Abstract
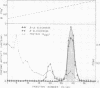
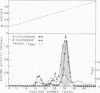
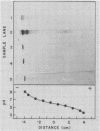
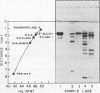
The fact that fungal glucans will stimulate soybeans to accumulate phytoalexins prompted an investigation of soybean cell β-1,3-glucanases and β-glucosidases, as well as the ability of these enzymes to hydrolyze the fungal glucans. Several β-1,3-glucanases and β-glucosidases can be solubilized from the walls of suspension-cultured soybean cells by treatment with 1.0 molar sodium acetate buffer. An enzyme, which has been termed β-glucosylase I, is the dominant β-1,3-glucanase in the cell wall extracts. Utilizing CM-Sephadex chromatography, hydroxylapatite chromatography, and affinity chromatography, β-glucosylase I has been purified 71-fold, with 39% recovery, from the mixture of cell wall enzymes. The affinity chromatography column material was prepared by covalently attaching p-aminophenyl-1-β-d-glucopyranoside, an analog of a β-glucosylase I substrate, to Sepharose. β-Glucosylase I, purified by this procedure, yields a single band on isoelectric focusing gels (pH 8.9). However, the purified β-glucosylase I yields a darkly-staining protein band at an apparent molecular weight of 69,000 and several lightly-staining protein bands in sodium dodecyl sulfate polyacrylamide gels. Additional purification procedures fail to remove these lightly-staining protein bands.
β-Glucosylase I will hydrolyze the β-glucan substrates, laminarin (3-linked) and lichenan (3- and 4-linked), and therefore, possesses β-glucanase activity. Studies of the progressive hydrolysis of laminarin by β-glucosylase I demonstrate that the enzyme hydrolyzes polysaccharide substrates in an exo manner. β-Glucosylase I will also hydrolyze a variety of low molecular weight β-glucosides including various β-linked diglucosides. Thus, β-glucosylase I also possesses β-glucosidase activity.
Several lines of evidence are presented that the β-glucanase and the β-glucosidase activities exhibited by purified β-glucosylase I preparations are catalyzed by the same enzyme. This evidence includes inhibition studies which indicate that the β-glucanase and the β-glucosidase activities of β-glucosylase I are catalyzed at the same active site. β-Glucosylase I will also catalyze glucosyl transfer. This catalytic activity is responsible for the observed ability of the enzyme to synthesize di- and trisaccharides from laminarin. The disaccharides formed by β-glucosylase I-catalyzed transglucosylation are the β-anomers of the 6-, 4-, 3-, and 2-linked diglucosides in the relative proportions of 10:1:1:1. The ability of β-glucosylase I to catalyze glucosyl transfer indicates that β-glucosylase I is biochemically more similar to previously studied β-glucosidases than to β-glucanases. This conclusion is supported by the observation that β-glucosylase I is strongly inhibited by 1,5-d-gluconolactone, an inhibitor of β-glucosidases but not of β-glucanases.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abeles F. B., Bosshart R. P., Forrence L. E., Habig W. H. Preparation and purification of glucanase and chitinase from bean leaves. Plant Physiol. 1971 Jan;47(1):129–134. doi: 10.1104/pp.47.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albersheim P., Valent B. S. Host-pathogen interactions in plants. Plants, when exposed to oligosaccharides of fungal origin, defend themselves by accumulating antibiotics. J Cell Biol. 1978 Sep;78(3):627–643. doi: 10.1083/jcb.78.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers A. R., Ebel J., Valent B., Albersheim P. Host-Pathogen Interactions: X. Fractionation and Biological Activity of an Elicitor Isolated from the Mycelial Walls of Phytophthora megasperma var. sojae. Plant Physiol. 1976 May;57(5):760–765. doi: 10.1104/pp.57.5.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Albersheim P. Host-Pathogen Interactions : XVII. HYDROLYSIS OF BIOLOGICALLY ACTIVE FUNGAL GLUCANS BY ENZYMES ISOLATED FROM SOYBEAN CELLS. Plant Physiol. 1981 Jul;68(1):221–228. doi: 10.1104/pp.68.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Wade M., Albersheim P. Host-Pathogen Interactions: XV. Fungal Glucans Which Elicit Phytoalexin Accumulation in Soybean Also Elicit the Accumulation of Phytoalexins in Other Plants. Plant Physiol. 1978 Dec;62(6):918–921. doi: 10.1104/pp.62.6.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darvill A. G., McNeil M., Albersheim P. Structure of Plant Cell Walls: VIII. A New Pectic Polysaccharide. Plant Physiol. 1978 Sep;62(3):418–422. doi: 10.1104/pp.62.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel J., Ayers A. R., Albersheim P. Host-Pathogen Interactions: XII. Response of Suspension-cultured Soybean Cells to the Elicitor Isolated from Phytophthora megasperma var. sojae, a Fungal Pathogen of Soybeans. Plant Physiol. 1976 May;57(5):775–779. doi: 10.1104/pp.57.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel J., Schaller-Hekeler B., Knobloch K. H., Wellman E., Grisebach H., Hahlbrock K. Coordinated changes in enzyme activities of phenylpropanoid metabolism during the growth of soybean cell suspension cultures. Biochim Biophys Acta. 1974 Oct 8;362(3):417–424. doi: 10.1016/0304-4165(74)90137-8. [DOI] [PubMed] [Google Scholar]
- HOFSTEE B. H. Non-inverted versus inverted plots in enzyme kinetics. Nature. 1959 Oct 24;184:1296–1298. doi: 10.1038/1841296b0. [DOI] [PubMed] [Google Scholar]
- Heyn A. N. Glucanase activity in coleoptiles of Avena. Arch Biochem Biophys. 1969 Jul;132(2):442–449. doi: 10.1016/0003-9861(69)90387-7. [DOI] [PubMed] [Google Scholar]
- Keegstra K., Albersheim P. The Involvement of Glycosidases in the Cell Wall Metabolism of Suspension-cultured Acer pseudoplatanus Cells. Plant Physiol. 1970 Jun;45(6):675–678. doi: 10.1104/pp.45.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee H. J., Wilson I. B. Enzymic parameters: measurement of V and Km. Biochim Biophys Acta. 1971 Sep 22;242(3):519–522. doi: 10.1016/0005-2744(71)90144-6. [DOI] [PubMed] [Google Scholar]
- Moore A. E., Stone B. A. A -I,3-glucan hydrolase from Nicotiana glutinosa. I. Extraction, purification and physical properties. Biochim Biophys Acta. 1972 Jan 20;258(1):238–247. doi: 10.1016/0005-2744(72)90982-5. [DOI] [PubMed] [Google Scholar]
- Moore A. E., Stone B. A. A -I,3-glucan hydrolase from Nicotiana glutinosa. II. Specificity, action pattern and inhibitor studies. Biochim Biophys Acta. 1972 Jan 20;258(1):248–264. doi: 10.1016/0005-2744(72)90983-7. [DOI] [PubMed] [Google Scholar]
- Reese E. T., Maguire A. H., Parrish F. W. Glucosidases and exo-glucanases. Can J Biochem. 1968 Jan;46(1):25–34. doi: 10.1139/o68-005. [DOI] [PubMed] [Google Scholar]
- Reid M. S., Bieleski R. L. A simple apparatus for vertical flat-sheet polyacrylamide gel electrophoresis. Anal Biochem. 1968 Mar;22(3):374–381. doi: 10.1016/0003-2697(68)90278-9. [DOI] [PubMed] [Google Scholar]
- Steers E., Jr, Cuatrecasas P., Pollard H. B. The purification of beta-galactosidase from Escherichia coli by affinity chromatography. J Biol Chem. 1971 Jan 10;246(1):196–200. [PubMed] [Google Scholar]
- Villa T. G., Notario V., Benítez T., Villanueva J. R. Purification of an exo-1,3-beta-glucanase from Candida utilis. Can J Biochem. 1976 Nov;54(11):927–934. doi: 10.1139/o76-134. [DOI] [PubMed] [Google Scholar]
- Wong Y. S., Maclachlan G. A. 1,3-beta-D-glucanases from Pisum sativum seedlings. I. Isolation and purification. Biochim Biophys Acta. 1979 Dec 7;571(2):244–255. doi: 10.1016/0005-2744(79)90095-0. [DOI] [PubMed] [Google Scholar]
- Wong Y. S., Maclachlan G. A. 1,3-beta-D-glucanases from Pisum sativum seedlings. II. Substrate specificities and enzymic action patterns. Biochim Biophys Acta. 1979 Dec 7;571(2):256–269. doi: 10.1016/0005-2744(79)90096-2. [DOI] [PubMed] [Google Scholar]
- York W. S., McNeil M., Darvill A. G., Albersheim P. Beta-2-linked glucans secreted by fast-growing species of Rhizobium. J Bacteriol. 1980 Apr;142(1):243–248. doi: 10.1128/jb.142.1.243-248.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]