The core domain of the eukaryotic ribosome assembly factor complex Rpf2–Rrs1 from A. nidulans was crystallized.
Keywords: ribosome assembly factors, Rpf2, Rrs1, Aspergillus nidulans
Abstract
Rpf2 and Rrs1 are essential proteins for ribosome biogenesis. These proteins form a complex (the Rpf2-subcomplex) with 5S rRNA and two ribosomal proteins (L5 and L11). This complex is recruited to the ribosome precursor (the 90S pre-ribosome). This recruitment is necessary for the maturation of 25S rRNA. Genetic depletion of Rpf2 and Rrs1 results in accumulation of the 25S rRNA precursor. In this study, Rpf2 and Rrs1 from Aspergillus nidulans were co-overexpressed in Escherichia coli, purified and crystallized. Subsequent analysis revealed that these crystals contained the central core region of the complex consisting of both N-terminal domains. X-ray diffraction data were collected to 2.35 Å resolution. Preliminary analysis revealed that the crystals belonged to space group P212121, with unit-cell parameters a = 54.1, b = 123.3, c = 133.8 Å. There are two complexes in the asymmetric unit. Structure determination using selenomethionine-labelled protein is in progress.
1. Introduction
In eukaryotes, the 80S ribosome is composed of four ribosomal RNAs (25S, 18S, 5.8S and 5S rRNAs) and approximately 80 ribosomal proteins. The biogenesis of this supracomplex starts with the transcription of 35S pre-rRNA (which spans the sequences of the 25S, 18S and 5.8S rRNAs) by RNA polymerase I and of 5S rRNA by RNA polymerase III in the nucleolus. They are assembled into a 90S ribonucleoprotein particle (the 90S pre-ribosome) with many ribosomal proteins and assembly factors. The external and internal spacers of 35S pre-rRNA are removed by the steps listed below to yield 27SA2 and 20S pre-rRNAs. Depending on these steps, 90S is divided into two pre-ribosomal particles: pre-60S and pre-40S. The 5′ cleavage of 27SA2 generates 27SB, which is further divided into 7S and 25.5S (the precursors of 5.8S and 25S rRNAs, respectively). After these processes, pre-60S and pre-40S exit from the nucleolus through the nucleoplasm to the cytoplasm to form mature functional subunits (Tschochner & Hurt, 2003 ▶).
Many steps in ribosome biogenesis have been elucidated by studies on Saccharomyces cerevisiae. More than 200 factors are required for these steps (Venema & Tollervey, 1999 ▶; Kressler et al., 1999 ▶). Rpf2 and Rrs1 are two of these factors. Genetic depletion of Rpf2 and Rrs1 results in accumulation of the 27SB intermediate. Therefore, these proteins are implicated in the processing of 27SB to 25S rRNA (Morita et al., 2002 ▶).
In addition, Rpf2 and Rrs1 have been found to be associated with the 5S RNP complex consisting of 5S rRNA, L5 and L11 (the Rpf2-subcomplex), which is recruited to the 90S pre-ribosome (Zhang et al., 2007 ▶). As the genetic depletion of Rpf2 or Rrs1 inhibited the recruitment of 5S rRNA and two ribosomal proteins (L5 and L11) to the 90S pre-ribosome, the subsequent processing of pre-rRNA and export were limited (Zhang et al., 2007 ▶; Miyoshi et al., 2004 ▶; Nariai et al., 2005 ▶). Direct interactions between each of Rpf2, Rrs1, L5 and L11 have also been confirmed by in vitro pull-down assays (Morita et al., 2002 ▶; Miyoshi et al., 2002 ▶; Nariai et al., 2005 ▶; Zhang et al., 2007 ▶). The formation of a subcomplex containing Rpf2 and Rrs1 was suggested by the stoichiometric behaviour of Rpf2 and Rrs1 in the wild-type and mutant strains (Zhang et al., 2007 ▶).
To shed light on ribosome biogenesis from a structural point of view, we initated the elucidation of the structures of the Rpf2 and Rrs1 proteins. Here, we report the expression, purification, crystallization and preliminary X-ray analysis of the Rpf2–Rrs1 complex.
2. Materials and methods
2.1. Macromolecule production
Because the C-terminal region of Rpf2 was predicted to be disordered, the genes containing the coding region for a C-terminally truncated form (Δ305–332) of Aspergillus nidulans (An) Rpf2(1–304) and full-length AnRrs1(1–218) were cloned into expression vectors. These two vectors were co-transformed into Escherichia coli B834 (DE3) pRARE2 cells (Novagen) and the obtained colonies were tested for the co-expression of both subunits (Table 1 ▶). Cells were grown at 310 K in LB medium to an OD600 of 0.6. The culture was then induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.25 mM and shifted to 298 K. After 16 h incubation, the cells were harvested by centrifugation at 4500g for 30 min at 277 K.
Table 1. Macromolecule-production information.
Additional residues are underlined.
| Source organism | A. nidulans FGSC A4 |
| DNA source | The genome of A. nidulans |
| Rpf2(1304) | |
| Expression vector | pET-28a (Novagen) |
| Cloning site | NdeIXhoI |
| Additional residues | Downstream box†, N-terminal His tag, TEV cleavage site |
| Expression host | E. coli B834 (DE3) pRARE2 |
| Complete amino-acid sequence of the construct produced | MNHKHHHHHHSSGENLYFQGHMLREVKPKNPRTARILKAKEPQLIEGAKRVLLLHGSKCPTPLHTVLKVFHSLTTPHSVLFHKKNENIHPFESTESLEFLANKNDCGMVIFGSSNKKRPNCLTIARIFDSKVLDMAELLLLPDANGEGIPEMNRLSMHVAIGLRPLMLFSGSAWDDTTSTTHTMLKSMLVDLFKGETSDKIDVEGLQYALMVGAEEPTAGLAPIIHLRWYKIVTKRSGHKLPRVELEEIGPKLDFKVGRIQEAPRDVMKEAMKQGKKPNEEIKNKKNIGIDLIGDKVGRVHLAKQDLGGLQTRKMKGLKRRAG |
| Rrs1(1218) (full length) | |
| Expression vector | pCDF Duet-1 (Novagen) |
| Cloning site | NdeINcoI |
| Additional residues | Downstream box† |
| Expression host | E. coli B834 (DE3) pRARE2 |
| Complete amino-acid sequence of the construct produced | MNHTMANSETEAKSTIKPKPERLPITVSKPTPYTFDLGHLLANDPNPLELPKSEPLNASLKATARDGVQSLLNQLLTTCPITSSQQGVLLTLPAPSTILPRHKPLPTPKPPTKWELFARKKGIGKYSNKPGAALADKERRKKLVYDEESGEWVPRWGYKGKNKDDDQWLVEVKEKDWKKEEEAAAKGSSIRGMSRAERKERIRRNERKMRANERNSRKSGGK |
The downstream box is a sequence element that enhances translation (Sprengart et al., 1996 ▶).
The cell pellets (from 3 l culture) were resuspended in suspension buffer [50 mM Tris–HCl pH 7.5, 600 mM NaCl, 10%(v/v) glycerol] with 0.1 mg ml−1 DNase, 0.5 mg ml−1 lysozyme and protease-inhibitor cocktail (cOmplete EDTA-free; Roche) and were disrupted by sonication. The lysate was centrifuged for 30 min at 40 000g and 277 K.
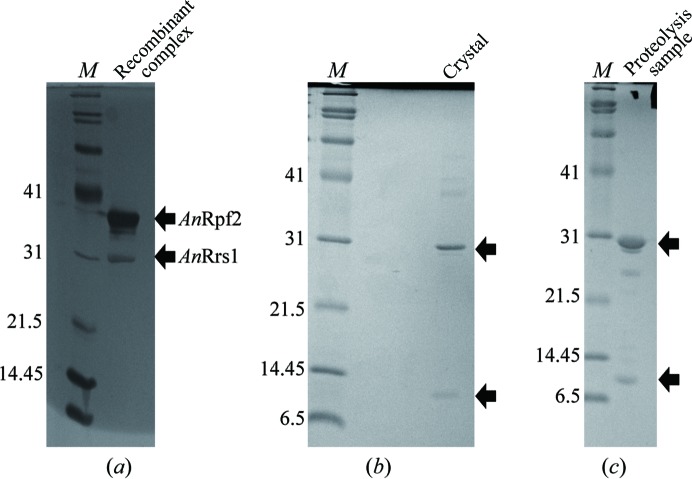
The supernatant was loaded onto a HisTrap HP column (GE Healthcare) pre-equilibrated with suspension buffer using an ÄKTApurifier system (GE Healthcare). The column was then subsequently washed with a 4% solution of the elution buffer [50 mM Tris–HCl pH 7.5, 600 mM NaCl, 10%(v/v) glycerol, 500 mM imidazole]. The protein was eluted with 50% elution buffer. The fractions containing the AnRpf2(1–304)–Rrs1 complex were pooled and diluted up to threefold with dilution buffer [50 mM Tris–HCl pH 7.5 containing 5%(v/v) glycerol]. The sample was further purified using a Resource S column (GE Healthcare) equilibrated with dilution buffer. The complex was eluted with a linear gradient of 0.2–2 M sodium chloride in the same buffer. Fractions containing the target complex were subsequently loaded onto a HiLoad 16/60 Superdex 200 prep-grade column (GE Healthcare) equilibrated with SEC buffer [20 mM HEPES–NaOH pH 7.5, 50 mM NaCl, 1%(v/v) glycerol] and eluted with the same buffer. The peak fractions were concentrated and stored at 190 K. The purity of the complex was assessed by SDS–PAGE (Fig. 1 ▶ a).
Figure 1.
15% SDS–PAGE gel stained with Coomassie Brilliant Blue. (a) Recombinant AnRpf2–Rrs1 produced in E. coli after purification. (b) Crystals. (c) Cleavage products by trypsin. Lane M contains molecular-mass marker (labelled in kDa).
2.2. Crystallization and characterization of the obtained crystals
Crystallization screening was performed using The JCSG Core Suites I–IV and The Classics Suite (Qiagen) using the sitting-drop vapour-diffusion method in 96-well plates (NeXtal Evolution μplate; Qiagen) with the additive reagent layered carboxylpropylamidephenylsilica (CPAPhS; Yao et al., 2008 ▶). Drops were formed by mixing 1 µl protein solution, 1 µl SEC buffer containing 45 ng µl−1 CPAPhS and 1 µl reservoir solution and were equilibrated against 0.1 ml reservoir solution at 293 K. Crystals were grown using The JCSG Core Suite I condition No. 34 [0.1 M HEPES–NaOH pH 6.5, 20%(w/v) PEG 6000 (final pH 7.0); Fig. 2 ▶] after 2–4 weeks. Isolated crystals were dissolved and analyzed by SDS–PAGE and MALDI–TOF MS. These results showed that the crystals had two fragments (about 30 and 10 kDa fragments).
Figure 2.
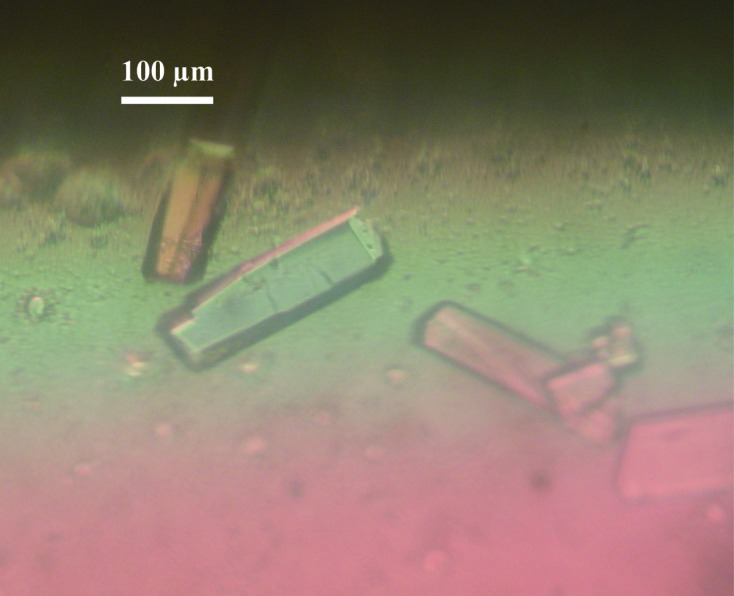
Crystals of the Rpf2–Rrs1 complex. The scale bar represents 100 µm.
To identify the two fragments, the 30 kDa band of the SDS–PAGE was transferred onto a polyvinylidene difluoride (PVDF) membrane and sequenced using automatic Edman degradation.
2.3. Limited proteolysis analysis
For further identification of the crystallized components, we performed limited proteolysis using trypsin. Following Ni-affinity chromatography, a pooled fraction of the AnRpf2(1–304)–Rrs1(1–218) complex was incubated with trypsin (Sigma) at a ratio of 100:1(w:w) AnRpf2(1–304)–Rrs1(1–218):trypsin in buffer consisting of 50 mM Tris–HCl pH 7.5, 350 mM NaCl, 100 mM imidazole, 7%(v/v) glycerol at 277 K for 1 h. To separate trypsin and cleavage products, the mixture was further purified on a HiTrap Heparin column (GE Healthcare) equilibrated with dilution buffer. The protein was eluted with a linear gradient of 0.2–1 M sodium chloride in the same dilution buffer. Fractions containing the target protein were then loaded onto a HiLoad 16/60 Superdex 200 prep-grade column equilibrated with the above-mentioned SEC buffer. The purified digestion mixtures were analyzed by SDS–PAGE. The gel was stained with Coomassie Brilliant Blue. The N-terminal sequence of the product in the 10 kDa band was determined using the above-mentioned method.
2.4. Data collection and processing
X-ray diffraction data were collected from this complex crystal on beamline BL41XU at SPring-8, Harima, Japan (proposal No. 2012A1494). Prior to the experiment, the crystals were cryoprotected by transfer into a solution containing 20%(v/v) glycerol for a few seconds and flash-cooled. A total of 360 images were collected with 0.5° oscillation and 1 s exposure time per image. For phasing by anomalous dispersion using Se atoms as scattering factors, we also produced SeMet-labelled Rpf2(18–254)–Rrs1(10–114) crystals. The optimized crystallization condition was 0.1 M HEPES–NaOH pH 8.0 containing 14%(w/v) PEG 3350 and 300 mM NaCl. The SeMet-labelled Rpf2(18–254)–Rrs1(10–114) crystals were cryoprotected with 20%(v/v) glycerol. A single-wavelength anomalous diffraction (SAD) data set was collected on beamline AR-NE3A at Photon Factory (PF), Tsukuba, Japan. All diffraction data were indexed, integrated, merged and scaled using XDS (Kabsch, 2010 ▶). Details of the data-collection and processing statistics are provided in Table 2 ▶.
Table 2. Statistics of data collection.
Values in parentheses are for the highest resolution shell.
| Native crystal | SeMet-labelled crystal | |
|---|---|---|
| Diffraction source | BL41XU, SPring-8 | NE-3A, PF |
| Wavelength () | 1.0000 | 0.9788 |
| Temperature (K) | 100 | 100 |
| Detector | Rayonix MX225HE | ADSC Quantum 270 CCD |
| Crystal-to-detector distance (mm) | 220 | 397.1 |
| Rotation range per image () | 0.5 | 0.8 |
| Total rotation range () | 180 | 180 |
| Exposure time per image (s) | 1 | 1.5 |
| Space group | P212121 | P41212 |
| a, b, c () | 54.1, 123.3, 133.8 | 128.9, 128.9, 57.5 |
| , , () | 90, 90, 90 | 90, 90, 90 |
| Mosaicity () | 0.152 | 0.219 |
| Resolution range () | 502.35 (2.492.35) | 503.51 (3.713.51) |
| Total No. of reflections | 274543 (43024) | 87626 (13867) |
| No. of unique reflections | 38131 (5930) | 11215 (1781) |
| Completeness (%) | 99.4 (97.6) | 99.9 (99.7) |
| Multiplicity | 7.20 (7.26) | 7.81 (7.56) |
| I/(I) | 14.51 (2.57) | 12.96 (1.95) |
| R meas † (%) | 9.1 (77.6) | 15.7 (108.3) |
| Overall B factor from Wilson plot (2) | 56.8 | 89.5 |
R
meas = 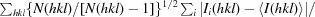
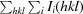 , where I(hkl) and N(hkl) are the mean intensity of a set of equivalent reflections and the multiplicity, respectively.
, where I(hkl) and N(hkl) are the mean intensity of a set of equivalent reflections and the multiplicity, respectively.
3. Results and discussion
We tried to crystallize several homologues of the Rpf2–Rrs1 complex such as those from Saccharomyces cerevisiae, Candida albicans and Chaetomium thermophilum, and finally obtained crystals of the Rpf2–Rrs1 complex from A. nidulans. SDS–PAGE analysis of the dissolved crystals suggested the presence of single species at around 30 and 10 kDa (Fig. 1 ▶ b). This result suggested that the crystals had two fragments (of about 30 and 10 kDa) which are not intact proteins. In addition, the dissolved crystals were analyzed by MALDI–TOF MS. The mass-spectrometric results revealed masses of 27 342 and 11 334 Da. The sequence of the 20 N-terminal amino-acid residues of the 30 kDa fragment was determined to be 18AKEPQLIEGAKRVLLLHGSK37 from AnRpf2, suggesting that the crystals contained degraded AnRpf2. On the other hand, the 10 kDa band could not be identified because the amount of the applied crystals was too small.
Limited proteolysis by trypsin showed that single species were present at around 30 and 10 kDa, suggesting that the composition of the trypsin-treated sample was similar to that of the obtained crystals (Fig. 1 ▶ c). The sequence of the eight N-terminal amino-acid residues using 10 kDa fragment was determined to be 10STIKPKPE17 from AnRrs1.
Taken together, molecular-weight determination using mass spectrometry and N-terminal amino-acid sequencing revealed that the crystallized components are the N-terminal domains of AnRpf2 (residues 18–262) and AnRrs1 (residues 10–113), and that the remaining C-terminal regions were degraded during the crystallization process. Consistent with this, the PSIPRED and DISOPRED2 programs for predicting secondary structure and disordered regions, respectively (McGuffin et al., 2000 ▶; Ward et al., 2004 ▶), suggested that residues 250–270 and the C-terminal region (residues 290–304) of AnRpf2 are disordered. In addition, secondary-structure prediction of Rrs1 suggested that it has N- and C-terminal domains connected by a central linker region (residues 88–108), implying that these N- and C-terminal regions might behave separately. Furthermore, we examined the limited proteolysis of the free Rpf2 molecule and found that the N-terminal domain is also fragmented (data not shown). These results indicate that the tight core region of the Rpf2–Rrs1 complex is crystallized after scission of the remaining residues that are not involved in the interactions. Such highly flexible terminal domains of both proteins may prevent the production of crystals of the full-length complex.
The SAD data from the SeMet-labelled crystal have been successfully phased. Phase calculation, density modification and preliminary model building were performed using phenix.autosol (Terwilliger et al., 2009 ▶). All ten selenium sites were found and used for phasing. Model building is currently in progress.
Acknowledgments
We are grateful to the beamline staff of SPring-8 and PF for their help in data collection. This work was supported by a Grant-in-Aid for Scientific Research (B) (No. 25291008 to MY) and Challenging Exploratory Research (No. 24657068 to MY) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kressler, D., Linder, P. & de La Cruz, J. (1999). Mol. Cell. Biol. 19, 7897–7912. [DOI] [PMC free article] [PubMed]
- McGuffin, L. J., Bryson, K. & Jones, D. T. (2000). Bioinformatics, 16, 404–405. [DOI] [PubMed]
- Miyoshi, K., Shirai, C., Horigome, C., Takenami, K., Kawasaki, J. & Mizuta, K. (2004). FEBS Lett. 565, 106–110. [DOI] [PubMed]
- Miyoshi, K., Tsujii, R., Yoshida, H., Maki, Y., Wada, A., Matsui, Y., Toh-E, A. & Mizuta, K. (2002). J. Biol. Chem. 277, 18334–18339. [DOI] [PubMed]
- Morita, D., Miyoshi, K., Matsui, Y., Toh-E, A., Shinkawa, H., Miyakawa, T. & Mizuta, K. (2002). J. Biol. Chem. 277, 28780–28786. [DOI] [PubMed]
- Nariai, M., Tanaka, T., Okada, T., Shirai, C., Horigome, C. & Mizuta, K. (2005). Nucleic Acids Res. 33, 4553–4562. [DOI] [PMC free article] [PubMed]
- Sprengart, M. L., Fuchs, E. & Porter, A. G. (1996). EMBO J. 15, 665–674. [PMC free article] [PubMed]
- Terwilliger, T. C., Adams, P. D., Read, R. J., McCoy, A. J., Moriarty, N. W., Grosse-Kunstleve, R. W., Afonine, P. V., Zwart, P. H. & Hung, L.-W. (2009). Acta Cryst. D65, 582–601. [DOI] [PMC free article] [PubMed]
- Tschochner, H. & Hurt, E. (2003). Trends Cell Biol. 13, 255–263. [DOI] [PubMed]
- Venema, J. & Tollervey, D. (1999). Annu. Rev. Genet. 33, 261–311. [DOI] [PubMed]
- Ward, J. J., McGuffin, L. J., Bryson, K., Buxton, B. F. & Jones, D. T. (2004). Bioinformatics, 20, 2138–2139. [DOI] [PubMed]
- Yao, K., You, X., Shi, L., Wan, W., Yu, F. & Chen, J. (2008). Langmuir, 24, 302–309. [DOI] [PubMed]
- Zhang, J., Harnpicharnchai, P., Jakovljevic, J., Tang, L., Guo, Y., Oeffinger, M., Rout, M. P., Hiley, S. L., Hughes, T. & Woolford, J. L. Jr (2007). Genes Dev. 21, 2580–2592. [DOI] [PMC free article] [PubMed]