The cystallization of human aquaporin 1 is described.
Keywords: aquaporin 1, water channel, baculovirus, crystal packing
Abstract
Aquaporin water channels (AQPs) are found in almost every organism from humans to bacteria. In humans, 13 classes of AQPs control water and glycerol homeostasis. Knockout studies have suggested that modulating the activity of AQPs could be beneficial for the treatment of several pathologies. In particular, aquaporin 1 is a key factor in cell migration and angiogenesis, and constitutes a possible target for anticancer compounds and also for the treatment of glaucoma. Here, a preliminary crystallographic analysis at 3.28 Å resolution of crystals of human aquaporin 1 (hAQP1) obtained from protein expressed in Sf9 insect cells is reported. The crystals belonged to the tetragonal space group I422, with unit-cell parameters a = b = 89.28, c = 174.9 Å, and contained one monomer per asymmetric unit. The hAQP1 biological tetramer is generated via the crystallographic fourfold axis. This work extends previous electron crystallographic studies that used material extracted from human red blood cells, in which the resolution was limited to approximately 3.8 Å. It will inform efforts to improve lattice contacts and the diffraction limit for the future structure-based discovery of specific hAQP1 inhibitors.
1. Introduction
Water transport through biological membranes is facilitated by the homotetrameric channels formed by aquaporins (AQPs; Benga et al., 1986 ▶; Denker et al., 1988 ▶; Preston et al., 1992 ▶). The human genome contains 13 different AQPs (Ishibashi et al., 2009 ▶) divided into the water-specific channels AQP0, AQP1, AQP2, AQP4, AQP5 and AQP8 (orthodox aquaporins) and the water or neutral solute facilitators AQP3, AQP7, AQP9 and AQP10 (aquaglyceroporins) (Hara-Chikuma & Verkman, 2006 ▶). AQP1, the first water channel to be characterized, was discovered in red blood cells and renal proximal tubules (Agre et al., 1993 ▶; Denker et al., 1988 ▶). AQP1 can establish a bidirectional transport of water in response to osmotic gradients, strictly excluding ions, including protons. Aquaporins are essential for the maintenance of water homeostasis (Agre, 2006 ▶; Agre et al., 2002 ▶). The biological importance as well as the potential for medical applications of AQP regulation has recently been reviewed (Huber et al., 2012 ▶; Verkman, 2008 ▶, 2012 ▶; Castle, 2005 ▶). Expression of AQP1, AQP4 and AQP9 in mammalian brain cells has been demonstrated (Badaut et al., 2007 ▶; Nielsen et al., 1993 ▶; Oshio et al., 2005 ▶) and several reports suggest that inhibitors of AQP-mediated water transport may constitute an alternative treatment for cerebral oedema. Accordingly, AQP4-null mice were resistant to the formation of cerebral oedema after either acute water intoxication or ischaemic stroke (Manley et al., 2004 ▶; Verkman, 2000 ▶). AQP inhibitors could also be useful for cancer treatment. Indeed, AQP1 is involved in tumour angiogenesis (Clapp & Martínez de la Escalera, 2006 ▶; Verkman et al., 2008 ▶; Saadoun et al., 2005 ▶), and AQP5 and AQP1 are up-regulated in lung cancer cells (Zhang et al., 2010 ▶). In the eye, several AQPs, including AQP1 and AQP4, protect the epithelium and regulate intraocular pressure (Verkman, 2003 ▶). Mice lacking AQP1 and AQP2 have reduced intraocular pressure (Zhang et al., 2002 ▶), and hence inhibition of water transport by AQP1 and AQP4 may provide an alternative treatment for glaucoma. Thus, specific inhibitors of AQPs are actively sought, and streamlined procedures to overexpress and obtain crystal structures of relevant human AQPs are required for informing structure-based drug discovery. Previously, the structure of human red blood cell AQP1 (hAQP1) has been described at 3.8 Å resolution using electron crystallographic data obtained from two-dimensional crystals (Murata et al., 2000 ▶). A crystal structure of the closely related bovine aquaporin 1 (bAQP1) that also used protein material extracted from bovine red blood cells (Sui et al., 2001 ▶) was solved at 2.2 Å resolution. Here, we expressed hAQP1 in Sf9 insect cells and report a preliminary crystallographic analysis at 3.28 Å resolution. This report forms a starting basis for crystal-engineering attempts to improve crystal quality.
2. Materials and methods
2.1. Protein production, purification and activity
2.1.1. Cloning and expression of hAQP1
The hAQP1 gene was cloned into the pFB-Lic-Bse vector (Addgene) with the forward primer TACTTCCAATCCATGGCCAGCGAGTTCAAGAAGAA and the reverse primer TATCCACCTTTACTGTCATTTGGGCTTCATCTCCACCC using ligation-independent cloning (Yokoyama, 2003 ▶). The construct contains an N-terminal hexahistidine tag and a TEV protease cleavage site. Bacmid production, Sf9 insect-cell transfection and virus production were performed as described previously (Table 1 ▶). Early passaged virus particles (P0) were amplified to P2 and were used to infect 2 l log-phase (3 × 106 cells ml−1) insect cells for recombinant protein expression. The multiplicity of infection (MOI) was kept between 2 and 3, and the culture was incubated at 27°C for 56 h at 140 rev min−1. The culture was spun down at 1500g for 30 min and the pellet was washed twice with 50 ml phosphate-buffered saline (PBS).
Table 1. Macromolecule-production information.
| Source organism | Human |
| DNA source | Commercial |
| Forward primer | ACTTCCAATCCATGGCCAGCGAGTTCAAGAAGAA |
| Reverse primer | TATCCACCTTTACTGTCATTTGGGCTTCATCTCCACCC |
| Cloning vector | pFB-Lic-Bse |
| Expression vector | pFB-Lic-Bse |
| Expression host | S. frugiperda Sf9 |
| Complete amino-acid sequence of the construct produced | MGHHHHHHSSGVDLGTENLYFQSMASEFKKKLFWRAVVAEFLATTLFVFISIGSALGFKYPVGNNQTAVQDNVKVSLAFGLSIATLAQSVGHISGAHLNPAVTLGLLLSCQISIFRALMYIIAQCVGAIVATAILSGITSSLTGNSLGRNDLADGVNSGQGLGIEIIGTLQLVLCVLATTDRRRRDLGGSAPLAIGLSVALGHLLAIDYTGCGINPARSFGSAVITHNFSNHWIFWVGPFIGGALAVLIYDFILAPRSSDLTDRVKVWTSGQVEEYDLDADDINSRVEMKPK |
2.1.2. Purification of hAQP1
Spodoptera frugiperda insect-cell pellets were resuspended in 5 ml lysis buffer [20 mM Tris–HCl pH 8, 300 mM NaCl, 2 mM β-mercaptoethanol, 10%(v/v) glycerol, cOmplete protease-inhibitor cocktail, benzonase and 2%(v/v) n-octyl-β-d-glucopyranoside (OG; Anagrade, Affymetrix)] per gram. Cell lysis was achieved by 10 min sonication followed by five passages through a microfluidizer. The mixture was then centrifuged at 40 000g for 30 min at 4°C. The supernatant was loaded onto Ni–NTA beads pre-equilibrated with lysis buffer. After overnight binding at 4°C, the resin was washed with 30 volumes of washing buffer [20 mM Tris–HCl pH 8, 300 mM NaCl, 25 mM imidazole, 2 mM β-mercaptoethanol, 10%(v/v) glycerol]. hAQP1 was eluted with five volumes of elution buffer [20 mM Tris–HCl pH 8, 300 mM NaCl, 300 mM imidazole, 2 mM β-mercaptoethanol, 10%(v/v) glycerol, 1%(v/v) OG] (Table 2 ▶). Eluted fractions were pooled and concentrated before being subjected to gel-filtration chromatography using a HiLoad 16/60 Superdex 200 preparation-grade column (GE Healthcare) at 4°C. The protein was eluted with 20 mM Tris–HCl pH 8, 300 mM NaCl, 1%(v/v) OG.
Table 2. Crystallization.
| Method | Hanging drop |
| Plate type | 24-well Linbro plate |
| Temperature (C) | 18 |
| Protein concentration (mgml1) | 10 |
| Buffer composition of protein solution | 20mM TrisHCl pH 8, 300mM NaCl, 1%(v/v) OG |
| Composition of reservoir solution | 0.05M glycine pH 9, 50%(v/v) PEG 400, 5% glycerol, 0.05%(v/v) CYMAL-6, 0.05%(v/v) n-nonyl--D-thiomaltopyranoside, 0.05%(v/v) dodecyl--D-maltoside |
| Volume and ratio of drop | 2l protein solution, 2:1 ratio |
| Volume of reservoir (l) | 500 |
2.1.3. Stopped-flow water-permeability assay
Liposomes were prepared using the film-rehydration method (Olson et al., 1979 ▶). Briefly, a thin lipid film was formed by drying the required amount of Escherichia coli lipids (Avanti Polar Lipids Inc.) pre-dissolved in chloroform under a nitrogen stream. The film was kept in a vacuum desiccator for at least 2 h, followed by the addition of purified hAQP1 protein and topping up with reconstitution buffer (20 mM Tris–HCl pH 8.0) to obtain a protein:lipid molar ratio of 1:400 in a final volume of 1 ml. The reconstitution mixture was incubated for 1 h at room temperature to allow proteoliposome formation, followed by extrusion through a 400 nm pore-size polycarbonate membrane using an Avestin extruder to obtain liposomes with a uniform size distribution. The diameter of the hAQP1 proteoliposomes was measured by dynamic light scattering (90 Plus Particle Size Analyzer, Brookhaven Instruments). An empty liposome control was prepared in the same way without addition of protein. The water permeability of proteoliposomes and empty liposomes was determined by following the light-scattering intensity changes in a SX20 stopped-flow spectrometer (Applied Photophysics) with a dead time of 1.1 ms. Experiments were carried out at 23°C. Shrinkage kinetics were measured from the time course of 90° scattered light intensity at 500 nm. For each experiment, five to ten replicates were averaged. Water permeability was measured by rapidly mixing with hyperosmolar PBS containing 500 mM sucrose as a nonpermeable osmolyte to establish a 250 mM inwardly directed osmotic gradient.
2.2. Crystallization of hAQP1
Small crystals with lenticular to oil-like droplet shapes were initially obtained at 20°C using the commercial crystallization kit Membrane Gold (Molecular Dimensions Inc., UK) in a mother liquor consisting of 0.05 M glycine pH 9, 50%(v/v) PEG 400, 5% glycerol, 0.05%(v/v) CYMAL-6, 0.05%(v/v) n-nonyl-β-d-thiomaltopyranoside, 0.05%(v/v) dodecyl-β-d-maltoside (DDM). A volume of 2 µl hAQP1 (10 mg ml−1) was mixed with 1 µl crystallization buffer using the hanging-drop vapour-diffusion technique. Crystals with sharper edges could be obtained within 2–4 d by incubation at 18°C. Crystals were retrieved from the crystallization drop using a nylon loop and were directly flash-cooled in liquid nitrogen.
2.3. Data collection and processing
2.3.1. Data collection, structure solution and preliminary refinement
Data sets were collected at a wavelength of 1.00 Å on the PXIII beamline at the Swiss Light Source, Paul Scherrer Intitute (SLS-PSI). Diffraction images from four crystals were collected on a PILATUS 2M detector at a distance of 410 mm using 4 s exposures and 1° oscillation. Reflections were indexed and integrated using XDS (Kabsch, 2010 ▶) in the tetragonal space group I422, with unit-cell parameters a = b = 89.28, c = 174.85 Å (Table 3 ▶). The solvent fraction was 56.1%. Diffraction intensities were subsequently scaled and merged using AIMLESS from the CCP4 suite (Evans, 2011 ▶).
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | PXIII, SLS-PSI |
| Wavelength () | 1.00 |
| Temperature (K) | 110 |
| Detector | PILATUS 2M |
| Crystal-to-detector distance (mm) | 410 |
| Rotation range per image () | 1 |
| Total rotation range () | 120 |
| Exposure time per image (s) | 4 |
| Space group | I422 |
| a, b, c () | 89.3, 89.3, 174.8 [93.3, 93.3, 180.5]† |
| , , () | 90.00, 90.00, 90.00 |
| Mosaicity () | 0.81 |
| Resolution range () | 20.03.28 (3.673.28) |
| Total No. of reflections | 140417 (25840) |
| No. of unique reflections | 5560 (1525) |
| Completeness (%) | 99.05 (99.05) |
| Multiplicity | 25.3 (16.9) |
| I/(I) | 22.1 (3.9)‡ |
| R r.i.m. | 0.115 (0.679) |
| Overall B factor from Wilson plot (2) | 103.1 |
The unit-cell parameters for the related crystal form of bAQP1 (PDB entry 1j4n) are given in square brackets.
The diffraction is anisotropic, extending to about 3.3 resolution along c* and to only 4.55 resolution in the perpendicular directions.
2.3.2. Structure-solution refinement and packing analysis
The structure was solved by molecular replacement using a monomer of bovine aquaporin 1 (PDB entry 1j4n) as a search probe (Sui et al., 2001 ▶). Iterative cycles of model building and structure refinement were performed using Coot (Emsley et al., 2010 ▶) and BUSTER (Bricogne et al., 2011 ▶) (Table 4 ▶). Analysis of hAQP1 tetramer formation and crystal packing was performed using PISA (http://www.ebi.ac.uk/pdbe/pisa/).
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| Resolution range () | 20.03.28 (3.673.28) |
| Completeness (%) | 99.05 (99.05) |
| Cutoff | 0 |
| No. of reflections, working set | 5555 (1445) |
| No. of reflections, test set | 259 (77) |
| Final R cryst | 0.2338 (0.2392) |
| Final R free | 0.2978 (0.2413) |
| Blow DPI | 0.538 |
| No. of non-H atoms | |
| Protein | 1722 |
| Water | 6 |
| Total | 1728 |
| R.m.s. deviations | |
| Bonds () | 0.010 |
| Angles () | 1.36 |
| Average B factors (2) | |
| Protein | 197.1 |
| Water | 81.1 |
| Ramachandran plot (%) | |
| Most favoured | 75.4 |
| Allowed | 20.1 |
| Generously allowed | 4.0 |
| Disallowed | 0.5 |
3. Results and discussion
3.1. Cloning and expression of hAQP1
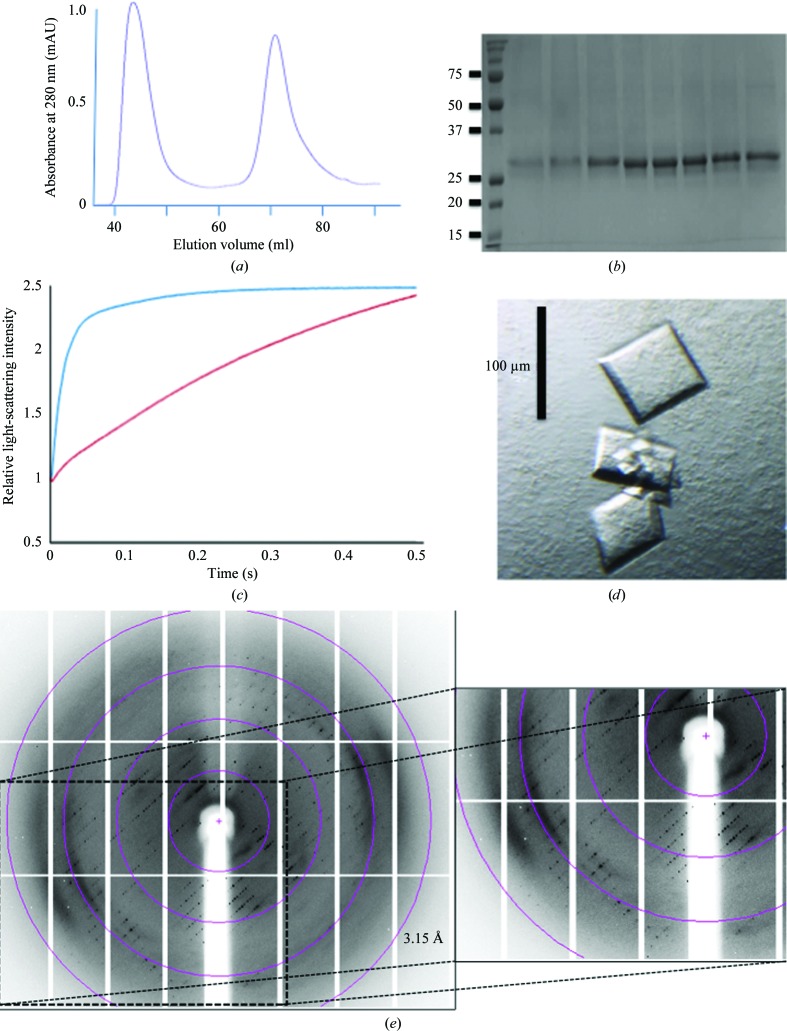
hAQP1 was cloned in frame with an N-terminal hexahistidine tag and a TEV protease cleavage site. Initial attempts to express the protein in E. coli using various constructs failed to produce good yields. The protein was therefore expressed in S. frugiperda Sf9 cells. Yeast expression systems such as Saccharomyces cerevisiae and Pichia pastoris have been shown to be adequate for heterologous expression of AQPs (Nyblom et al., 2007 ▶; Bomholt et al., 2013 ▶). We found that Sf9 cells constitute a suitable system for the expression of hAQP1 and are consistently capable of generating 1–3 mg purified active protein per litre of expression medium (Fig. 1 ▶).
Figure 1.
(a) Gel-filtration chromatogram of hAQP1, which elutes at 72 ml, corresponding to a homotetramer. The first peak that elutes at 47 ml contains hAQP1 aggregates. (b) 12% SDS–PAGE of hAQP1 from the tetramer fraction after gel-filtration chromatography. (c) hAQP1 water-permeability functional assay. Light-scattering intensity changes of hAQP1-embedded proteoliposomes (blue curve) compared with the control empty liposome (red curve), plotted as a function of time. (d) Tetragonal-shaped crystals of hAQP1. (e) Diffraction image from one of the four hAQP1 data sets used to assemble a complete highly redundant data set. The inset shows a magnified view of the anisotropic diffraction pattern that extends to approximately 3.3 Å resolution along c*, while diffraction is weak beyond 6.0 Å resolution in the perpendicular directions. Circles indicating 12.9, 6.4, 4.2 and 3.1 Å resolution limits are shown. Significant diffuse scattering was also evident from the diffraction images.
3.2. hAQP1 purification and activity
Purification of hAQP1 was achieved by a two-step purification protocol without ultracentrifugation. Based on previous reports (Sui et al., 2001 ▶), we used OG for protein extraction followed by an Ni–NTA metal-affinity column and a gel-filtration chromatography step, in which hAQP1 elutes as a tetramer and higher oligomers (Fig. 1 ▶ a). Asn42 has a putative N-linked glycosylation site. Protein glycosylation is present when hAQP1 is purified from erythrocytes (Zeidel et al., 1992 ▶), but not when it is purified from S. cerevisiae (Laizé et al., 1997 ▶) or P. pastoris (Emami et al., 2013 ▶). From an SDS–PAGE analysis, no evidence of glycosylation was found for hAQP1 expressed in Sf9 cells (Fig. 1 ▶ b). PNGase treatment resulted in only a slight decrease of the molecular mass from 31 623 to 31 531 Da as measured by mass spectrometry (theoretical mass of 31 136 Da), also suggesting that the present hAQP1 was devoid of glycosylation. To verify that the purified recombinant hAQP1 protein was functional, we performed stopped-flow permeability assays using liposomes. In response to osmotic shock, liposomes enriched with a 1:400 aquaporin:lipid molar ratio displayed an exponential rate of water transport (Fig. 1 ▶ c) compared with control liposomes, demonstrating that the recombinant hAQP1 protein is active.
3.3. Protein crystallization, data collection and structure solution
Refinement of the initial crystallization conditions was achieved by varying the concentration of precipitant and adding two detergents to the crystallization mixture, as described in §2, leading to larger tetragonal-shaped crystals that were suitable for data collection (Fig. 1 ▶ d). Most crystals tested on a synchrotron beamline, however, diffracted to only 5–10 Å resolution. Data from four crystals that diffracted anisotropically to 3.2 Å resolution along c* but only to about 4.5–5 Å resolution in the perpendicular directions were merged to obtain a highly redundant data set to 3.28 Å resolution that was used for structure determination (Table 3 ▶). Residues 1–235 of the polypeptide chain are visible in the electron-density map, including all six transmembrane helices. Although present in the crystallized protein, the C-terminal end (residues 236–269) is not visible, presumably owing to its high mobility. This mobility considerably restricts the stability of the lattice (see below) and is presumably responsible for the poorer diffraction of the present hAQP1 crystal form compared with bAQP1 crystals (Sui et al., 2001 ▶). The loop spanning residues 39–48 that includes the putative glycosylation site at Asn42 is also flexible and poorly defined. Overall, helices located near the crystallographic fourfold at the centre of the tetramer are well ordered with most of the side chain visible, whilst the quality of the electron-density map decreases radially, with only backbone atoms of helices clearly visible at the periphery of the tetramer.
3.4. Monomer structure
Not surprisingly, the overall three-dimensional structure of hAQP1 is conserved with respect to bAQP1, with which it shares 92% amino-acid sequence identity: 248 residues are identical over 271 aligned residues (Figs. 2 ▶ and 3 ▶). A superposition of the hAQP1 and the bovine AQP1 monomers (PDB entry 1j4n; Sui et al., 2001 ▶; Murata et al., 2000 ▶) returns an r.m.s. deviation of 1.16 Å for 916 main-chain atoms. The monomer structure can be broken up into three regions: the transmembrane (TM), cytoplasmic and extracellular regions (Verkman & Mitra, 2000 ▶). The hAQP1 monomer comprises six TM α-helices, two non-membrane-spanning helices and two characteristic loops (Borgnia et al., 1999 ▶) located on opposite sides of the membrane that partially penetrate the bilayer and interact through the conserved Asn-Pro-Ala or ‘NPA’ motifs. Both extramembrane regions form an inner duct that penetrates into the TM region and form a pore that restricts its diameter at the centre of the TM region, allowing the transit of water molecules. Helices M3 and M7 are embedded in the core of the protein facing each other, building a pseudo-TM helix. Both helices contain the conserved ‘NPA’ sequence motif that is present throughout AQPs and contributes to the formation of the pore-regulatory site.
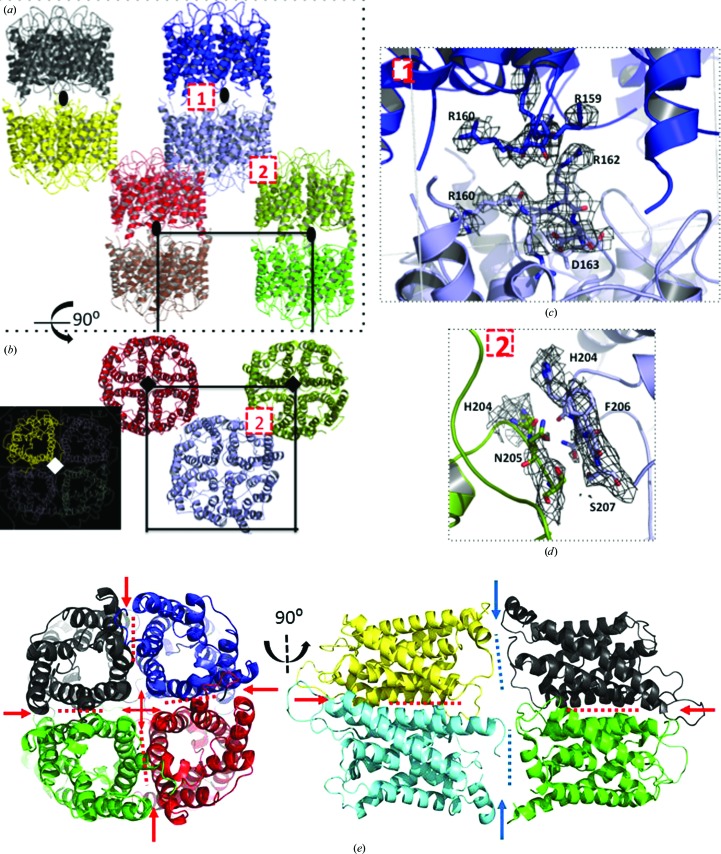
Figure 2.
Crystal packing. Two perpendicular views of the hAQP1 crystal packing are displayed: (a) along the crystallographic dyad (black ovals) that relates two tetramers in a head-to-tail fashion and (b) along the fourfold c* axis (black and white squares) that generates the hAQP1 tetramer (inset). The unit-cell boundaries are indicated. The magnified views in (c) and (d) show the two main protein–protein contacts that generate this crystal form. (e) Corresponding perpendicular views of the bAQP1 tetramers. Blue arrows indicate the large interface formed between the N-terminal and C-terminal regions of the head-to tail bQAQP1 which is missing in the hAQP1 lattice. Figures were prepared using PyMOL (v.1.3r1; Schrödinger).
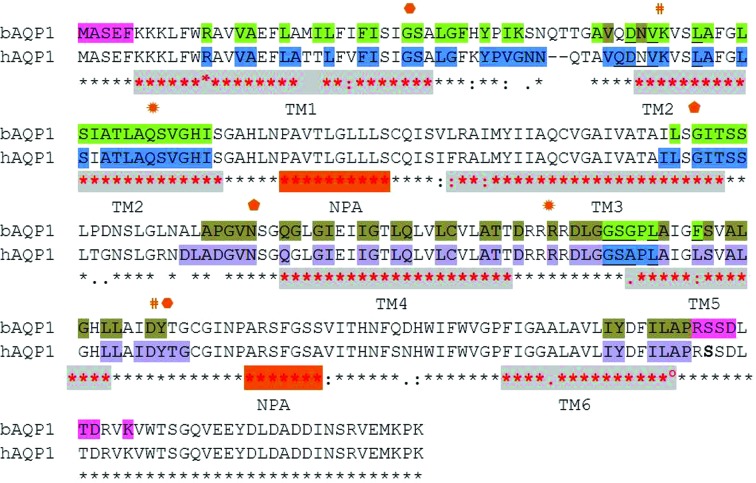
Figure 3.
Sequence alignment and crystal packing of bAQP1 and hAQP1 (using ClustalW; http://www.ebi.ac.uk/Tools/msa/clustalw2/). Conserved residues are marked with stars below the sequences. Red stars and grey shading indicate the six transmembrane (TM) helices. The two ‘NPA’ motifs are shaded in orange and labelled. Residues involved in tetramer and crystal-packing formation are highlighted with the following colour code: blue and purple for hAQP1 and green and tan for bAQP1. Residues in pink (at the N-terminal and C-terminal ends of bAQP1) are involved in the crystal contact found only in the bAQP1 lattice (see text). The symbols denote residues involved in either conserved hydrogen-bond (hexagons, pentagons and sun symbols) or salt-bridge (hashes) interactions.
3.5. Tetramer formation and crystal packing
Orthogonal views of the hAQP1 crystal lattice are shown in Fig. 2 ▶. The hAQP1 tetramer is formed via the crystallographic fourfold axis (Figs. 2 ▶ a and 2 ▶ b). Hydrophobic residues projecting from TM helices M2 and M6 (Fig. 3 ▶) contribute to tetramer formation. In the extracellular half of hAQP1, the hydrophobic patch contributed by residues Val50, Leu54 and Leu58 of TM helix M2 and Leu170 and Leu174 (Phe in bAQP1) of TM helix M6 from each monomer interact with equivalent residues related by the crystallographic fourfold. In turn, in the cytoplasmic half of the protein, TM helix M6 contributes residues Leu172 and Phe176 to the tetrameric interface. Two tetramers related by a crystallographic dyad are oriented head-to tail along the crystallographic c axis (Fig. 2 ▶ a). Contacts between the two tetramers are limited to the arginine-rich loops 158–162 (conserved in bAQP1; see Fig. 3 ▶) that are brought into close proximity via a crystallographic dyad (Fig. 2 ▶ c). Finally, the hAQP1 lattice is stabilized via a set of contacts between two sets of AQP1 tetramers involving Phe206 and Asn205 (Fig. 2 ▶ d). We compared the crystal lattices formed by hAQP1 (this work) and bAQP1 (PDB entry 1j4n; Sui et al., 2001 ▶) both by visual inspection and using the PDBePISA server (Krissinel & Henrick, 2007 ▶). The crucial difference between the bAQP1 and hAQP1 lattices is the presence of an additional packing interface of about 400 Å2 that is established between the N-terminal region of one bAQP1 tetramer and the C-terminal region of the tetramer related by a crystallographic dyad. This crystal contact is only present in the bAQP1 lattice and is stabilized via the formation of six hydrogen bonds (Figs. 2 ▶ e and 3 ▶). In the crystal structure of bAQP, these residues (shaded in pink in Fig. 3 ▶) stabilize the head-to-tail intermolecular interface. This interface is absent in the hAQP1 crystal lattice owing to the disorder of the C-terminal polypeptide chain and leads to a slight tilt of the tetramers, leading to the observed variation in unit-cell parameters (Table 3 ▶). This reduction in important lattice contacts is consistent with the medium resolution of the diffraction pattern, with the pattern of diffuse scattering (Fig. 1 ▶ e) and also with the high overall Wilson temperature factor of this crystal form (Table 4 ▶). Since both the human and bovine proteins share the same N-terminal and C-terminal amino-acid sequences (Fig. 3 ▶), it is possible that the presence of the N-terminal hexahistidine tag in hAQP1 interferes with the formation of this crystal contact, and efforts are under way to cleave the hAQP1 histidine tag. In order to promote stronger intermolecular contacts and to stabilize the crystal lattice for improvement of the diffraction properties, the basic loop that establishes contacts around the crystallographic dyad could also be targeted by mutagenesis, for instance by mutating Arg162 to an acidic residue to promote the formation of an intermolecular salt bridge with Arg159 (Fig. 2 ▶ c).
4. Conclusion
In conclusion, this work demonstrates the overall structural conservation between hAQP1 and bAQP1, identifies a crucial crystal contact for lattice stabilization and suggests some routes to improve the diffraction properties of hAQP1 crystals for future structure-based drug discovery.
Acknowledgments
We thank colleagues at the PPP for helpful discussions during the course of this study. We acknowledge the provision of excellent beamtime and support by the staff of SLS, Villigen, Switzerland. This project was supported by grant CRP2008 from the National Research Foundation to the JL and JT laboratories.
References
- Agre, P. (2006). Proc. Am. Thorac. Soc. 3, 5–13. [DOI] [PMC free article] [PubMed]
- Agre, P., King, L. S., Yasui, M., Guggino, W. B., Ottersen, O. P., Fujiyoshi, Y., Engel, A. & Nielsen, S. (2002). J. Physiol. 542, 3–16. [DOI] [PMC free article] [PubMed]
- Agre, P., Preston, G. M., Smith, B. L., Jung, J. S., Raina, S., Moon, C., Guggino, W. B. & Nielsen, S. (1993). Am. J. Physiol. 265, F463–F476. [DOI] [PubMed]
- Badaut, J., Brunet, J.-F. & Regli, L. (2007). Metab. Brain Dis. 22, 251–263. [DOI] [PubMed]
- Benga, G., Popescu, O., Borza, V., Pop, V. I., Muresan, A., Mocsy, I., Brain, A. & Wrigglesworth, J. M. (1986). Eur. J. Cell Biol. 41, 252–262. [PubMed]
- Bomholt, J., Hélix-Nielsen, C., Scharff-Poulsen, P., Pedersen, P. A. & Hong, W. (2013). PLoS One, 8, e56431. [DOI] [PMC free article] [PubMed]
- Borgnia, M., Nielsen, S., Engel, A. & Agre, P. (1999). Annu. Rev. Biochem. 68, 425–458. [DOI] [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). BUSTER v.2.11.2. Cambridge: Global Phasing Ltd.
- Castle, N. A. (2005). Drug Discov. Today, 10, 485–493. [DOI] [PubMed]
- Clapp, C. & Martínez de la Escalera, G. (2006). Trends Endocrinol. Metab. 17, 1–2. [DOI] [PubMed]
- Denker, B. M., Smith, B. L., Kuhajda, F. P. & Agre, P. (1988). J. Biol. Chem. 263, 15634–15642. [PubMed]
- Emami, S., Fan, Y., Munro, R., Ladizhansky, V. & Brown, L. S. (2013). J. Biomol. NMR, 55, 147–155. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Hara-Chikuma, M. & Verkman, A. S. (2006). Cell. Mol. Life Sci. 63, 1386–1392. [DOI] [PMC free article] [PubMed]
- Huber, V. J., Tsujita, M. & Nakada, T. (2012). Mol Aspects Med. 33, 691–703. [DOI] [PubMed]
- Ishibashi, K., Hara, S. & Kondo, S. (2009). Clin. Exp. Nephrol. 13, 107–117. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Laizé, V., Ripoche, P. & Tacnet, F. (1997). Protein Expr. Purif. 11, 284–288. [DOI] [PubMed]
- Manley, G. T., Binder, D. K., Papadopoulos, M. C. & Verkman, A. S. (2004). Neuroscience, 129, 983–991. [DOI] [PubMed]
- Murata, K., Mitsuoka, K., Hirai, T., Walz, T., Agre, P., Heymann, J. B., Engel, A. & Fujiyoshi, Y. (2000). Nature (London), 407, 599–605. [DOI] [PubMed]
- Nielsen, S., Smith, B. L., Christensen, E. I. & Agre, P. (1993). Proc. Natl Acad. Sci. USA, 90, 7275–7279. [DOI] [PMC free article] [PubMed]
- Nyblom, M., Öberg, F., Lindkvist-Petersson, K., Hallgren, K., Findlay, H., Wikström, J., Karlsson, A., Hansson, Ö., Booth, P. J., Bill, R. M., Neutze, R. & Hedfalk, K. (2007). Protein Expr. Purif. 56, 110–120. [DOI] [PubMed]
- Olson, F., Hunt, C. A., Szoka, F. C., Vail, W. J. & Papahadjopoulos, D. (1979). Biochim. Biophys. Acta, 557, 9–23. [DOI] [PubMed]
- Oshio, K., Watanabe, H., Song, Y., Verkman, A. S. & Manley, G. T. (2005). FASEB J. 19, 76–78. [DOI] [PubMed]
- Preston, G. M., Carroll, T. P., Guggino, W. B. & Agre, P. (1992). Science, 256, 385–387. [DOI] [PubMed]
- Saadoun, S., Papadopoulos, M. C., Hara-Chikuma, M. & Verkman, A. S. (2005). Nature (London), 434, 786–792. [DOI] [PubMed]
- Sui, H., Han, B.-G., Lee, J. K., Walian, P. & Jap, B. K. (2001). Nature (London), 414, 872–878. [DOI] [PubMed]
- Verkman, A. S. (2000). Curr. Opin. Nephrol. Hypertens. 9, 517–522. [DOI] [PubMed]
- Verkman, A. S. (2003). Exp. Eye Res. 76, 137–143. [DOI] [PubMed]
- Verkman, A. S. (2008). Expert Rev. Mol. Med. 10, e13. [DOI] [PMC free article] [PubMed]
- Verkman, A. S. (2012). Annu. Rev. Med. 63, 303–316. [DOI] [PMC free article] [PubMed]
- Verkman, A. S., Hara-Chikuma, M. & Papadopoulos, M. C. (2008). J. Mol. Med. 86, 523–529. [DOI] [PMC free article] [PubMed]
- Verkman, A. S. & Mitra, A. K. (2000). Am. J. Physiol. Renal Physiol. 278, F13–F28. [DOI] [PubMed]
- Yokoyama, S. (2003). Curr. Opin. Chem. Biol. 7, 39–43. [DOI] [PubMed]
- Zeidel, M. L., Ambudkar, S. V., Smith, B. L. & Agre, P. (1992). Biochemistry, 31, 7436–7440. [DOI] [PubMed]
- Zhang, D., Vetrivel, L. & Verkman, A. S. (2002). J. Gen. Physiol. 119, 561–569. [DOI] [PMC free article] [PubMed]
- Zhang, Z., Chen, Z., Song, Y., Zhang, P., Hu, J. & Bai, C. (2010). J. Pathol. 221, 210–220. [DOI] [PubMed]