation of toxic ions and other solutes in the vacuole (Hamilton et al., 2001). ACC synthase was increased in leaves of wheat by the accumulation of Cd and TCB (Ge et al., 2009). The induction of ACC synthase can stimulate ethylene production (Wong et al., 2001). Ethylene, as an intermediary of the stressful signal, plays an key defense role such as regulating plant growth and development, inducing the expression of various defensive genes in response to Cd and TCB stresses (O'Donnell et al., 1996).
Copper (Cu) is one of the most hazardous pollutants in agricultural fields. Environmental levels of Cu are rapidly increasing due to excessive use of phosphate fertilizer and irrigation with sewage and water polluted by activities of the metallurgy industry. Cu not only reduces crop yields, but also accumulates in the edible parts of plants, allowing it to easily enter the food chain and cause adverse effects on human health (Cobbett and Goldsbrough, 2002). The results of morphological and physiological studies have demonstrated that plant height, root length, fresh and dry weight, and pigment content of leaves (carotenoid and chlorophyll) are significantly decreased during stress. In addition, the malondialdehyde (MDA) content of roots and leaves increases significantly while Cu contents increased gradually, whereas they having a strong correlation during Cu stress (Li et al., 2013). 2-DE analysis of wheat proteins under copper stress led to the identification of 93 proteins, which were involved in signal transduction, carbohydrate metabolism, protein metabolism, energy production, and transportation, in roots and leaves. Moreover, a number of proteins related to carbohydrate metabolism, protein metabolism, and photosynthesis were specifically changed in the roots or leaves, and included lipoxygenase 2.1 in the leaves and 5, 10- methylenetetrahydrofolate reductase in the roots (Li et al., 2013).
Prospects and challenges
For plants to respond optimally to the ever-changing environment, precise modulation of the proteome is essential for adaptive responses in metabolism and development. Plant adaptive responses to stress need to be decoded in terms of the specific, underlying biological processes. A proteomic approach is useful not only for reconstruction of the plant response to various stresses as a whole, but is also to able to allow different stress responsive pathways to be individually dissected. Most wheat proteomic studies published to date have mainly focused on the effects and plant responses to individual stresses, and proteomic changes in response to multiple stresses remain to be elucidated. However, such an approach would allow a greater understanding of the cross-talk that occurs between different stress signal pathways.
Conventional gel-based proteomic approaches, and gel free-mass spectrometry (MS)-based methods involving label-based and label-free protein quantification have been extensively used with their own advantages and disadvantages for characterization of stress-responsive proteins in wheat (Kong et al., 2010; Oh et al., 2014; Zhang et al., 2014). Gel based approaches are widely used for their simplicity, reproducibility, wide molecular weight coverage, and detection of post-translational modifications (Ghosh and Xu, 2014). Nonetheless, 2-DE approach has some known limitations like issues related to reproducibility, identifying low-abundance and hydrophobic proteins, exceedingly large or small proteins, as well as basic proteins and co-migration of proteins (Hossain et al., 2013). However, for investigation of membrane proteins interactions, blue native PAGE (BN-PAGE), a gel-based approach has become a method of choice for its simplicity and suitability for lipophilic entities. A systematic strategy involving blue native polyacrylamide gel electrophoresis (BN-PAGE) and SDS-PAGE has been designed for the proteomic analysis of wheat chloroplast protein complexes (Meng et al., 2011). Using this method, chloroplast envelopes, thylakoids, and stromata were enriched effectively, and more than 18 complexes were obtained by BN-PAGE.
The inherent limitations of 2-DE can be overcome by the multidimensional protein identification technique (MudPIT), a non-gel method of shotgun proteomics approach. Quantification in MudPIT analysis is usually performed through in vitro labeling techniques such as isotope tags for relative and absolute quantitation (iTRAQ) and isotope coupled affinity tags (ICAT). Exploiting the ICAT labeling method, Islam et al. (2003) successfully unraveled the expression of wheat seed proteins, which were reported to be difficult through 2-DE approach due to co-synthesis of proteins by genes belong to three different genomes namely, A, B, and D. Moreover, metabolic seed protein fraction in durum wheat were analyzed through gel-free shotgun proteomics, that allowed identifying over 380 proteins exhibiting greater molecular weight diversity ranging from 7 to 258 kDa (Fercha et al., 2013).
The accuracy of proteomic data is highly dependent on the sample preparation and analysis techniques, including the isolation, separation, visualization, and identification of the full complement of proteins. Despite recent advances, more emphasis needs to be given to protein extraction protocols, particularly for very low abundance proteins such as transcription factors, kinases, and transporters. Fluorophore-tagged protein immune-precipitation and label-free MS-based quantification techniques have been shown to be superior to the classical 2-DE-based proteomic approach for the accurate identification of low-abundance signaling and regulatory protein complexes (Smaczniak et al., 2012). In addition, advanced tools such as laser-capture micro-dissection (Dembinsky et al., 2007) for tissue proteomics could be adapted for the identification of tissue- and cell-specific proteins involved in plant stress tolerance mechanisms. Moreover, the selected reaction monitoring technique is an emerging MS technique for the targeted and highly sensitive identification and accurate quantitation of very low abundance proteins and modified peptides in complex biological mixtures (Calvo et al., 2011; Picotti and Aebersold, 2012).
The development of effective strategies for post-proteomics or functional proteomics analyses requires an accurate understanding of the effects of post-translational modifications on individual proteins and the interactions between modified proteins and their associated metabolic pathways, in addition to how these pathways are integrated into the cellular metabolic network. In particular, the post-translational protein modifications such as phosphorylation, glycosylation, and oxidation, which are thought to play key roles in stress signaling, require greater understanding at the molecular level. MS-based phosphoproteomic technology has become an invaluable tool for the identification of phosphoproteins and mapping of phosphorylation sites. Nevertheless, identification of the in-vivo protein phosphorylation sites, which is required for the functional characterization of target proteins, is a difficult challenge for any phosphoproteomic study. Immobilized metal affinity chromatography (IMAC) and immunoprecipitation using antibodies against phosphorylated amino acids are the two most common pre-fractionation techniques used in MS analysis (Zhang et al., 2014). Significant recent progress has been made in the quantitative and dynamic analyses of mapped phosphorylation sites. The isolation of phosphopeptides by IMAC followed by MS/MS or MS(n) analysis has enabled the detection of hundreds of new in-vivo phosphorylation sites (Bentem et al., 2006). The more advanced iTRAQ and stable isotope labeling by/with amino acids (SILAC) labeling techniques label peptides in vitro just prior to MS or label proteins during cell growth, respectively, and allow for changes in individual phosphorylation sites to be measured over time in response to stress.
In addition to the development of functional proteomics, a greater focus on subcellular proteomics of wheat is warranted, as organelle proteome studies generate detailed information on the intrinsic mechanisms of plant abiotic stress responses. Isolation of the target organelle from total tissue extracts is one of the most challenging aspects of subcellular proteomics. Conventional methods of subcellular fractionation typically involve differential and density-gradient centrifugation, which use a series of centrifugation steps to separate different cellular compartments or organelles from cell homogenates based on mass and/or density. In contrast, free-flow electrophoresis fractionates organelles based on their net global isoelectric charges or electrophoretic mobilities. Moreover, immunoaffinity purification is a more advanced technique that has been used successfully for the isolation of organelles with high specificity and yields (Ackermann and Berna, 2007). Despite the application of these advanced proteomics techniques, many organelle proteins, including both stress-induced and house-keeping proteins, remain unclassified. Future initiatives aimed at identifying and characterizing organellar proteins are expected to aid in the global proteomic profiling of environmentally stressed plants.
The combination of these proteomic approaches will facilitate characterization of target regulatory proteins and aid in their molecular cloning to address fundamental questions about plant physiology under abiotic stress. Finally, integration of wheat “Omics” data, such as genomics, transcriptomics, proteomics, metabolomics, and interactomics, with rapidly evolving bioinformatics tools and interactive databases is essential to construct mathematical models that will provide a more comprehensive picture about the mechanisms underlying plant performance under adverse conditions.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Glossary
Abbreviations
- 2-DE
two-dimensional polyacrylamide gel electrophoresis
- IEF
isoelectric focusing
- pI
isoelectric point
- IPG
immobilized pH gradient
- iTRAQ
isobaric tags for relative and absolute quantitation
- LC
liquid chromatography
- MS
mass spectrometry
- SILAC
stable isotope labeling by amino acids in cell culture
- RuBisCO
ribulose 1,5-bisphosphate carboxylase/oxygenase
- OEE
oxygen-evolving enhancer protein
- ROS
reactive oxygen species
- LEA
late-embryogenesis abundant
- ABA
abscisic acid.
References
- Abas L., Luschnig C. (2010). Maximum yields of microsomal-type membranes from small amounts of plant material without requiring ultracentrifugation. Anal Biochem. 401, 217–227. 10.1016/j.ab.2010.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi F. M., Komatsu S. (2004). A proteomic approach to analyze salt-responsive proteins in rice leaf sheath. Proteomics 4, 2072–2081. 10.1002/pmic.200300741 [DOI] [PubMed] [Google Scholar]
- Ackermann B. L., Berna M. J. (2007). Coupling immunoaffinity techniques with MS for quantitative analysis of low abundance protein biomarkers. Exp. Rev. Proteomics 4, 175–186. 10.1586/14789450.4.2.175 [DOI] [PubMed] [Google Scholar]
- Adamis P. D., Gomes D. S., Pinto M. L., Panek A. D., Eleutherio E. C. (2004). The role of glutathione transferases in cadmium stress. Toxicol. Lett. 154, 81–88. 10.1016/j.toxlet.2004.07.003 [DOI] [PubMed] [Google Scholar]
- Ahsan N., Nakamura T., Komatsu S. (2012). Differential responses of microsomal proteins and metabolites in two contrasting cadmium (Cd)-accumulating soybean cultivars under Cd stress. Amino Acids 42, 317–327. 10.1007/s00726-010-0809-7 [DOI] [PubMed] [Google Scholar]
- Ali G. M., Komatsu S. (2006). Proteomic analysis of rice leaf sheath during drought stress. J. Proteome Res. 5, 396–403. 10.1021/pr050291g [DOI] [PubMed] [Google Scholar]
- Alvarez S., Roy Choudhury S., Pandey S. (2014). Comparative quantitative proteomics analysis of the ABA response of roots of drought-sensitive and drought-tolerant wheat varieties identifies proteomic signatures of drought adaptability. J. Proteome Res. 13, 1688–1701. 10.1021/pr401165b [DOI] [PubMed] [Google Scholar]
- Andrews D. L., MacAlpine D. M., Johnson J. R., Kelley P. M., Cobb B. G., Drew M. C. (1994). Differential induction of mRNAs for the glycolytic ethanolic fermentative pathways by hypoxis and anoxia in maize seedlings. Plant Physiol. 106, 1575–1582. 10.1104/pp.106.4.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghari R., Ebrahimzadeh H. (2006). Drought stress increases the expression of wheat leaf ribulose-1, 5- bisphosphate carboxylase/oxyenase protein. Iranian J. Sci. Tech. Trans. A 30, 1–7. 11368350 [Google Scholar]
- Ashraf M., Harris P. J. C. (2013). Photosynthesis under stressful environments: an overview. Photosynthetica 51, 163–190 10.1007/s11099-013-0021-6 [DOI] [Google Scholar]
- Baginsky S., Gruissem W. (2002). Endonucleolytic activation directs dark-induced chloroplast mRNA degradation. Nucleic Acids Res. 30, 4527–4533. 10.1093/nar/gkf561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancel E., Rogniaux H., Debiton C., Chambon C., Branlard G. (2010). Extraction and proteome analysis of starch granule-associated proteins in mature wheat kernel (Triticum aestivum L.). J. Proteome Res. 9, 3299–3310. 10.1021/pr9010525 [DOI] [PubMed] [Google Scholar]
- Basu A., Basu U., Taylor G. J. (1994). Induction of microsomal membrane proteins in roots of an aluminum-resistant cultivar of triticum aestivum L. under conditions of aluminum stress. Plant Physiol. 104, 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentem S., Roitinger E., Anrather D., Csaszar E., Hirt H. (2006). Phosphoproteomics as a tool to unravel plant regulatory mechanisms. Physiol. Planta. 126, 110–119 10.1111/j.1399-3054.2006.00615.x [DOI] [Google Scholar]
- Bota J., Medrano H., Flexas J. (2004). Is photosynthesis limited by decreased Rubisco activity and RuBP content under progressive water stress? New Phytol. 162, 671–681. 10.1111/j.1469-8137.2004.01056.x [DOI] [PubMed] [Google Scholar]
- Brenchley R., Spannagl M., Pfeifer M., Barker G. L., D'Amore R., Allen A. M., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491, 705–710. 10.1038/nature11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G., Vasquez-Tello A., Danyluk J., Sarhan F. (2002). Two novel intrinsic annexins accumulate in wheat membranes in response to low temperature. Plant Cell Physiol. 41, 177–184. 10.1093/pcp/41.2.177 [DOI] [PubMed] [Google Scholar]
- Budak H., Akpinar B. A., Unver T., Turktas M. (2013). Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI–MS/MS. Plant Mol. Biol. 83, 89–103. 10.1007/s11103-013-0024-5 [DOI] [PubMed] [Google Scholar]
- Bueno P., Piqueras A. (2002). Effect of transition metals on stress, lipid peroxidation and antioxidant enzyme activities in tobacco cell cultures. Plant Growth Regul. 36, 161–167 10.1023/A:1015044705137 [DOI] [Google Scholar]
- Burks E. A., Bezerra P. P., Le H., Gallie D. R., Browning K. S. (2001). Plant initiation factor 3 subunit composition resembles mammalian initiation factor 3 and has a novel subunit. J. Biol. Chem. 276, 2122–2131. 10.1074/jbc.M007236200 [DOI] [PubMed] [Google Scholar]
- Caldas T. D., El Yaagoubi A., Richarme G. (1998). Chaperone properties of bacterial elongation factor EF-Tu. J. Biol. Chem. 273, 11478–11482. 10.1074/jbc.273.19.11478 [DOI] [PubMed] [Google Scholar]
- Calvo E., Camafeita E., Fernández-Gutiérrez B., López J. A. (2011). Applying selected reaction monitoring to targeted proteomics. Expert. Rev. Proteomics 8, 165–173. 10.1586/epr.11.11 [DOI] [PubMed] [Google Scholar]
- Cantero A., Barthakur S., Bushart T. J., Chou S., Morgan R. O., Fernandez M. P., et al. (2006). Expression profiling of the Arabidopsis annexin gene family during germination, de-etiolation and abiotic stress. Plant Physiol. Biochem. 44, 13–24. 10.1016/j.plaphy.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Carrari F., Coll-Garcia D., Schauer N., Lytovchenko A., Palacios-Rojas N., Balbo I., et al. (2005). Deficiency of a plastidial adenylate kinase in Arabidopsis results in elevated photosynthetic amino acid biosynthesis and enhanced growth. Plant Physiol. 137, 70–82. 10.1104/pp.104.056143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso G., Cavaliere C., Foglia P., Gubbiotti R., Samperi R., Laganà A. (2009). Analysis of drought responsive proteins in wheat (Triticum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant Sci. 177, 570–576 10.1016/j.plantsci.2009.08.007 [DOI] [Google Scholar]
- Chakrabarty A. M. (1998). Nucleoside diphosphate kinase: role in bacterial growth, virulence, cell glycerald and polysaccharide synthesis. Mol. Microbiol. 28, 875–882. 10.1046/j.1365-2958.1998.00846.x [DOI] [PubMed] [Google Scholar]
- Chaves M. M., Flexas J., Pinheiro C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103, 551–560. 10.1093/aob/mcn125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Warren C. R., Adams M. A. (2000). Separation of Rubisco in extracts of plant leaves by capillary electrophoresis with sieving polymers. Anal. Lett. 33, 579–587 10.1080/00032710008543075 [DOI] [Google Scholar]
- Chinnusamy V., Schumaker K., Zhu J. K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 55, 225–236. 10.1093/jxb/erh005 [DOI] [PubMed] [Google Scholar]
- Cho J. H., Hwang H., Cho M. H., Kwon Y. K., Jeon J. S., Bhoo S. H., et al. (2008). The effect of DTT in protein preparations for proteomic analysis: removal of a highly abundant plant enzyme, ribulose bisphosphate carboxylase/oxygenase. J. Plant Biol. 51, 297–301 10.1007/BF03036130 [DOI] [Google Scholar]
- Clemens S. (2006). Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie 88, 1707–1719. 10.1016/j.biochi.2006.07.003 [DOI] [PubMed] [Google Scholar]
- Cobbett C., Goldsbrough P. (2002). Phytochelatins and metallothioneins: roles in heavy metal detoxification and homeostasis. Annu. Rev. Plant Biol. 53, 159–182. 10.1146/annurev.arplant.53.100301.135154 [DOI] [PubMed] [Google Scholar]
- Crafts-Brandner S. J., Salvucci M. E. (2000). RuBisCO activase constrains the photosynthetic potential of leaves at high temperature and CO2. Proc. Natl. Acad. Sci. U.S.A. 97, 13430–13435. 10.1073/pnas.230451497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cushman J. C. (1993). Molecular cloning and expression of chloroplast NADP-malate dehydrogenase during Crassulacean acid metabolism induction by salt stress. Photosynth. Res. 35, 15–27. 10.1007/BF02185408 [DOI] [PubMed] [Google Scholar]
- D'Amici G. M., Huber G. C., Zolla L. (2009). Separation of thylakoid membrane proteins by sucrose gradient ultracentrifuge or blue native-SDS-PAGE two-dimensional electrophoresis, in Membrane Proteomics: Methods and Protocols, eds Peirce M. J., Waits R. (New York, NY: Springer; ), 61–67. [DOI] [PubMed] [Google Scholar]
- Dai Z. M. (2010). Activities of enzymes involved in starch synthesis in wheat grains differing in starch content. Russ. J. Plant Physiol. 57, 74–78 10.1134/S1021443710010103 [DOI] [Google Scholar]
- Davis D. G., Swanson H. R. (2001). Activity of stress-related enzymes in the perennial weed leafy spurge (Euphorbia esula L.). Env. Exp. Bot. 46, 95–108 10.1016/S0098-8472(01)00081-8 [DOI] [Google Scholar]
- Dechassa D., Khairy S., Robert T., Zachary S. (2011). Proteomic analysis of soybean roots under aluminum stress. Intl. J. Plant Genomics 2011:282531. 10.1155/2011/282531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delisle G., Champoux M., Houde M. (2001). Characterization of oxalate oxidase and cell death in Al-sensitive and tolerant wheat roots. Plant Cell Physiol. 42, 324–333. 10.1093/pcp/pce041 [DOI] [PubMed] [Google Scholar]
- Dembinsky D., Woll K., Saleem M., Liu Y., Fu Y., Borsuk L. A., et al. (2007). Transcriptomic and proteomic analyses of pericycle cells of the maize primary root. Plant Physiol. 145, 575–588. 10.1104/pp.107.106203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirevska K., Simova-Stoilova L., Vassileva V., Vaseva I., Grigorova B., Feller U. (2008). Drought-induced leaf protein alterations in sensitive and tolerant wheat varieties. Gen. Appl. Plant Physiol. 34, 79–102. [Google Scholar]
- Demirevska K., Zasheva D., Dimitrov R., Simova-Stoilova L., Stamenova M., Feller U. (2009). Drought stress effects on RuBisCO in wheat: changes in the RuBisCO large subunit. Acta Physiol. Plant. 31, 1129–1138 10.1007/s11738-009-0331-2 [DOI] [Google Scholar]
- Ding Y., Ma Q. H. (2004). Characterization of a cytosolic malate dehydrogenase cDNA which encodes an isozyme toward oxaloacetate reduction in wheat. Biochemie 86, 509–518. 10.1016/j.biochi.2004.07.011 [DOI] [PubMed] [Google Scholar]
- Drummond R. D., Guimarães C. T., Felix J., Ninamango-Cárdenas F. E., Carneiro N. P., Paiva E., et al. (2001). Prospecting sugarcane genes involved in aluminum tolerance. Genet. Mol. Biol. 24, 221–230 10.1590/S1415-47572001000100029 [DOI] [Google Scholar]
- Edwards R., Dixon D. P., Walbot V. (2000). Plant glutathione Stransferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 5, 193–198. 10.1016/S1360-1385(00)01601-0 [DOI] [PubMed] [Google Scholar]
- Erdmann B., Hoffmann P., Wiedenroth E. M. (1986). Changes in the root system of wheat seedlings following root anaerobiosis I. anatomy and respiration in Triticum aestivum L. Ann. Bot. 58, 597–605. [Google Scholar]
- Feiz L., Irshad M., Pont-Lezica R. F., Canut H., Jamet E. (2006). Evaluation of cell wall preparation for proteomics: a new procedure for purifying cell walls from Arabidopsis hypocotyls. Plant Methods 2:10. 10.1186/1746-4811-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller U., Anders I., Mae T. (2008). Rubiscolytics: fate of Rubisco after its enzymatic function in a cell is terminated. J. Exp. Bot. 59, 1615–1624. 10.1093/jxb/erm242 [DOI] [PubMed] [Google Scholar]
- Fercha A., Capriotti A. L., Caruso G., Cavaliere C., Gherroucha H., Samperi R., et al. (2013). Gel-free proteomics reveal potential biomarkers of priming-induced salt tolerance in durum wheat. J. Proteomics 91, 486–499. 10.1016/j.jprot.2013.08.010 [DOI] [PubMed] [Google Scholar]
- Flexas J., Ribas-Carbó M., Bota J., Galmés J., Henkle M., Martínez-Cañellas S., et al. (2006). Decreased RuBisCO activity during water stress is not induced by decreased relative water content but related to conditions of low stomatal conductance and chloroplast CO2 concentration. New Phytol. 172, 73–82. 10.1111/j.1469-8137.2006.01794.x [DOI] [PubMed] [Google Scholar]
- Ford K. L., Cassin A., Bacic A. (2011). Quantitative proteomic analysis of wheat cultivars with differing drought stress tolerance. Front. Plant Sci. 2:44. 10.3389/fpls.2011.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukayama H., Abe R., Uchida N. (2010). SDS-dependent proteases induced by ABA and its relation to Rubisco and Rubisco activase contents in rice leaves. Plant Physiol. Biochem. 48, 808–812. 10.1016/j.plaphy.2010.08.002 [DOI] [PubMed] [Google Scholar]
- Fukuda A., Chiba K., Maeda M., Nakamura A., Maeshima M., Tanaka Y. (2004). Effect of salt and osmotic stresses on the expression of genes for the vacuolar H+-pyrophosphatase, H+-ATPase subunit A, and Na+/H+ antiporter from barley. J. Exp. Bot. 55, 585–594. 10.1093/jxb/erh070 [DOI] [PubMed] [Google Scholar]
- Fukuda T., Saito A., Wasaki J., Shinano T., Osaki M. (2007). Metabolic alterations proposed by proteome in rice roots grown under low P and high Al concentration under pH. Plant Sci. 172, 1157–1165 10.1016/j.plantsci.2007.02.020 [DOI] [Google Scholar]
- Gao L., Yan X., Li X., Guo G., Hu Y., Ma W., et al. (2011). Proteome analysis of wheat leaf under salt stress by two-dimensional difference gel electrophoresis (2D-DIGE). Phytochemistry 72, 1180–1191. 10.1016/j.phytochem.2010.12.008 [DOI] [PubMed] [Google Scholar]
- Ge C., Ding Y., Wang Z., Wan D., Wang Y., Shang Q., et al. (2009). Responses of wheat seedlings to cadmium, mercury and trichlorobenzene stresses. J. Environ. Sci. 21, 806–813. 10.1016/S1001-0742(08)62345-1 [DOI] [PubMed] [Google Scholar]
- Ghosh D., Xu J. (2014). Abiotic stress responses in plant roots: a proteomics perspective. Front. Plant Sci. 5:6. 10.3389/fpls.2014.00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill B. S., Appels R., Botha-Oberholster A., Buell C. R., Bennetzen J. L., Chalhoub B., et al. (2004). A workshop report on wheat genome sequencing: international genome research on wheat consortium. Genetics 168, 1087–1096. 10.1534/genetics.104.034769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong P., Zhang J., Li H., Yang C., Zhang C., Zhang X., et al. (2010). Transcriptional profiles of drought-responsive genes in modulating transcription signal transduction, and biochemical pathways in tomato. J. Exp. Bot. 61, 3563–3575. 10.1093/jxb/erq167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattan S. R., Grieve C. M. (1992). Mineral element acquisition and growth response of plants grown in saline environments. Agr. Ecosyst. Environ. 38, 275–300 10.1016/0167-8809(92)90151-Z [DOI] [Google Scholar]
- Gupta P. K., Mir R. R., Mohan A., Kumar J. (2008). Wheat genomics: present status and future prospects. Int. J. Plant Genomics 2008:896451. 10.1155/2008/896451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge S., Gatenby A. A. (1995). Rubisco synthesis, assembly, mechanism, and regulation. Plant Cell 7, 809–819. 10.1105/tpc.7.7.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisalihoglu G., Hart J. J., Wang Y. H., Cakmak I., Kochian L. V. (2003). Zinc efficiency is correlated with enhanced expression and activity of Cu/Zn superoxide dismutase and carbonic anhydrase in wheat. Plant Physiol. 131, 595–602. 10.1104/pp.011825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton C. A., Good A. G., Taylor G. J. (2001). Induction of vacuolar ATPase and mitochondrial ATP synthase by aluminium in aluminium-resistant cultivar of wheat. Plant Physiol. 125, 2068–2077. 10.1104/pp.125.4.2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock J. T., Henson D., Nyirenda M., Desikan R., Harrison J., Lewis M., et al. (2005). Proteomic identification of glyceraldehydes 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiol. Bioch. 43, 828–835. 10.1016/j.plaphy.2005.07.012 [DOI] [PubMed] [Google Scholar]
- Haque M. E., Abe F., Mori M., Oyanagi A., Komatsu S., Kawaguchi K. (2014). Characterization of a wheat pathogenesis-related protein, TaBWPR-1.2, in seminal roots in response to waterlogging stress. J. Plant Physiol. 171, 602–609. 10.1016/j.jplph.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Haque M. E., Kawaguchiand K., Komatsu S. (2011). Analysis of proteins in aerenchymatous seminal roots of wheat grown in hypoxic soils under waterlogged conditions. Protein Pept. Lett. 18, 912–924. 10.2174/092986611796011455 [DOI] [PubMed] [Google Scholar]
- Hashimoto M., Komatsu S. (2007). Proteomic analysis of rice seedlings during cold stress. Proteomics 7, 1293–1302. 10.1002/pmic.200600921 [DOI] [PubMed] [Google Scholar]
- Herman E. M., Helm R. M., Jung R., Kinney A. J. (2003). Genetic modification removes an immunodominant allergen from soybean. Plant Physiol. 132, 36–43. 10.1104/pp.103.021865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges M., Flesch V., Gálvez S., Bismuth E. (2003). Higher plant NADP dependent isocitrate dehydrogenases, ammonium assimilation and NADPH production. Plant Physiol. Biochem. 41, 577–585 10.1016/S0981-9428(03)00062-7 [DOI] [Google Scholar]
- Hossain Z., Khatoon A., Komatsu S. (2013). Soybean proteomics for unraveling abiotic stress response mechanism. J. Proteome Res. 12, 4670-4684. 10.1021/pr400604b [DOI] [PubMed] [Google Scholar]
- Huang B., Chu C. H., Chen S. H., Juan H. F., Chen Y. M. (2006). A proteomics study of the mung bean glycerald regulated by brassinosteroids under conditions of chilling stress. Cell. Mol. Biol. Lett. 11, 264–278. 10.2478/s11658-006-0021-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo C. M., Zhao B. C., Ge R. C., Shen Y. Z., Huang Z. J. (2004). Proteomic analysis of the salt tolerance mutant of wheat under salt stress. Yi Chuan Xue Bao (Chinese) 31, 1408–1414. [PubMed] [Google Scholar]
- Hurkman W. J., Vensel W. H., Tanaka C. K., Whitehand L., Altenbach S. B. (2009). Effect of high temperature on albumin and globulin accumulation in the endosperm proteome of the developing wheat grain. J. Cereal Sci. 49, 12–23 10.1016/j.jcs.2008.06.014 [DOI] [Google Scholar]
- Hurt E., Hauska G. (1981). A cytochrome f/b6 complex of five polypeptides with plastoquinol-plastocyanin-oxidoreductase activity from spinach chloroplasts. Eur. J. Biochem. 117, 591–595. 10.1111/j.1432-1033.1981.tb06379.x [DOI] [PubMed] [Google Scholar]
- Ishida H., Nishimori Y., Sugisawa M., Makino A., Mae T. (1997). The large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is fragmented into 37-kda and 16-kda polypeptides by active oxygen in the lysates of chloroplasts from primary leaves of wheat. Plant Cell Physiol. 38, 471–479. 10.1093/oxfordjournals.pcp.a029191 [DOI] [PubMed] [Google Scholar]
- Islam N., Tsujimoto H., Hirano H. (2003). Wheat proteomics: relationship between fine chromosome deletion and protein expression. Proteomics 3, 307–316. 10.1002/pmic.200390044 [DOI] [PubMed] [Google Scholar]
- Jacoby R. P., Millar A. H., Taylor N. L. (2010). Wheat mitochondrial proteomes provide new links between antioxidant defense and plant salinity tolerance. J. Proteome Res. 9, 6595–6604. 10.1021/pr1007834 [DOI] [PubMed] [Google Scholar]
- Jambunathan N., Mahalingam R. (2006). Analysis of Arabidopsis growth factor gene 1 (GFG1) encoding a nudix hydrolase during oxidative signaling. Planta 224, 1–11. 10.1007/s00425-005-0183-y [DOI] [PubMed] [Google Scholar]
- Jamet E., Albenne C., Boudart G., Irshad M., Canut H., Pont-Lezica R. (2008). Recent advances in plant cell wall proteomics. Proteomics 8, 93–98. 10.1002/pmic.200700938 [DOI] [PubMed] [Google Scholar]
- Jensen R. G., Bahr J. T. (1977). Ribulose 1,5-bisphosphate carboxylase-oxygenase. Ann. Rev. Plant Physiol. 28, 379–400 10.1146/annurev.pp.28.060177.002115 [DOI] [Google Scholar]
- Jiang S. S., Liang X. N., Li X., Wang S. L., Lv D. W., Ma C. Y., et al. (2012). Wheat drought-responsive grain proteome analysis by linear and nonlinear 2-DE and MALDI-TOF mass spectrometry. Int. J. Mol. Sci. 13, 16065–16083. 10.3390/ijms131216065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. Q., Yang B., Harris N. S., Deyholos M. K. (2007). Comparative proteomics analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58, 3591–3607. 10.1093/jxb/erm207 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Cobb B. G., Drew M. C. (1989). Hypoxic induction of anoxia tolerance in root tips of Zea mays L. Plant Physiol. 91, 837–841. 10.1104/pp.91.3.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A. H., Cho K., Choi J. S., Bae K. H., Komatsu S., Uozumi N., et al. (2013b). The wheat chloroplastic proteome. J. Proteomics 93, 326–342. 10.1016/j.jprot.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Kamal A. H., Cho K., Choi J. S., Jin Y., Park C. S., Lee J. S., et al. (2013a). Patterns of protein expression in water-stressed wheat chloroplasts. Biol. Plant. 57, 305–312 10.1007/s10535-012-0290-0 [DOI] [Google Scholar]
- Kamal A. H., Cho K., Kim D. E., Uozumi N., Chung K. Y., Lee S. Y., et al. (2012a). Changes in physiology and protein abundance in salt-stressed wheat chloroplasts. Mol. Biol. Rep. 39, 9059–9074. 10.1007/s11033-012-1777-7 [DOI] [PubMed] [Google Scholar]
- Kamal A. H., Cho K., Komatsu S., Uozumi N., Choi J. S., Woo S. H. (2012b). Towards an understanding of wheat chloroplasts: a methodical investigation of thylakoid proteome. Mol. Biol. Rep. 39, 5069–5083. 10.1007/s11033-011-1302-4 [DOI] [PubMed] [Google Scholar]
- Kamal A. H., Rashid H., Sakata K., Komatsu S. (2015). Gel-free quantitative proteomic approach to identify cotyledon proteins in soybean under flooding stress. J. Proteomics 112C, 1–13. 10.1016/j.jprot.2014.08.014 [DOI] [PubMed] [Google Scholar]
- Kim K. H., Kamal A. H., Shin K. H., Choi J. S., Heo H. Y., Woo S. H. (2010). Large-scale proteome investigation in wild relatives (A, B, and D genomes) of wheat. Acta Biochim. Biophys. Sin. 42, 709–716. 10.1093/abbs/gmq079 [DOI] [PubMed] [Google Scholar]
- Kim S. T., Cho K. S., Jang Y. S., Kang K. Y. (2001). Two-dimensional electrophoretic analysis of rice proteins by polyethylene glycol fractionation for protein arrays. Electrophoresis 22, 2103–2109. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Furuta Y., Ohno T., Hara T., Koyama H. (2005). Quantitative trait loci controlling aluminum tolerance in two accessions of Arabidopsis thaliana (Landsberg erecta and Cape Verde Islands). Plant Cell Environ. 28, 1516–1524 10.1111/j.1365-3040.2005.01388.x [DOI] [Google Scholar]
- Kobayashi Y., Hoekenga O. A., Itoh H., Nakashima M., Satio S., Shaff J. E., et al. (2007). Characterization AtALMT1 expression in aluminuminducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 145, 843–852. 10.1104/pp.107.102335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian L. V. (1995). Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 237–260 10.1146/annurev.pp.46.060195.001321 [DOI] [Google Scholar]
- Komatsu S., Wada T., Abalea Y., Nouri M. Z., Nanjo Y., Nakayama N., et al. (2009). Analysis of plasma membrane proteome in soybean and application to flooding stress response. J. Proteome Res. 8, 4487–4499. 10.1021/pr9002883 [DOI] [PubMed] [Google Scholar]
- Komatsu S., Yanagawa Y. (2013). Cell wall proteomics of crops. Front. Plant Sci. 4:17. 10.3389/fpls.2013.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F. J., Oyanagi A., Komatsu S. (2010). Cell wall proteome of wheat roots under flooding stress using gel-based and LC MS/MS-based proteomics approaches. Biochim. Biophys. Acta 1804, 124–136. 10.1016/j.bbapap.2009.09.023 [DOI] [PubMed] [Google Scholar]
- Krishnan H. B., Natarajan S. S. (2009). A rapid method for depletion of Rubisco from soybean (Glycine max) leaf for proteomic analysis of lower abundance proteins. Phytochemistry 70, 1958–1964. 10.1016/j.phytochem.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Lane B. G., Dunwell J. M., Bay J. A., Schmitt M. R., Cuming A. C. (1993). German, a marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268, 12239–12242. [PubMed] [Google Scholar]
- Lanubile R., Piro G., Dalessandro G. (1997). Effect of Brefeldin A on the synthesis and transport of cell wall polysaccharides and proteins in pea root seedlings. J. Exp. Bot. 48, 1925–1933 10.1093/jexbot/48.316.1925 [DOI] [Google Scholar]
- Laurell C. B. (1966). Quantitative estimation of proteins by electrophoresis in agarose gel containing antibodies. Anal. Biochem. 15, 45–52 10.1016/0003-2697(66)90246-6 [DOI] [PubMed] [Google Scholar]
- Lee E. J., Matsumura Y., Soga K., Hoson T., Koizumi N. (2007). Glycosyl hydrolases of cell wall are induced by sugar starvation in Arabidopsis. Plant Cell Physiol. 48, 405–413. 10.1093/pcp/pcm009 [DOI] [PubMed] [Google Scholar]
- Leech R. M., Marrison J. L. (1996). Immunofluorescent quantitation of chloroplast proteins. Plant J. 10, 1169–1175. 10.1046/j.1365-313X.1996.10061169.x [DOI] [PubMed] [Google Scholar]
- Li G., Peng X., Xuan H., Wei L., Yang Y., Guo T., et al. (2013). Proteomic analysis of leaves and roots of common wheat (Triticum aestivum L.) under copper-stress conditions. J. Proteome Res. 12, 4846–4861. 10.1021/pr4008283 [DOI] [PubMed] [Google Scholar]
- Li W., Zhang C., Lu Q., Wen X., Lu C. (2011). The combined effect of salt stress and heat shock on proteome profiling in Suaeda salsa. J. Plant Physiol. 168, 1743–1752. 10.1016/j.jplph.2011.03.018 [DOI] [PubMed] [Google Scholar]
- Majoul T., Bancel E., Triboï E., Ben Hamida J., Branlard G. (2003). Proteomic analysis of the effect of heat stress on hexaploid wheat grain: characterization of heat-responsive proteins from total endosperm. Proteomics 3, 175–183. 10.1002/pmic.200390026 [DOI] [PubMed] [Google Scholar]
- Mansour M. M. F., Salama K. H. A., Al-Mutawa M. M. (2003). Transport proteins and salt tolerance in plants. Plant Sci. 164, 891–900 10.1016/S0168-9452(03)00109-2 [DOI] [Google Scholar]
- Martin P. K., Koebner R. M. D. (1995). Sodium and chloride ions contribute synergistically to salt toxicity in wheat. Biol. Plant. 37, 265–271 10.1007/BF02913224 [DOI] [Google Scholar]
- Matthews R. G., Sheppard C., Goulding C. (1998). Methylenetetrahydro folate reductase and methionine synthase: biochemistry and molecular biology. Eur. J. Pediatr. 157, 54–59 10.1007/PL00014305 [DOI] [PubMed] [Google Scholar]
- McLennan A. G. (2006). The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63, 123–143. 10.1007/s00018-005-5386-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memon A. R. (2004). The role of ADP-ribosylation factor and SAR1 in vesicular trafficking in plants. Biochim. Biophys. Acta 1664, 9–30. 10.1016/j.bbamem.2004.04.005 [DOI] [PubMed] [Google Scholar]
- Meng Q., Rao L., Xiang X., Zhou C., Zhang X., Pan Y. (2011). A systematic strategy for proteomic analysis of chloroplast protein complexes in wheat. Biosci. Biotech. Biochem. 75, 2194–2199. 10.1271/bbb.110479 [DOI] [PubMed] [Google Scholar]
- Miflin B. J., Habash D. Z. (2002). The role of glutamine synthetase and glutamate dehydrogenase in nitrogen assimilation and possibilities for improvement in the nitrogen utilization of crops. J. Exp. Bot. 53, 979–987. 10.1093/jexbot/53.370.979 [DOI] [PubMed] [Google Scholar]
- Millar A. H., Sweetlove L. J., Giege P., Leaver C. J. (2001). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727. 10.1104/pp.010387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S., Tripathi R. D., Srivastava S., Dwivedi S., Trivedi P. K., Dhankher O. P., et al. (2009). Thiol metabolism play significant role during cadmium detoxification by Ceratophyllum demersum L. Bioresour. Technol. 100, 2155–2161. 10.1016/j.biortech.2008.10.041 [DOI] [PubMed] [Google Scholar]
- Mock H. P., Grimm B. (1997). Reduction of uroporphyrinogen decarboxylase by antisense RNA expression affects activities of other enzymes involved in tetra-pyrrole biosynthesis and leads to light dependent necrosis. Plant Physiol. 113, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi P. P., Moieni A., Hiraga S., Komatsu S. (2012). Organ-specific proteomic analysis of drought-stressed soybean seedlings. J. Proteomics 75, 1906-1923. 10.1016/j.jprot.2011.12.041 [DOI] [PubMed] [Google Scholar]
- Murota K. I., Ohshita Y., Watanabe A., Aso S., Sato F., Yamada Y. (1994). Changes related to salt tolerance in thylakoid membranes of photoautotrophically cultured green tobacco cells. Plant Cell Physiol. 35, 107–113. [Google Scholar]
- Nandi D., Tahiliani P., Kumar A., Chandu D. (2006). The ubiquitin-proteasome system. J. Biosci. 31, 137–155 10.1007/BF02705243 [DOI] [PubMed] [Google Scholar]
- Ndimba B. K., Chivasa S., Simon W. J., Slabas A. R. (2005). Identification of Arabidopsis salt and osmotic stress responsive proteins using twodimensional difference gel electrophoresis and mass spectrometry. Proteomics 5, 4185–4196. 10.1002/pmic.200401282 [DOI] [PubMed] [Google Scholar]
- Nishikawa M., Hashida M., Takakura Y. (2009). Catalase delivery for inhibiting ROS-mediated tissue injury and tumor metastasis. Adv. Drug Del. Rev. 61, 319–326. 10.1016/j.addr.2009.01.001 [DOI] [PubMed] [Google Scholar]
- Nowitzki U., Gelius-Dietrich G., Schwieger M., Henze K., Martin W. (2004). Chloroplast phosphoglycerate kinase from Euglena gracilis: endosymbiotic gene replacement going against the tide. Eur. J. Biochem. 271, 4123–4131. 10.1111/j.1432-1033.2004.04350.x [DOI] [PubMed] [Google Scholar]
- O'Donnell P. J., Calvert C., Atzorn R., Wasternack C., Leyser H. M. O., Bowles D. J. (1996). Ethylene as a signal mediating the wound response of tomato plants. Science 274, 1914–1917. [DOI] [PubMed] [Google Scholar]
- Oh M. W., Roy S. K., Kamal A. H., Cho K., Cho S. W., Park C. S., et al. (2014). Proteome analysis of roots of wheat seedlings under aluminum stress. Mol. Biol. Rep. 41, 671–681. 10.1007/s11033-013-2905-8 [DOI] [PubMed] [Google Scholar]
- Oyanagi A. (2008). Relationship of growth variability, with ground level and soil water content in a large paddy field in Inashiki city, Ibaraki-prefecture in 2007-Wet injury of wheat. Jpn. J. Crop. Sci. (Japanese) 77, 511–515 10.1626/jcs.77.511 [DOI] [Google Scholar]
- Palmgren M. G., Harper J. F. (1999). Pumping with plant P-type ATPases. J. Exp. Bot. 50, 883–893 10.1093/jxb/50.Special_Issue.883 [DOI] [Google Scholar]
- Pang Q., Chen S., Dai S., Chen Y., Wang Y., Yan X. (2010). Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J. Proteome Res. 9, 2584–2599. 10.1021/pr100034f [DOI] [PubMed] [Google Scholar]
- Parker R., Flowers T. J., Moore A. L., Harpham N. V. J. (2006). An accurate and reproducible method for proteome profiling of the effects of salt stress in the rice leaf lamina. J. Exp. Bot. 57, 1109–1118. 10.1093/jxb/erj134 [DOI] [PubMed] [Google Scholar]
- Peng Z., Wang M., Li F., Lv H., Li C., Xia G. (2009). A proteomic study of the response to salinity and drought stress in an lntrogression strain of bread wheat. Mol. Cell. Proteomics 8, 2676–2686. 10.1074/mcp.M900052-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picotti P., Aebersold R. (2012). Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat. Methods 9, 555–566. 10.1038/nmeth.2015 [DOI] [PubMed] [Google Scholar]
- Pradet A., Raymond P. (1983). Adenine nucleotide ratios and adenylate energy charge in energy metabolism. Ann. Rev. Plant Physiol. 34, 199–224 10.1146/annurev.pp.34.060183.001215 [DOI] [Google Scholar]
- Qu Z. L., Zhong N. Q., Wang H. Y., Chen A. P., Jian G. L., Xia G. X. (2006). Ectopic expression of the cotton non-symbiotic hemoglobin gene GhHbd1 triggers defense responses and increases disease tolerance in Arabidopsis. Plant Cell Physiol. 47, 1058–1068. 10.1093/pcp/pcj076 [DOI] [PubMed] [Google Scholar]
- Reynolds M., Bonnett D., Chapman S. C., Furbank R. T., Manès Y., Mather D. E., et al. (2010). Raising yield potential of wheat. I. Overview of a consortium approach and breeding strategies. Exp. Bot. 62, 439–452. 10.1093/jxb/erq311 [DOI] [PubMed] [Google Scholar]
- Ribeiro M., Nunes-Miranda J. D., Branlard G., Carrillo J. M., Rodriguez-Quijano M., Igrejas G. (2013). One hundred years of grain omics: identifying the glutens that feed the world. J. Proteome Res. 12, 4702–4716. 10.1021/pr400663t [DOI] [PubMed] [Google Scholar]
- Rizhsky L., Liang H., Mittler R. (2002). The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 130, 1143–1151. 10.1104/pp.006858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer S., Hugueney P., Bouvier F., Camara B., Kuntz M. (1993). Expression of the genes encoding the early carotenoid biosynthetic enzymes in Capsicum annuum. Biochem. Biophy. Res. Commun. 196, 1414–1421. 10.1006/bbrc.1993.2410 [DOI] [PubMed] [Google Scholar]
- Safár J., Simková H., Kubaláková M., Cíhalíková J., Suchánková P., Bartos J., et al. (2010). Development of chromosome-specific BAC resources for genomics of bread wheat. Cytogenet. Genome Res. 129, 211–223. 10.1159/000313072 [DOI] [PubMed] [Google Scholar]
- Salekdeh G. H., Komatsu S. (2007). Crop proteomics: aim at sustainable agriculture of tomorrow. Proteomics 7, 2976–2996. 10.1002/pmic.200700181 [DOI] [PubMed] [Google Scholar]
- Sanità di Toppi L., Gabbrielli R. (1999). Response to cadmium in higher plants. Environ. Exp. Bot. 41, 105–130 10.1016/S0098-8472(98)00058-6 [DOI] [Google Scholar]
- Schafleitner R., Wilhelm E. (2002). Isolation of wound-responsive genes from chestnut (Castanea sativa) microstems by mRNA display and their differential expression upon wounding and infection with the chestnut blight fungus (Chryphonectria parasitica). Physiol. Mol. Plant Pathol. 61, 339–348 10.1006/pmpp.2003.0448 [DOI] [Google Scholar]
- Schwartz A., Ilan N., Schwarz M., Scheaffer J., Assmann S. M., Schroeder J. I. (1995). Anion-channel blockers inhibit S-type anion channels and abscisic acid responses in guard cells. Plant Physiol. 109, 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann J. R., Critchley C. (1985). Effects of salt stress on the growth, ion content, stomatal glyceral and photosynthetic capacity of a salt-sensitive species, Phaseolus vulgaris L. Planta 164, 151–162. 10.1007/BF00396077 [DOI] [PubMed] [Google Scholar]
- Seo H. S., Song J. T., Cheong J. J., Lee Y. H., Lee Y. W., Hwang I., et al. (2001). Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. U.S.A. 98, 4788–4793. 10.1073/pnas.081557298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S., Ishikawa T., Tamoi M., Miyagawa Y., Takeda T., Yabuta Y., et al. (2002). Regulation and function of ascorbate peroxidase isoenzymes. J. Exp. Bot. 53, 1305–1319. 10.1093/jexbot/53.372.1305 [DOI] [PubMed] [Google Scholar]
- Shin K. H., Kamal A. H. M., Cho K., Choi J. S., Jin Y., Paek N. C., et al. (2011). Defense proteins are induced in wheat spikes exposed to Fusarium graminearum. Plant Omics J. 4, 270–277. [Google Scholar]
- Siggaard-Andersen M., Kauppinen S., von Wettstein-Knowles P. (1991). Primary structure of a cerulenin-binding β-ketoacyl-[acyl carrier protein] synthase from barley chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 88, 4114–4118. 10.1073/pnas.88.10.4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar P., Sharmila P., Saradhi P. P. (2000). Proline alleviates salt-stress-induced enhancement in ribulose-l, 5-bisphosphate oxygenase activity. Biochem. Biophys. Res. Commun. 279, 512–515. 10.1006/bbrc.2000.4005 [DOI] [PubMed] [Google Scholar]
- Smaczniak C., Li N., Boeren S., America T., van Dongen W., Goerdayal S. S., et al. (2012). Proteomics-based identification of low-abundance signaling and regulatory protein complexes in native plant tissues. Nat. Protoc. 7, 2144–2158. 10.1038/nprot.2012.129 [DOI] [PubMed] [Google Scholar]
- Soussi M., Ocaña A., Lluch C. (1998). Effect of salt stress on growth, photosynthesis and nitrogen fixation in chick-pea (Cicer arietinum L.). J. Exp. Bot. 325, 1329–1337. [Google Scholar]
- Spreitzer R. J., Savucci M. E. (2002). Rubisco: structure, regulatory interactions and possibilities for a better enzme. Annu. Rev. Plant Biol. 53, 449–485. 10.1146/annurev.arplant.53.100301.135233 [DOI] [PubMed] [Google Scholar]
- Sugihara K., Hanagata N., Dubinsky Z., Baba S., Karube I. (2000). Molecular characterization of cDNA encoding oxygen evolving enhancer protein 1 increased by salt treatment in the mangrove Bruguiera gymnorrhiza. Plant Cell Physiol. 41, 1279–1285. 10.1093/pcp/pcd061 [DOI] [PubMed] [Google Scholar]
- Sun-Wada G. H., Wada Y., Futai M. (2003). Vacuolar H+ pumping ATPases in luminal acidic organelles and extracellular compartments: common rotational mechanism and diverse physiological roles. J. Bioen. Biomem. 35, 347–358. 10.1023/A:1025780932403 [DOI] [PubMed] [Google Scholar]
- Suping Z., Roger S., Theodore W. T. (2009). Proteome changes induced by aluminum stress in tomato roots. J. Exp. Bot. 60, 1849–1857. 10.1093/jxb/erp065 [DOI] [PubMed] [Google Scholar]
- Tezara W., Mitchell V. J., Driscoll S. D., Lawlor D. W. (1999). Water stress inhibits plant photosynthesis by decreasing coupling factor and ATP. Nature 401, 914–917 10.1038/44842 [DOI] [Google Scholar]
- Tezara W., Mitchell V. J., Driscoll S. D., Lawlor D. W. (2002). Effects of water deficit and its interaction with CO2 supply on the biochemistry and physiology of photosynthesis in sunflower. J. Exp. Bot. 53, 1781–1791. 10.1093/jxb/erf021 [DOI] [PubMed] [Google Scholar]
- Thornton S., Anand N., Purcell D. (2003). Not just for housekeeping: protein initiation and elongation factors in cell growth and tumorigenesis. J. Mol. Med. 81, 536–548. 10.1007/s00109-003-0461-8 [DOI] [PubMed] [Google Scholar]
- Trebitsh T., Danon A. (2001). Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proc. Natl. Acad. Sci. U.S.A. 98, 12289–12294. 10.1073/pnas.211440698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugane K., Kobayashi K., Niwa Y., Ohba Y., Wada K., Kobayashi H. (1999). A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 11, 1195–1206. 10.1105/tpc.11.7.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda A., Kathiresan A., Bennett J., Takabe T. (2006). Comparative transcriptome analyses of barley and rice under salt stress. Theor. Appl. Genet. 112, 1286–1294. 10.1007/s00122-006-0231-4 [DOI] [PubMed] [Google Scholar]
- Vaseva I. I., Grigorova B. S., Simova-Stoilova L. P., Demirevska K. N., Feller U. (2010). Abscisic acid and late embryogenesis abundant protein profile changes in winter wheat under progressive drought stress. Plant Biol. (Stuttg) 12, 698–707. 10.1111/j.1438-8677.2009.00269.x [DOI] [PubMed] [Google Scholar]
- Vassilev A., Iordanov I., Chakalova E., Kerin V. (1995). Effect of cadmium stress on growth and photosynthesis of young barley (H. vulgare L.) plants. II. Structural and functional changes in the photosynthetic apparatus. Bulg. J. Plant Physiol. 21, 12–21. [Google Scholar]
- Veeranagamallaiah G., Jyothsnakumari G., Thippeswamy M., Chandra Obul Reddy P., Surabhi G. K., Sriranganayakulu G., et al. (2008). Proteomic analysis of salt stress responses in foxtail millet (Setaria glycer L. cv. Prasad) seedlings. Plant Sci. 175, 631–641 10.1016/j.plantsci.2008.06.017 [DOI] [Google Scholar]
- Vij S., Tyagi A. K. (2006). Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genomics 276, 565–575. 10.1007/s00438-006-0165-1 [DOI] [PubMed] [Google Scholar]
- Wahid A., Gelani S., Ashraf M., Foolad M. R. (2007). Heat tolerance in plants: an overview. Environ. Exp. Bot. 61, 199–223 10.1016/j.envexpbot.2007.05.011 [DOI] [Google Scholar]
- Wang C., Hu H., Xu Y., Li X. X., Zhang H. J. (2011). Differential proteomic analysis of cadmium-responsive proteins in wheat leaves. Biol. Plant 55, 586–590 10.1007/s10535-011-0131-6 [DOI] [Google Scholar]
- Wang M. C., Peng Z. Y., Li C. L., Li F., Liu C., Xia G. M. (2008). Proteomic analysis on a high salt tolerance introgression strain of Triticum aestivum/Thinopyrum ponticum. Proteomics 8, 1470–1489. 10.1002/pmic.200700569 [DOI] [PubMed] [Google Scholar]
- Wardley T. M., Bhalla P. L., Dalling M. J. (1984). Changes in the number and composition of chloroplasts during senescence of mesophyll cells of attached and detached leaves of wheat (Triticum aestivum L.). Plant Physiol. 75, 421–424. 10.1104/pp.75.2.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren C. R., Adams M. A., Chen Z. (2000). Is photosynthesis related to concentrations of nitrogen and Rubisco in leaves of Australian native plants? Aust. J. Plant Physiol. 27, 407–416 10.1071/PP98162 [DOI] [Google Scholar]
- Watson B. S., Lei Z., Dixon R. A., Sumner L. W. (2004). Proteomics of Medicago sativa cell walls. Phytochemistry 65, 1709–1720. 10.1016/j.phytochem.2004.04.026 [DOI] [PubMed] [Google Scholar]
- Watson H. C., Walker N. P., Shaw P. J., Bryant T. N., Wendell P. L., Fothergill L. A., et al. (1982). Sequence and structure of yeast phosphoglycerate kinase. EMBO J. 1, 1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss W., Gorg A. (2007). Two-dimensional electrophoresis for plant proteomics. Met. Mol. Biol. 355, 121–143. 10.1385/1-59745-227-0:121 [DOI] [PubMed] [Google Scholar]
- Welch R. M., Norvell W. A. (1999). Mechanisms of cadmium uptake, translocation and deposition in plants, in Cadmium in Soils and Plants, eds McLaughlin M. J., Singh B. R. (Dordrecht: Kluwer Academic Publishers; ), 125–150. [Google Scholar]
- Widjaja I., Naumann K., Roth U., Wolf N., Mackey D., Dangl J. L., et al. (2009). Combining subproteome enrichment and Rubsico depletion enables identification of low abundance proteins differentially regulated during plant defense. Proteomics 9, 138–147. 10.1002/pmic.200800293 [DOI] [PubMed] [Google Scholar]
- Wong W. S., Li G. G., Ning W., Xu Z. F., Hsiao W. L. W., Zhang L. Y., et al. (2001). Repression of chilling-induced ACC accumulation in transgenic citrus by over-production of antisense 1-aminocyclopropane-1-carboxylate synthase RNA. Plant Sci. 161, 969–977 10.1016/S0168-9452(01)00505-2 [DOI] [Google Scholar]
- Woo E. J., Dunwe J. M., Goodenough P. W., Marvier A. C., Pickersgill R. W. (2000). Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Biol. 7, 1036–1040. 10.1038/80954 [DOI] [PubMed] [Google Scholar]
- Worden A. Z., Lee J. H., Mock T., Rouzé P., Simmons M. P., Aerts A. L., et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 324, 268–272. 10.1126/science.1167222 [DOI] [PubMed] [Google Scholar]
- Xu J., Li Y., Sun J., Du L., Zhang Y., Yu Q., et al. (2013). Comparative physiological and proteomic response to abrupt low temperature stress between two winter wheat cultivars differing in low temperature tolerance. Plant Biol. (Stuttg). 15, 292–303. 10.1111/j.1438-8677.2012.00639.x [DOI] [PubMed] [Google Scholar]
- Yamamoto Y., Kobayashi Y., Devi S. R., Rikiishi S., Matsumoto H. (2002). Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 128, 63–72. 10.1104/pp.010417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S. P., Zhang Q. Y., Tang Z. C., Su W. A., Sun W. N. (2006). Comparative proteome analysis provides new insights into chilling stress response in rice. Mol. Cell. Proteomics 5, 235–244. 10.1002/pmic.200400853 [DOI] [PubMed] [Google Scholar]
- Yang F., Jørgensen A. D., Li H., Søndergaard I., Finnie C., Svensson B., et al. (2011). Implications of high-temperature events and water deficits on protein profiles in wheat (Triticum aestivum L. cv. Vinjett) grain. Proteomics 11, 1684–1695. 10.1002/pmic.201000654 [DOI] [PubMed] [Google Scholar]
- Yang Q., Wang Y., Zhang J., Shi W., Qian C., Peng X. (2007). Identification of aluminum-responsive proteins in rice roots by a proteomic approach: cysteine synthase as a key player in Al response. Proteomics 7, 737–749. 10.1002/pmic.200600703 [DOI] [PubMed] [Google Scholar]
- Yang T., Poovaiah B. W. (2002). Hydrogen peroxide homeostasis: activation of plant catalase by calcium calmodulin. Proc. Natl. Acad. Sci. U.S.A. 99, 4097–4102. 10.1073/pnas.052564899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C. M., Wen J., Motes C. M., Sparks J. A., Blancaflor E. B. (2008). A class I ADP-ribosylation factor GTPase-activating protein is critical for maintaining directional root hair growth in Arabidopsis. Plant Physiol. 147, 1659–1674. 10.1104/pp.108.119529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssefian S., Nakamura M., Sano H. (1993). Tobacco plants transformed with the O-acetylserine (thiol) lyase gene of wheat are resistant to toxic levels of hydrogen sulphide gas. Plant J. 4, 759–769. 10.1046/j.1365-313X.1993.04050759.x [DOI] [PubMed] [Google Scholar]
- Zang X., Komatsu S. (2007). A proteomics approach for identifying osmotic-stress-related proteins in rice. Phytochemistry 68, 426–437. 10.1016/j.phytochem.2006.11.005 [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu D., Zhang J., Jiang W., Luo G., Yang W., et al. (2013). Novel insights into the composition, variation, organization, and expression of the low-molecular-weight glutenin subunit gene family in common wheat. J. Exp. Bot. 64, 2027–2040. 10.1093/jxb/ert070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Lv D., Ge P., Bian Y., Chen G., Zhu G., et al. (2014). Phosphoproteome analysis reveals new drought response and defense mechanisms of seedling leaves in bread wheat (Triticum aestivum L.). J. Proteomics 109C, 290–308. 10.1016/j.jprot.2014.07.010 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Shirley N., Lahnstein J., Fincher G. B. (2005). Characterization and expression patterns of UDP-D-Glucuronate decarboxylase genes in barley. Plant Physiol. 138, 131–141. 10.1104/pp.104.057869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Sauve R. (2009). Salt-induced and salt-suppressed proteins in tomato leaves. J. Amer. Soc. Hort. Sci. 134, 289–294. [Google Scholar]
- Zhou S., Sauve R., Thannhauser T. W. (2009). Proteome changes induced by aluminum stress in tomato roots. J. Exp. Bot. 60, 1849–1857. 10.1093/jxb/erp065 [DOI] [PubMed] [Google Scholar]
- Zhu J., Mickelson S. M., Kaeppler S. M., Lynch J. P. (2006). Detection of quantitative trait loci for seminal root traits in maize (Zea mays L.) seedlings grown under differential phosphorus levels. Theor. Appl. Genet. 113, 1–10. 10.1007/s00122-006-0260-z [DOI] [PubMed] [Google Scholar]
- Zörb C., Herbst R., Forreiter C., Schubert S. (2009). Short-term effects of salt exposure on the maize chloroplast protein pattern. Proteomics 9, 4209–4220. 10.1002/pmic.200800791 [DOI] [PubMed] [Google Scholar]