Abstract
Glioblastomas (GBMs) are lethal brain cancers that are resistant to current therapies. We investigated the cytotoxicity of human allogeneic NK cells against patient-derived GBM in vitro and in vivo, as well as mechanisms mediating their efficacy. We demonstrate that KIR2DS2 immunogenotype NK cells were more potent killers, notwithstanding the absence of inhibitory killer Ig–like receptor (KIR)-HLA ligand mismatch. FACS-sorted and enriched KIR2DS2+ NK cell subpopulations retained significantly high levels of CD69 and CD16 when in contact with GBM cells at a 1:1 ratio and highly expressed CD107a and secreted more soluble CD137 and granzyme A. In contrast, KIR2DS2− immunogenotype donor NK cells were less cytotoxic against GBM and K562, and, similar to FACS-sorted or gated KIR2DS2− NK cells, significantly diminished CD16, CD107a, granzyme A, and CD69 when in contact with GBM cells. Furthermore, NK cell–mediated GBM killing in vitro depended upon the expression of ligands for the activating receptor NKG2D and was partially abrogated by Ab blockade. Treatment of GBM xenografts in NOD/SCID mice with NK cells from a KIR2DS2+ donor lacking inhibitory KIR-HLA ligand mismatch significantly prolonged the median survival to 163 d compared with vehicle controls (log-rank test, p = 0.0001), in contrast to 117.5 d (log-rank test, p = 0.0005) for NK cells with several inhibitory KIR-HLA ligand mismatches but lacking KIR2DS2 genotype. Significantly more CD56+CD16+ NK cells from a KIR2DS2+ donor survived in nontumor-bearing brains 3 wk after infusion compared with KIR2DS2− NK cells, independent of their proliferative capacity. In conclusion, KIR2DS2 identifies potent alloreactive NK cells against GBM that are mediated by commensurate, but dominant, activating signals.
Introduction
Glioblastoma (GBM) is the most frequent and malignant brain tumor in adults (1) and is treated by surgical debulking, followed by concurrent temozolomide chemotherapy and fractionated radiotherapy. Despite this aggressive multimodal treatment, the median survival of patients remains only 14.6 mo (2) and emphasizes the need for developing novel and more effective adjuvant treatments.
Several studies investigated the efficacy of adoptively transferred autologous lymphokine-activated killer cells (3) but a meager few investigated purified NK cells (4) against GBM. Moreover, the T cell component of lymphokine-activated killer cells can inhibit NK cells’ activity due to the development of regulatory T cells (5). We recently demonstrated elevated expression of MHC class I molecules on patient GBM cells that may render tolerant autologous NK cells (6). Therefore, we hypothesized that adoptive transfer of purified allogeneic NK cells that are killer Ig–like receptor (KIR)-HLA class I ligand mismatched to the patient’s tumor might be more efficient. NK cells are innate lymphoid cells that are specialized to recognize and kill tumor- and virus-infected cells through germline-encoded receptors that transmit activating or inhibitory signals (7). Natural cytotoxicity receptor (NCRs) are activating receptors that recognize a variety of viral and bacterial molecules expressed on infected cells, as well as ligands expressed on cancer cells (8). However, many of these ligands have not been identified. Ligation of NKG2D, a member of the NK group 2 (NKG2) receptors, to stress-induced molecules, which are expressed most frequently on cancer cells, results in NK cell activation (9, 10). In contrast, NKG2A receptor mediates inhibitory signals upon recognition of HLA-E molecules that are often upregulated on cancer cells to escape the immune system (11). KIRs recognize classical MHC class I (HLA-ABC in humans). KIRs with short cytoplasmic domains are activating receptors, whereas ligation of the KIR with long cytoplasmic domains to their cognate HLA class I ligands transmits a cascade of inhibitory signals that mediates NK cell tolerance to self cells (12). A number of malignancies have been associated with particular KIR genotypes [e.g., reduced frequency of KIR2DS2 was reported in pre-B acute lymphoblastic leukemia (13), as well as KIR2DL2 and KIR2DS2 in autologous NK cells from chronic myeloid leukemia patients (14)]. Limited literature exists on the predictive potential and functionality of KIR immunogenotypes in solid tumors (15, 16), in particular GBM. NK cells display cytotoxicity against altered self cells that downregulate HLA class I molecules as a result of the “missing self” mechanism (17). However, KIR-HLA class I ligand mismatch can occur in an allogeneic setting when the donor’s NK cells express KIRs that do not recognize the recipient’s HLA class I molecules, resulting in diminished inhibitory signals for NK cells (18). In addition, when activating KIR, NKG2D and NCRs are ligated to their cognate ligands, the balance may be tipped towards activating signals, and this ultimately determines cytotoxic potency (19) and production of proinflammatory cytokines and chemokines. Thus, the therapeutic potential of NK cells against cancer was explored in vitro (20–22) and in vivo (23, 24). Allogeneic NK cells were demonstrated to mediate the graft-versus-leukemia effect through KIR-HLA class I ligand mismatch in patients receiving hematopoietic stem cell transplantation (HSCT) (18). In the current study, we aimed to elucidate the potential of allogeneic NK cells against GBM. Interaction of NKG2D receptor with its ligands expressed on tumor cells, as well as the presence of KIR2DS2 receptor, had greater impact on GBM lysis than did the inhibitory KIR-HLA class I mismatch. KIR2DS2+ NK cell subsets exhibited a functional activation advantage, cytokine production, propensity for degranulation, and greater persistence in vivo compared with KIR2DS2− NK cells. To our knowledge, this is the first report demonstrating efficacy of allogeneic NK cells against patient-derived solid tumor in vitro and in vivo mediated by alloreactive subpopulations with dominant functional activation signals.
Materials and Methods
NK cell isolation and culture
This study was approved by the Regional Ethical Committee of Western Norway, Bergen (2013/96) and informed consent to participate was obtained from all donors. PBMCs were obtained from the blood of 10 healthy donors using Leucosep 50-ml tubes with a high-density polyethylene membrane (Greiner Bio-One, Frickenhausen, Germany) and Lymphocyte Separation Medium (Lonza, Lysaker, Norway). NK cells were purified using a human NK Cell Isolation Kit (Miltenyi Biotec, Bergisch Gladbach, Germany), according to the manufacturer’s protocol. The NK cells were cultured in CellGro GMP Serum-free Stem Cell Growth Medium (CellGenix, Freiburg, Germany) supplemented with 10% FCS (PAA Laboratories, Pasching, Austria), 500 U/ml IL-2 (R&D Systems, Abingdon, U.K.), and the NK Cell Expansion/Activation Kit containing anti-CD2 and anti-CD335 (NKp46) Abs (Miltenyi Biotec), according to the manufacturer’s instructions. After 2 wk of culture, NK cells for in vivo experiments were washed, and beads were removed using a magnetic separator (Miltenyi Biotec).
K562 and patient GBM cell culture
GBM cell lines were established from patient biopsies collected during surgical procedures at Haukeland University Hospital. The Regional Ethical Committee of Western Norway, Bergen and the Norwegian Data Protection Agency approved the collection of tumor tissue. Patients gave their informed consent to specimen collection for research purposes, and their samples were analyzed anonymously. Tumor tissue was mechanically dissociated as previously described (6). The human chronic myelogenous leukemia cell line K562 (American Type Culture Collection, Manassas, VA) was cultured in suspension with RPMI 1640 medium supplemented with 10% FCS, 5% penicillin-streptomycin, and 1 mM HEPES at 37°C and 5% CO2. GBM primary cultures obtained from patients P3 (GBM), 2012-018 (GBM with gliosarcoma differentiation), and 2011-20-I (GBM) were propagated in Neural Basal medium (Invitrogen, Hämeenlinna, Finland) supplemented with 1% GlutaMAX (Invitrogen), 2% B-27 (Invitrogen), 5% penicillin-streptomycin (Lonza), 10 μg/ml epidermal growth factor, and fibroblast growth factor (PeproTech) at 37°C and 5% CO2. Cells in exponential growth phase were used for all assays.
GBM spheroid culture
For in vivo experiments, P3 GBM cells were propagated in vitro as spheroids in DMEM (Sigma-Aldrich, Oslo, Norway) supplemented with 10% FCS (PAA Laboratories), 100 U/ml penicillin-streptomycin (Lonza), and 5 μg/ml plasmocin (InvivoGen, Toulouse, France) in (30% v/v) agar-coated flasks. Standardized spheroids containing 104 cells/spheroid were cultured in 4% v/v methylcellulose-DMEM in round-bottom 96-well plates over 3 wk.
DNA isolation, KIR typing, HLA typing, and KIR-HLA mismatch
DNA was isolated from GBM cells and from the NK cell donors’ blood using DNeasy Blood & Tissue Kit (QIAGEN, Germantown, MD), according to the manufacturer’s protocol. The GBM cells’ HLA class I (loci A, B, and C) was genotyped by ProImmune (Oxford, U.K.) using the PCR-based sequence-specific oligonucleotide probe hybridization technique. High-resolution HLA typing (with PCR sequence-specific primers or PCR sequencing) was performed for the HLA-C locus on 2012-018 cells. The expression of HLA-G and HLA-E was evaluated by flow cytometry. In addition, the surface expression of selected HLA class I epitopes was evaluated by flow cytometry using anti–HLA-Bw4, A9, A32, and anti–HLA-A3 Abs (Abcam, Cambridge, U.K.). KIR typing was performed using the KIR Typing kit (Miltenyi Biotec), according to the manufacturer’s protocol. KIR-HLA class I mismatch was defined as the absence of inhibitory signal resulting from absence of the receptor gene in the donor’s DNA, whereas the ligands’ genes were present in the GBM cells’ DNA.
In vitro cytotoxicity assays
In vitro cytotoxicity assays were performed with NK cells cultured for 2 wk against GBM cells and the K562 chronic myelogenous leukemia cell line as a positive control. Target cells were washed and labeled with CFSE (Sigma-Aldrich). Following the washing steps, NK cells were cocultured with target cells at E:T ratios of 2:1, 5:1, 10:1, and 20:1 for 4 h at 37°C and 5% CO2 in RPMI 1640 medium (Invitrogen) supplemented with 10% FCS (PAA Laboratories), 50 U/ml penicillin-streptomycin (Lonza), and 1 mM HEPES (Invitrogen). Culture of target cells alone was used as a negative control. In six cytotoxicity assays we blocked the NK cell receptors using the following Abs: anti-NKG2D and anti-KIR2DS4 at final concentration of 5 μg/106 effector cells and anti-KIR2DS2 at a final concentration of 3 μg/106 effector cells (Supplemental Table I). Isotype-control mouse IgG1 (R&D Systems), mouse IgG2a (R&D Systems), and rabbit IgG (Abcam) were used at the same concentration as the Ab to which they corresponded. Each experimental condition was performed in three replicates. Then cells were washed with PBS containing 0.5% BSA (Miltenyi Biotec) and labeled with a Live/Dead Fixable Near-IR Dead Cell Stain Kit (Invitrogen), according to the manufacturer’s protocol, or with TO-PRO3 (Invitrogen) for 1 min before data acquisition on a BD LSR Fortessa flow cytometer (BD Biosciences, Trondheim, Norway). The data were analyzed using BD FACSDiva v6.2 software (BD Biosciences). The target cells were identified as CFSE+, and effector cells were identified as CFSE−. The dead target cells were identified as CFSE+TO-PRO3+ or CFSE+Live/Dead+. Spontaneous death was defined as the proportion of dead target cells cultured alone (negative control), and this value was subtracted from the proportion of dead target cells cocultured with effector cells. Each cytotoxicity assay was repeated in at least 3 or 4 independent experiments.
KIR2DS2 cell sorting and activation assays
FACS sorting of KIR2DS2+ and KIR2DS2− NK cells was carried out to study the functional features of KIR2DS2+ NK cell subsets. Purified NK cells from KIR2DS2+ donors were sorted after expansion for 2 wk. NK cells were stained with anti-KIR2DS2 Ab, as described below (“Flow cytometry phenotyping”), and 106 cells with the highest fluorescence intensity and 106 negative cells were sorted using a FACSAria II (BD Biosciences); cells with intermediate fluorescence intensity were discarded (see gating strategy in Fig. 2Af). Cells stained with isotype control and stained KIR2DS2 immunogenotype–negative cells were used as controls for gating. Data were recorded and analyzed using BD FACSDiva v6.2 software (BD Biosciences) and FlowJo v10 software (TreeStar, Ashland, OR). After sorting, KIR2DS2+ and KIR2DS2− subsets were expanded for two additional weeks to obtain greater cell numbers. Then, NK cells were plated in either monoculture 1:0 or in coculture with GBM cells at an E:T ratio of 1:1 for 4 h at 37°C and 5% CO2 in serum-free RPMI 1640 medium without cytokine supplements. Following the 4-h incubation, supernatants were collected and stored at −80°C pending cytokine analysis. The cells were washed, immunostained, and phenotyped for surface and intracellular markers.
Cytokine analysis
Supernatants from two independent activation assays (above) were collected and analyzed in duplicate for diverse cytokines, granzymes, and perforin using a Luminex-based immunoassay and a kit that detected 17 analytes. This Milliplex Human Magnetic Cytokine Bead Panel (cat. no. HCD8MAG15K-17PMX; Merck Millipore, Darmstadt, Germany) was used according to the manufacturer’s instructions.
In vivo NK cell therapy for GBM-bearing mice
All animal procedures were performed in accordance with the European Convention for the Protection of Vertebrates Used for Scientific purposes, and the protocols were approved by The Norwegian Animal Research Authority (Oslo, Norway). A total of 40 7-wk-old NOD/SCID mice (Taconic) of both genders were implanted intracranially with P3 tumor spheroids. Each animal was anesthetized with sevoflurane (Abbott Laboratories) and secured in a stereotactic frame (Kopf Instruments, Tujunga, CA). A burr hole was made with a microdrill (Kopf Instruments) 0.5 mm posterior to the bregma and 2 mm to the right of the sagittal suture. Five spheres/animal were injected 2 mm below the brain surface with a Hamilton syringe. One week after implantation, mice were subjected to either of the following treatments administered intracranially using the same coordinates as the previously implanted tumor spheroids: 1) 10 μl PBS containing 4.4 g/l glucose (n = 5) or 2) anti-CD2 and anti-NKp46 Abs at a concentration of 0.05 μg/ml in 10 μl PBS (without conjugated beads) used as control for NK cell expansion (Miltenyi Biotec) (n = 6) and 3) 106 NK cells from donor 4 or donor 3 diluted in 10 μl PBS containing 4.4 g/l glucose (n = 14 and n = 16, respectively). Two weeks after implantation, mice were injected intracranially with a second 10-μl dose of PBS containing glucose (n = 5), 10 μl Abs (without conjugated beads) used for NK cell expansion (Miltenyi Biotec) (n = 6), or 106 NK cells from donor 4 or donor 3 diluted in 10 μl PBS containing glucose (n = 7 and n = 8, respectively), as described above. Animals were monitored for survival and inspected daily. Animals were sacrificed by CO2 inhalation and dislocation of the neck when they developed neurologic symptoms, such as rotational behavior, reduced activity, or delayed grooming and dome head. Their brains were fixed in formalin for H&E and immunohistochemistry (IHC) staining.
In vivo off-target NK cell therapy
A total of 45 7-wk-old NOD/SCID mice (Taconic, Denmark) of both genders was treated intracranially with 10 μl PBS containing 4.4 g/l glucose (n = 3) or 106 NK cells from donor 4 (KIR2DS2−) or donor 3 (KIR2DS2+) diluted in 10 μl PBS containing 4.4 g/l glucose (n = 18 each) administered into the same sterotaxic brain coordinates as described in the section above. One week later, some mice were injected intracranially with a second dose of 10 μl PBS containing 4.4 g/l glucose (n = 3) or 106 NK cells from donor 4 or donor 3 diluted in 10 μl PBS containing 4.4 g/l glucose (n = 13 each), as described above. Three animals from each group were sacrificed at 1, 3, and 5 wk after the second NK cell dose, and their brains were dissociated into single-cell suspensions using a Neural Tissue Dissociation Kit containing papain (Miltenyi Biotec), according to the manufacturer’s protocol, and analyzed by flow cytometry. The remaining animals were monitored for survival, inspected daily, and sacrificed when they developed neurologic symptoms, as described above. Their brains were fixed in formalin for H&E and IHC staining.
Flow cytometry phenotyping
PBMCs, NK cells, GBM cells, and cells obtained from the dissociated mice brains were counted and washed with PBS containing 0.5% BSA. The cells were stained with a Live/Dead Fixable Near-IR or a Yellow Dead Cell Stain Kit (Invitrogen) and fluorochrome-conjugated Abs (Supplemental Table I) or isotype controls for 20 min at 4°C protected from light. Then, cells were washed, fixed, and permeabilized with Cytofix/Cytoperm (BD Biosciences), followed by intracellular staining with fluorochrome-conjugated Abs (Supplemental Table I) or isotype controls for 20 min at 4°C, protected from light. Cells were washed, and data were acquired on a BD LSR Fortessa (BD Biosciences) flow cytometer. The data were analyzed using BD FACSDiva v6.2 software (BD Biosciences) and FlowJo v10 software (TreeStar). The proportion of specifically stained cells indicated on the graphs was calculated as the percentage of marker-positive cells subtracting the percentage of isotype control–positive cells. Debris and dead cells were excluded from the analysis based on a forward scatter (FSC) versus side scatter (SSC) plot and Live/Dead staining. Doublets were excluded based on an SSC-A versus SSC-H plot. In the in vivo experiments, human NK cells were defined as CD45+CD3−CD56+ cells, and human tumor cells were defined as nestin+CD45− cells. Murine immune cells were gated as CD45+ cells by mouse-specific staining. Microglia and macrophages were defined as CD11b+CD45low and CD11b+CD45high populations, respectively. In the in vitro experiments, human NK cell subpopulations were gated based on CD56, CD16 expression, and lack of CD3. Thereafter, the gated populations were analyzed for expression of CD4, CD8, NKG2D (CD314), NKp46 (CD335), NKG2A, CD69, CD107a, Ki-67, IFN-γ, and KIRs—KIR2DL4, KIR2DL3, KIR2DL1, KIR3DL1, KIR3DL2/1, KIR2DL2/3, KIR2DS4, and KIR2DS2 (Supplemental Table I). To assess the KIR2DS2+ subpopulation, KIR2DS2+ and KIR2DS2− cells were gated (see gating strategy in Fig. 2Ab) and analyzed for the expression of CD16, CD69, NKG2D, KIR2DL1, KIR3DL1, KIR3DL2/1, and KIR2DL2/3. Importantly, for good separation of KIR3DL2 and KIR3DL1, as well as KIR2DL2 and KIR2DL3, two-step staining was performed. NK cells were preincubated for 20 min with anti-KIR2DL1 and anti-KIR3DL1 Abs before incubation with anti-KIR3DL2/1 and anti-KIR2DL2/3 Abs.
Immunohistochemistry
Formalin-fixed paraffin-embedded sections of the animals’ brains were subjected to IHC staining using standard procedures (25). Following unmasking of antigenic determinants, the sections were labeled with the following primary Abs: mouse anti-human nestin (Millipore, Oslo, Norway), mouse anti-human STEM121 (StemCells, Cambridge, U.K.), mouse anti-human NKp46 (BioLegend, San Diego, CA), mouse anti-human Ki-67 (Dako, Glostrup, Denmark), and rat anti-mouse CD31 (Dianova, Hamburg, Germany). After the washing steps, endogenous peroxidase blocking was applied, and sections were incubated with secondary Abs. Staining was visualized with the avidin-biotin-peroxidase complex method with 3,3′-diaminobenzidine and H2O2 (DCS, Hamburg, Germany), according to the manufacturer’s protocol (VECTASTAIN; Vector Laboratories, Burlingame, CA). Thereafter, the sections were counterstained with Harris hematoxylin. The percentages of Ki-67+ cells (Ki-67 labeling index) and NKp46+ cells were quantified in five randomly selected high-power (400× magnification) microscopic fields in all animals in the study group. Detection of apoptotic cells was performed with the TUNEL assay, according to the manufacturer’s instructions (Roche Applied Bioscience, Mannheim, Germany), and apoptotic cells were quantified as previously described (25). Quantification of CD31+ microvascular density and area fraction of CD31+ cells was performed as previously described (25).
Magnetic resonance imaging
Magnetic resonance imaging (MRI) was performed with a 7-Tesla Bruker PharmaScan system (Bruker, Ettlingen, Germany) and linear-volume radiofrequency coil with an internal diameter of 23 mm. T1-weighted MRI was obtained using a rapid-acquisition relaxation enhancement sequence with relaxation time = 900 ms, excitation time = 9.00 ms, rare factor = 4, a slice thickness of 1.0 mm, and resolution of 0.0078 cm/pixel. T2-weighted MRI was obtained using a rapid-acquisition relaxation enhancement sequence with relaxation time = 3500 ms, excitation time = 35.22 ms, rare factor = 8, a slice thickness of 1.0 mm, and resolution of 0.0078 cm/pixel. MRI scanning was performed 12, 19, and 26 d after tumor spheroid implantation (pretreatment, posttreatment 1 and posttreatment 2, respectively), and MRI scanning longitudinally followed the same three animals/group. Each measurement involved a tripilot scan, a T2-weighted scan, a precontrast T1-weighted scan, and a postcontrast T1-weighted scan, with 0.1 ml Dotarem contrast agent (gadoteric acid 279.3 mg/ml; Guerbet, Roissy, France) injected s.c.
Statistical analyses
Survival data were analyzed using the Kaplan–Meier method with the Mantel–Cox log-rank test. The phenotyping data obtained by flow cytometry and IHC quantification were analyzed using a nonparametric Mann–Whitney U one-way ANOVA with Dunn multiple-comparison post hoc test or two-way ANOVA with Bonferroni multiple-comparison post hoc test when more than two categorical and dependent variables were analyzed. All analyses were performed with Prism statistical software, version 5.0 (GraphPad, La Jolla, CA). The p values < 0.05 were considered statistically significant.
Results
Expression of GBM stem cell–like markers
Previous studies demonstrated that GBM cells propagated in growth factor–defined stem cell medium retained tumor heterogeneity, resembled glioma stem–like cell (GSC) phenotypes, and were more susceptible to NK cell lysis (26). Therefore, we characterized the GBM short-term cultures for their expression of immature differentiation markers and ligands for NK cell receptors (Table I, Supplemental Fig. 1). A high percentage (87–100%) of P3 and 2011-20-I GBM cells expressed nestin (Table I, Supplemental Fig. 1H and 1I, respectively), as well as vimentin and glial fibrillary acidic protein (GFAP) intermediate filaments (Table I, Supplemental Fig. 1K and 1L, respectively). A total of 40% of 2012-018 gliosarcoma cells expressed A2B5 (Supplemental Fig. 1S), 80% expressed GFAP (Supplemental Fig. 1M), and 80.4% expressed vimentin, consistent with their gliosarcoma differentiation (Table I). Prominin-1/CD133, which was reported to identify GSCs (27), was expressed by 39% of P3 cells, 31.5% of 2011-20-I cells, and 21.1% of 2012-018 GBM cells (Table I, Supplemental Fig. 1N–P, respectively). A total of 40.7% of 2012-018 GBM cells expressed CD15 (Table I), which was reported to be enriched on CD133− GSCs (28). P3 and 2011-20-I GBM cells did not express CD31, whereas 35.2% of 2012-018 gliosarcoma cells expressed it (Table I). Taken together, the GBM short-term cultures differentially expressed markers associated with the immature GSC phenotype, as opposed to more differentiated cells.
Table I. Expression of GBM stem–like markers and NK cell ligands.
| Ligand/Marker | P3 (% of Cells) | 2011-20-I (% of Cells) | 2012-018 (% of Cells) | P3 (MFI) | 2011-20-I (MFI) | 2012-018 (MFI) |
|---|---|---|---|---|---|---|
| ULBP-1 | 68.23 | 41 | 33.2 | 23,923 | 36,621 | 19,524 |
| ULBP-2/5/6 | 95.22 | 90.18 | 71.2 | 183,413 | 67,147 | 58,897 |
| ULBP-3 | 63.03 | 50 | 41.4 | 28,371 | 19,871 | 29,928 |
| MICA | 91.03 | 95.16 | 91.65 | 371 | 344 | 234 |
| MICB | 6.91 | 13.13 | 41.22 | 11,482 | 43,564 | 13,626 |
| HLA-A,B,C | 99.28 | 99.7 | 99.17 | 1,571 | 724 | 9,456 |
| HLA-DR,DP,DQ | 0 | 19.5 | 63.42 | 0 | 466 | 900 |
| HLA-E | 0 | 0 | 25.2 | 0 | 0 | 415 |
| HLA-G | 0 | 0 | 0.1 | 0 | 0 | 971 |
| GFAP | 98.8 | 100 | 79.98 | 1,131 | 1,123 | 466 |
| Vimentin | 95.6 | 87.6 | 80.4 | 1,399 | 532 | 3,206 |
| Nestin | 97.6 | 99.7 | 96.81 | 6,148 | 2,115 | 1,176 |
| CD31 | 0 | 0 | 35.23 | 0 | 0 | 677 |
| CD133 | 39 | 31.5 | 21.12 | 172 | 102 | 419 |
| A2B5 | 42.6 | 77.3 | 40.04 | 250 | 489 | 877 |
| CD15 | 0.25 | 0 | 40.72 | 0 | 0 | 679 |
| HLA-A3 | — | — | 98.1 | 0 | 0 | 5,584 |
| HLA-Bw4 | 97.5 | 77.4 | 97.3 | 2,354 | 621 | 4,802 |
The proportion of specifically stained cells was calculated by subtracting the percentage of isotype-control–positive cells from the percentage of marker-positive cells.
—, HLA-A3 not present on genotype and not expressed on protein level.
Expression of HLA class I and II molecules on GBM cells
KIR-HLA class I mismatch was reported to improve the outcome of leukemia patients receiving HSCT (18) and to play a role in the NK cells’ cytotoxicity against solid tumors in vitro (21, 29). To investigate the impact of KIR-HLA class I mismatch on NK cell potency against GBM, we genotyped the HLA class I alleles and determined their expression at the protein level. At least 99% of all GBM cells expressed the classical HLA-A, -B, and -C molecules. None expressed the nonclassical HLA-G, whereas 25.2% of 2012-018 GBM cells expressed HLA-E (Table I, Supplemental Fig. 1G). A total of 19.5% of 2011-20-I GBM and 63.4% of 2012-018 cells expressed HLA class II molecules, P3 did not express them (Table I). HLA-class I genotyping of GBM cells was also performed (Supplemental Table II), and the surface expression of HLA-A and -B molecules was confirmed by flow cytometry. A total of 98.1% of 2012-018 cells expressed HLA-A3 (Table I, Supplemental Fig. 1D), and 97.5, 77.4, and 97.3 of P3, 2011-20-I, and 2012-018 cells expressed HLA-Bw4, respectively (Table I, Supplemental Fig. 1A–C, respectively). The 2012-018 cells expressed HLA-A, HLA-B, HLA-C, and HLA-Bw4 at the highest density, as indicated by mean fluorescence intensity (MFI) data (Table I). The Ab that we used to detect HLA-Bw4 also recognizes HLA-A9 and HLA-A39 molecules; however, all cell lines were negative for these alleles at the genomic level (Supplemental Table II). Moreover, a number of HLA-A alleles, as well as HLA-B alleles, including HLA-B*44 and HLA-B*27 present in the genome of P3, HLA-B*44 present in the genome of 2011-20-I, and HLA-B*37 present in the genome of 2012-018, belong to a group of alleles with the HLA-Bw4 epitope (Supplemental Table II). Thus, all of the GBM cells differentially expressed ligands for KIRs.
Purified and ex vivo–expanded NK cells efficiently kill GBM cells in vitro but are less potent against gliosarcoma cells
We purified NK cells from 10 healthy donors and analyzed phenotype, morphology of freshly isolated (resting) and cultured (activated) NK cells, and expansion rates of NK cells in culture. As expected, freshly isolated NK cells (CD56+CD3−) were predominantly CD56dimCD16+ under steady-states and had a rounded phase-bright morphology (Supplemental Fig. 1T and 1V, respectively). After 2 wk in culture, the NK cells acquired a CD56brightCD16bright phenotype and a ramified, phase-dark morphology (Suppl. Fig. 1U and 1W, respectively). Inhibitory KIRs, NKG2D, and NKp46 were not significantly affected by culture conditions (Supplemental Fig. 1X). However, NKG2A receptor expression was attenuated on activated NK cells (p = 0.0073, Supplemental Fig. 1X). Although there was a donor-dependent variability, the purified NK cells were successfully expanded 8.73 ± 4.22-fold in culture (mean ± SEM, range 5.07–10.82; data not shown). Activated NK cells from all donors were able to kill the GBM cells; however, the efficiency was donor dependent (Supplemental Fig. 1Y). 2012-018 cells were significantly less susceptible to NK cell–mediated lysis compared with P3 and 2011-20-I cells at all E:T ratios (p < 0.001 and p < 0.0001, respectively, at E:T ratio of 5:1; p < 0.0001 at an E:T ratio of 10:1; and p < 0.05 and p < 0.0001, respectively, at an E:T ratio of 20:1, Kruskal–Wallis test, Supplemental Fig. 1Y). No significant difference was observed between P3 and 2011-20-I cells (Supplemental Fig. 1Y). All donor NK cells efficiently killed control K562 cells (p < 0.001 and p < 0.0001 compared with GBM cells, Supplemental Fig. 1Y).
Ligation of NKG2D-activating receptor to stress-induced ligands is important for NK cell lysis of GBM cells
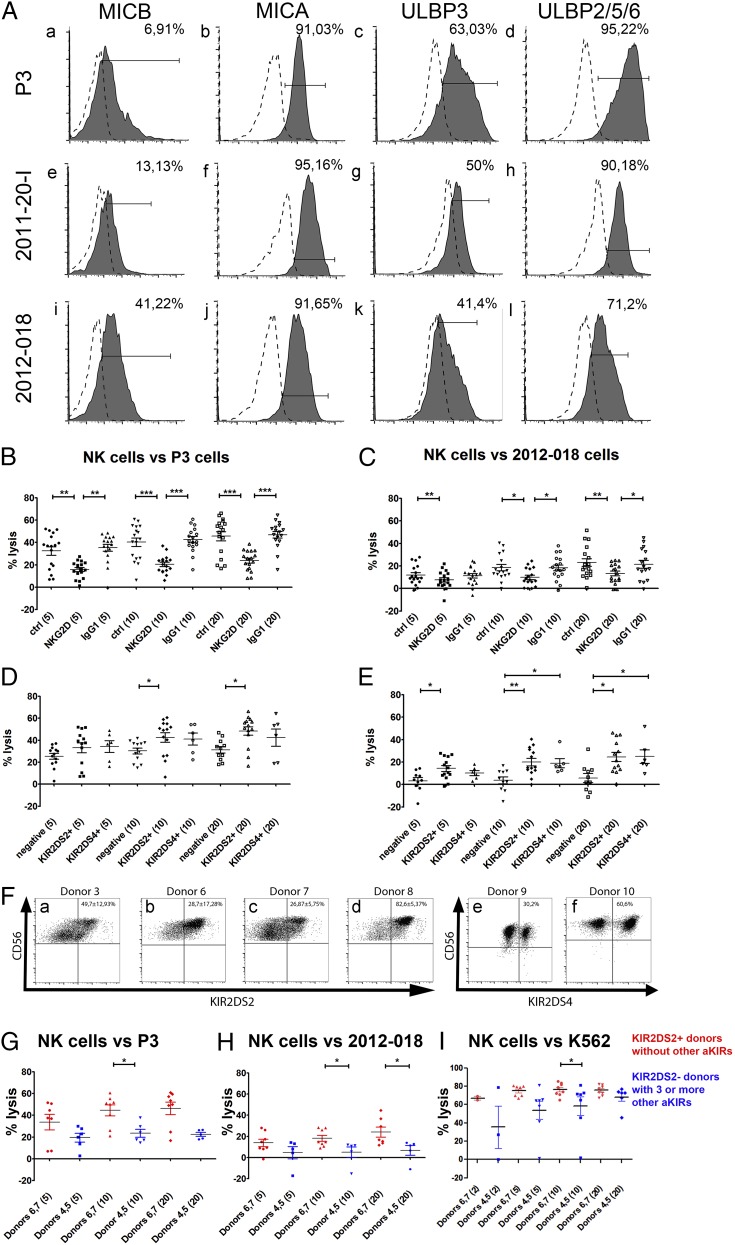
To investigate the impact of NKG2D on the differential susceptibility of GBM cells to NK lysis, we determined the expression of its ligands—MHC class I–related chain A (MICA), MHC class I–related chain B (MICB), and UL16-binding proteins (ULBPs) by GBM cells. A total of 41.2% of 2012-018 gliosarcoma cells expressed MICB (Fig. 1Ai, Table I), whereas 6.9 and 13.1% of P3 and 2011-20-I cells, respectively, expressed it (Fig. 1Aa, 1Ae, respectively, Table I). High percentages (91–95%) of all GBM cells expressed MICA (Fig. 1Ab, 1Af, 1Aj, Table I). Various proportions of all three GBMs expressed ULBP-1, ULBP-3 (Fig. 1Ac, 1Ag, 1Ak), and ULBP-2/5/6 (Fig. 1Ad, 1Ah, 1Al) (Table I). P3 was the GBM with the highest proportion of ULBP-1+, ULBP-3+ (Fig. 1Ac), and ULBP-2/5/6+ (Fig. 1Ad) cells (68.2, 63, and 95.2%, respectively, Table I), followed by 2011-20-I (Fig. 1Ag, 1Ah, respectively; 41, 50, and 90.2%, Table I), whereas 2012-018 had the lowest percentages of positive cells of all three GBMs (Fig. 1Ak, 1Al, respectively; 33.2, 41.4, and 71.2%, Table I). 2012-018 cells also expressed ULBP-1, ULBP-2/5/6, and MICA at the lowest density compared with P3 and 2011-20-I cells, as indicated by the MFI data (Table I).
Next, we performed in vitro cytotoxicity assays with activated NK cells obtained from six healthy donors against P3 and 2012-018 cells and observed that all donor NK cells efficiently killed the GBM cells from both sources, although the percentage of lysed 2012-018 cells was lower than P3 cells. Blocking NKG2D receptor significantly attenuated NK cell–mediated killing of P3 cells at all E:T ratios, whereas isotype-control IgG1 Abs had no effect (p = 0.0006 at E:T ratio of 5:1; p < 0.0001 at E:T ratio of 10:1; and p < 0.0001 at E:T ratio 20:1; Friedman test, NKG2D blocking compared with controls [no Ab and isotype control], Fig. 1B). Similar results were obtained for 2012-018 cells (p = 0.0031 at E:T ratio of 5:1; p = 0.0114 at E:T ratio of 10:1; and p = 0.0048 at E:T ratio of 20:1; Friedman test, NKG2D blocking compared with control [no Ab and isotype control], Fig. 1C). Taken together, these data indicate that activating signals via NKG2D are important for NK lysis of GBM cells. However, donor dependency of NK cell lysis of GBM and the GBM cells' partial rescue after NKG2D blockade indicated that other mechanisms contributed to NK cell efficacy.
FIGURE 1.
Ligation of NKG2D to stress-induced ligands and the activating KIR2DS2 receptor are important for efficient NK cell–mediated cytotoxicity of GBM cells. (A) Surface expression of ligands for NKG2D receptors MICB, MICA, ULBP3, and ULBP 2/5/6 on P3 (Aa–Ad), 2011-20-I (Ae–Ah), and 2012-018 (Ai–Al) cells. Dashed graphs represent negative controls; filled graphs represent staining. The proportion of specifically stained cells indicated was calculated by subtracting the percentage of isotype-control–positive cells from the percentage of marker-positive cells. The percentage lysis by NK cells of P3 (B) and 2012-018 (C) GBM cells at E:T ratios of 5:1, 10:1, and 20:1, with and without addition of blocking NKG2D or IgG1 isotype-control Abs. Mean percentage (± SEM) of lysis of P3 (D) and 2012-018 (E) GBM cells mediated by KIR2DS2+ or KIR2DS4+ NK cells compared with NK cells lacking both receptors (negative) at E:T ratios of 5:1, 10:1, and 20:1. Data are mean ± SEM of three or four independent experiments/donor (n = 6 donors). The x-axis shows experimental condition and E:T ratio (in parentheses). *p < 0.05, **p = 0.01, ***p < 0.001. (F) CD56 versus KIR2DS2 dot plots represent the gating strategy of KIR2DS2+ subpopulations of donors 3 (Fa), 6 (Fb), 7 (Fc), and 8 (Fd). CD56 versus KIR2DS4 dot plots represent the gating strategy for KIR2DS4+ expression on NK cell subpopulations of donors 9 (Fe) and 10 (Ff). Data are mean percentage (± SEM) of cells expressing KIR2DS2 or KIR2DS4 receptors. Mean percentage (± SEM) of lysis of P3 GBM cells (G), 2012-018 GBM cells (H), and K562 cells (I) mediated by NK cells from donors 6 and 7 (red: possessing KIR2DS2 gene and no other activating KIR genes) compared with NK cells from donors 4 and 5 (blue: lacking KIR2DS2 gene, but possessing three or more genes of other activating KIRs) at E:T ratios of 2:1, 5:1, 10:1, and 20:1. Data are mean ± SEM of three or four independent experiments/donor. *p < 0.05.
Activating KIR2DS2 receptor is important for NK cell potency against GBM cells in vitro, independent of inhibitory KIR repertoire
To investigate the impact of KIR–HLA interactions between NK cells and GBM cells, genomic KIR typing of the donors and HLA class I typing of GBM cells were performed (Table II, Supplemental Table II). 2012-018 cells expressed all HLA class I alleles that are known ligands for KIRs (Table I, Supplemental Fig. 1C, 1D, Supplemental Table II). P3 and 2011-20-I cells did not express any known ligands for KIR3DL2 (Table I, Supplemental Table II). Next, we compared the mean percentage of GBM cells lysed by NK cells isolated from donors expressing all inhibitory KIRs with lysis by donors missing one inhibitory KIR and lysis by donors missing two or three inhibitory KIRs. No significant difference in lysis for any of the GBM cells was observed (data not shown), indicating that KIR-HLA ligand mismatch was unlikely to be a major mechanism contributing to GBM killing.
Table II. The KIR genotypes of donors’ NK cells and the HLA genotype of P3 GBM cells, 2011-20-I GBM cells, and 2012-018 gliosarcoma.
 |
Legend colors: red, – NK or GBM cells are positive for KIR or HLA ligand, respectively; green, - NK or GBM cells are negative for KIR or HLA ligand, white, – HLA ligand is unknown; yellow highlights, donors used in in vivo experiments. Mismatch is denoted by two different colors from group HLA and KIR, match is denoted by two red colors.
C2 epitope recognized by KIR2DL2/3.
+, NK cells positive for KIR or HLA gene; –, sample negative for KIR or HLA gene; +del, NK cells have partial deletion KIR2DS4.
Therefore, we investigated the impact of activating KIR genes and found that NK cells possessing the KIR2DS2 immunogenotype (KIR2DS2+) were significantly more efficient against P3 cells at an E:T ratio of 10:1 (p = 0.0373, Kruskal–Wallis test with Dunn multiple-comparison test, Fig. 1D) and an E:T ratio of 20:1 (p = 0.0202, Kruskal–Wallis test, Fig. 1D), as well as against 2012-018 cells at an E:T ratio of 5:1 (p = 0.0331, Kruskal–Wallis test, Fig. 1E), an E:T ratio of 10:1 (p = 0.0056; Kruskal–Wallis test, Fig. 1E), and an E:T ratio of 20:1 (p = 0.0099, Kruskal–Wallis test, Fig. 1E), compared with KIR2DS2−/KIR2DS4− donors (negative). No significant difference was observed for 2011-20-I tumor cells (data not shown). NK cells from KIR2DS4+ donors were significantly more cytotoxic against 2012-018 cells at an E:T ratio of 10:1 (p = 0,0056, Kruskal–Wallis test, Fig. 1E) and an E:T ratio of 20:1 (p = 0.0099, Kruskal–Wallis test, Fig. 1E) compared with negative donors. These results were independent of the expression of inhibitory KIRs, because similar levels of expression were observed in these three groups (data not shown). The surface expression of KIR2DS2 or KIR2DS4 on cultured NK cells possessing the KIR2DS2 or KIR2DS4 immunogenotype was determined by flow cytometry. A total of 36.39 ± 19.35% (range: 14.1–86.4%, n = 4) of NK cells from donors with the KIR2DS2+ genotype expressed KIR2DS2 on the cell surface (Fig. 1Fa–d). KIR2DS4+ subsets accounted for 45.4 ± 21.5% in the two donors possessing the KIR2DS4 immunogenotype (Fig. 1Fe, 1Ff). In addition, we compared the cytotoxicity of NK cells from donors 4 and 5, who lacked the KIR2DS2 gene but possessed three or more other activating KIR genes, with NK cells from donors 6 and 7, who possessed only the KIR2DS2-activating KIR gene (Table II). The latter NK cells were significantly more cytotoxic against P3 cells at an E:T ratio of 10:1 (p = 0.0127, Mann–Whitney U test, Fig. 1G) and against 2012-018 cells at an E:T ratio of 10:1 (p = 0.0451, Mann–Whitney U test, Fig. 1H) and an E:T ratio of 20:1 (p = 0.0177, Mann–Whitney U test, Fig. 1H).
KIR2DS2 immunogenotype confers a functional activation advantage
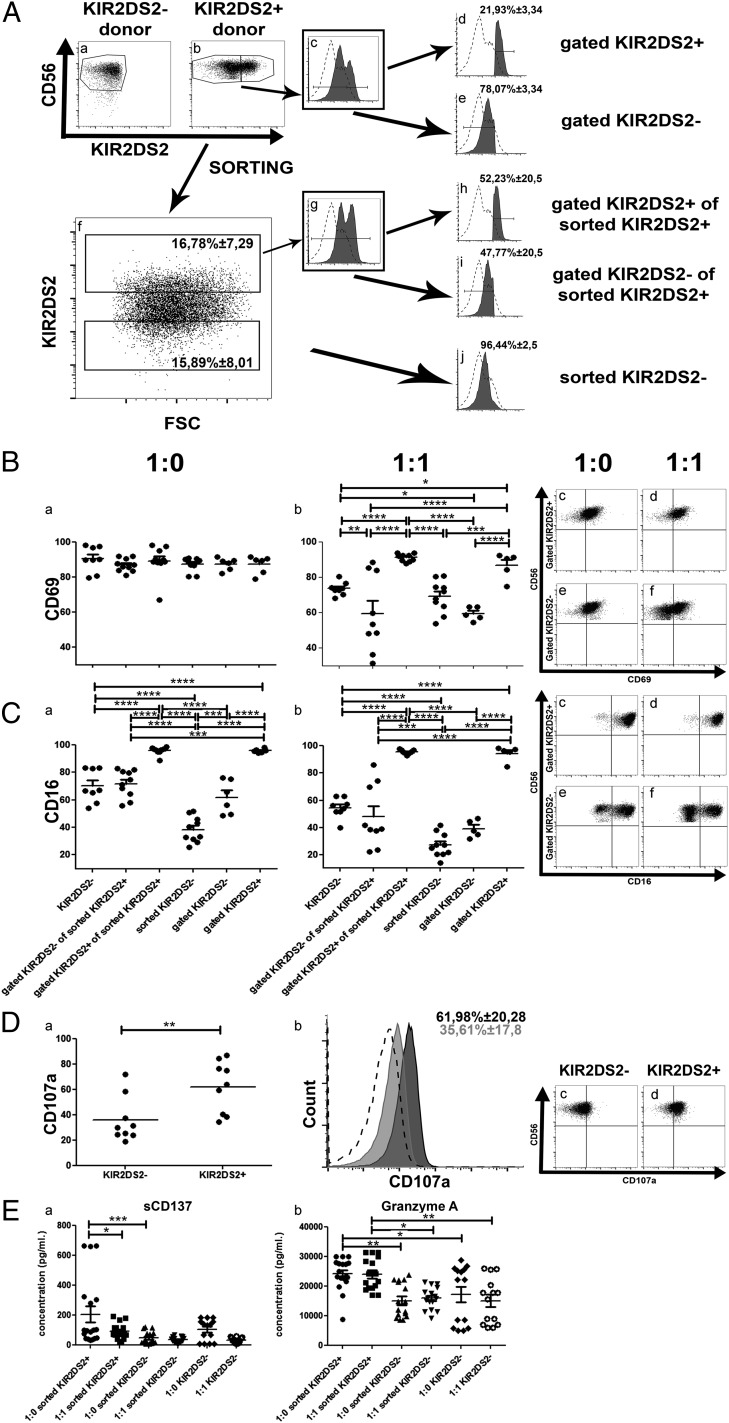
HLA-A*11:01 was recently identified as a ligand for KIR2DS2 (30). Despite the absence of the HLA-A*11:01 gene on the genotype of the P3, 2012-018, and 2011-20-I cells (Table II, Supplemental Table II), NK cells from donors with a KIR2DS2+ genotype killed P3 and 2012-018 cells more efficiently than did NK cells from donors without the KIR2DS2 gene (Fig. 1D, 1E). Indeed, NK cells with a KIR2DS2+ immunogenotype killed K562 cells lacking HLA ligands significantly better than did KIR2DS2− cells (p = 0.0345, Kruskal–Wallis test, Fig. 1I), pointing against the KIR–HLA ligand interaction as a mechanism for a cytotoxic effect. To investigate whether KIR2DS2 conferred an intrinsic functional advantage over other subpopulations, we investigated their activation, proliferation, and cytokine production in vitro, as well as their survival capabilities in vivo in mice without tumors. We used heterogeneous NK cells isolated from donors possessing the KIR2DS2 gene (Fig. 2Ab, 2Ac) for in vitro assays. Subsequently, we gated the KIR2DS2+ and KIR2DS2− subpopulations (Fig. 2Ad, 2Ae) and compared them with KIR2DS2 immunogenotype–negative NK cells (Fig. 2Aa). Alternatively, KIR2DS2+ and KIR2DS2− NK cell subsets were sorted using FACS (Fig. 2Af), expanded for 2 wk (Fig. 2Ag) prior to use in assays as gated KIR2DS2+ of sorted KIR2DS2+ cells, gated KIR2DS2− of sorted KIR2DS2+ cells, or sorted KIR2DS2− cells (Fig. 2Ah–j, respectively), and compared with KIR2DS2− NK cells (Fig. 2Aa). In monoculture, all NK subpopulations expressed similar levels of CD69, an early activation Ag (31) (p > 0.05, Fig. 2Ba). Upon coculture with GBM cells at an E:T ratio of 1:1, only the sorted KIR2DS2+ or gated KIR2DS2+ NK cells retained high levels of CD69 (p < 0.0001, two-way ANOVA with Bonferroni multiple-comparisons test), whereas KIR2DS2− donor NK cells and KIR2DS2− sorted or gated subsets significantly downregulated CD69 (Fig. 2Bb, representative dot plots Fig. 2Bc–f). Importantly, the sorted or gated KIR2DS2− subsets diminished CD69 to a greater extent (p < 0.001 and p < 0.05, respectively) than did immunogenotype-negative donor NK cells, indicating that the KIR2DS2+ fractions were the activated subsets (Fig. 2Bb). In contrast to CD69, only the monocultured sorted KIR2DS2+ or gated KIR2DS2+ NK cells expressed significantly high levels of CD16 (p < 0.0001, two-way ANOVA with Bonferroni multiple-comparisons test, Fig. 2Ca), which also was retained in coculture with GBM cells (p < 0,0001, Fig. 2Cb, representative dot plots Fig. 2Cc–f). Immunogenotype-negative donor NK cells and KIR2DS2− subsets did not differ significantly in their low CD16 expression levels before and after coculture with GBM (p > 0.05, Fig. 2C). Furthermore, greater proportions of heterogeneous KIR2DS2+ NK cells significantly expressed LAMP-1 (CD107a) (p = 0.0040, Mann–Whitney U test, Fig. 2D), indicating that they were activated for degranulation (32). There was no difference in the expression of NKG2D, NKp46, and inhibitory KIRs in the KIR2DS2+ and KIR2DS2− NK cell subsets (data not shown). There also was no statistically significant difference in proliferation based on BrdU incorporation, Ki-67 labeling index, and expansion rates between the KIR2DS2+ and KIR2DS2− NK cell subsets (data not shown).
FIGURE 2.
Activating KIR2DS2 receptor contributes to a functional advantage for NK cells. (A) Gating strategy of KIR2DS2 subpopulations from NK cell donors. (Aa) CD56 versus KIR2DS2 dot plot showing NK cell gating of KIR2DS2− donors. (Ab) CD56 versus KIR2DS2 dot plot showing NK cell gating of KIR2DS2+ donors. KIR2DS2− and KIR2DS2+ subpopulations are gated. (Ac) KIR2DS2 graph of the KIR2DS2+ donors. KIR2DS2− and KIR2DS2+ subpopulations are gated. (Ad) KIR2DS2 graph of the KIR2DS2+ gated subpopulation of KIR2DS2+ donors. (Ae) KIR2DS2 graph of the KIR2DS2− gated subpopulation of KIR2DS2+ donors. (Af) KIR2DS2 versus FSC dot plot representative of the KIR2DS2+ and KIR2DS2− sorting procedure of KIR2DS2+ donors. KIR2DS2− and KIR2DS2+ subpopulations are gated. Data are mean percentage (± SEM) of cells. (Ag) KIR2DS2 graph of KIR2DS2+ sorted cells, obtained after 2 wk in culture following the initial sorting. KIR2DS2− and KIR2DS2+ subpopulations are gated. (Ah) KIR2DS2 graph of KIR2DS2+ gated subpopulation of KIR2DS2+ sorted cells. (Ai) KIR2DS2 graph of KIR2DS2− gated subpopulation of KIR2DS2+ sorted cells. (Aj) KIR2DS2 graph of KIR2DS2− sorted cells, obtained after 2 wk in culture following the initial sorting. Data are mean percentage (± SEM) of cells. Dashed histograms represent negative control (immunogenotype KIR2DS2− donor stained for KIR2DS2) and filled black histograms represent gated or sorted population stained for KIR2DS2. (B) Percentage of cells from each of the subpopulation described above expressing the CD69 surface receptor when cultured as monoculture 1:0 (Ba) or cocultured with P3 GBM (1:1) (Bb). Dot plots of CD56 versus CD69 for gated KIR2DS2+ (Bc and Bd) and gated KIR2DS2− (Be and Bf) subpopulations when cultured as monoculture 1:0 or cocultured with P3 (1:1). (C) Percentage of cells of each subpopulation described above expressing the CD16 surface receptor when cultured as monoculture 1:0 (Ca) or cocultured with P3 (1:1) (Cb). Dot plots of CD56 versus CD16 of the gated KIR2DS2+ (Cc and Cd) and gated KIR2DS2− (e and f) subpopulations when cultured as monoculture 1:0 or cocultured with P3 (1:1). (Da) Percentage of cells of the KIR2DS2− and KIR2DS2+ donors expressing the CD107a surface receptor. (Db) Representative CD107a graph of KIR2DS2− donors (filled gray), KIR2DS2+ donors (filled black), and negative controls (dashed line). Data are mean percentage (± SEM) of positive cells. Dot plots of CD56 versus CD107a of the KIR2DS2− (Dc) and KIR2DS2+ (Dd) donors. (E) Concentration (pg/ml) of sCD137 (Ea) and granzyme A (Eb) in the supernatant of cultures of different subpopulations of NK cells in monocultures 1:0 or in cocultures with P3 (1:1) for 4 h in serum-free RPMI 1640 without cytokine supplement. Data are mean ± SEM of three or four independent experiments/donor. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Activated KIR2DS2 immunogenotype NK cells preferentially express soluble CD137 and granzyme A lytic granules
Cytokine analyses revealed no significant difference in IFN-γ, TNF-α, perforin, or granzyme B release between KIR2DS2+ and KIR2DS2− NK cells in monoculture or coculture (Supplemental Fig. 2). However, release of the splice variant of costimulatory receptor soluble CD137 (sCD137; ILA) (33), a member of the TNFR family, was significantly enhanced in the KIR2DS2+ sorted NK cell subsets in monoculture compared with KIR2DS2− sorted subsets (p < 0.001, one-way ANOVA, Fig. 2Ea), as well as in 1:1 cocultures with GBM cells (p < 0.05, one-way ANOVA with Bonferroni multiple-comparisons test, Fig. 2Ea); however, it was not significantly different from immunogenotype KIR2DS2− donors. Although levels of perforin and granzyme B cytolytic granules could not differentiate the NK cells from immunogenotype KIR2DS2− donors and FACS-sorted subsets from KIR2DS2+ donors (Supplemental Fig. 2), granzyme A release was significantly elevated in the KIR2DS2+ sorted subsets compared with KIR2DS2− sorted subsets and in NK cells from immunogenotype KIR2DS2− donors in monoculture, as well as in 1:1 coculture with GBM cells (p < 0.05 and p < 0.01 respectively, one-way ANOVA with Bonferroni multiple-comparisons test, Fig. 2Eb). These data collectively indicate that the KIR2DS2+ subset is distinct from the KIR2DS2− subset, as well as KIR2DS2− NK cells from immunogenotype-negative donors, with regard to intrinsic activation states, degranulation, and cytotoxic capacity.
Treatment with a single dose of NK cells significantly increases survival of GBM-bearing animals
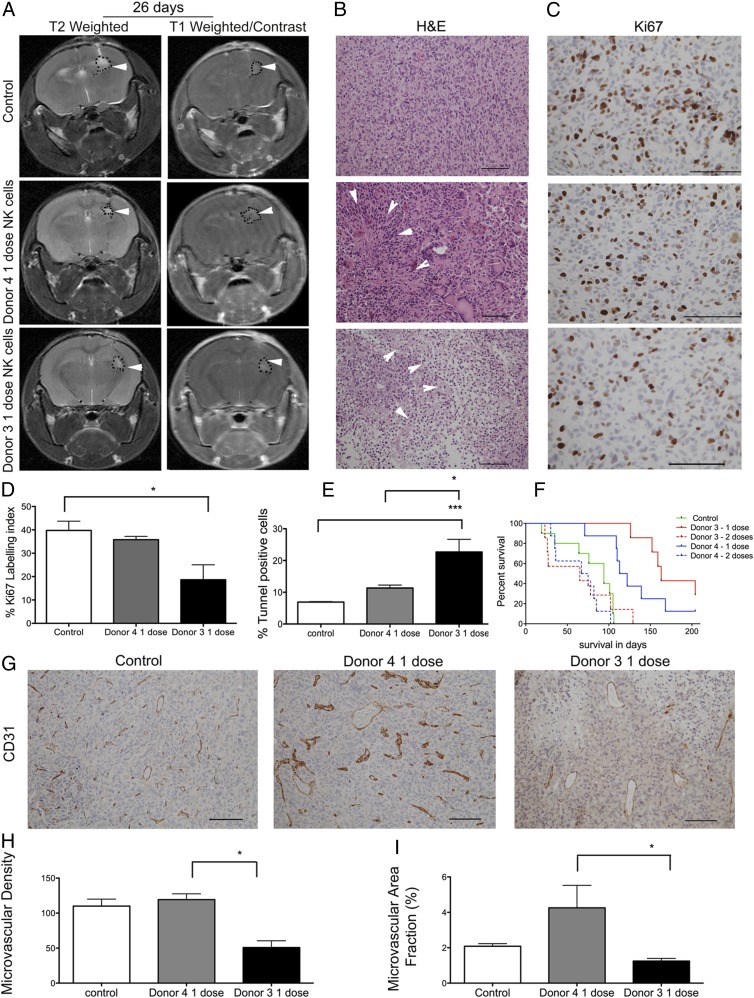
Next, we asked whether NK cells’ efficacy against solid GBM in vivo is dependent on the KIR repertoire. Thus, we treated mice bearing human GBM with activated NK cells isolated from donors 3 and 4, who possessed different inhibitory and activating KIR gene repertoires (Table II). Donor 3 NK cells possessed a KIR2DS2+ genotype but had no inhibitory KIR-HLA ligand mismatch. In contrast, donor 4 NK cells had three inhibitory KIR-HLA ligand mismatches but lacked the KIR2DS2 gene. MRI performed 26 d after implantation of the GBM tumor spheres (12 d after injection of the first dose of NK cells) revealed contrast-enhancing small tumor lesions in the right hemisphere on postcontrast T1-weighted images from control mice and mice that received a single dose of NK cells from donor 3 or donor 4 (Fig. 3A, right panels). In all mice scanned, T2-weighted sequences displayed high signal intensities in the same areas, consistent with localized edema formation indicative of tumor development (Fig. 3A, left panels). H&E staining performed post mortem showed diffuse infiltrative tumors with high cellular density in the control group (Fig. 3B) and large regions of necrotic cell death in the treatment groups (Fig. 3B). The Ki-67–labeling index revealed greater tumor cell proliferation in untreated control tumors compared with tumors treated with one dose of NK cells from donor 3 or donor 4 (Fig. 3C, 3D). Donor 3’s NK cells significantly reduced tumor proliferation compared with controls (p = 0.02, one-way ANOVA, Kruskal–Wallis 7.53, df = 2, Fig. 3D). Larger areas of apoptotic cells were seen in the tumors treated with donor 3’s NK cells compared with vehicle-treated control tumors (p = 0.0004, one-way ANOVA, Kruskal Wallis 15.55, df = 2, Fig. 3E) and with tumors treated with donor 4’s NK cells (p = 0.04, one-way ANOVA, Kruskal–Wallis 15.55, df = 2, Fig. 3E). There was no significant difference in the survival of animals in control groups treated with PBS vehicle only and animals treated with anti-CD2 and anti-NKp46 Abs, which are components of NK cell expansion in the Miltenyi Biotec kit and were used as controls (data not shown). Therefore, we merged these animals into one control group. Mice treated with a single dose of 106 NK cells obtained from donor 4 or donor 3 lived significantly longer than did untreated control animals, with a median survival of 117.5 d (p = 0.0005, log-rank 15.62, df = 1), 163 d (p = 0.0001, log-rank 12.24, df = 1), and 94 d, respectively (Fig. 3F). There was no significant difference in the survival of animals treated with one dose of NK cells obtained from donor 4 and donor 3 (p = 0.11, log-rank 2.45, df = 1, Fig. 3F). Moreover, repeated doses of NK cells from both donor 3 and donor 4 significantly diminished the survival benefit compared with a single dose of NK cells from donor 4 (p = 0.002, log-rank 8.89, df = 1, Fig. 3F) or from donor 3 (p = 0.0004, log-rank 12.34, df = 1, Fig. 3F). Next, we investigated angiogenesis as an indicator of tumor growth and metabolism. Treatment with a single dose of NK cells from donor 3 significantly reduced tumor microvascular densities compared with NK cells from donor 4 (p = 0.04, one-way ANOVA, Kruskal Wallis 6.0, df = 2, Fig. 3G, 3H). The microvessels of tumors treated with donor 4 NK cells were dysregulated and dilated, indicating florid angiogenesis (Fig. 3G), as revealed by a significantly larger fraction of CD31+ vessels compared with tumors treated with donor 3 NK cells (p = 0.01, one-way ANOVA, Kruskal–Wallis 8.0, df = 3, Fig. 3I).
FIGURE 3.
Treatment with a single dose of NK cells significantly increases animal survival. (A) T2-weighted and postcontrast T1-weighted images of NOD/SCID mice bearing P3 tumors treated with one dose of NK cells from donor 4 or donor 3 or controls. Tumors are indicated by arrowheads. (B) H&E staining of tumors treated with donor 3–derived or donor 4–derived NK cells. Necrosis is indicated by arrowheads (scale bar, 100 μm; original magnification ×200). (C) Ki-67+ cells (scale bar, 100 μm; original magnification ×400). Percentage Ki-67–positive (D) and cell death as represented by the percentage of TUNEL+ apoptotic/necrotic cells (E). Data are mean ± SEM. (F) Kaplan–Meier survival curves of animals treated with vehicle PBS/CD2 and NKp46 Abs (n = 10 animals) or with one or two doses of NK cells from donor 4 or donor 3 (n = 7 animals/group). (G) CD31 immunostaining from control mouse and mice that received one dose of NK cells from donor 4 or donor 3 (scale bar, 100 μm; original magnification ×200). Microvascular density (H) and percentage of area positive for CD31 microvessels (I). *p < 0.05, ***p < 0.001.
Increased proinflammatory macrophages/microglia within the tumor after one dose of NK cells
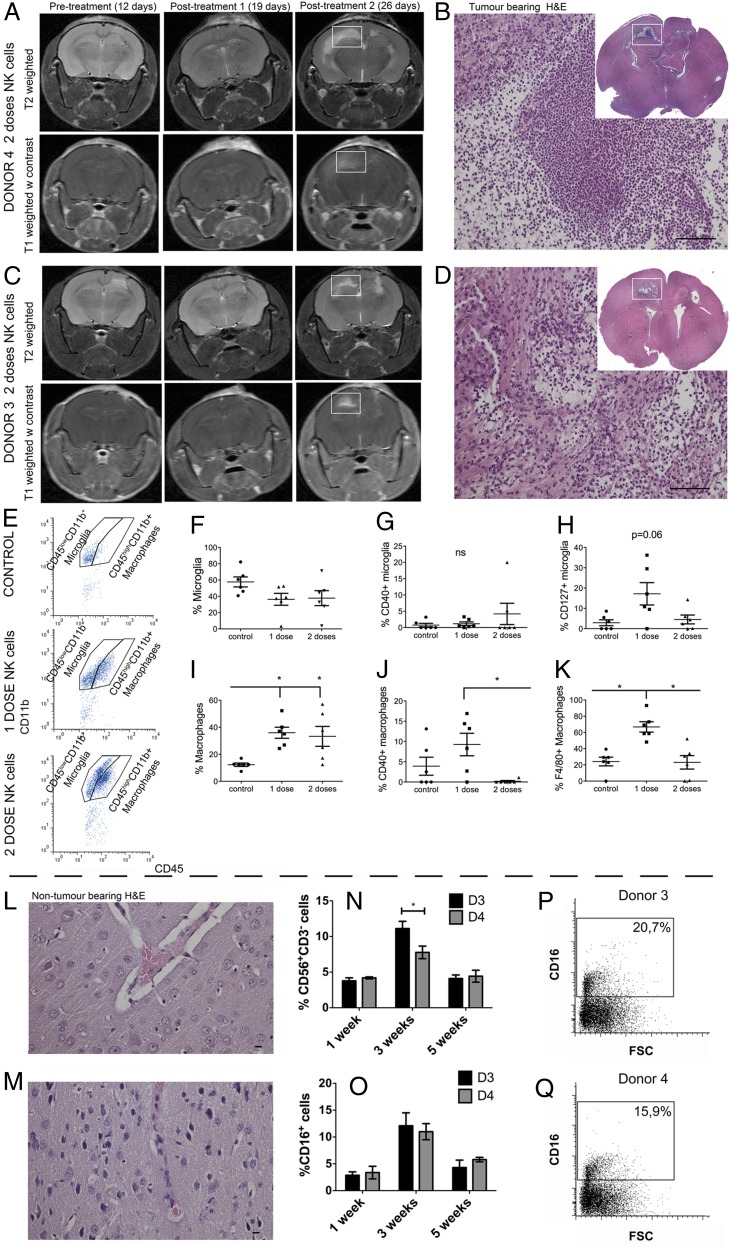
We hypothesized that the repeated NK cell treatments reduced animal survival as the result of increased inflammation. Longitudinal T1- and T2-weighted MRI follow-up revealed radiological changes that appeared after the second dose of NK cells from both donors (Fig. 4A, 4C). Histological analysis revealed numerous immune cells at the ipsilateral sites of tumor implantation and in the contralateral hemisphere, destruction of the white matter tracts of the corpus callosum, and high numbers of foamy cells (Fig. 4B, 4D). To characterize the cell types involved, flow cytometry was performed on brain tissue dissociated into a single-cell suspension. We distinguished macrophages from microglia based on the relative expression of CD45 and CD11b (34); the CD45highCD11b+ cellular populations represented macrophages, and CD45lowCD11b+ cells were microglia (Fig. 4E). Although proportions of microglia remained unchanged between controls and tumors treated with one or two doses of NK cells (p > 0.05, Fig. 4F), macrophages were significantly recruited to the tumors after treatment with one or two doses of NK cells (p = 0.005, one-way ANOVA, Kruskal–Wallis 10.26, df = 2, Fig. 4I). The macrophages significantly increased costimulatory CD40 molecules after one dose of NK cell treatment compared with two doses of treatment (p = 0.02, one-way ANOVA, Kruskal–Wallis 7.46, df = 2, Fig. 4J), whereas this was not the case for microglia (Fig. 4G). Single-dose NK cell treatment significantly increased F4/80 expression on macrophages compared with the control and two doses of NK cells (p = 0.0045, one-way ANOVA, Kruskal–Wallis 10.79, df = 2, Fig. 4K). Microglia showed a tendency toward elevated IL-7R (CD127) expression compared with the control group and the group that received two doses of NK cells (p = 0.061, one-way ANOVA, Kruskal–Wallis 5.5, df = 2, Fig. 4H). Off-target experiments with the two-dose NK cell regimen from immunogenotype KIR2DS2+ (donor 3) and KIR2DS2− NK cells (donor 4) in a separate cohort of nontumor-bearing mice demonstrated that NK cells were well tolerated, because the animals were alive at 4 mo post transplantation (p = 0.38, log-rank 4.16, df = 4, data not shown). Histological analyses of brains revealed normal histoarchitecture and regenerating tissue in areas of NK cell injection (Fig. 4L, 4M). Significantly more CD56+CD3− NK cells that were CD16+ from KIR2DS2+ donor 3 persisted in the brain (p = 0,03; two-way ANOVA with Bonferroni multiple-comparisons test) at 3 wk compared with the KIR2DS2− donor 4 NK cells (Fig. 4N–Q). There was no difference in proliferation, as indicated by the Ki-67–labeling index (data not shown), at 1, 3, and 5 wk posttransplant.
FIGURE 4.
Increased proinflammatory macrophages/microglia within the tumor after one-dose NK cell treatment. (A) Longitudinal T2-weighted (upper panels) and postcontrast T1-weighted (lower panels) images of NOD/SCID mice bearing P3 tumors before (12 d) and after treatment with one or two doses (19 and 26 d, respectively) of NK cells from donor 4. (B) H&E staining of tumor-bearing mouse brain after two doses of donor 3 NK cells. Immune infiltrates in the contralateral hemisphere region as indicated by the white box in (A) (scale bar, 100 μm; original magnification ×200). Inset shows a whole-brain histological section stained with H&E from the white box region on MR images from a representative animal treated with donor 4–derived NK cells. (C) Longitudinal T2-weighted (upper panels) and postcontrast T1-weighted (lower panels) images of NOD/SCID mice bearing P3 tumors before (12 d) and after treatment with one or two doses (19 and 26 d, respectively) of NK cells from donor 3. (D) H&E staining of tumor-bearing mouse brain after two doses of donor 4 NK cells. Immune infiltrates in the contralateral hemisphere region as indicated by the white box in (C) (scale bar, 100 μm; original magnification ×200). Inset shows a whole-brain histological section stained with H&E from the white box region on MR images from a representative animal treated with donor 3–derived NK cells. (E) Representative examples of flow cytometry gating of microglia (CD45lowCD11b+) and macrophage (CD45highCD11b+) populations of the control group (top panel) and the groups that received one dose (middle panel) or two doses (bottom panel) of NK cells. (F) Proportions of microglia within all immune (CD45+) cells as evaluated by flow cytometry. Data are mean ± SEM (n = 6/group). Proportions of microglia expressing CD40 (G) or CD127 (H), as evaluated by flow cytometry. Data are mean ± SEM (n = 6/group). (I) Proportions of macrophages within all immune (CD45+) cells, as evaluated by flow cytometry. Data are mean ± SEM (n = 6/group). Proportions of macrophages expressing CD40 (J) or F4/80 (K), as evaluated by flow cytometry. Data are mean ± SEM (n = 6/group). Histological H&E staining of nontumor-bearing mouse brain after two doses of donor 4 NK cells (L) or two doses of donor 3 NK cells (M) (scale bar, 10 μm; original magnification ×400). Percentage of donor 3 and donor 4 NK cells (CD56+ and CD3−) (N) and CD16+ NK cells (O) in nontumor–bearing mouse brains 1, 3, and 5 wk after NK cell treatment. Dot plots of CD16 versus FSC showing the percentage of CD16+ cells present 3 wk after donor 3 (P) or donor 4 (Q) NK cell treatment. *p < 0.05.
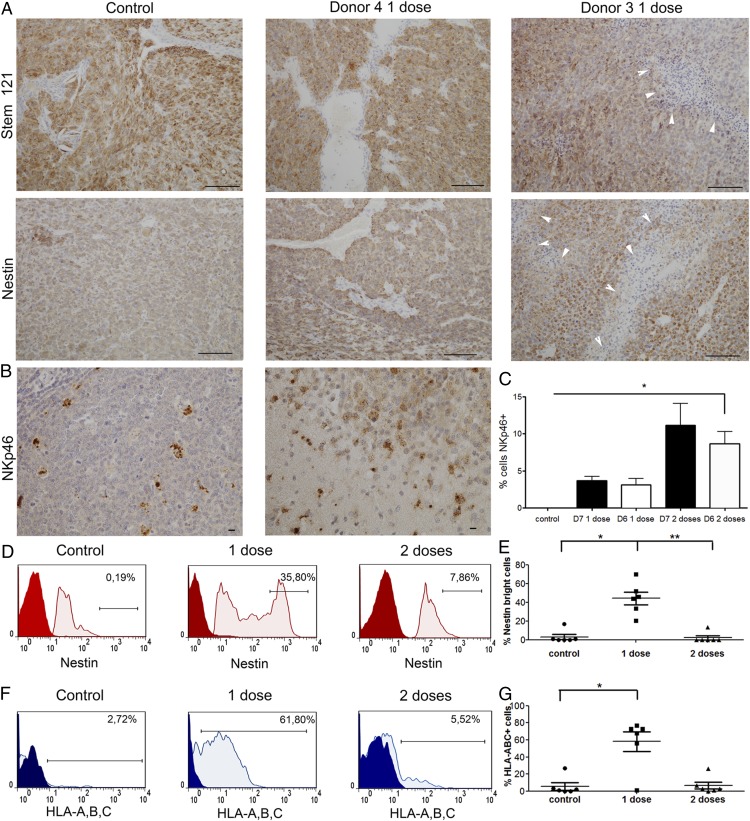
Single-dose NK cell treatment upregulates nestin and HLA-ABC expression on tumor cells
We identified the human cells engrafted in the mouse brains using human-specific STEM121, which labeled highly cellular control tumors, donor 4 NK cell–treated tumors, and donor 3 NK cell–treated tumors with extensive areas of tumor necrosis (Fig. 5A). Tumor cells were confirmed by their expression of human-specific nestin (Fig. 5A). NKp46+ NK cells could be visualized in the tumor tissues (Fig. 5B, 5C) from all treatment groups. Flow cytometric phenotyping of the mouse brains revealed that one dose of NK cells upregulated the expression of nestin, which manifested as a higher proportion of nestinbright tumor cells, compared with controls and mice treated with a double dose of NK cells (41.0 ± 9.6% versus 3.1 ± 2.8%, p > 0.05, and versus 2.3 ± 2.3%, p < 0.05, respectively, n = 6, Fig. 5D, 5E). Tumor cells from mice treated with one dose of NK cells upregulated the expression of HLA-A, HLA-B, and HLA-C compared with the control group (58.0 ± 11.78% versus 5.5 ± 4.3%, p < 0.05, n = 6, Fig. 5F, 5G) and compared with mice treated with a double dose of NK cells (58.0 ± 11.78% versus 6.6 ± 4.1%, p > 0.05, n = 6, Fig. 5F, 5G), thus potentially providing ligands for KIRs. Tumor cells from all treatment groups expressed low levels of HLA-E (<4%) and HLA-G (<9%, data not shown), thereby lacking ligands for the inhibitory/activating receptors NKG2A and KIR2DL4, respectively, expressed on NK cells.
FIGURE 5.
Single-dose NK cell treatment induces nestin, HLA-A, HLA-B, and HLA-C expression on tumor cells. (A) Human-specific STEM121+ cells in P3 tumor-bearing mice treated with vehicle controls or one dose of NK cells from donor 4 or donor 3 (scale bar, 100 μm; original magnification ×200). Nestin expression by IHC in tumors treated with one dose of NK cells from donor 4 or donor 3 compared with controls post mortem (scale bar, 100 μm; original magnification ×200). White arrowheads in (A) indicate regions of necrotic tumor tissue. (B) Human-specific NKp46 staining demonstrates infiltration of the injected NK cells into the tumor in vivo (right panel) and staining of human tonsil used as a positive control (left panel) (scale bar, 10 μm; original magnification ×400). (C) Percentage of NKp46+ cells (mean ± SEM) in tumor tissue. (D) Representative graphs showing nestin expression on tumor cells from control animals and animals treated with one or two doses of NK cells, as evaluated by flow cytometry. The gate represents the “nestinbright” population within all tumor (nestin+) cells (red: negative control; pink: nestin). (E) Proportion of nestinbright population within all tumor (nestin+) cells, as evaluated by flow cytometry. Data are mean ± SEM (n = 6/group). (F) Representative graphs showing HLA-A,B,C expression on tumor cells from control animals and animals treated with one or two doses of NK cells, as evaluated by flow cytometry. The gate represents the HLA-A,B,C+ population within all tumor (nestin+) cells (blue: isotype control; pale blue: HLA-A,B,C). (G) Proportions of HLA-A,B,C+ population within all tumor (nestin+) cells, as evaluated by flow cytometry. Data are mean ± SEM (n = 6/group). *p < 0.05, **p < 0.01.
Discussion
NK cells play an important role in anticancer immunity because of their multiple mechanisms of recognizing and killing transformed cells. The NCRs NKp30, NKp44, and NKp46 were demonstrated to induce NK cell activation via recognition of tumor-associated Ags (9, 35, 36). Overexpression of stress-induced ligands for NKG2D-activating receptor on tumor cells also was shown to trigger NK cell–mediated tumor cell lysis (37). The above-mentioned mechanisms determine NK cell potency in a tumor-dependent manner, because NCRs and NKG2D are constitutively expressed on NK cells in most individuals, whereas different tumors variably express tumor-associated Ags and may display different expression levels of NKG2D ligands.
In the current study, we investigated the cytotoxicity of human allogeneic NK cells against patient-derived GBM and gliosarcoma cells. These tumor cells were propagated in stem cell medium to maintain the genomic and phenotypic heterogeneity, as well as the cancer-initiating properties of the original tumor in situ (27, 38). These aggressive, therapy-resistant GSCs were demonstrated to be more susceptible to NK cell–mediated lysis (26). Our patient-derived GBMs and gliosarcoma cells displayed phenotypes consistent with immature GSCs, because they expressed the stem cell markers CD133 and A2B5, propagated as neurospheres, and derived tumors in vivo in mice, features that are consistent with other reports (26, 27). Furthermore, nearly 100% of the GBM cells expressed HLA-A, HLA-B, and HLA-C molecules, whereas 25.2% of gliosarcoma cells also expressed nonclassical HLA-E, confirming our previous analyses of patients’ GBM biopsies in situ (6). Purified and ex vivo–cultured NK cells obtained from 10 unrelated donors were cytotoxic against all GBMs, as well as control K562 cells that lack HLA ligands. However, the gliosarcoma cells were less susceptible to NK cell–mediated lysis, likely as a result of higher HLA levels and lower expression of most of the ligands for NKG2D-activating receptor. This is supported by our findings demonstrating that blocking the NKG2D receptor significantly diminished cytotoxicity against both P3 and 2012-018 cells. However, the NK cells’ cytotoxicity against GBM cells was donor dependent, so we hypothesized that this was due to their different KIR repertoires. The KIR-HLA class I mismatch was reported to mediate the graft-versus-leukemia effect (18), improve the outcome of patients receiving HSCT (18), and play an important role in the NK cells’ cytotoxicity against solid tumors in vitro (21, 29). However, we found no correlation between NK cell potency and the degree of inhibitory KIR-HLA class I ligand mismatch. A caveat to these interpretations is our small donor/tumor sample size, which may not represent the diversity of the NK repertoire (39), as well as the fact that we did not examine allelic polymorphisms that were demonstrated to influence the impact of KIR-HLA ligand mismatch on hematological malignancies (40). Nevertheless, we investigated the possible impact of activating KIRs on the efficacy of NK cell–mediated lysis of GBM cells. Although activating KIRs often recognize the same ligands as the inhibitory KIRs, they do so with lower affinity. KIR2DS2 does not bind well to HLA-C, and only substitution of a tyrosine residue at position 45 of KIR2DS2 restores some binding affinity to HLA-C (30). Therefore, the physiological function of activating KIRs has remained elusive (41); until recently, activating KIRs were not considered to play a major role in cancer. Our data indicate that the KIR2DS2 gene may identify alloreactive NK cell subsets with higher potency against GBM, independently of the repertoire of inhibitory or other activating KIRs. KIR2DS4+ NK cells were more potent against GBM than were KIR2DS2−/KIR2DS4− NK cells, but they were not significantly more potent than KIR2DS2+ NK cells. Recently, HLA-A*11:01 was demonstrated to bind KIR2DS2 (30); however, the KIR2DS2+ NK cells exhibited greater cytotoxic potency against all GBMs without HLA-A*11:01 ligands, as well as K562 cells lacking all HLA ligands. These findings might indicate that an intrinsic cytotoxic potency, rather than KIR–HLA ligand interactions, is the mechanism of potency associated with the KIR2DS2 immunogenotype. However, the existence of other, unidentified ligands cannot be ruled out. We were unable to unequivocally demonstrate the impact of KIR2DS2 and KIR2DS4 because KIR2DS2 and KIR2DS4 receptors were expressed at the protein level on only a minor subpopulation of NK cells in contrast to NKG2D, which is constitutively expressed on all NK cells; the anti-KIR2DS2 and anti-KIR2DS4 Abs that we used are not known to have a blocking, neutralizing, or stimulating effect; and high linkage disequilibrium between KIR2DS2 and KIR2DL2 genes—only 4 aa residues distinguish them at the ligand-binding domain–means that Abs against KIR2DS2 might also recognize KIR2DL2. Generating highly specific neutralizing monoclonal anti-KIR2DS2 and anti-KIR2DS4 Abs, as well as an NK cell culture protocol that allows the selective expansion of KIR2DS2+ or KIR2DS4+ cells, might enable functional investigations of the role of KIR2DS2 and KIR2DS4 receptors in NK cell–mediated lysis of tumor cells. Despite these shortcomings, we demonstrated functionally that KIR2DS2+-enriched NK cell subsets retained significantly higher levels of CD69 and CD16 and secreted significantly more sCD137, which might regulate their activation. The increased cytotoxic potential of KIR2DS2+ NK cells was consistent with significantly higher intrinsic levels of CD107a+ cytolytic lysosomes and increased granzyme A degranulation, when in contact with GBM cells, compared with KIR2DS2− NK cell subsets and NK cells from immunogenotype KIR2DS2−. Human granzyme A is a serine protease with trypsin-like activity; it cleaves basic arginine and lysine residues to induce rapid caspase-independent cell death through ssDNA breaks (42, 43). Thus, tumor cells that are resistant to caspase-mediated cell death, including GBMs that overexpress Bcl-2, might be sensitive to the apoptotic effects of granzyme A. More KIR2DS2+ NK cells also persisted in vivo at 3 wk, consistent with increased sCD137 costimulatory factors.
Furthermore, we investigated the impact of KIR repertoires on the NK cells’ potency against solid GBM in vivo and demonstrated that donor 3’s NK cells, which possessed the KIR2DS2+ gene, prolonged median survival by 72 d compared with control and by 46 d compared with donor 4’s KIR2DS2− NK cells, and manifested as decreased tumor proliferation, reduced angiogenesis, and increased apoptosis. We reported recently that early structural, contrast-enhanced MRI imaging fails to distinguish tumor volumes in immunotherapy strategies (44); thus, we used it in this study to visualize tumor establishment. Based on these in vivo findings and the in vitro findings, we speculate that the difference in the outcome of the animals partially corresponds to the alloreactive KIR2DS2+ NK cells. Despite less inhibitory KIR-HLA class I mismatch, donor 3’s NK cells demonstrated better therapeutic efficacy, indicating that the inhibitory signal resulting from interactions of inhibitory KIR with their ligands may be overridden by activating signals mediated by KIR2DS2, in addition to other activating receptors, such as NCRs and NKG2D. Moreover, NK cells from donor 4 were less efficient, despite the presence of three other activating KIR genes; this may suggest that the KIR2DS2 genotype represents a dominant activation signal. However, this finding requires confirmation in larger cohorts of donor/tumor samples for which the KIR diversity is more extensively accounted. Nevertheless, our study suggests that KIR2DS2 may constitute an important clinical criterion for NK cell donor selection with regard to the development of effective cell-based anticancer therapies.
The lack of extended survival in double dose–treated, tumor-bearing animals may reflect an “on-target” adverse effect that occurs as the result of extensive inflammation due to host macrophage recruitment, in addition to tumor-dependent factors. Off-target experiments that treated naive nontumor-bearing mice with the same NK cell schedule showed that it was well tolerated; it did not result in dose-dependent toxicity. Compared with double dose–treated and control animals, a higher proportion of macrophages from single dose–treated tumor-bearing animals expressed CD40, a costimulatory receptor that interacts with the CD154 ligand that is reported to be expressed on NK cells (45). The microglia of single dose–treated animals upregulated the expression of CD127, the IL-7R that mediates inflammation (46, 47); thus, the increased expression of its receptor may suggest an induced proinflammatory response of host microglia that, in addition to host CD40-expressing macrophages, might contribute to the beneficial effect of single-dose treatment. In our previous study using a rat glioma model, we proposed a mechanism of NK cell–mediated immune modulation of microglia to proinflammatory phenotypes through IFN-γ secretion and recruitment of M1-like macrophages from the periphery (48). In that study, combination treatment with NK cells and mAb9.2.27 against NG2/CSPG4 induced the functional switch of microglia from tumor supportive, anti-inflammatory M2-like cells to proinflammatory M1-like cells that effectively reduced GBM growth in vivo. We speculate that a similar cross-talk between macrophages and microglia, in response to NK cell–derived factors, occurred in the current study. In addition, nestin expression was upregulated on the tumor cells of animals treated with a single dose of NK cells compared with double dose–treated animals and controls. The nestin intermediate filament is expressed on neural progenitor cells under physiological conditions and is attenuated upon differentiation of neural stem cells. However, nestin is overexpressed in glioma cells, reactive astrocytes (49), and it can be re-expressed during other pathological conditions, such as injury- or infection-related inflammation (50, 51). Thus, we speculate that the increased nestin expression on tumor cells in single dose–treated animals is associated with the proinflammatory effects mediated by the adoptively transferred NK cells. Tumor cells in single dose–treated animals upregulated the expression of HLA class I molecules compared with controls. Reddy et al. (52) demonstrated that glioma cell lines upregulated HLA class I expression in response to IFN-β and IFN-γ treatment in vitro. Therefore, the effect that we observed in vivo might be due to IFN-γ release by activated NK cells.
To investigate the influence of KIR–HLA class I interactions on the therapeutic effect, we chose to use NOD/SCID mice bearing human tumor and human NK cells isolated from healthy donors. Xenograft models using patient-derived tumor material are considered to be more representative, and lack of functional host NK cells in NOD/SCID mice eliminates the possible influence of host NK cells on the study results. However, the disadvantage of this model is the inability to assess the full impact of the host immune system, which is of great importance in the context of allogeneic cellular therapy. Therefore, further studies are needed to evaluate whether allogeneic NK cell therapy is applicable for GBM patients with respect to immune-conditioning regimens, mode of delivery, dose escalation, maximum tolerated doses to the brain, and management of acute local inflammation after NK cell infusions. To summarize, the possibility of using NK cells against solid tumors is a promising strategy that has just started to be realized (53, 54). To our knowledge, our study is the first to demonstrate that KIR2DS2+ NK cells may represent an alloreactive subset with potent cytotoxicity against patient-derived solid GBM tumors in vivo mediated by elevated activation levels and effective granzymes against GBM. The impact of KIR2DS2 should be investigated further in a larger cohort, because it may identify donors with more potent NK cells. Selective expansion of NK cells with the desired phenotype could enhance anticancer therapeutics.
Supplementary Material
Acknowledgments
We thank Bodil B. Hansen, Tove Johannsen, Ingrid Sandvik Gavlen, Kristin Paulsen Rye, Endy Spriet, Krishna M. Talasila, and Tina Pavlin for efficient technical assistance. We thank the neurosurgical department (Haukeland University Hospital) for providing tumor tissue, and we are grateful to the volunteers who donated blood for the experiments and the patients who consented to donate tumor tissue for research. Flow cytometry and MRI were performed at the Molecular Imaging Center, University of Bergen.
This work was supported by grants from The Bergen Medical Research Foundation, The Norwegian Research Council, The Meltzer Fund, and The Norwegian Cancer Society.
A.G.N. and J.K. designed and performed experiments, monitored animals, conducted in vivo experiments, analyzed and interpreted the data, and wrote the manuscript. L.L. collected blood from healthy donors, sorted the KIR2DS2+ NK cells using FACS, monitored animals, and conducted in vivo experiments. M.Z. performed experiments and contributed to data analysis. A.E. designed and performed experiments and contributed to data analysis. Ø.B. contributed to cytokine analysis and manuscript editing. J.Z. contributed to data interpretation and manuscript editing. P.Ø.E. contributed to ethical approvals, in vivo experiment design and surgery, data interpretation, and manuscript editing. M.C. conceived of and designed the research, analyzed and interpreted the data, coordinated project activities, wrote the manuscript, and provided funding.
The online version of this article contains supplemental material.
- FSC
- forward scatter
- GBM
- glioblastoma
- GFAP
- glial fibrillary acidic protein
- GSC
- glioma stem–like cell
- HSCT
- hematopoietic stem cell transplantation
- IHC
- immunohistochemistry
- KIR
- killer Ig–like receptor
- MFI
- mean fluorescence intensity
- MICA
- MHC class I–related chain A
- MICB
- MHC class I–related chain B
- MRI
- magnetic resonance imaging
- NCR
- natural cytotoxicity receptor
- NKG2
- NK group 2
- sCD137
- soluble CD137
- SSC
- side scatter
- ULBP
- UL16-binding protein.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Louis D. N., Ohgaki H., Wiestler O. D., Cavenee W. K., Burger P. C., Jouvet A., Scheithauer B. W., Kleihues P. 2007. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 114: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R., Mason W. P., van den Bent M. J., Weller M., Fisher B., Taphoorn M. J., Belanger K., Brandes A. A., Marosi C., Bogdahn U., et al. European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups.; National Cancer Institute of Canada Clinical Trials Group 2005. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352: 987–996. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa E., Takano S., Ohno T., Tsuboi K. 2012. Adoptive cell transfer therapy for malignant gliomas. Adv. Exp. Med. Biol. 746: 109–120. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa E., Tsuboi K., Takano S., Uchimura E., Nose T., Ohno T. 2004. Intratumoral injection of IL-2-activated NK cells enhances the antitumor effect of intradermally injected paraformaldehyde-fixed tumor vaccine in a rat intracranial brain tumor model. Cancer Sci. 95: 98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghiringhelli F., Ménard C., Martin F., Zitvogel L. 2006. The role of regulatory T cells in the control of natural killer cells: relevance during tumor progression. Immunol. Rev. 214: 229–238. [DOI] [PubMed] [Google Scholar]
- 6.Kmiecik J., Poli A., Brons N. H., Waha A., Eide G. E., Enger P. O., Zimmer J., Chekenya M. 2013. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J. Neuroimmunol. 264: 71–83. [DOI] [PubMed] [Google Scholar]
- 7.Lanier L. L. 2003. Natural killer cell receptor signaling. Curr. Opin. Immunol. 15: 308–314. [DOI] [PubMed] [Google Scholar]
- 8.Koch J., Steinle A., Watzl C., Mandelboim O. 2013. Activating natural cytotoxicity receptors of natural killer cells in cancer and infection. Trends Immunol. 34: 182–191. [DOI] [PubMed] [Google Scholar]
- 9.Moretta A., Bottino C., Vitale M., Pende D., Cantoni C., Mingari M. C., Biassoni R., Moretta L. 2001. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu. Rev. Immunol. 19: 197–223. [DOI] [PubMed] [Google Scholar]
- 10.Bottino C., Castriconi R., Moretta L., Moretta A. 2005. Cellular ligands of activating NK receptors. Trends Immunol. 26: 221–226. [DOI] [PubMed] [Google Scholar]
- 11.Mittelbronn M., Simon P., Löffler C., Capper D., Bunz B., Harter P., Schlaszus H., Schleich A., Tabatabai G., Goeppert B., et al. 2007. Elevated HLA-E levels in human glioblastomas but not in grade I to III astrocytomas correlate with infiltrating CD8+ cells. J. Neuroimmunol. 189: 50–58. [DOI] [PubMed] [Google Scholar]
- 12.Campbell K. S., Purdy A. K. 2011. Structure/function of human killer cell immunoglobulin-like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 132: 315–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almalte Z., Samarani S., Iannello A., Debbeche O., Duval M., Infante-Rivard C., Amre D. K., Sinnett D., Ahmad A. 2011. Novel associations between activating killer-cell immunoglobulin-like receptor genes and childhood leukemia. Blood 118: 1323–1328. [DOI] [PubMed] [Google Scholar]
- 14.Middleton D., Diler A. S., Meenagh A., Sleator C., Gourraud P. A. 2009. Killer immunoglobulin-like receptors (KIR2DL2 and/or KIR2DS2) in presence of their ligand (HLA-C1 group) protect against chronic myeloid leukaemia. Tissue Antigens 73: 553–560. [DOI] [PubMed] [Google Scholar]
- 15.Carrington M., Wang S., Martin M. P., Gao X., Schiffman M., Cheng J., Herrero R., Rodriguez A. C., Kurman R., Mortel R., et al. 2005. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J. Exp. Med. 201: 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naumova E. S., Sukhotina N. N., Naumov G. I. 2004. Molecular-genetic differentiation of the dairy yeast Kluyveromyces lactis and its closest wild relatives. FEMS Yeast Res. 5: 263–269. [DOI] [PubMed] [Google Scholar]
- 17.Vivier E., Ugolini S., Blaise D., Chabannon C., Brossay L. 2012. Targeting natural killer cells and natural killer T cells in cancer. Nat. Rev. Immunol. 12: 239–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruggeri L., Capanni M., Urbani E., Perruccio K., Shlomchik W. D., Tosti A., Posati S., Rogaia D., Frassoni F., Aversa F., et al. 2002. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 295: 2097–2100. [DOI] [PubMed] [Google Scholar]
- 19.Köhler K., Xiong S., Brzostek J., Mehrabi M., Eissmann P., Harrison A., Cordoba S. P., Oddos S., Miloserdov V., Gould K., et al. 2010. Matched sizes of activating and inhibitory receptor/ligand pairs are required for optimal signal integration by human natural killer cells. PLoS ONE 5: e15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tallerico R., Todaro M., Di Franco S., Maccalli C., Garofalo C., Sottile R., Palmieri C., Tirinato L., Pangigadde P. N., La Rocca R., et al. 2013. Human NK cells selective targeting of colon cancer-initiating cells: a role for natural cytotoxicity receptors and MHC class I molecules. J. Immunol. 190: 2381–2390. [DOI] [PubMed] [Google Scholar]
- 21.Igarashi T., Wynberg J., Srinivasan R., Becknell B., McCoy J. P., Jr., Takahashi Y., Suffredini D. A., Linehan W. M., Caligiuri M. A., Childs R. W. 2004. Enhanced cytotoxicity of allogeneic NK cells with killer immunoglobulin-like receptor ligand incompatibility against melanoma and renal cell carcinoma cells. Blood 104: 170–177. [DOI] [PubMed] [Google Scholar]
- 22.Koh J., Lee S. B., Park H., Lee H. J., Cho N. H., Kim J. 2012. Susceptibility of CD24(+) ovarian cancer cells to anti-cancer drugs and natural killer cells. Biochem. Biophys. Res. Commun. 427: 373–378. [DOI] [PubMed] [Google Scholar]
- 23.Swift B. E., Williams B. A., Kosaka Y., Wang X. H., Medin J. A., Viswanathan S., Martinez-Lopez J., Keating A. 2012. Natural killer cell lines preferentially kill clonogenic multiple myeloma cells and decrease myeloma engraftment in a bioluminescent xenograft mouse model. Haematologica 97: 1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jachimowicz R. D., Fracasso G., Yazaki P. J., Power B. E., Borchmann P., Engert A., Hansen H. P., Reiners K. S., Marie M., von Strandmann E. P., Rothe A. 2011. Induction of in vitro and in vivo NK cell cytotoxicity using high-avidity immunoligands targeting prostate-specific membrane antigen in prostate carcinoma. Mol. Cancer Ther. 10: 1036–1045. [DOI] [PubMed] [Google Scholar]
- 25.Wang J., Svendsen A., Kmiecik J., Immervoll H., Skaftnesmo K. O., Planagumà J., Reed R. K., Bjerkvig R., Miletic H., Enger P. O., et al. 2011. Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS ONE 6: e23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avril T., Vauleon E., Hamlat A., Saikali S., Etcheverry A., Delmas C., Diabira S., Mosser J., Quillien V. 2012. Human glioblastoma stem-like cells are more sensitive to allogeneic NK and T cell-mediated killing compared with serum-cultured glioblastoma cells. Brain Pathol. 22: 159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. 2004. Identification of human brain tumour initiating cells. Nature 432: 396–401. [DOI] [PubMed] [Google Scholar]
- 28.Bari R., Bell T., Leung W. H., Vong Q. P., Chan W. K., Das Gupta N., Holladay M., Rooney B., Leung W. 2009. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood 114: 5182–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Re F., Staudacher C., Zamai L., Vecchio V., Bregni M. 2006. Killer cell Ig-like receptors ligand-mismatched, alloreactive natural killer cells lyse primary solid tumors. Cancer 107: 640–648. [DOI] [PubMed] [Google Scholar]
- 30.Liu J., Xiao Z., Ko H. L., Shen M., Ren E. C. 2014. Activating killer cell immunoglobulin-like receptor 2DS2 binds to HLA-A*11. Proc. Natl. Acad. Sci. USA 111: 2662–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ziegler S. F., Ramsdell F., Hjerrild K. A., Armitage R. J., Grabstein K. H., Hennen K. B., Farrah T., Fanslow W. C., Shevach E. M., Alderson M. R. 1993. Molecular characterization of the early activation antigen CD69: a type II membrane glycoprotein related to a family of natural killer cell activation antigens. Eur. J. Immunol. 23: 1643–1648. [DOI] [PubMed] [Google Scholar]
- 32.Aktas E., Kucuksezer U. C., Bilgic S., Erten G., Deniz G. 2009. Relationship between CD107a expression and cytotoxic activity. Cell. Immunol. 254: 149–154. [DOI] [PubMed] [Google Scholar]
- 33.Setareh M., Schwarz H., Lotz M. 1995. A mRNA variant encoding a soluble form of 4-1BB, a member of the murine NGF/TNF receptor family. Gene 164: 311–315. [DOI] [PubMed] [Google Scholar]
- 34.Fux M., van Rooijen N., Owens T. 2008. Macrophage-independent T cell infiltration to the site of injury-induced brain inflammation. J. Neuroimmunol. 203: 64–72. [DOI] [PubMed] [Google Scholar]
- 35.Brandt C. S., Baratin M., Yi E. C., Kennedy J., Gao Z., Fox B., Haldeman B., Ostrander C. D., Kaifu T., Chabannon C., et al. 2009. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J. Exp. Med. 206: 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sivori S., Parolini S., Marcenaro E., Castriconi R., Pende D., Millo R., Moretta A. 2000. Involvement of natural cytotoxicity receptors in human natural killer cell-mediated lysis of neuroblastoma and glioblastoma cell lines. J. Neuroimmunol. 107: 220–225. [DOI] [PubMed] [Google Scholar]
- 37.Cooper M. A., Fehniger T. A., Turner S. C., Chen K. S., Ghaheri B. A., Ghayur T., Carson W. E., Caligiuri M. A. 2001. Human natural killer cells: a unique innate immunoregulatory role for the CD56(bright) subset. Blood 97: 3146–3151. [DOI] [PubMed] [Google Scholar]
- 38.Kenney-Herbert E. M., Ball S. L., Al-Mayhani T. M., Watts C. 2011. Glioblastoma cell lines derived under serum-free conditions can be used as an in vitro model system to evaluate therapeutic response. Cancer Lett. 305: 50–57. [DOI] [PubMed] [Google Scholar]
- 39.Horowitz, A., D. M. Strauss-Albee, M. Leipold, J. Kubo, N. Nemat-Gorgani, O. C. Dogan, C. L. Dekker, S. Mackey, H. Maecker, G. E. Swan, et al. 2013. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med. 5: 208ra145. [DOI] [PMC free article] [PubMed]
- 40.Bari R., Rujkijyanont P., Sullivan E., Kang G., Turner V., Gan K., Leung W. 2013. Effect of donor KIR2DL1 allelic polymorphism on the outcome of pediatric allogeneic hematopoietic stem-cell transplantation. J. Clin. Oncol. 31: 3782–3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.López-Botet M., Bellón T., Llano M., Navarro F., García P., de Miguel M. 2000. Paired inhibitory and triggering NK cell receptors for HLA class I molecules. Hum. Immunol. 61: 7–17. [DOI] [PubMed] [Google Scholar]
- 42.Beresford P. J., Xia Z., Greenberg A. H., Lieberman J. 1999. Granzyme A loading induces rapid cytolysis and a novel form of DNA damage independently of caspase activation. Immunity 10: 585–594. [DOI] [PubMed] [Google Scholar]
- 43.Lieberman J., Fan Z. 2003. Nuclear war: the granzyme A-bomb. Curr. Opin. Immunol. 15: 553–559. [DOI] [PubMed] [Google Scholar]
- 44.Rygh, C. B., J. Wang, M. Thuen, A. Gras Navarro, E. M. Huuse, F. Thorsen, A. Poli, J. Zimmer, O. Haraldseth, S. A. Lie, et al. 2014. Dynamic Contrast Enhanced MRI Detects Early Response to Adoptive NK Cellular Immunotherapy Targeting the NG2 Proteoglycan in a Rat Model of Glioblastoma. PloS One 9: e108414. [DOI] [PMC free article] [PubMed]
- 45.Scott M. J., Hoth J. J., Stagner M. K., Gardner S. A., Peyton J. C., Cheadle W. G. 2004. CD40-CD154 interactions between macrophages and natural killer cells during sepsis are critical for macrophage activation and are not interferon gamma dependent. Clin. Exp. Immunol. 137: 469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fry T. J., Mackall C. L. 2002. Interleukin-7: from bench to clinic. Blood 99: 3892–3904. [DOI] [PubMed] [Google Scholar]
- 47.Hartgring S. A., van Roon J. A., Wenting-van Wijk M., Jacobs K. M., Jahangier Z. N., Willis C. R., Bijlsma J. W., Lafeber F. P. 2009. Elevated expression of interleukin-7 receptor in inflamed joints mediates interleukin-7-induced immune activation in rheumatoid arthritis. Arthritis Rheum. 60: 2595–2605. [DOI] [PubMed] [Google Scholar]
- 48.Poli A., Wang J., Domingues O., Planagumà J., Yan T., Rygh C. B., Skaftnesmo K. O., Thorsen F., McCormack E., Hentges F., et al. 2013. Targeting glioblastoma with NK cells and mAb against NG2/CSPG4 prolongs animal survival. Oncotarget 4: 1527–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin X., Jin X., Jung J. E., Beck S., Kim H. 2013. Cell surface Nestin is a biomarker for glioma stem cells. Biochem. Biophys. Res. Commun. 433: 496–501. [DOI] [PubMed] [Google Scholar]
- 50.Jin J. K., Jeong B. H., Na Y. J., Kim Y. S., Carp R. I., Wie M. B., Moon C., Shin T. 2004. Increased expression of the embryonic intermediate filament, nestin, in the brains of scrapie-infected mice. Neurosci. Lett. 367: 254–258. [DOI] [PubMed] [Google Scholar]
- 51.Ha Y., Choi J. U., Yoon D. H., Cho Y. E., Kim T. S. 2002. Nestin and small heat shock protein expression on reactive astrocytes and endothelial cells in cerebral abscess. Neurosci. Res. 44: 207–212. [DOI] [PubMed] [Google Scholar]
- 52.Reddy P. G., Graham G. M., Datta S., Guarini L., Moulton T. A., Jiang H. P., Gottesman M. M., Ferrone S., Fisher P. B. 1991. Effect of recombinant fibroblast interferon and recombinant immune interferon on growth and the antigenic phenotype of multidrug-resistant human glioblastoma multiforme cells. J. Natl. Cancer Inst. 83: 1307–1315. [DOI] [PubMed] [Google Scholar]
- 53.Mimura K., Kamiya T., Shiraishi K., Kua L. F., Shabbir A., So J., Yong W. P., Suzuki Y., Yoshimoto Y., Nakano T., et al. 2014. Therapeutic potential of highly cytotoxic natural killer cells for gastric cancer. Int. J. Cancer 135: 1390–1398. [DOI] [PubMed] [Google Scholar]
- 54.Geller M. A., Knorr D. A., Hermanson D. A., Pribyl L., Bendzick L., McCullar V., Miller J. S., Kaufman D. S. 2013. Intraperitoneal delivery of human natural killer cells for treatment of ovarian cancer in a mouse xenograft model. Cytotherapy 15: 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.