Abstract
Tubulin protomers undergo an extensive array of post-translational modifications to tailor microtubules to specific tasks. One such modification, the acetylation of lysine-40 of α-tubulin, located in the lumen of microtubules, is associated with stable, long-living microtubule structures. MEC-17 was recently identified as the acetyltransferase that mediates this event. We have determined the crystal structure of the catalytic core of human MEC-17 in complex with its cofactor acetyl-CoA at 1.7 Å resolution. The structure reveals that the MEC-17 core adopts a canonical Gcn5-related N-acetyltransferase (GNAT) fold that is decorated with extensive surface loops. An enzymatic analysis of 33 MEC-17 surface mutants identifies hot-spot residues for catalysis and substrate recognition. A large, evolutionarily conserved hydrophobic surface patch is identified that is critical for enzymatic activity, suggesting that specificity is achieved by interactions with the α-tubulin substrate that extend outside of the modified surface loop. An analysis of MEC-17 mutants in C. elegans shows that enzymatic activity is dispensable for touch sensitivity.
Keywords: X-ray crystallography, Tubulin acetyltransferase, Substrate recognition, C. elegans, Mechanosensation
Introduction
Microtubules are cytoskeletal components with a diverse range of essential cellular functions. Cytoplasmic microtubules mediate cell motility, transport, and shape, while specialized microtubule based structures include the mitotic spindle, required for chromosome segregation, and the axoneme, the core component of cilia. All microtubule filaments share a common architecture. Heterodimers of α- and β-tubulin assemble in a head-to-tail fashion into linear polymers termed protofilaments, which associate laterally to form a cylindrical filament. Polymerization of tubulin heterodimers into microtubules will occur spontaneously in vitro in the presence of GTP.1
Cells have adopted several means to tailor the common microtubule structure to distinct cellular functions. These include the activity of microtubule-associated proteins, and the expression of various α- and β-tubulin isoforms.2 Microtubules are also subject to a wide range of evolutionarily conserved post-translational modifications. A majority of these post-translational modifications occur on the flexible C-terminal tails of α- and β-tubulin that protrude from the microtubule. These include the removal and addition of the C-terminal tyrosine of α-tubulin, the removal of the penultimate C-terminal glutamate of α-tubulin, and the addition of poly-glycine and poly-glutamate chains to a number of glutamate residues near the C-termini of both α- and β-tubulin.3-5 An additional microtubule post-translational modification, the acetylation of lysine-40 (K40) of α-tubulin, is believed to uniquely occur in the lumen of the microtubule filament.3,4
Acetylation of α-tubulin K40 has long been associated with stable microtubule structures, such as those in the cytoplasmic array of neuronal axons, and in the ciliary axoneme.4 However, a coherent picture of the effects of this acetylation event on microtubule function has yet to emerge. Genetic manipulations abrogating α-tubulin K40 acetylation appear to cause no detectable phenotype in Tetrahymena thermophila, a loss of touch sensation in Caenorhabditis elegans, anatomical abnormalities in Danio rerio, and delayed cilia assembly in Homo sapiens.6-8 Additional reports have shown that Kinesin-1 preferentially travels on acetylated microtubule tracks, the endoplasmic reticulum preferentially slides along acetylated microtubules, acetylated microtubules bind to and inhibit the Na+/K+ pump, and microtubule acetylation regulates protofilament number in native microtubules.9-15
Recent biochemical and cell biological studies have identified MEC-17, or αTAT-1, as the α-tubulin K40 acetyltransferase.6,8 MEC-17 is highly conserved among ciliated organisms, and displays strong substrate specificity, with no discernable activity towards histone tails. To further aid in the understanding of MEC-17 and the effects of α-tubulin K40 acetylation, we have determined the crystal structure of the catalytic domain of human MEC-17 in complex with acetyl-CoA. Structure-guided mutagenesis was used to determine hot-spot residues that abolish enzymatic activity. Strikingly, a large evolutionarily conserved hydrophobic patch is required for enzymatic activity, suggesting that substrate specificity is established by an extensive interaction with α-tubulin, extending beyond the typical peptide substrate interaction. A mutational analysis in C. elegans establishes an additional, non-catalytic function of MEC-17 that is responsible for touch sensitivity.
Results
Structure determination
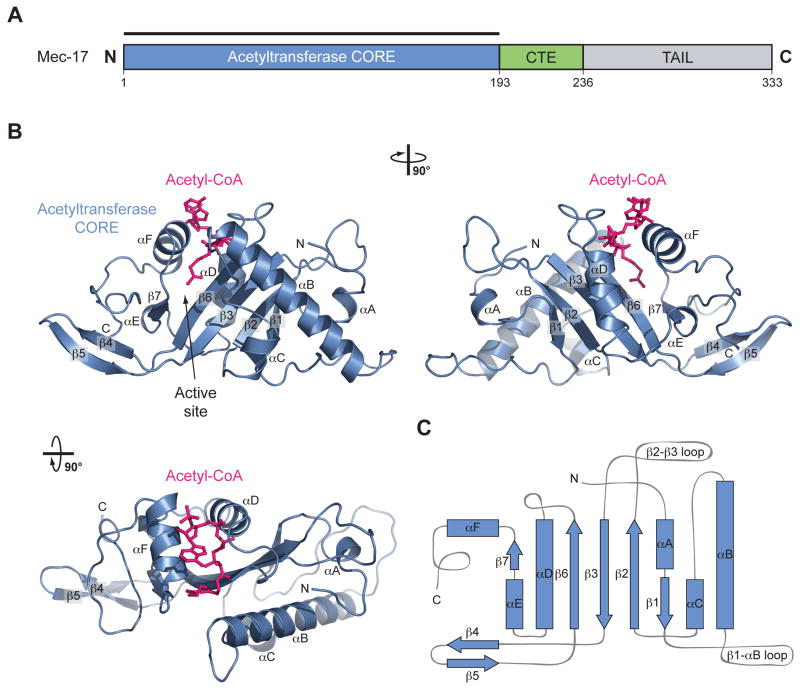
Using secondary structure prediction and sequence conservation analyses, we designed a series of C-terminal deletion constructs of H. Sapiens MEC-17 for expression in E. coli (Table S1). Of these, a stable and well expressing fragment encompassing the core catalytic domain was identified (residues 1 to 193, hereafter referred to as MEC-17CORE). Crystals of MEC-17CORE in the presence of acetyl-CoA were obtained in the orthorhombic space group P21212. The structure was solved by single anomalous dispersion (SAD) using X-ray diffraction data obtained from selenomethionine-labeled crystals. The final model contains residues 1 to 193 and was refined to Rwork and Rfree values of 17.8 % and 21.3 %, respectively. For the details of the data collection and refinement statistics, see Table 1.
Table 1. Crystallographic analysis.
| Crystal 1 SeMet | |
|---|---|
| Data collection | |
| Synchrotron | SSRL |
| Beamline | BL12-2 |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 41.7, 122.2, 37.5 |
| α, β, γ (°) | 90, 90, 90 |
| Se Peak | |
|
|
|
| Wavelength (Å) | 0.97940 |
| Resolution (Å) | 50.0 – 1.7 |
| No. observations | 345,965 |
| No. unique reflections | 39,430 (3,277) |
| Rsym | 7.5 (70.4) |
| <I / σI> | |
| Completeness (%) | 97.2 (81.6) |
| Redundancy | 8.8 (6.4) |
| Refinement | |
| Resolution (Å) | 40.0 – 1.7 |
| No. reflections | |
| total | 37,288 |
| test set | 3,500 (9.4%) |
| Rwork / Rfree | 17.8 / 21.3 |
| No. atoms | 1,826 |
| Protein | 1,567 |
| Ligand/ion | 52 |
| Water | 209 |
| B-factors | 36.2 |
| Protein | 35.9 |
| Ligand/ion | 22.5 |
| Water | 42.4 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.1 |
Highest-resolution shell is shown in parentheses.
SSRL, Stanford Synchrotron Radiation Lightsource
Highest-resolution shell is shown in parentheses
As determined by Molprobity34
Structural Overview
The MEC-17CORE•acetyl-CoA crystal structure is shown in Figure 1 and supplementary movies S1 and S2. MEC-17CAT adopts a globular α/β fold that features a central five-stranded anti-parallel β-sheet, composed of β1-3, β6, and β7. An additional β-hairpin insertion between β3 and β6, β4 and β5, packs against a C-terminal unstructured tail marked by three conserved phenylalanines (F183, F186, and F190). The central β-sheet is decorated on either side by six α-helices of varying lengths (αA-F). The central β-sheet along with αD form a structural element that, despite weak sequence identity, is strongly reminiscent of Gcn5 and other GNAT family members.16,17
Fig. 1. Structural Overview of MEC-17.
(a) Domain structure. Blue, acetyltransferase domain; Green, C-terminal extension (CTE); Gray, unstructured C-terminal tail. (b) Cartoon representation of the MEC-17CORE•acetyl-CoA structure, shown rotated 90° to the right and bottom. (c) Schematic representation of the architecture of MEC-17CORE with secondary structure elements indicated.
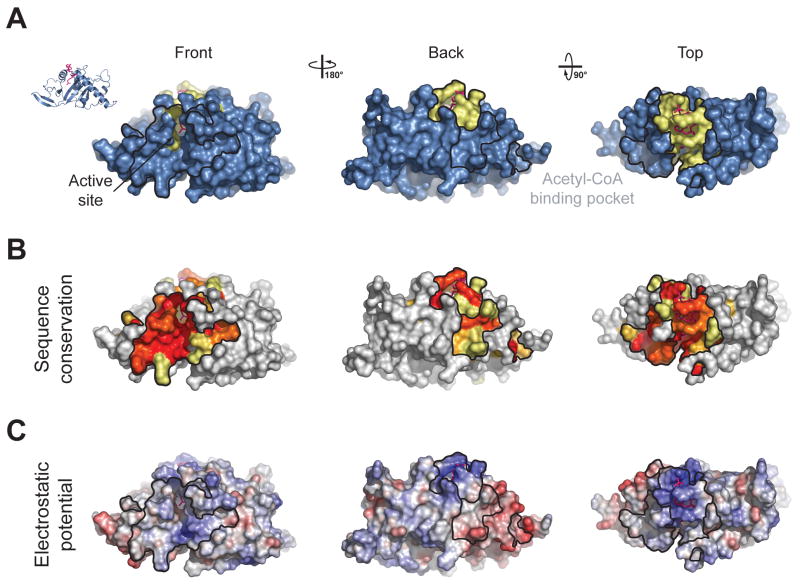
Acetyl-CoA is encapsulated in a highly conserved, predominantly basic pocket on the surface of MEC-17CORE, between αD, αF and β6. A total of 18 residues contact the cofactor, constituting a buried surface area of ∼ 1440 Å2. Two invariant residues (R132 and K162) flank the adenine ring of CoA. R132 and K169 form hydrogen bonds with the 3′-ADP phosphate moiety, while three main chain amides (G134, G136, and R137) form hydrogen bonds with the pyrophosphate. The acetyl-CoA pantetheine arm is bound by a combination of hydrophobic van-der-Waals contacts and hydrogen bonds primarily with F124, Q131, and S160. As with the MEC-17CORE protein, acetyl-CoA adopts an overall conformation similar to that seen in complex with other GNAT family members, particularly in the acetyl and pantetheine moieties.
The donor acetyl group is oriented by a hydrogen bond with the backbone amide of F124, and sits at the base of the acetyl-CoA binding pocket, proximal to a groove which must bind the incoming lysine-40 substrate residue of α-tubulin and its surrounding residues. This groove is also predominantly basic (Fig. 2), and could mediate binding to several acidic residues, which surround lysine-40 on α-tubulin. Our crystal structure is in agreement with previously published structural data from several other groups,18-20 (add reference-Ding) as indicated by RMSDs of ∼0.6 Å2 over 193 Cα atoms for available human MEC-17 structures.
Fig. 2. Surface Properties of MEC-17.
(a) Surface representation colored according to identified binding regions, with the acetyl-CoA binding pocket in yellow. (b) Surface representation of MEC-17CORE colored according to a multispecies sequence alignment. The conservation at each position is mapped onto the surface and is shaded in a color gradient from white (below 60 % identity) to light yellow (60 % identity) to dark red (100 % identity). (c) Surface representation of MEC-17CORE colored according to electrostatic potential. The electrostatic potential is plotted onto the surface and colored in a gradient from red (-10 kBT/e) to blue (10 kBT/e).
Mutational Analysis
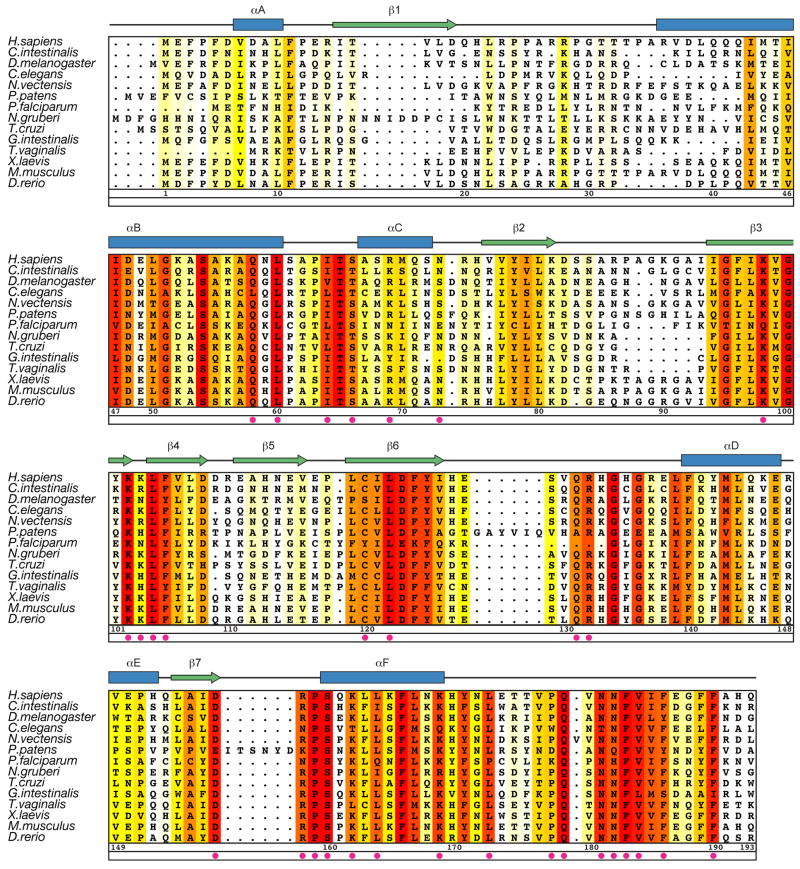
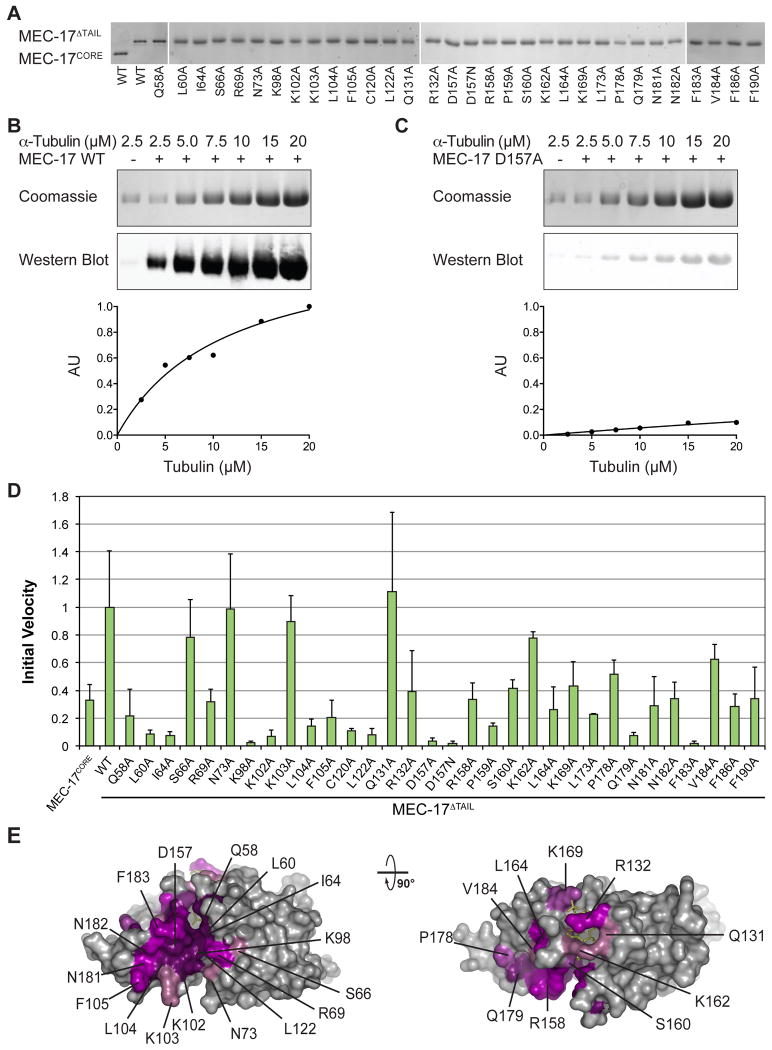
To identify key residues for catalytic activity and substrate recognition by MEC-17, the crystal structure and a sequence conservation analysis (Fig. 3) were used to guide the design of two C-terminal truncations and 33 surface mutations. These variants were tested in an in vitro enzymatic assay, using purified MEC-17 proteins, α-tubulin, and an acetylated K40 α-tubulin specific antibody, which allowed the determination of the initial velocity, Michaelis-Menten constant, and turnover number. All mutants examined were purified to homogeneity in milligram quantities and are indistinguishable from the wildtype protein in their behavior on a gel-filtration column, demonstrating their proper folding. The results of these experiments are summarized in Figure 4 and Table 2.
Figure 3. Multi-species Sequence Alignment of MEC-17 Homologues.
The secondary structure is indicated above the sequence as blue cylinders (α helices), green arrows (β sheets), and gray lines (coil regions). The numbering below the alignment is relative to human MEC-17. Overall sequence conservation at each position is shaded in a color gradient from white (below 60 % identity) to yellow (60 % identity) to dark red (100 % identity). Magenta dots indicate mutated residues in our biochemical and in vivo analyses.
Fig. 4. Mutational Analysis.
(a) WT and mutant forms of MEC-17 used in in vitro biochemical assays visualized on SDS-PAGE gels. (b) Representative biochemical data for wildtype MEC-17. Reaction volumes separated by SDS-PAGE stained with Coomassie brilliant blue, or immunoblotted with an acetylated K40 α-tubulin specific antibody (top). Quantification of immunoblots using an Odyssey Imaging System (bottom). (c) The same as in (b), for a catalytically dead mutant (D157A). (d) Normalized initial velocities of all tested MEC-17CORE mutants. Error bars represent standard error. All experiments were repeated at least 3 times. (e) Surface representation colored according to relative acetyltrasferase activity of alanine mutants in a gradient from light purple (>80 % activity) to dark purple (<20 % activity). A 90° rotated view is shown on the right.
Table 2. Biochemical analysis of MEC-17 Mutants.
The Michaelis-Menten Km and Vmax values were determined by direct fitting of linear initial velocities determined at substrate concentrations of 1-20 μM. Error values represents standard error.
| Biochemical Analysis of MEC-17 Mutants | |||
|---|---|---|---|
|
| |||
| Mutation | Km (μM) | kcat ×10-2 (sec-1) | kcat / Km ×103 (sec-1M-1) |
|
| |||
| WT (1-236) | 13.64±2.70 | 3.79±.39 | 2.78±.84 |
| WT (1-193) | 45.37±55.76 | 3.24±3.03 | 0.71±1.53 |
| Q58A | N/A | N/A | N/A |
| L60A | N/A | N/A | N/A |
| I64A | N/A | N/A | N/A |
| S66A | 84.26±66.24 | 9.23±6.16 | 1.10±1.59 |
| R69A | 98.42±48.78 | 2.76±1.18 | 0.28±0.26 |
| N73A | 23.39±3.24 | 4.91±.43 | 2.10±0.47 |
| K98A | N/A | N/A | N/A |
| K102A | N/A | N/A | N/A |
| K103A | 31.24±18.55 | 2.79±1.15 | 0.89±0.61 |
| L104A | N/A | N/A | N/A |
| F105A | N/A | N/A | N/A |
| C120A | N/A | N/A | N/A |
| L122A | N/A | N/A | N/A |
| Q131A | 14.64±4.06 | 2.75±1.2 | 1.88±1.34 |
| R132A | 52.03±29.31 | 3.32±1.46 | 0.63±0.92 |
| D157A | N/A | N/A | N/A |
| D157N | N/A | N/A | N/A |
| R158A | 79.48±67.3 | 4.81±3.42 | 0.61±0.94 |
| P159A | 36.88±18.73 | .97±.35 | 0.26±0.23 |
| S160A | 50.94±22.51 | 3.67±1.26 | 0.72±0.56 |
| K162A | 48.4±13.66 | 6.49±1.46 | 1.34±0.68 |
| L164A | 3.45±5.25 | .56±.12 | 1.62±2.81 |
| K169A | 43.28±20.78 | 3.45±1.25 | 0.80±0.67 |
| L173A | 36.75±54.19 | 1.99±2.12 | 0.54±1.38 |
| P178A | 18.8±8.39 | 2.39±.63 | 1.27±0.90 |
| Q179A | N/A | N/A | N/A |
| N181A | N/A | N/A | N/A |
| N182A | 58.17±37.87 | 4.14±2.15 | 0.71±0.83 |
| F183A | N/A | N/A | N/A |
| V184A | 47.12±55.95 | 5.77±5.24 | 1.22±2.57 |
| F186A | 15.23±15.17 | .77±.43 | 0.51±0.79 |
| F190A | 107.3±99.03 | 5.97±4.82 | 0.56±0 |
We first tested the activity of the crystallized MEC-17CORE (residues 1 to 193), as well as a slightly longer fragment (residues 1 to 236, hereafter referred to as MEC-17ΔTAIL) that encompasses the MEC-17CORE and an additional evolutionarily conserved C-terminal extension (CTE). In agreement with earlier work MEC-17ΔTAIL exhibits increased activity.8 Specifically, while both MEC17 fragments possess similar turnover numbers, the MEC-17CORE exhibits a three times higher KM value than MEC-17ΔTAIL (∼45 vs. ∼15 μM). While the CTE is absent in our structure, these findings suggest that it forms additional contacts with α-tubulin. Full-length MEC-17 severely degraded during purification, consistent with the predicted lack of secondary structure elements in the remainder of the protein (Fig. 1A), and therefore was not tested. Based on these results, we used MEC-17ΔTAIL for our mutational analyses. Our experimentally determined KM value for MEC-17ΔTAIL (∼15 μM) was in the range of those previously reported (∼2 μM and ∼30 μM), while our turnover number (∼4 × 10-2 s-1) was ∼50 fold higher (∼6 × 10-4 s-1 and ∼20 × 10-4 s-1).8,20
Mutations in the acetyl-CoA binding pocket, including those that interact with the adenine ring (R132A and K162A), ADP-phosphate (K169A), and panthenine arm (Q131A and S160A), all have a mild effect on enzymatic activity. Together, these results suggest a very robust protein-cofactor interaction that is insensitive to individual mutations.
The MEC-17 active site sits at the base of the cofactor-binding pocket. GNAT family members use a general base, a glutamate in the case of Gcn5, to activate a water molecule to deprotonate the substrate lysine, priming it for nucleophilic attack on the cofactor acetyl-group.21,22 Potential general bases in MEC-17 include Q58, C120 and D157, which are located ∼5 Å away from the donor acetyl group. Mutation of each of these residues to alanine severely compromises enzymatic activity. D157 forms a salt bridge with R158, but could potentially undergo structural rearrangements upon substrate binding. To test whether the carboxylate moiety of D157 is required for enzymatic activity, we analyzed a D157N mutant and found that D157N had no enzymatic activity. Additional conserved residues in the active site (L60, I64, and R158) are presumed to position the substrate lysine and their mutation to alanine severely reduced the catalytic activity, as expected. Below the acetyl-CoA binding pocket and active site αC, β3, β6, and connecting loops contribute to a conserved surface groove. Basic residues (R69, K98 and K102) lining this groove prove indispensible for catalysis, and could interact with acidic residues adjacent to the substrate lysine, as previously suggested.18,19,23 The active site architecture and identified catalytic residues are in agreement with the findings in recent publications.18-20
MEC-17 displays two additional highly conserved surface patches which are adjacent to the substrate-binding groove (Fig. 2B). Whereas mutations in one of them (S66A and N73A) had no discernable effect on catalytic activity, mutations in the second patch show a pronounced effect on MEC-17 catalytic activity (L104A, F105A, L164A, L173A, P178A, Q179A, F183A, V184A, F186A, and F190A). These ten residues are located in the β4-β5 hairpin, helix αF, or a C-terminal region following helix αF, are primarily hydrophobic, and constitute approximately 1400 Å2 of solvent accessible surface area on MEC-17CORE. The size of this patch and its distance from the active site, support the conclusion that MEC-17 engages not only the loop that harbors K40, but makes extensive contacts with α-tubulin. We explored this possibility by testing the ability of MEC-17 to acetylate or bind to a 20-residue α-tubulin peptide centered on K40, consistent with earlier work.23 Indeed, the peptide proved insufficient for either enzymatic activity or interaction in vitro (Fig. S1).
In summary, our data suggest that the substrate specificity of MEC17 is the result of numerous contacts between regions of α-tubulin that extend outside the acetylated K40 loop and a large evolutionarily conserved surface patch on MEC-17CORE, as well as a C-terminal extension following the MEC-17CORE. Further atomic details of the substrate recognition of MEC-17 await additional structural characterization.
In vivo Analysis
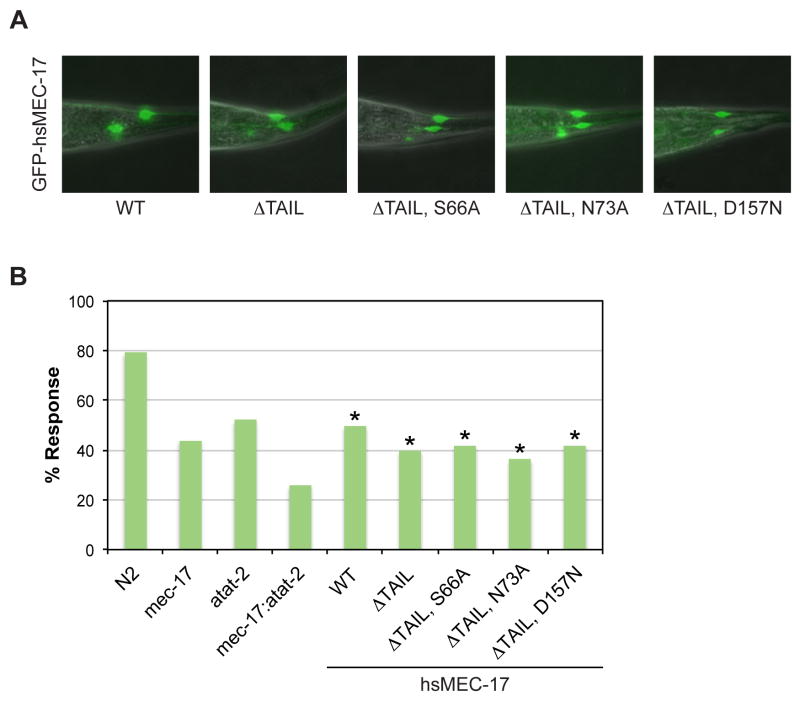
MEC-17 and its paralog ATAT-2 acetylate K40 on α-tubulin and are required for touch sensitivity in C. elegans that is mediated by the touch receptor neurons.6,8,24 However, it is currently unclear whether their catalytic activity is required for mechanosensation, as different groups have found a catalytically inactive MEC-17 mutant to be either sufficient or insufficient for rescuing touch sensitivity of a mec-17; atat-2 double null mutant.8,14 To address these contradicting results, we tested the ability of MEC-17 variants to restore touch sensitivity of a mec-17(ok2109); atat-2(ok2415) double null mutant C. elegans strain. The expression of all MEC-17 variants was verified by the fluorescence signal from a C-terminal GFP-tag, and all worms tested contained marked and comparable levels of GFP fluorescence (Fig. 5A). The results are summarized in Figure 5 and Supplementary Movies S3 and S4.
Fig. 5. Functional Analysis of Human MEC-17 Variants in C. elegans.
(a) Merged differential interference contrast (DIC) and fluorescent images of representative transgenic worms expressing GFP-tagged human MEC-17 variants. (b) Touch sensitivity of C. elegans strains, represented as percentage of positive responses. Asterisks indicate statistical significance based on Fisher's Exact Test. N2 denotes the wild-type strain, mec-17 the single null mutant mec-17(ok2109) strain, atat-2 the single null mutant atat-2(ok2415) strain, and mec-17; atat-2 the double null mutant mec-17(ok2109); atat-2(ok2415) strain. Remaining strains are the mec-17; atat-2 double null mutant strain with the indicated hsMEC-17 variant present as a transgene. All hsMEC-17 variants, irrespective of catalytic activity, rescue the touch sensitivity of the double mutant.
We first tested full-length wild-type human MEC-17, and MEC-17ΔTAIL which both restore touch sensitivity to the same levels as the atat-2(ok2415) single null mutant strain (∼50 and ∼40 % of worms, respectively; Fig. 5B). These results demonstrate that the C-terminal tail of human MEC-17, which is not present in the C. elegans protein, is dispensable for mechanosensation. We next introduced three mutants into the truncated H. sapiens MEC-17 fragment: a catalytically dead active site mutant (D157N) and two catalytically active surface mutants (S66A and N73A). We found that the catalytically dead D157N mutant (42 %) and the catalytically active S66A (42 %) and N73A (36 %) mutants all rescued touch sensitivity in approximately the same percentage of worms. These data are consistent with previous work, despite the use of different genetic backgrounds and expression systems.14 Together, these results support the conclusion that MEC-17 promotes mechanosensation in C. elegans through a non-catalytic function.
Discussion
Microtubules are extensively post-translationally modified on flexible C-terminal tails that protrude from the filament. One prominent exception is the highly conserved acetylation of the lumenal lysine-40 of α-tubulin. To shed further light on this post-translational modification, we have determined the crystal structure of the H. sapiens tubulin acetyltransferase MEC-17 in complex with acetyl-CoA. Our structural data were used to guide a series of biochemical and in vivo experiments to determine residues required for tubulin acetylation and the functional significance of MEC-17 activity in C. elegans.
The detailed catalytic mechanism of MEC-17 remains open for debate. GNAT family members employ a catalytic base to activate a water molecule to deprotonate the substrate lysine.22 Different groups have proposed that Q58, D157, or D157 and C120 participate as general bases in MEC-17.19,20,23 Our data cannot currently distinguish among these potential mechanisms. Each of these residues is an appropriate distance from the acetyl-CoA in our crystal structure. Alanine mutations of Q58, C120 and D157 severely compromise enzymatic activity. Notably, C120 does not exhibit the same level of conservation as the other two potential general bases. Additional studies are required for a comprehensive understanding of catalysis by MEC-17.
Our mutational analysis, along with those of others, point to a basic groove below the active site that interacts with acidic residues surrounding lysine-40 of α-tubulin providing significant substrate specificity relative to histone tails.19,23 However, our further results suggest a more extensive interaction with the substrate that goes beyond the typical recognition of an extended peptide segment, as is common for other acetyltransferases.17,25 In particular, our biochemical data establish an extensive surface patch, particularly in the β4-β5 hairpin and C-terminal loop, that while distal from the active site is necessary for efficient catalysis. While previous results suggested the importance of the β4-β5 hairpin,20 our work greatly expands upon the size of this patch as well as its sensitivity to mutation. Additional evidence for this notion is provided by the inability of MEC-17 to either bind or acetylate a 20-residue α-tubulin peptide centered on K40 in vitro. A more detailed understanding of this recognition awaits further structural exploration of MEC-17 in complex with α-tubulin.
Pioneering genetic studies identified a role for MEC-17 in C. elegans mechanosensation more than thirty years ago.24 Two different groups have examined the role of MEC-17 catalytic function in vivo.8,14. Both studies provide strong evidence that tubulin acetylation (of specialized tubulin MEC-12) requires MEC-17 catalytic activity, but differ in their observations of the extent to which the acetyltransferase activity of MEC-17 is required for whole animal touch sensitivity. Our results with human protein in C. elegans are more consistent with a dispensable function of the catalytic activity for the whole animal behavior. Close examination of the published data as well as our data leads us to question utility of this whole animal behavioral assay for a quantitative assessment of touch receptor neuronal function. The touch assay has been stunningly successful in identifying genes with strong effects on touch receptor neuron function. The double acetyltransferase mutant in everyone's hands still displays some response to light mechanical stimulation, raising the possibility of another acetylation pathway, if you hold with acetylation being crucial for the behavior, or consistent with the view that acetylation helps but is not essential for whole animal mechanosensation. Future work will be required to elucidate the mechanistic basis for the identified non-catalytic activities of MEC-17 in mechanosensation. In particular, cell based assays, for example using genetically encoded calcium sensors, might provide a more accurate indication of the role of acetylation. Our structural and functional analyses presented here provide the framework for these studies and will assist in further research into MEC-17 and tubulin acetylation as a whole.
Materials and Methods
Protein expression and purification
The DNA fragments of H. Sapiens MEC-17 were amplified by PCR (Addgene plasmid 27099) and cloned into a modified pET28a vector (Novagen) that contained a PreScission protease site after the N-terminal hexahistidine tag.8,26 MEC-17 mutants were generated using QuikChange mutagenesis (Stratagene), and confirmed by DNA sequencing. Details of the bacterial expression constructs can be found in Table S1.
All proteins were expressed in E. coli BL21-CodonPLus(De3)-RIL cells (Stratagene) grown in LB media supplemented with appropriate antibiotics. Seleno-L-methionine-labeled protein was produced with a methionine pathway inhibition protocol.27 Protein expression was induced at OD600 of ∼0.6 with 500 μM IPTG for 16-18 hours at 18 °C. Cells were harvested by centrifugation, resuspended in a buffer containing 20 mM Tris, pH 7.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 2 μM bovine lung aprotinin (Sigma), and complete EDTA-free protease inhibitor cocktail (Roche) and flash frozen in liquid nitrogen.
Thawed cells were lysed with a cell disrupter (Avestin) and the lysate was centrifuged for 60 minutes at 40,000 × g. The cleared lysate was applied to a Ni-NTA column (Qiagen) and eluted via an imidazole gradient. Pooled Ni-NTA fractions were cleaved with PreScission protease for 12 hours and further purified over a HiLoad16/60 Superdex 200 column (GE Healthcare) equilibrated in 20 mM Tris, pH 7.0, 150 mM NaCl, and 5 mM DTT. Pooled fractions were concentrated to 12 mg/ml and used in subsequent crystallization trials or for biochemical activity assays.
Structure determination and refinement
Crystals of MEC-17CORE were grown at 21 °C in hanging drops containing 1 μl of protein, which contained a two-fold molar excess of acetyl-CoA (Sigma), and 1 μl of a reservoir solution consisting of 26 % (w/v) PEG 3350, 100 mM Bis-Tris, pH 6.5, and 200 mM NaCl. Crystals grew in the space group P21212 and reached a maximum size of approximately 100 μm × 100 μm × 100 μm within a week. For cryo-protection, crystals were stabilized in 30 % (w/v) PEG 3350, 100 mM Bis-Tris, pH 6.5, 200 mM NaCl and 20 % glycerol (added in 5 % increments). X-ray diffraction data were collected at 100K at beamline 12-2 at the Stanford Synchrotron Radiation Lightsource (SSRL). X-ray intensities were processed using the HKL2000 denzo/scalepack package.28 Phases from a SeMet SAD data set were obtained with SHARP,29 which following density modification by DM30 yielded an electron density map of high quality. ARP/wARP was used for initial automated model building, followed by manual model building in Coot.30,31 Refinement was carried out in Phenix and the final model contains residues 1-193 and was refined to Rfree and Rwork values of 21.3 and 17.8%, respectively.32 The stereochemical quality of the model was assessed with PROCHECK and MolProbity.33,34 For details of the data collection and refinement statistics, see Table 1.
MEC-17 Activity Assays
In vitro MEC-17 acetyltransferase reactions were initiated by the addition of 1 μM recombinant MEC-17 to varying concentrations of porcine brain tubulin (Cytoskeleton), in 10 μl of a buffer containing 80 mM PIPES, pH 6.9, 0.5 mM MgSO4, 0.8 mM EGTA, 1 mM GDP, 5 mM DTT, 10 % glycerol, and 30 μM acetyl-CoA. Reactions were carried out at room temperature for 30 minutes, in the previously demonstrated linear range of MEC-17 activity8 and stopped by the addition of SDS loading buffer. Reaction volumes were separated on a 12.5 % SDS-PAGE gel, transferred to PVDF membranes, and immunoblotted with a mouse anti-acetylated K40 α-tubulin antibody (Sigma, 1:3,000 dilution) followed by an IRDye-800 conjugated goat anti-mouse antibody (Li-Cor, 1:15,000 dilution). Bound IRDye-800 conjugated antibody was detected and quantified using the Odyssey Imaging System (Li-Cor). Previously demonstrated levels of porcine brain tubulin K40 acetylation were used to estimate concentrations from measured absorbance units.35 All initial velocity measurements were the average of at least three independent experiments. Km and kcat values were determined by fitting the concentration dependence of initial velocities using GraphPad Prism software 6.0.
In vivo analysis
C. elegans was handled as described previously.36 All strains used are derivatives of C. elegans N2 Bristol strain. The following mutations were used: LGIV: mec-17(ok2109); LGX: atat-2(ok2415). DNA fragments of H. Sapiens and C. elegans MEC-17 were amplified by PCR and cloned into Fire vector pPD95.75. To drive gene expression in the TRNs, C-terminally GFP-tagged MEC-17 was then fused to 850 bp of the promoter region of mec-7 using PCR fusion.37 The construct was injected with a Pmyo-2∷dsRed co-injection marker.38,39 All worms analyzed were selected for comparable levels of GFP fluorescence. All touch assays were performed as previously described.24 Briefly, nematode touch sensitivity was determined by stroking an eyebrow hair attached to a toothpick across the anterior or posterior half of an adult animal. Movement away from the eyebrow stroke was considered a touch-sensitive response. Results indicate the percentage of worms that responded to a single touch, and all trials included at least 100 worms.
For fluorescence microscopy, animals were mounted on 2 % agarose pads in M9 buffer, containing 10 mM sodium azide (Sigma-Aldrich), and examined with a 40×, 0.75 numerical aperture oil-immersion objective (UPLFLN, Olympus) on a standard epifluorescence microscope (IX71, Olympus). Images were captured with an electron-multiplying CCD camera (C9100-13, Hamamatsu).
Illustration and figures
The sequence alignment of MEC-17 was generated using ClustalX and colored with Alscript.40,41 Figures were generated using PyMOL (www.pymol.org). Electrostatic potential was calculated with APBS.42
Supplementary Material
Highlights.
Crystal structure of the catalytic core of human MEC-17 in complex with acetyl-CoA
Enzymatic analysis of mutants identifies hot-spot residues for catalysis and substrate recognition
Large, conserved surface patch that is critical for enzymatic activity suggests extensive interactions with α-tubulin
Analysis of mutants in C. elegans establishes that enzymatic activity of MEC-17 is dispensable for touch sensitivity
Acknowledgments
We thank Daniel H. Lin, Alina Patke, Tobias Stuwe, Karsten Thierbach, and Yunji Wu for critical reading of the manuscript, David King for mass spectrometry analysis, Maxene Nachury and Irini Topalidou for providing material, Jens Kaiser and the scientific staff of SSRL beamline 12-2 for their support with X-ray diffraction measurements. We acknowledge the Gordon and Betty Moore Foundation for their support of the Molecular Observatory at the California Institute of Technology. The operations at the SSRL are supported by the Department of Energy and by the National Institutes of Health. AMD and PJM are supported by a National Institutes of Health Research Service Award (5 T32 GM07616). AH was supported by the Albert Wyrick V Scholar Award of the V Foundation for Cancer Research, the 54th Mallinckrodt Scholar Award of the Edward Mallinckrodt, Jr. Foundation, and a Kimmel Scholar Award of the Sidney Kimmel Foundation for Cancer Research. Research supported in part by the Howard Hughes Medical Institute, with which P.W.S. is an investigator.
Footnotes
Supplementary Data: Supplementary data to this article can be found online at http://dx.doi.org/xx.xxxx/j.jmb.xxxx.xx.xxx
Accession Numbers: The atomic coordinates and structure factors of the H. sapiens MEC-17 have been deposited in the Protein Data Bank under accession code 4IF5.
References
- 1.Nogales E. Structural insights into microtubule function. Annu Rev Biochem. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 2.Luduena RF. Multiple forms of tubulin: different gene products and covalent modifications. Int Rev Cytol. 1998;178:207–75. doi: 10.1016/s0074-7696(08)62138-5. [DOI] [PubMed] [Google Scholar]
- 3.Garnham CP, Roll-Mecak A. The chemical complexity of cellular microtubules: Tubulin post-translational modification enzymes and their roles in tuning microtubule functions. Cytoskeleton (Hoboken) 2012 doi: 10.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–86. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 5.Szyk A, Deaconescu AM, Piszczek G, Roll-Mecak A. Tubulin tyrosine ligase structure reveals adaptation of an ancient fold to bind and modify tubulin. Nat Struct Mol Biol. 2011;18:1250–8. doi: 10.1038/nsmb.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. MEC-17 is an alpha-tubulin acetyltransferase. Nature. 2010;467:218–22. doi: 10.1038/nature09324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaertig J, Cruz MA, Bowen J, Gu L, Pennock DG, Gorovsky MA. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J Cell Biol. 1995;129:1301–10. doi: 10.1083/jcb.129.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc Natl Acad Sci USA. 2010;107:21517–22. doi: 10.1073/pnas.1013728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casale CH, Alonso AD, Barra HS. Brain plasma membrane Na+,K+-ATPase is inhibited by acetylated tubulin. Mol Cell Biochem. 2001;216:85–92. doi: 10.1023/a:1011029125228. [DOI] [PubMed] [Google Scholar]
- 10.Cueva JG, Hsin J, Huang KC, Goodman MB. Posttranslational acetylation of alpha-tubulin constrains protofilament number in native microtubules. Curr Biol. 2012;22:1066–74. doi: 10.1016/j.cub.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman JR, Webster BM, Mastronarde DN, Verhey KJ, Voeltz GK. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–75. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–72. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 13.Santander VS, Bisig CG, Purro SA, Casale CH, Arce CA, Barra HS. Tubulin must be acetylated in order to form a complex with membrane Na(+),K (+)-ATPase and to inhibit its enzyme activity. Mol Cell Biochem. 2006;291:167–74. doi: 10.1007/s11010-006-9212-9. [DOI] [PubMed] [Google Scholar]
- 14.Topalidou I, Keller C, Kalebic N, Nguyen KC, Somhegyi H, Politi KA, Heppenstall P, Hall DH, Chalfie M. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr Biol. 2012;22:1057–65. doi: 10.1016/j.cub.2012.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zampar GG, Chesta ME, Carbajal A, Chanaday NL, Diaz NM, Casale CH, Arce CA. Acetylated tubulin associates with the fifth cytoplasmic domain of Na(+)/K(+)-ATPase: possible anchorage site of microtubules to the plasma membrane. Biochem J. 2009;422:129–37. doi: 10.1042/BJ20082410. [DOI] [PubMed] [Google Scholar]
- 16.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vetting MW, LP SdC, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–26. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 18.Kormendi V, Szyk A, Piszczek G, Roll-Mecak A. Crystal structures of tubulin acetyltransferase reveal a conserved catalytic core and the plasticity of the essential N-terminus. J Biol Chem. 2012 doi: 10.1074/jbc.C112.421222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taschner M, Vetter M, Lorentzen E. Atomic resolution structure of human alpha-tubulin acetyltransferase bound to acetyl-CoA. Proc Natl Acad Sci USA. 2012;109:19649–54. doi: 10.1073/pnas.1209343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friedmann DR, Aguilar A, Fan J, Nachury MV, Marmorstein R. Structure of the alpha-tubulin acetyltransferase, alphaTAT1, and implications for tubulin-specific acetylation. Proc Natl Acad Sci USA. 2012;109:19655–60. doi: 10.1073/pnas.1209357109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanner KG, Trievel RC, Kuo MH, Howard RM, Berger SL, Allis CD, Marmorstein R, Denu JM. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–60. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 22.Rojas JR, Trievel RC, Zhou J, Mo Y, Li X, Berger SL, Allis CD, Marmorstein R. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–8. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 23.Li W, Zhong C, Li L, Sun B, Wang W, Xu S, Zhang T, Wang C, Bao L, Ding J. Molecular basis of the acetyltransferase activity of MEC-17 towards alpha-tubulin. Cell Res. 2012;22:1707–11. doi: 10.1038/cr.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chalfie M, Sulston J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev Biol. 1981;82:358–70. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 25.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–95. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 26.Hoelz A, Nairn AC, Kuriyan J. Crystal structure of a tetradecameric assembly of the association domain of Ca2+/calmodulin-dependent kinase II. Mol Cell. 2003;11:1241–51. doi: 10.1016/s1097-2765(03)00171-0. [DOI] [PubMed] [Google Scholar]
- 27.Doublie S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol. 1997;276:523–30. [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–324. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.La Fortelle E, DB G. Maximum-likelihood heavy-atom parameteter refinement in the MIR and MAD methods. Methods Enzymol. 1997;276:476–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 30.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski RA, M MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 34.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–5. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 36.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–30. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- 38.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–82. [PubMed] [Google Scholar]
- 39.Shaham S, Horvitz HR. Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev. 1996;10:578–91. doi: 10.1101/gad.10.5.578. [DOI] [PubMed] [Google Scholar]
- 40.Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–5. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 41.Barton GJ. ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 42.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.