Abstract
Based on a bioinformatics analysis of the Middle East respiratory syndrome coronavvirus (MERS-CoV) S protein, we synthesized a panel of peptides coupled to keyhole limpet haemocyanin and used them to raise antibodies in rabbits. In addition, the recombinant receptor-binding domain (RBD) was used to raise polyclonal antibodies in mice. All of the antibodies raised by S-peptide immunisation were specific and sensitive for S protein expressed in transfected cells in the indirect immunofluorescence assay or Western blotting. The RBD efficiently elicited neutralizing antibodies against MERS-CoV by blocking viral entry at the binding step. Furthermore, we found that the SP3 peptide, corresponding to amino-acid residues 736–761 of the S protein, elicited robust neutralizing activities by blocking viral entry at the postbinding and membrane fusion steps. We conclude that amino-acid residues 736–761 of the S protein carry neutralizing epitopes that may be used in the development of vaccines and antiviral agents against MERS-CoV.
Introduction
Middle East respiratory syndrome coronavvirus (MERS-CoV), a novel β coronavirus, was recently identified as the causative agent of a severe acute respiratory illness in humans in the Middle East (24). This finding has raised serious concerns about the possibility of a pandemic infection similar to that caused by severe acute respiratory syndromecoronavirus (SARS-CoV) (2,20). As of 4 July 2014, there have been 827 confirmed cases of infection with MERS-CoV, and 287 of the affected people died (www.who.org). Cases have been linked to many regions of Asia, Africa, Europe, and America. According to recent data, people with a mild respiratory illness may be infected with MERS-CoV; in some cases, the infected people have no respiratory symptoms (5,20,21,23). Patients with a chronic disease or compromised immune system have a higher risk of becoming infected and/or developing complications (2,5,20,21,26). There have been small clusters of infection in several countries, suggesting that person-to-person transmission is possible when close contact occurs (16,21). The rapid identification of effective therapeutics is a high priority, because there is currently no specific therapy or vaccine for MERS-CoV and the resulting disease is severe with a high case-fatality rate.
MERS-CoV belongs to the genus Betacoronavirus, in the Family Coronaviridae, as SARS-CoV does. However, they do not use the same host cell receptor for infection (14,19). Complete genome sequencing indicated that this new virus is the first lineage C Betacoronavirus species known to infect humans (22). CoVs are positive-strand RNA viruses (4). The virion comprises a nucleocapsid (N) core surrounded by an envelope containing three membrane proteins: spike (S), membrane, and envelope. The S protein of MERS-CoV, a 1353-amino-acid type I membrane glycoprotein, is known to be responsible for receptor binding (9,15,19,22), membrane fusion (9a), and the induction of neutralising antibodies (7–9,18). Although the S protein of MERS-CoV shares little amino-acid identity with that of other CoVs (<30%) (22), it shares common structural features with the S proteins of other CoVs (11,15,22,23a). Its two components are S1, which contains the receptor-binding domain (RBD) (7–9,15,18), and S2, which contains the fusion peptide (9a). Dipeptidyl peptidase 4 (also known as CD26) was identified as a functional receptor for MERS-CoV, and the structural basis of S/receptor engagement has been explored (15,19,20,21,23a). A recent report, indeed, showed the presence of S-specific neutralizing antibodies in MERS-CoV-infected patients (20,21,26). Therefore, the S protein is recognized as the primary target of neutralizing antibodies. Knowledge of the antigenic determinants that can elicit neutralizing antibodies could be beneficial for the development of a protective vaccine.
In this study, we aimed at identifying neutralizing epitopes in the MERS-CoV S protein that may be used for the development of a vaccine or therapeutic agents against MERS-CoV infection. Although a properly folded RBD could be the most import target for neutralizing antibodies, as demonstrated for SARS-CoV (6,10,13), the identification of other neutralizing epitopes in the S could assist in the development of a vaccine and therapeutics against MERS-CoV infection. We synthesized peptides from different regions of the MERS-CoV S protein based on a bioinformatics analysis and used them to raise antibodies in rabbits. Recombinant RBD (rRBD) was used to raise polyclonal antibodies in mice. The antisera were then tested in terms of their ability to bind S protein derived from the transfection of the codon-optimized S gene and their capacity to neutralize MERS-CoV using an in vitro neutralizing assay based on lentiviral pseudotyped particles expressing full-length MERS-CoV S protein. We confirmed that the RBD could efficiently elicit neutralizing antibodies against MERS-CoV and is an essential target for vaccine development. A novel neutralizing epitope corresponding to amino-acid residues 736–761 of the S protein was also identified.
Materials and Methods
Cell lines and plasmids
BHK-21, Huh-7, and 293FT were cultured in Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 10% heat-inactivated fetal bovine serum (HyClone), penicillin (100 U/mL), streptomycin (100 g/mL), nonessential amino acids (0.1 mM), and L-glutamine (2 mM; Life Technologies).
The codon-optimized S gene of MERS-CoV derived from the published sequence (GenBank accession number: JX869059) was chemically synthesized (Qingke Bio-Tech Engineering Service Co., Ltd.). The S expression plasmid was constructed by inserting the full-length S gene into pVRC (a gift from Dr. Gary Nabel, VRC, NIH, USA) under the control of the CMV promoter to produce pVRC-SY (nCoV).
pNL4-3R-.E-Luc (encoding a provirus containing luciferase and HIV gag-pol), pVRC8304 (encoding the spike glycoprotein of SARS-CoV), and pM.D (encoding the VSV-G glycoprotein) were used to generate pseudoviruses and have been described elsewhere (9a,10,18,24).
Synthesis of the rRBD and peptide panel
The amino-acid sequence of the MERS-CoV S protein was downloaded from NCBI GenBank, and immunogenic regions containing potential human B-cell epitopes were predicted using BepiPred and ABCPred B (Table 1). The designated panel of SP2 (-1 and -2), SP3, and NP peptides was synthesized with a cysteine residue added to the C terminal using the solid-phase method (ZhongKeYaGuang Co.) and dissolved in 5% dimethyl sulfoxide at 2 mg/mL. In addition, the peptides were coupled to keyhole limpet haemocyanin (KLH) at the N-terminus by the method of Lee et al. (15a) and treated as previous described (21a). All peptides were analyzed by high-performance liquid chromatography and mass spectroscopy to verify their identity. The information of the S and N peptides used as the immunogens are listed in Table 2.
Table 1.
Design of the Synthesized Peptides Based on Data from a Previous Bioinformatics Analysis
| Method | Position | Epitope | Score |
|---|---|---|---|
| BCPred | 509 | DDRTEVPQLVNANQYSPCVS | 0.979 |
| 530 | VPSTVWEDGDYYRKQLSPLE | 0.848 | |
| 738 | LPDTPSTLTPRSVR | 1 | |
| ABCred | 509 | DDRTEVPQLVNANQYS | 0.8 |
| 532 | STVWEDGDYYRKQLSP | 0.97 | |
| 736 | CALPDTPSTLTPRSVR | 0.89 | |
| 748 | RSVRSVPGEMRLAS | 0.9 |
Table 2.
Immunogens and Groupings Used to Raise Antisera in Mice or Rabbits
| Group | Immunogens | Domain or location of epitopes | Antiserum |
|---|---|---|---|
| 1 | RBD (MERS-CoV) | 367–606 aa of S | Anti-RBD (EMC), from mice |
| 2 | RBD (SARS-CoV) | 318–510 aa of S | Anti-SARS (W624), from mice |
| 3 | P24 (HIV-1) | Anti P24, from mice | |
| 4 | SP2-1 (MERS-CoV) coupled to KLH; | 509–530 aa of S | Anti SP2, from rabbit |
| SP2-2 (MERS-CoV) coupled to KLH | 529–554 aa of S | ||
| 5 | SP3 (MERS-CoV) coupled to KLH | 731–761 aa of S | Anti SP3, from rabbit |
| 6 | NP1 (MERS-CoV) coupled to KLH | 201–218 aa of N | Anti NP1, from rabbit |
rRBD was prepared based on the coding sequence of MERS-CoV RBD (GenBank accession number: JX869059; spike residues 367–606) using the Bac-to-Bac Baculovirus Expression System (Life Technologies) and purified on a Superdex 200 column (GE Healthcare) as previously described. The purified RBD of SARS-CoV (covering spike residues 318–510 and expressed in HEK293T cells; a gift from Dr. He Yuxian, Institute of Pathogen Biology, CAMS, Beijing) was used for mouse immunization as a control (Table 2).
Generation of polyclonal antibodies against immunogens in mice or rabbits
All animal experiments were conducted in accordance with the Guidelines for Animal Experiments described and approved by the Institutional Animal Care and Use Committee of the Chinese Center for Disease Control and Prevention.
The purified Rrbd (5 microgram per dose) was used to immunize female Balb/c mice (6–8 weeks old). The rRBD was supplemented with complete Freund's adjuvant (CFA; Sigma) for the first immunization and incomplete Freund's adjuvant (Sigma) for the second and third immunization. The mice were immunized thrice at 3-week intervals and monitored for seroconversion to the rRBD using an enzyme-linked immunosorbent assay (ELISA). Purified HIV-1 P24 protein was also used to immunize mice following the same protocol, to produce antisera of non-CoV as an irrelative control to test the background of assay.
One milligram of each of the S peptides was mixed with an equal volume of CFA and used as the formulation for immunization. One New Zealand White rabbit (female Balb/c, 6–8 weeks old) per group was immunized with the formulations to raise antibodies against the respective S peptides. The rabbits were given triple booster injections at 3 weeks after the initial immunization. Incomplete Freund's adjuvant instead of CFA was used in the formulations for booster injections at 2-week intervals. The sera were sampled for testing at 2 weeks after the last immunization.
Western blot analysis
293FT cells were transfected with individual S-expression plasmids using Lipofectamine 2000 reagent (Life Technologies). At 36 h post-transfection, the cells were lysed in ice-cold RIPA buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 0.5% sodium deoxycholate) supplemented with a protease inhibitor mixture (Sigma). The lysates were kept on ice for 10 min, centrifuged, and resolved by 6% SDS-PAGE. The proteins were then transferred to a PVDF membrane (Bio-Rad), blocked with 5% skim milk in PBST for 1 h, and then probed with the indicated primary antibodies at an appropriate dilution overnight at 4°C. The next day, the membrane was incubated with corresponding IRD Flour 800-labeled IgG secondary antibodies (LI-COR Biosciences) and were scanned by the Odyssey Infrared Imaging System (LI-COR Biosciences).
Indirect immunofluorescence assay
An immunofluorescence assay (IFA) was used to detect nCoV S glycoprotein expression in BHK cells. Briefly, BHK cells cultured on coverslips were fixed in 4% formaldehyde, permeabilized in 0.5% Triton X-100, blocked in 5% bovine serum albumin in phosphate-buffered saline, and then probed with indicated primary antibodies for 1 h at room temperature. After a wash, cells were incubated with their respective Alexa Fluor 488-conjugated secondary antibodies for another 1 h. The cells were then washed and stained with 4, 6-diamidino-2-phenylindole (DAPI) to detect nuclei. Fluorescence images were obtained and analyzed using an LSM 510 laser-scanning confocal microscope (Carl Zeiss).
Enzyme-linked immunosorbent assay
Testing for immunogen reactivity was performed using the rRBD or various peptides as the antigen in an ELISA. The ELISA procedure was carried out as previously described (10,24).
Neutralization assay
The LV-CoVpps (MERS-CoVpp and SARS-CoVpp) were produced by the transfection of human embryonic kidney 293FT cells using Fugen HD reagent (Roche) with a combination of pNL4-3R-E-Luc and pVRC-SY (nCoV) or pVRC8304. As a control, pVRC-SY (nCoV) was replaced with pMD.G. The LV-CoVpps were harvested at 48 h post-transfection, and the virus titer (presented as the HIV p24 antigen concentration) was determined using the Vironostika HIV-1 Antigen Microelisa System (BioMérieux).
To evaluate the neutralizing antibodies raised by the various peptides in our panel, we produced LV-CoVpps that harbored a luciferase reporter gene. To map the entry steps targeted by the polyclonal antisera, we established two LV-CoVpp-based neutralization assays to distinguish between antibody-mediated interference during binding and postbinding events by adapting a previously published method. In Protocol I, serially diluted sera after purification on a protein G column were incubated with LV-CoVpps at 37°C for 1 h before binding to the target cells, and the mixtures were then added to Huh7 cells for infection. After 48 h, the cells were lysed for a luciferase activity assay. In Protocol II, LV-CoVpps were incubated with Huh7 cells for 4 h at 4°C, and the antisera were then added to the inoculum. In this assay, the antibody was added after the binding of LV-CoVpp to the target cells, thus enabling us to study the effect of the antibodies on viral entry at the postbinding and fusion steps.
Normalized neutralizations were calculated as follows:
(relative luciferase units of pps with mock sera–relative luciferase units of pps with immune serum in a given dilution)/relative luciferase units of pps with mock sera.
We performed a neutralization assay using 96-well plates, and all experiments were carried out in triplicate.
Statistical analysis
Significant differences between the experimental and control groups were evaluated by a one-way analysis of variance using SPSS software (release 12.1; SPSS, Inc.). Differences were considered significant at p<0.05.
Results
Generation of antisera against the rRBD and panel of peptides
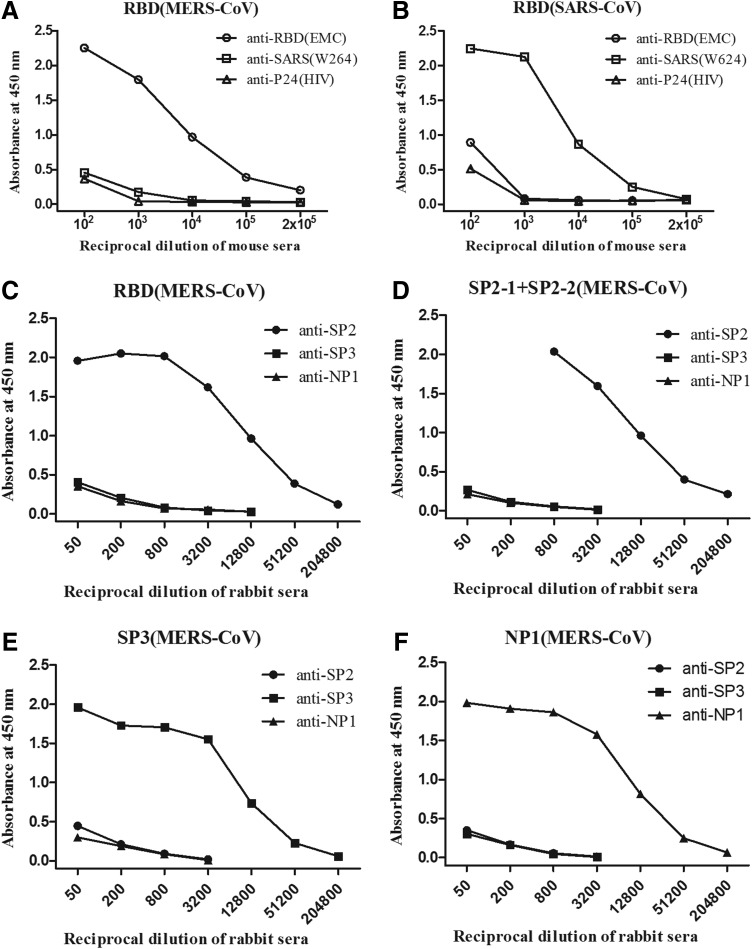
To determine the neutralizing epitopes on the S of MERS-CoV, the rRBD and panel of S peptides (SP2-1, SP2-2, and SP3) covering different regions were used as antigens (Fig. 1) to raise polyclonal antibodies in mice and rabbits, respectively. The RBD of SARS-CoV and two peptides from the N of MERS-CoV were included as control antigens (Table 2). Multiple booster injections were performed in an attempt to increase the antibody titers and specificity. All immunogens elicited high titers of IgG antibodies specific for the respective antigens as detected by ELISA (Fig. 2). The mean end-point titer of anti-RBD IgG in the mice reached >1×105, while that of the anti-S or -N peptide IgGs in the rabbits reached 1×104 after the last immunization. The antisera against the SP2 peptides in immunized mice showed a very high binding capacity with the RBD of MERS-CoV. No cross-reactivity was observed between the antisera against the RBD of MERS-CoV and the RBD of SARS-CoV, or vice versa. In addition, the MERs-CoV S-specific antiserum did not cross-react with the N peptides.
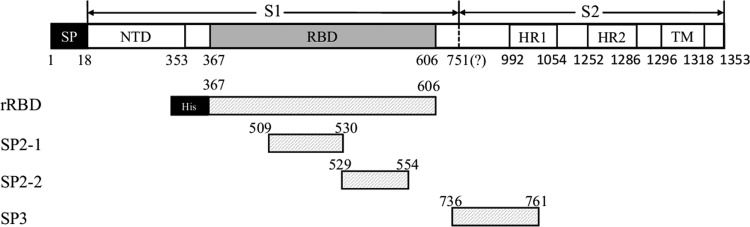
FIG. 1.
Schematic diagram of the Middle East respiratory syndrome coronavvirus (MERS-CoV) S protein and location of the immunogens (rRBD and panel of peptides) used in this study. The codon-optimized full-length S coding gene was inserted into the expression vector pVRC. SP, signal peptide (residues 1–18); RBD, receptor-binding domain (residues 367–606); 751?, S1/S2 cleavage site; HR1, heptad repeat 1 (residues 992–1054); HR2, heptad repeat 2 (residues 1252–1286); TM, transmembrane domain (residues 1296–1353); rRBD, recombinant RBD from the Bac-to-Bac system (indicated by a gray box). The S peptides used in this study were SP2-1 (residues 509–530), SP2-2 (residues 529–554), and SP3 (residues 736–761) and are indicated by gray boxes.
FIG. 2.
The immunization of mice with the RBD or rabbits with peptide-keyhole limpet hemocyanin (KLH) conjugates induced immunogen-specific serum IgG antibodies. The results are presented as the mean A450±standard error (SE) of triplicate experiments per group as detected by an ELISA using serial dilutions of the sera and indicated antigens. The antigen-specific IgG binding ability of pooled serum samples was determined by an rRBD ELISA (A–C), SP2 peptide ELISA (D), SP3 peptide ELISA (E), and N peptide ELISA (F).
Characterization of full-length S expression in transfected cells by Western blotting and IFA
The specificity of the sera induced by the rRBD and SP peptides was further confirmed by Western blotting and IFA using recombinant S proteins from 293FT and BHK cells transfected with pVRC-SY (nCoV) (Fig. 3).
FIG. 3.
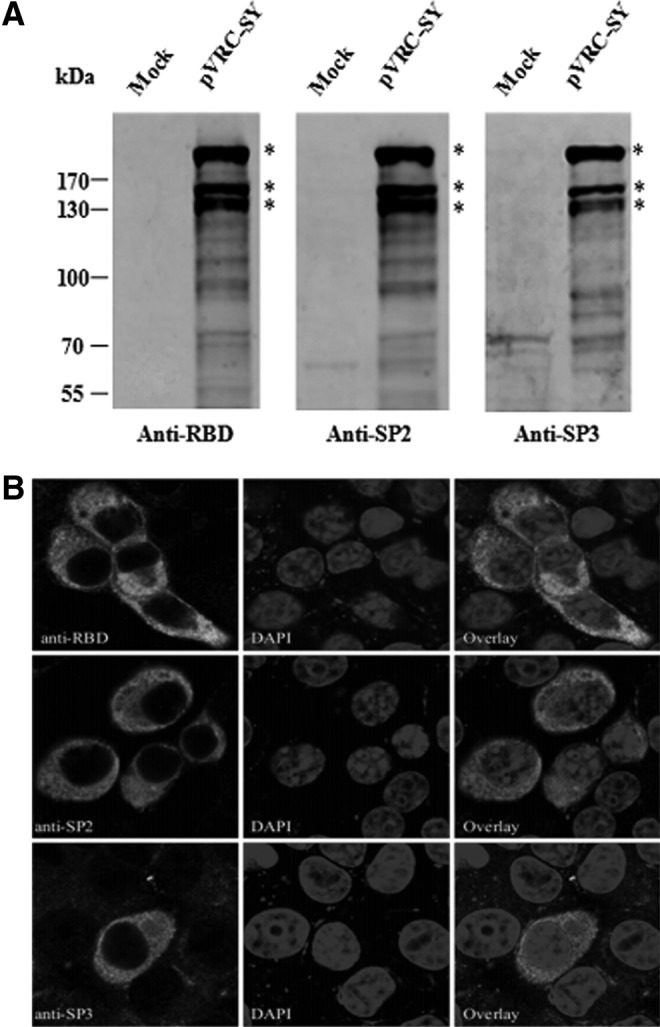
Expression of the MERS-CoV S protein in 293FT and BHK cells using antisera against the immunogens used in this study. Samples were derived from 293FT or BHK cells transiently transfected with pVRC-SY or pVRC8304 (indicated as Mock). (A) Western blot showing S-protein expression. 293FT cells were transfected with pVRC-SY(nCoV). Thirty-six hours later, cell lysates were prepared and Western blots were performed by using the indicated antiserum. Asterisks indicate the correct bands of S protein. (B) Immunofluorescence assay (IFA) of pVRC-SY(nCoV)-transfected BHK cells. Cells were fixed, permeabilized, and stained for either the S protein using the indicated antibody, or chromatin using a 4, 6-diamidino-2-phenylindole (DAPI) stain. The first row represents S protein, and the second row represents the chromatin. Cells were analyzed by confocal microscopy using a 100× objective, and representative images are shown.
All antisera raised against the rRBD or SP2 and SP3 peptides recognized a dominant band with a size of ∼200 kDa in pVRC-SY (nCoV)-transfected 293FTcell lysates, as expected from the full-length monomeric S protein (Fig. 3A). In addition, two major bands with a size of ∼130–170 kDa were detected by the anti-RBD and anti-SP sera, most likely representing the unglycosylated form of the S protein and glycosylated form of S1, demonstrating that the MERS-CoV S may be cleaved to S1 and S2. Our data also showed several smaller low-molecular-weight products, suggesting that S1 or S2 was further cleaved. These were S-specific bands, because they were not detected in mock-transfected cells. The antiserum from N-peptide immunization in rabbits was used for Western blotting, but no specific band was observed (data not shown). This result indicates that all antibodies raised against the rRBD and S peptides bound specifically to the S of MERS-CoV.
To further investigate whether these polyclonal antibodies could specifically bind to the native S protein and to identify the location of S expression, a confocal IFA was used to analyze S expression in BHK cells transfected with pVRC-SY. Binding of the antisera to S protein in the cytoplasm and on the cell surface was detected, suggesting that these polyclonal antibodies recognized the properly folded, mature form of MERS-CoV S.
Neutralizing activity against MERS-CoV was elicited by the rRBD and peptide targeting amino-acid residues 736–761 of the S protein
After determining that all of the polyclonal antibodies against the rRBD and S peptides could recognize the native form of MERs-CoV S protein, we further tested their neutralizing activities.
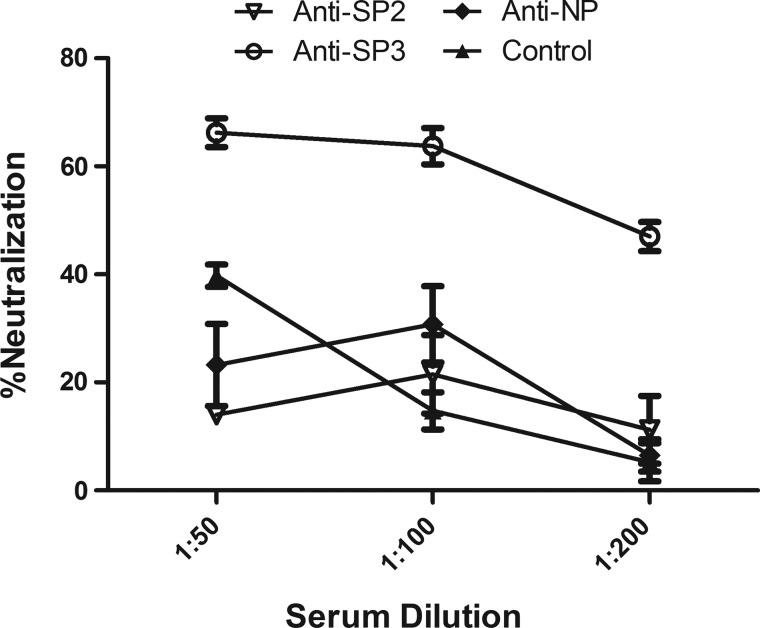
To assess the neutralizing capacity of the antisera after the last bleed from rabbits injected with SP2 (SP2-1 and -2, targeting residues 509–554 of the S protein), SP3 (targeting residues 731–761 of the S protein), and NP (targeting residues of the N protein), MERS-CoVpp was premixed 1:1 with the indicated dilution of antiserum before inoculation into Huh-7 cells. Luciferase activity was then examined at 48 h postinfection (Fig. 4). Only the antiserum from rabbits injected with SP3 showed neutralizing activity, and the titer of NT50 reached 50% neutralization at a 1:100 dilution. A recent report indicated that the RBD could be an important target for neutralizing antibodies; we confirmed that rRBD elicited robust neutralizing antibodies against MERS-CoV in mice (Fig. 5), reaching 100% neutralization at a 1:100 dilution. However, the rabbit antiserum against SP2 (located at the C-terminus of the RBD) did not show neutralizing activity. In addition, a CD analysis of the SP2-1 and -2 peptides showed that they were linear (data not shown).
FIG. 4.
Neutralization assay to screen the rabbit antisera raised by peptide immunization. MERSpp was premixed 1:1 with the indicated dilution of sera at 37°C before inoculation into Huh-7 cells, and the infection efficacy was monitored at 48 h postinfection (Protocol I). The normalized neutralization was calculated as described in Materials and Methods. The experiment was carried out in triplicate; data from one representative case are shown. Error bars indicate the SE of the mean.
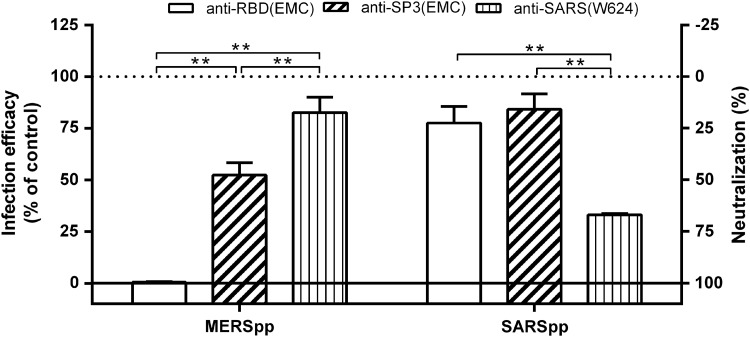
FIG. 5.
Specificity of the neutralization activity of the antisera used in this study. MERSpp or SARSpp was premixed 1:1 with the indicated dilution of antisera at 37°C before inoculation into Huh-7 cells, and the infection efficacy was monitored at 48 h postinfection (Protocol I). The normalized neutralization was calculated as described in Materials and Methods. The experiment was carried out in triplicate; data from one representative case are shown. Error bars indicate the SE of the mean.
To test the specificity of the neutralizing antibodies elicited by the rRBD and SP3, we obtained the antiserum (W624) induced by the RBD of SARS-CoV-immunized mice and evaluated the cross-reactivity between the antiserum and two CoVpps (MERSpp and SARSpp). Our results indicate that the antiserum against the rRBD and SP3 of MERS-CoV could effectively neutralize an MERSpp infection in vitro, while it did not neutralize an SARSpp infection (Fig. 5). In contrast, the antiserum (W624) from the SARS-CoV RBD effectively neutralized an SARSpp infection in vitro, while it did not neutralize an MERSpp infection. These results suggest that the neutralizing antibodies induced by the rRBD and SP3 of MERS-CoV were specific and did not cross-neutralize SARS-CoV.
The neutralizing antibodies induced by the rRBD and SP3 targeted MERS-CoVpp entry at different steps
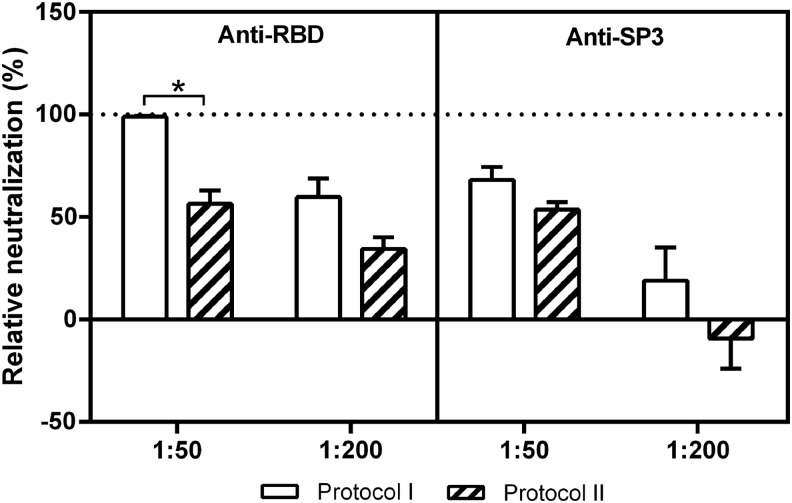
To map the entry steps targeted by the neutralizing antisera, we developed two MERSpp-based neutralization assays to allow us to distinguish between the binding and postbinding stages of infection. As described in the Materials and Methods, inhibitory antibodies were added before the binding of MERSpp to the target cells in Protocol I while antibodies were added after MERSpp binding to the target cells in Protocol II. As expected for antibodies targeting entry mediated by receptor-binding events, the relative neutralizing activity of anti-rRBD at a 1:50 dilution when added before binding (Protocol I) was significantly higher than that when added postbinding (Protocol II). However, no marked difference in the relative neutralizing activity of anti-SP3 at a 1:50 dilution was shown between Protocols I and II (Fig. 6). These data suggest that the anti-SP3 antibodies inhibited MERSpp entry during the postbinding and fusion process, in contrast to the anti-RBD antibodies.
FIG. 6.
Inhibition of an MERSpp infection by anti-RBD and -SP3 neutralizing antibodies at different steps. The experiment was performed as described in Materials and Methods. Briefly, antisera were added before the binding of MERSpp to Huh-7 cells in Protocol I, while antisera were added after MERSpp binding to target cells in Protocol II. The mean values±SE of a representative experiment performed in triplicate are shown. Statistically significant differences (p<0.01, t-test) in neutralization between Protocols I and II are indicated (*).
Discussion
The development of an effective vaccine and therapeutics against MERS-CoV infection is urgently needed (12,20). Neutralizing antibody responses often provide a first-line adaptive defense against infection by limiting virus entry and spread (3,12,20). The S protein of coronaviruses is not only the main determinant of tropism (14,19), but also the major antigenic target for neutralizing antibody induction (3,6–10,12,13). Therefore, it has been used as an immunogen for vaccine development. So far, little is known about the neutralizing epitopes and capacity of the MERS-CoV S protein. In this study, we found that the RBD and amino acids 736–761 of the MERS-CoV S protein induced neutralizing antibodies, representing potential targets for the inhibition of MERS-CoV entry at different steps. These findings have important implications not only for understanding the process and mechanism of neutralization, but also for the design of novel strategies to block MERS-CoV infection.
We first assessed the immunogenicity and reactive specificity of the RBD and panel of peptide-KLH conjugates as immunogens in mice and rabbits. An ELISA-based analysis of the serum IgG responses against the immunogens revealed that the RBD was more immunogenic in BALB/c mice than were the peptide-KLH conjugates in rabbits. Robust immunogen-specific IgG responses were obtained after several immunizations. Western blotting and confocal IFA indicated that the antiserum might bind most efficiently to the MERS-CoV S protein expressed in the cytoplasm and on the membrane; the size of the full-length monomeric S and S1 proteins detected is in agreement with our expectations and published data (18,25). This study also suggests that the MERS-CoV S protein undergoes stepwise cleavage to generate S1 and S2. However, further study is needed to identify the exact cleavage site and to determine whether the cleavage is cell dependent and tissue specific.
As shown in previous reports, the CoVs S protein is critical for target cell entry and harbors both linear and conformational epitopes for neutralization (1,6,9,10,13). However, the exact site and precise entry step targeted by neutralizing antibodies has not yet been defined, especially for MERS-CoV. The RBD of MERS-CoV is a key functional domain in the S protein, which is responsible for virus binding to receptors on target cells and that may contain neutralizing epitopes (8,9,13,15,18,20). We found that the rRBD (covering residues 367–605 of the S) elicited robust neutralizing antibodies against MERS-CoV in immunized mice by blocking viral entry at the binding step. These data are consistent with those in recent reports and confirm that the RBD of MERS-CoV is an ideal immunogen for vaccine development (7–9,12,18). We also found that antiserum induced by the SP2 peptide (targeting residues 509–554 of the S), which is located at the binding interface of the RBD according to recent crystallization data (15,23a), did not elicit potent neutralizing antibodies in immunized rabbits; however, high titers of binding antibodies to the RBD were observed. We suggest that the lack of neutralizing activity of the antiserum against SP2 could be due to the linear conformation of these immunogens. These data also suggest that the conformation of the RBD is essential for the induction of neutralizing activity, and that non-neutralizing immunodominant epitopes exist in the RBD of the MERS-CoV S protein.
This is the first linear neutralizing epitope located outside of the RBD in the S protein of MERS-CoV to be reported. This novel epitope targeting residues 736–761 is located at a possible proteocleavage site on the MERS-CoV S protein (15,20,22). We further demonstrated that the neutralizing activity induced by the SP3 peptide was specific for the postbinding and membrane fusion steps of MERS infection. Our data are consistent with a previous report on SARS-CoV (17), which indicated that human monoclonal antibody (5H10) directed to proteolytic cleavage site (791–805 amino acid) in S neutralized the virus in a rhesus macaque SARS model. Since multiple neutralizing epitopes have been identified on the S protein of SARS-CoV and envelope of HIV-1 (3,10,13,17), it is not unusual to find more than one neutralizing epitope on the MERS-CoV S protein.
In summary, a panel of S peptides based on bioinformatics predictions was confirmed to be immunogenic in this study. The RBD of the S protein of MERS-CoV could efficiently elicit neutralizing antibodies that targeted viral binding. Furthermore, we found a novel linear epitope that targeted amino-acid residues 736–761 of the S protein and which elicited robust neutralization by blocking viral entry at the postbinding and membrane fusion steps. We conclude that the amino-acid residues 736–761 of the S protein carry neutralizing epitopes that may be used to develop vaccines and antiviral agents against MERS-CoV.
Acknowledgments
The authors thank Dr Yuxian He (Institute of Pathogen Biology, Chinese Academy of Medical Sciences) for providing rRBD protein of SARS-CoV, and Dr. Gary Nabel (VRC, NIH, USA) for providing the plasmid of pVRC8304. This study was supported by China Mega-Project for Infectious Diseases Control and Prevention (2013ZX10004601, 2014ZX1000401).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Belouzard S, Millet JK, Licitra BN, and Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 2012;4: 1011–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breban R, Riou J, and Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet 2013;382:694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coughlin MM, Babcook J, and Prabhakar BS. Human monoclonal antibodies to SARS-coronavirus inhibit infection by different mechanisms. Virology 2009;394:39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Groot RJ, Cowley JA, Enjuanes L, et al. . Order of nidovirales. In: King A, Adams M, Carstens E, and Lefkowitz EJ, eds. Virus Taxonomy. The 9th report of the International Committee on Taxonomy of Viruses. San Diego, Academic Press, 2012:785–795 [Google Scholar]
- 5.de Groot RJ, Baker SC, Baric RS, et al. . Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol 2013;87:7790–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du L, He Y, Zhou Y, Liu S, Zheng BJ, and Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol 2009;7:226–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du L, Kou Z, Ma C, et al. . A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One 2013;8:e81587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du L, Zhao G, Kou Z, et al. . Identification of receptor-binding domain in S protein of the novel human coronavirus MERS-CoV as an essential target for vaccine development. J Virol 2013;87:9939–9942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Gao J, Lu G, Qi J, et al. . Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J Virol 2013;87:13134–13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gierer S, Bertram S, Kaup F, et al. . The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 2013;87:5502–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y, Lu H, Siddiqui P, Zhou Y, and Jiang S. Receptor-binding domain of severe acute respiratory syndrome coronavirus spike protein contains multiple conformation-dependent epitopes that induce highly potent neutralizing antibodies. J Immunol 2005;174:4908–4915 [DOI] [PubMed] [Google Scholar]
- 11.Holmes KV, and Dominguez SR. The new age of virus discovery: genomic analysis of a novel human betacoronavirus isolated from a fatal case of pneumonia. mBio 2013;4:e00548-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hotez PJ, Bottazzi ME, Tseng CT, et al. . Calling for rapid development of a safe and effective MERS vaccine. Microbes Infect 2014;pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang S, Bottazzi ME, Du L, et al. . Roadmap to developing a recombinant coronavirus S protein receptor-binding domain vaccine for severe acute respiratory syndrome. Expert Rev Vaccines 2012;11:1405–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn JH, Li W, Choe H, and Farzan M. Angiotensin-converting enzyme 2: a functional receptor for the SARS coronavirus. Cell Mol Life Sci 2004;61:2738–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a.Lee A, Powell JE, Tregear GW, Niall HD, and Stevens VC. A method for preparing fl-hCG COOH peptide-carrier conjugates of predictable composition. Mol Immunol 1980;17:749–756 [DOI] [PubMed] [Google Scholar]
- 15.Lu G, Hu Y, Wang Q, et al. . Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 2013;500:227–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, and Stephens GM. Family cluster of Middle East respiratory syndrome coronavirus infections. N Engl J Med 2013;368:2487–2494 [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi-Akiyama T, Ishida I, Fukushi M, et al. . Fully human monoclonal antibody directed to proteolytic cleavage site in severe acute respiratory syndrome (SARS) coronavirus S protein neutralizes the virus in a rhesus macaque SARS model. J Infect Dis 2011;203:1574–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mou H, Raj VS, van Kuppeveld FJ, Rottier PJ, Haagmans BL, and Bosch BJ. The receptor binding domain of the new MERS coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol 2013;87:9379–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raj VS, Mou H, Smits SL, et al. . Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013;495:251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raj VS, Osterhaus AD, Fouchier RA, and Haagmans BL. MERS: emergence of a novel human coronavirus. Curr Opin Virol 2014;5:58–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Snijders A, Benaissa-Trouw BJ, Oosterlaken TA, et al. . Identification of linear epitopes on Semliki Forest virus E2 membrane protein and their effectiveness as a synthetic peptide vaccine. J Gen Virol 1991;72(Pt 3):557–565 [DOI] [PubMed] [Google Scholar]
- 21.The Who Mers-Cov Research Group. State of Knowledge and Data Gaps of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in Humans. PLoS Curr 2013;5:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Boheemen S, de Graaf M, Lauber C, et al. . Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio 2012;3:e00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Wang N, Shi X, Jiang L, et al. . Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 2013;23:986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.World Health Organization. Novel coronavirus infection—update. www.who.int/csr/don/2014_07_04_mers/en/, 2014
- 24.Yan K, Tan W, Wang H, et al. . SARS-CoV spike proteins expressed by the vaccinia virus Tiantan strain: secreted sq protein induces robust neutralization antibody in mice. Viral Immunol 2009;22:57–66 [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Zhang L, Geng H, et al. . The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 2013;4:951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, and Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012;367:1814–1820 [DOI] [PubMed] [Google Scholar]