The tyrosine phosphatase AmsI from E. amylovora was cloned, overexpressed in E. coli, purified and crystallized. X-ray diffraction data were collected to a resolution of 1.57 Å.
Keywords: Erwinia, exopolysaccharides, amylovoran, tyrosine phosphatase, AmsI
Abstract
The Gram-negative bacterium Erwinia amylovora is a destructive pathogen of plants belonging to the Rosaceae family. Amongst its pathogenicity factors, E. amylovora produces the exopolysaccharide amylovoran, which contributes to the occlusion of plant vessels, causing wilting of shoots and eventually resulting in plant death. Amylovoran biosynthesis requires the presence of 12 genes (from amsA to amsL) clustered in the ams region of the E. amylovora genome. They mostly encode glycosyl transferases (AmsG, AmsB, AmsD, AmsE, AmsJ and AmsK), proteins involved in amylovoran translocation and assembly (AmsH, AmsL and AmsC), and also a tyrosine kinase (AmsA) and a tyrosine phosphatase (AmsI), which are both involved in the regulation of amylovoran biosynthesis. The low-molecular-weight protein tyrosine phosphatase AmsI was overexpressed as a His6-tagged protein in Escherichia coli, purified and crystallized. X-ray diffraction data were collected to a maximum resolution of 1.57 Å in space group P3121.
1. Introduction
Erwinia amylovora is a Gram-negative bacterium belonging to the Enterobacteriaceae family. It causes the devastating disease called fire blight in Rosaceae such as apple, pear, raspberry, cotoneaster and pyracantha (Vanneste, 2000 ▶). E. amylovora infects the host plants during spring, primarily in the flowers through the nectarthodes, or during summer, attacking the succulent tissues of shoots through wounds caused by hail, wind, storms and insects. Once established in the plant, the bacteria move through the vascular system of the plant, spreading the infection to other tissues and escaping the plant defence with the help of a capsular exopolysaccharide (EPS), which also represents one of the most important pathogenicity factors of E. amylovora (Vrancken et al., 2013 ▶). The EPS causes occlusion of the plant vessels, resulting in the characteristic wilting of fire blight, and eventually leading to plant death (Gross et al., 1992 ▶). The EPS of E. amylovora contains two different types of sugar polymers, amylovoran and levan, that are essential to biofilm formation, which in turn protects the bacteria and helps to spread the disease. Δams mutants defective in the synthesis of amylovoran were nonpathogenic, while Δlsc mutants defective in the synthesis of levan did not colonize xylem vessels and only moved slowly into apple shoots (Koczan et al., 2009 ▶). Levan is a fructo-oligosaccharide synthesized by the action of a single enzyme, levansucrase (Geier & Geider, 1993 ▶; Caputi, Cianci et al., 2013 ▶; Caputi, Nepogodiev et al., 2013 ▶), while the biosynthesis of the long-chain amylovoran requires a complicated enzymatic system. Amylovoran is a repetition of oligosaccharide units of galactose, glucose, glucuronic acid and pyruvate (Nimtz et al., 1996 ▶). EPS biosynthesis, regulation and secretion involves the action of 12 genes clustered in the 16 kb ams region (from amsA to amsL) in the E. amylovora genome (Bugert & Geider, 1995 ▶). Most of them are glycosyl transferases (AmsG, AmsB, AmsD, AmsE, AmsJ and AmsK); the other proteins are involved in amylovoran translocation and assembly (AmsH, AmsL and AmsC) and include a tyrosine kinase (AmsA) and a tyrosine phosphatase (AmsI) that are both involved in regulating the production of amylovoran (Langlotz et al., 2011 ▶). The biosynthesis of amylovoran is regulated by a network of proteins which includes the phosphorelay system RcsB, RcsC and RcsD (Wang et al., 2009 ▶, 2011 ▶) and the recently characterized negative regulator of amylovoran production AmyR (Wang et al., 2012 ▶).
AmsI, a protein tyrosine phosphatase (EC 3.1.3.48), was initially proposed to be a low-molecular-weight phosphatase (144 amino acids, molecular mass of 15 772.2 Da as calculated by ProtParam from ExPASy) used to dephosphorylate lipid carrier diphosphates during amylovoran assembly (Bugert & Geider, 1997 ▶). AmsI was later proposed to act in concert with AmsA, a tyrosine kinase associated with the production of exopolysaccharide (Ilan et al., 1999 ▶) and located downstream of the AmsI gene in the ams operon (Bugert & Geider, 1995 ▶). AmsA and AmsI are homologues of Wzc and Wzb, respectively, in Escherichia coli, which regulate colanic acid production. In E. coli the interplay between the autokinase Wzc and Wzb regulates colanic acid synthesis by changing the phosphorylation state of Wzc by the action of Wzb. The bacteria only produced colanic acid when the autophosphorylated Wzc is dephosphorylated by Wzb (Vincent et al., 2000 ▶). The structure of Wzb was solved by NMR and provided the evidence for a substrate-recognition mechanism that differs from that of eukaryotic tyrosine phosphatases, suggesting that Wzb could represent an ideal target for structure-based drug design to fight pathogenic bacteria (Lescop et al., 2006 ▶). We cloned, overexpressed, purified and crystallized AmsI. The structural data collected to 1.57 Å resolution will help towards a better understanding of the mechanism of its interaction with AmsA and drive the development of inhibitors able to eradicate fire blight.
2. Materials and methods
2.1. Cloning, overexpression and purification of E. amylovora AmsI
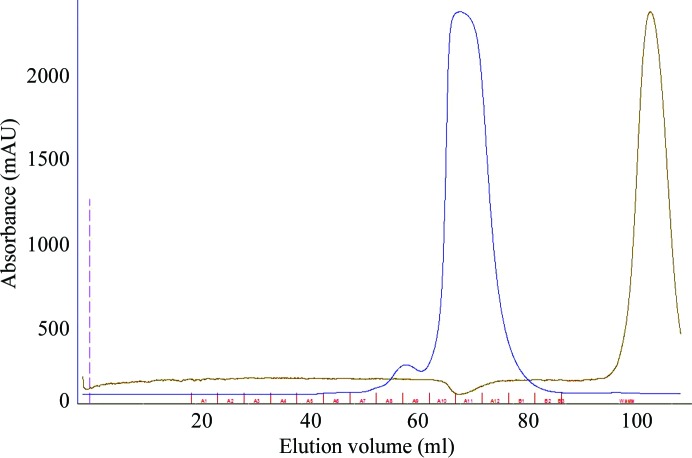
The amsI gene (UniProt Q46630) was PCR-amplified from genomic DNA isolated from E. amylovora strain Ea273 (ATCC 49946). The forward and reverse primers included NcoI and EcoRI restriction sites, respectively (underlined in Table 1 ▶). The PCR product was purified, digested for 3 h at 310 K and ligated into pETM-11 vector (Dümmler et al., 2005 ▶). The obtained plasmid (pETM-11::AmsI) was propagated in E. coli NovaBlue cells (EMD4Biosciences, Germany). E. coli BL21 (DE3) cells were transformed with pETM-11::AmsI for expression of the recombinant His6-tagged protein. Table 1 ▶ reports the details of the cloning procedure. After initial small-scale expression trials, which demonstrated good levels of protein expression, the transformed cells were grown overnight in 10 ml 2×YT medium containing kanamycin (30 µg ml−1) at 310 K. The culture was used to inoculate 1 l medium (1:100 dilution) and grown at 310 K to an OD600 of 0.8. The temperature was then decreased to 293 K and after 1 h the culture was induced with 1 mM IPTG and grown for a further 16 h. The cells were harvested by centrifugation at 4500g for 15 min at 277 K, washed with 100 ml ice-cold 50 mM sodium phosphate pH 7.4, 250 mM NaCl, 0.5 mM TCEP (buffer A) and centrifuged again, yielding 6 g of wet cell paste. The cells were resuspended in 50 ml buffer A containing 0.2 mg ml−1 lysozyme and a protease-inhibitor tablet (Roche, Switzerland), stirred for 30 min at room temperature and lysed by sonication (Soniprep, MSE, UK) on ice for 2 min using 10 s cycles (15.6 MHz) and a 9.5 mm diameter probe. Cell debris was removed by centrifugation at 18 000g for 20 min at 277 K. After filtration through a 0.45 µm cellulose acetate filter, the cell extract was loaded onto a HisTrap HP 5 ml column (GE Healthcare, Sweden) previously equilibrated with buffer A at a flow rate of 2 ml min−1. The column was then washed with buffer A supplemented with 20 mM imidazole until the A 280 reached the baseline. An imidazole gradient from 20 to 500 mM in buffer A for 20 column volumes (100 ml) was applied and the enzyme eluted at a concentration of 150 mM imidazole. The fractions containing the enzyme were pooled, concentrated by ultrafiltration using a Vivaspin 20 (Sartorius) and loaded onto a Sephadex S75 16/60 column (1.6 × 60 cm; GE Healthcare, Sweden) equilibrated with 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM TCEP (buffer B) at a flow rate of 1 ml min−1 (Fig. 1 ▶). All purification steps were carried out at room temperature. Protein purity was confirmed by SDS–PAGE.
Table 1. Macromolecule-production information.
| Source organism | E. amylovora strain Ea273 (ATCC 49946) |
| DNA source | Genomic |
| Forward primer | 5-CGATCACCATGGTCAATTCAATCTTAGTGG |
| Reverse primer | 5-CGAGAATTCCTATCGGCTTAATGCG |
| Cloning vector | pETM-11 |
| Expression vector | pETM-11 |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced† | MKHHHHHHPMSDYDIPTTENLYFQGAMVNSILVVCIGNICRSPTGERLLKAALPERKIASAGLKAMVGGSADETASIVANEHGVSLQDHVAQQLTADMCRDSDLILVMEKKHIDLVCRINPSVRGKTMLFGHWINQQEIADPYKKSRDAFEAVYGVLENAAQKWVNALSR |
The AmsI sequence is underlined.
Figure 1.
Elution profile of the size-exclusion chromatography carried out as a second step of the purification using a Sephadex S75 16/60 column (1.6 × 60 cm; GE Healthcare, Sweden). The injection volume was 5 ml protein solution (the blue line represents the UV absorption at 280 nm). This step was used as a desalting step as well (the brown line represents the conductivity of the solution).
2.2. Crystallization
The protein was crystallized using the microbatch-under-oil method in 96-well MRC plates. The crystallization wells were protected from drying using adhesive ClearView sheets (Molecular Dimensions). Drops of 1 µl precipitant solution from the crystallization kits [PACT premier HT-96 and JCSG-plus HT-96, CSS1 and CSS2 (Brzozowski & Walton, 2001 ▶); Molecular Dimensions] were added to 15 µl volatile oil (Molecular Dimensions) using a multichannel pipette, immediately followed by 1 µl protein solution. A shower of very small crystals appeared within a week in condition A7 of the CSS2 screen (2.7 M ammonium sulfate, 100 mM sodium acetate pH 5.5). The conditions were replicated using a grid of different ammonium sulfate concentrations and crystals grew to a maximum size of about 0.2 × 0.2 × 0.2 mm in 1.7 M ammonium sulfate, 100 mM sodium acetate pH 5.5. A summary of crystallization is provided in Table 2 ▶. A crystal was scooped from the mother liquor using a cryoloop and flash-cooled at the beamline by quickly mounting it onto the goniometer straight into the nitrogen stream of the cryocooling system to test its diffraction properties. The high-salt mother liquor and the tiny layer of surrounding oil around the crystal provided good cryoprotection, allowing the collection of a complete high-quality data set (Table 3 ▶).
Table 2. Crystallization.
| Method | Microbatch under oil |
| Plate type | 96-well MRC plates |
| Temperature (K) | 293 |
| Protein concentration (mgml1) | 15 |
| Buffer composition of protein solution | 20mM TrisHCl pH 7.5, 150mM NaCl, 0.5mM TCEP |
| Composition of reservoir solution | 1.7M ammonium sulfate, 100mM sodium acetate pH 5.5 |
| Volume and ratio of drop | 1l, 1:1 |
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Diffraction source | EMBL P13, PETRA III |
| Wavelength () | 1.033 |
| Temperature (K) | 100 |
| Detector | PILATUS 6M |
| Crystal-to-detector distance (mm) | 222.6 |
| Rotation range per image () | 0.1 |
| Total rotation range () | 360 |
| Exposure time per image (s) | 0.1 |
| Space group | P3121 |
| a, b, c () | 66.03, 66.03, 72.27 |
| , , () | 90, 90, 120 |
| Mosaicity () | 0.08 |
| Resolution range () | 72.271.57 (1.601.57) |
| Total No. of reflections | 510981 (25345) |
| No. of unique reflections | 25949 (1253) |
| Completeness (%) | 100 (100) |
| Multiplicity | 19.7 (20.2) |
| I/(I) | 23.2 (5.5) |
| R r.i.m. † | 0.066 (0.598) |
| Overall B factor from Wilson plot (2) | 16.85 |
Estimated by multiplying the conventional R merge value by the factor [N/(N 1)]1/2, where N is the data multiplicity [R merge = 0.064 (0.583), data multiplicity = 19.7 (20.2)].
2.3. Data collection and processing
Diffraction data were collected at 100 K on the EMBL P13 beamline at the PETRA III storage ring, DESY, Hamburg, Germany. The wavelength was set to 1.033 Å using a Si(III) crystal monochromator (FMB Oxford). Data were collected with a focused beam collimated with a 70 µm diameter aperture using a PILATUS 6M detector (DECTRIS) and an MD2 goniometer (Maatel-EMBL) with a horizontal spindle axis. The data were processed using XDS (Kabsch, 2010 ▶), the space group was determined by POINTLESS (Evans, 2011 ▶) and the data were scaled with AIMLESS (Evans, 2006 ▶). The crystal diffracted to 1.57 Å resolution in space group P3121. Data-collection statistics are reported in Table 3 ▶.
3. Results and discussion
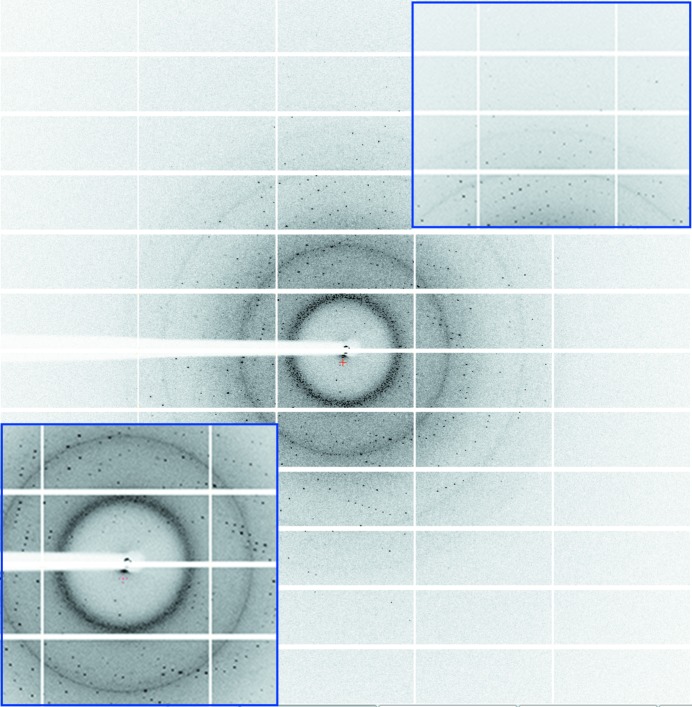
The E. amylovora AmsI gene was amplified from genomic DNA and ligated into pETM-11 expression vector. His6-tagged AmsI was overexpressed in E. coli, yielding 80 mg protein per litre of culture. Crystals of AmsI grew from a two-step protein-purification protocol without removing the His6 tag from the protein and using microbatch under oil, thus facilitating their transport to the beamline. The crystal of AmsI used for data collection belonged to space group P3121 and diffracted to 1.57 Å resolution (Fig. 2 ▶) without addition of cryoprotectant to the microbatch-under-oil crystallization conditions. The asymmetric unit contains one molecule of AmsI with a solvent content of 55.36% and a Matthews coefficient of 2.75 Å3 Da−1. The structure will be solved by molecular replacement using the model of Wzb already available in the PDB (Hagelueken et al., 2009 ▶). The results obtained will help us towards a better understanding of the regulation of amylovoran biosynthesis modulated by the interplay between AmsA and AmsI, and may possibly help in the design of molecules able to selectively inhibit bacterial tyrosine phosphatases.
Figure 2.
Diffraction image of AmsI collected on a PILATUS 6M detector. The insets show magnifications of the low-resolution and high-resolution regions.
Acknowledgments
We thank Dr M. Malnoy (Fondazione Edmund Mach, S. Michele all’Adige, Trento, Italy) for providing the genomic DNA from E. amylovora strain Ea273. We thank Dr M. Salomone-Stagni for crystal handling at the beamline. Plasmid pETM-11 was obtained from the European Molecular Biology Laboratory under a signed Material Transfer Agreement. Data were collected under European Molecular Biology Laboratory beam time award No. MX-152. This work was supported by the Autonomous Province of Bolzano (project: ‘A structural genomics approach for the study of the virulence and pathogenesis of Erwinia amylovora’).
References
- Brzozowski, A. M. & Walton, J. (2001). J. Appl. Cryst. 34, 97–101.
- Bugert, P. & Geider, K. (1995). Mol. Microbiol. 15, 917–933. [DOI] [PubMed]
- Bugert, P. & Geider, K. (1997). FEBS Lett. 400, 252–256. [DOI] [PubMed]
- Caputi, L., Cianci, M. & Benini, S. (2013). Acta Cryst. F69, 570–573. [DOI] [PMC free article] [PubMed]
- Caputi, L., Nepogodiev, S. A., Malnoy, M., Rejzek, M., Field, R. A. & Benini, S. (2013). J. Agric. Food Chem. 61, 12265–12273. [DOI] [PubMed]
- Dümmler, A., Lawrence, A.-M. & de Marco, A. (2005). Microb. Cell Factories, 4, 34. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. R. (2011). Acta Cryst. D67, 282–292. [DOI] [PMC free article] [PubMed]
- Geier, G. & Geider, K. (1993). Physiol. Mol. Plant Pathol. 42, 387–404.
- Gross, M., Geier, G., Rudolph, K. & Geider, K. (1992). Physiol. Mol. Plant Pathol. 40, 371–381.
- Hagelueken, G., Huang, H., Mainprize, I. L., Whitfield, C. & Naismith, J. H. (2009). J. Mol. Biol. 392, 678–688. [DOI] [PMC free article] [PubMed]
- Ilan, O., Bloch, Y., Frankel, G., Ullrich, H., Geider, K. & Rosenshine, I. (1999). EMBO J. 18, 3241–3248. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Koczan, J. M., McGrath, M. J., Zhao, Y. & Sundin, G. W. (2009). Phytopathology, 99, 1237–1244. [DOI] [PubMed]
- Langlotz, C., Schollmeyer, M., Coplin, D. L., Nimtz, M. & Geider, K. (2011). Physiol. Mol. Plant Pathol. 75, 163–169.
- Lescop, E., Hu, Y., Xu, H., Hu, W., Chen, J., Xia, B. & Jin, C. (2006). J. Biol. Chem. 281, 19570–19577. [DOI] [PubMed]
- Nimtz, M., Mort, A., Domke, T., Wray, V., Zhang, Y., Qiu, F., Coplin, D. & Geider, K. (1996). Carbohydr. Res. 287, 59–76. [DOI] [PubMed]
- Vanneste, J. L. (2000). Fire Blight: the Disease and its Causative Agent, Erwinia amylovora. Wallingford: CABI.
- Vincent, C., Duclos, B., Grangeasse, C., Vaganay, E., Riberty, M., Cozzone, A. J. & Doublet, P. (2000). J. Mol. Biol. 304, 311–321. [DOI] [PubMed]
- Vrancken, K., Holtappels, M., Schoofs, H., Deckers, T. & Valcke, R. (2013). Microbiology, 159, 823–832. [DOI] [PubMed]
- Wang, D., Korban, S. S., Pusey, P. L. & Zhao, Y. (2011). Phytopathology, 101, 710–717. [DOI] [PubMed]
- Wang, D., Korban, S. S., Pusey, P. L. & Zhao, Y. (2012). PLoS One, 7, e45038. [DOI] [PMC free article] [PubMed]
- Wang, D., Korban, S. S. & Zhao, Y. (2009). Mol. Plant Pathol. 10, 277–290. [DOI] [PMC free article] [PubMed]