Abstract
Chronic inflammation is highly prevalent in maintenance hemodialysis (MHD) patients, and it has been shown to be a strong predictor of morbidity and mortality. Mitochondrial DNA (mtDNA) released into circulation after cell damage can promote inflammation in patients and animal models. However, the role and mechanisms of circulatory mtDNA in chronic inflammation in MHD patients remain unknown. Sixty MHD patients and 20 health controls were enrolled in this study. The circulatory mtDNA was detected by quantitative real-time PCR assay. Plasma interleukin 6 (IL-6) and tumor necrosis factor α (TNF-α) were quantitated by ELISA assay. Dialysis systems in MHD patients and in vitro were used to evaluate the effect of different dialysis patterns on circulatory mtDNA. Circulatory mtDNA was elevated in MHD patients comparing to that of health control. Regression analysis demonstrated that plasma mtDNA was positively associated with TNF-α and the product of serum calcium and phosphorus, while negatively associated with hemoglobin and serum albumin in MHD patients. MtDNA induced the secretion of IL-6 and TNF-α in the THP-1 cells. Single high-flux hemodialysis (HF-HD) and on line hemodiafiltration (OL-HDF) but not low-flux hemodialysis (LF-HD) could partially reduce plasma mtDNA in MHD patients. In vitro, both HD and hemofiltration (HF) could fractional remove mtDNA. Collectively, circulatory mtDNA is elevated and its level is closely correlated with chronic inflammation in MHD patients. HF-HD and HDF can partially reduce circulatory mtDNA in MHD patients.
Introduction
Chronic inflammation is one of the characteristics of maintenance hemodialysis (MHD) patients, which has been shown to be a strong predictor of morbidity and mortality in this population [1]. It was demonstrated that chronic inflammation played an important role in the pathogenesis of cardiovascular disease (CVD) in MHD patients [2]. The increased levels of specific cytokines (interleukin 6 and tumor necrosis factor α) or acute phase proteins (C-reactive protein) have been found to be associated with CVD in MHD patients [3], [4]. Inflammation in MHD patients may be related to processes associated with renal failure itself such as uremic toxins retention, oxidative stress and dialysis processes such as dialysis membranes and dialysis fluid [5]–[8]. However, the mechanism of chronic inflammation state has not been clarified clearly.
Containing the proinflammatory CpG DNA repeats is an improtant strcture feature for mitochondrial DNA(mtDNA) [9]. The proinflammatory role of mtDNA has been confirmed in several diseases such as severe trauma [10]. MtDNA escaped from autophagy in the cardiomyocytes by external hemodynamic stress could lead to Toll Like Receptor (TLR)-9 mediated inflammatory responses and is capable of inducing myocarditis [11]. Plasma DNA is also elevated in the septic patients, which has potential use for predicting outcome in septic patients arriving at the emergency room [12]. Post-dialysis cell-free DNA (cf-DNA) level might be an independent predictor of all-cause mortality in MHD patients [13]. It has been described that apoptotic cf-DNA was increased in the plasma of MHD patients, which selectively stimulated the production of pro-inflammatory cytokines from human monocyte [14]. Thus far, the change of circulatory mtDNA in MHD patients and the mechanism in the formation of inflammatory state has not yet be concerned.
In this study, the expression of circulatory mtDNA and several inflammatory markers were determined in MHD patients. The association among plasma mtDNA, inflammatory cytokines and clinical characteristics of patients was analyzed. To clarify the removal effect of different hemodialysis patterns on circulatory mtDNA, the clearance was evaluated in MHD patients and in vitro respectively.
Materials and Methods
Ethics statement
All of the following details were approved by the responsible ethics committee of Nanjing Medical University before the study. The Permit Number of this study was KY026. All patients and healthy controls provided written informed consent before any study-related procedure was performed.
Study population
From Oct 2012 to Jan 2013, sixty MHD patients from the Center of Kidney Disease of 2nd Affiliated Hospital of Nanjing Medical University and 20 healthy controls from the Medical Examination Center of the hospital were recruited. MHD was defined as patients undergoing regular hemodialysis prescription, three times a week, for 4 h per session at least 6 months. All patients were in a stable condition and free from infection for at least 3 months. Patients were divided into three groups according to the dialysis patterns: Group A: low-flux hemodialysis (LF-HD) with PMMA membrane (B3-1.3A, TORAY, JAPAN); Group B: high-flux hemodialysis (HF-HD) with polysulfone membrane (TS-1.3A, TORAY, JAPAN); Group C: on line hemodiafiltration (OL-HDF) with polysulfone filter (TS-1.3U, TORAY, JAPAN). All these groups with a blood rate flow of 250 to 280 ml/min and a dialysis rate flow of 500 ml/min and HDF with an additional dosage of substitution 30L. Each group of 20 patients received the treatment as indicated.
Blood sampling
Two blood samples were collected for each patient: one before a single hemodialysis another at the end of it. Peripheral blood (2 ml) was collected into EDTA-containing tubes in each puncture. To ensure cell-free plasma collection, EDTA–blood were initially centrifuged for 10 min at 3000 rpm, followed by separation into a 1.5 ml clear polypropylene tube taking care not to disturb the buffer coat layer. The newly separated aliquot was centrifuged for a further 10 min at 10,000 rpm, after which the upper portion of plasma was removed and placed into a further clear tube and was frozen at −80°C prior to extraction [15]. DNA was extracted from 200 µl plasma samples with the use of a QIAamp DNA Mini and Blood Mini kit (Qiagen, Frederick, MD) using the “blood and body fluid protocol” as recommended by the manufacturer [16].
Isolation of mitochondria and mtDNA preparation
Mitochondria of human skeletal muscle tissue which was from surgical trauma by traffic accident were isolated as described previously [17]. MtDNA was extracted from the isolated mitochondria using DNeasy Blood & Tissue kit (Qiagen, Frederick, MD), which was prepared under sterile conditions. The concentration of mtDNA was then determined spectrophotometrically. No protein contamination was found and nuclear DNA was less than 0.001% by quantitative polymerase chain reaction (PCR) [10]. The purified mtDNA was diluted into serial concentrations which were used as standards of real-time PCR protocols. Part of the purified mtDNA was used in the simulated dialysis systems.
Real-time quantitative PCR protocols
Primers for human cytochrome B (Cyt B) (forward 5′-ATGACCCCAATACGCAAAAT-3′ and reverse 5′- CGAAGTTTCATCATGCGGAG-3′), human cytochrome C oxidase subunit III (COX III) (forward 5′-ATGACCCACCAATCACATGC-3′ and reverse 5′-ATCACATGGCTAGGCCGGAG-3′) were chosen for study because they are unique among mitochondrial molecules [18]. Primer sequences have no significant homology with DNA found in any bacterial species published on BLAST. Real-time PCR was performed using previously described primers. The PCR reaction mixture consisted of 1 ul of DNA, 12.5 µl of 2× SYBR Green PCR Master Mix (Roche Diagnostics GmbH Mannheim, Germany), 1 µl of forward and reverse primer, and 9.5 µl of nuclease-free water. PCR was carried out with an ABI Prism 7300 Sequence Detection System at 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min by amplifying serial dilutions of a known quantity of amplicons. Concentrations are expressed as nanograms per milliliter (ng/ml).
Cell line culture, differentiation and stimulation
The role of mtDNA on inflammation was studied on the human acute monocytic leukemia cell line—THP-1 cell line, which was cultured as previously described [19]. Cells were maintained as a continuous culture in RPMI1640 medium supplemented with 10% fetal bovine serum (Invitrogen) in a humidified atmosphere of 5% CO2 in air at 37°C. The medium was refreshed every 3 to 4 days. Initially, the THP-1 cells (106/ml) were differentiated toward the monocyte/macrophage phenotype in the presence of phorbol-12-myristate-13-acetate (PMA) 100 ng/ml for 36 hours. THP-1 cells were then incubated with purified mtDNA in different dosage from 0 hour to 32 hours to evaluate the effect on inflammatory markers.
IL-6 and TNF-α ELISA
The amount of IL-6 and TNF-α in the supernatant of THP-1 cells and in the plasma of MHD patients and controls were determined by ELISA (R&D Systems, Wiesbaden, Germany) following the protocol recommended by the manufacturer.
Simulated dialysis systems in vitro
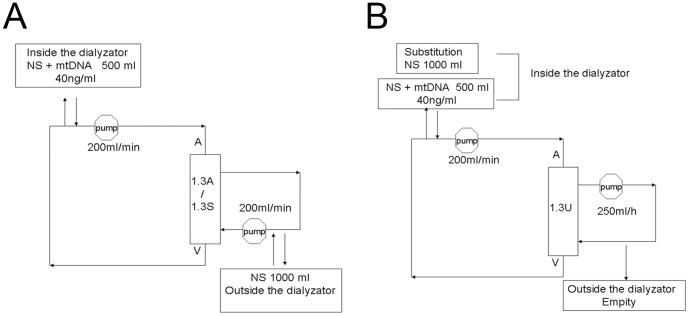
Three simulated dialysis patterns, LF-HD, HF-HD and hemofiltration (HF) were established in vitro. The simulated dialysis system was shown schematically in Fig. 1. In LF-HD and HF-HD, saline 500 ml with 40 ng/ml mtDNA was used as blood and saline 1000 ml was used as dialysate (Fig. 1 A). For the HF system, saline 500 ml with 40 ng/ml mtDNA was used as blood and saline 1000 ml was used as substitution (Fig. 1 B). The samples 500 µl were taken from the blood and dialysate side after 4 hours for the detection of mtDNA. After completion of the simulated dialysis, the dialyzer and filter were soaked with saline 20 ml overnight followed by DNA extraction. Ten fibers were taken from each dialyzer and filter, incubated with tissue buffer lysis 5 ml at 37°C overnight.
Figure 1. Three different dialysis systems, LF-HD, HF-HD and HF were established in vitro.
A. The schematic diagram of LF-HD and HF-HD in vitro. B. The schematic diagram of HF in vitro.
Statistic analysis
The data were analyzed using SPSS for windows version 18.0. Clinical data are expressed as the mean ± SD or median and range or frequency, as appropriate. Comparisons of continuous data of two groups used the independent t test. Compared t test was used in the data before and after hemodialysis. Logistic regression analysis was performed to describe the relationship between plasma mtDNA and clinical characteristics. A P value less than 0.05 was considered to represent a statistically significant difference.
Results
The clinical characteristics of MHD patients and controls
Baseline characteristics of participants were summarized in Table 1. MHD patients had a higher BMI (22.1±3.2 kg/m2) compared with controls (21.3±1.5 kg/m2, P = 0.001). Blood pressure especially systolic blood pressure (SBP) was higher in MHD patients (138.1±25.6 mmHg) than that in controls (106.5±10.6 mmHg, P = 0.005). The duration of hemodialysis was 57.2±45.6 months. MHD patients were more likely to have diabetes mellitus and the history of heart failure in recent one year. The hemoglobin (HGB) was lower in MHD patients than that in health controls, with 105.9±14.5 g/L vs 132.5±7.9 g/L (P = 0.034). Serum albumin (ALB) was 39.5±4.3 g/L in MHD patients, which was 46.9±3.1 g/L in controls. ALB adjusted Ca was 2.36±0.21 mmol/L in MHD patients and 2.28±0.13 mmol/L in controls (P = 0.060). Serum P and parathyroid hormone (PTH) were both higher in MHD patients than in controls, with serum P 1.71±0.55 mmol/L in MHD patients and 1.00±0.09 mmol/L in controls (P = 0.000); ln-PTH 4.60±1.38 ng/L in MHD patients and 3.57±0.38 ng/L in controls (P = 0.001). The product of Ca and P was also elevated in MHD patient.
Table 1. The clinical characteristics of MHD patients and health controls.
| Clinical characteristics | Controls | MHD patients | P value |
| No. of Cases | N = 20 | N = 60 | - |
| Age(years) | 41.0±11.7 | 57.7±11.3 | 0.685 |
| Male gender n (%) | 12(60.0) | 34(56.7) | 0.546 |
| BMI (kg/m2) | 21.3±1.5 | 22.1±3.2 | 0.001 |
| SBP (mmHg) | 106.5±10.6 | 138.1±25.6 | 0.005 |
| DBP (mmHg) | 69.3±8.3 | 79.3±11.8 | 0.127 |
| Duration of HD (months) | - | 57.2±45.6 | - |
| DM, n (%) | 0(0) | 9(15.0) | 0.026 |
| HF, n (%) | 0(0) | 8(13.3) | 0.091 |
| Smoking, n (%) | 6(30.0) | 19(31.7) | 0.386 |
| HGB (g/L) | 132.5±7.9 | 105.9±14.5 | 0.034 |
| Alb(g/L) | 46.9±3.1 | 39.5±4.3 | 0.209 |
| Ca(mmol/L) | 2.40±0.11 | 2.35±0.20 | 0.012 |
| Alb adjusted Ca (mmol/L) | 2.28±0.13 | 2.36±0.21 | 0.060 |
| Phosphate (mmol/L) | 1.00±0.09 | 1.71±0.55 | 0.000 |
| Product of Ca and P (mg/dl)2 | 27.3±2.8 | 48.6±16.8 | 0.000 |
| Ln-Intact-PTH (ng/L) | 3.57±0.38 | 4.60±1.38 | 0.001 |
| CHOL (mmol/L) | 4.28±0.48 | 4.28±0.77 | 0.095 |
| LDL-C (mmol/L) | 2.25±0.10 | 2.31±0.60 | 0.636 |
| CRP (mg/L) | 0.5–4.0 | 0.5–18.5 | 0.001 |
| IL-6 (pg/ml) | 43.8±8.8 | 69.1±53.4 | 0.177 |
| TNF-α(pg/ml) | 37.0±2.2 | 62.5±36.7 | 0.001 |
| mtDNA (ng/ml) | 42.7±8.5 | 129.1±101.3 | 0.000 |
Abbreviations: BMI: body mass index; SBP: systolic blood pressure; DBP: diastolic blood pressure; DM: diabetes mellitus; HF: heart failure; HGB: hemoglobin; Alb: albumin; Ca: calcium; P: phosphorus; PTH: parathyroid hormone; CHOL: cholesterol; LDL-C: low density lipoprotein-cholesterol; CRP: C-reaction protein; IL-6: interleukin 6; TNF-α: tumor necrosis factor α.
The circulatory mtDNA was significantly elevated in MHD patients
Circulatory mtDNA was evaluated from plasma of all 60 MHD patients before hemodialysis and 20 controls. Cyt B and COX III were applied to evaluate the quantity of mtDNA [18]. Cyt B was used as the main marker of mtDNA and COX III was used to validate the accuracy of Cyt B. It was shown that there was a strong correlation between the two mtDNA markers (Correlation coefficient 0.962, P = 0.000). In the 20 health controls, plasma mtDNA could be detected at a low concentration of 42.7±8.5 ng/ml, while plasma mtDNA was higher in MHD patients before hemodialysis with the concentration of 129.1±101.3 ng/ml. It was shown that circulatory mtDNA was significantly elevated in MHD patients (P<0.001) (Table 1).
Circulatory inflammatory markers were elevated in MHD patients
Plasma CRP levels (mg/L) were higher in MHD patients (0.5 to 18.5, median 4.0) than in controls (0.5 to 4.0, median 2.0). Plasma TNF-α levels (pg/ml) were significantly higher in MHD patients (62.5±36.7) than in controls (37.0±2.2) (P = 0.001). Circulatory IL-6 levels (pg/ml) were also evaluated in MHD patients (69.1±53.4) and controls (43.8±8.8), although there was no significant difference between two groups (P = 0.177) (Table 1). It was demonstrated that circulatory inflammatory markers were elevated in MHD patients.
Circulatory mtDNA was correlated with inflammatory parameters in MHD patients
MHD patients were stratified by the median plasma mtDNA concentration, one group with mtDNA>100 ng/ml, and the other group with mtDNA<100 ng/ml. In the univariate logistic regression analysis, plasma mtDNA was negatively correlated with HGB, serum ALB (P<0.005 for both) and was positively correlated with serum P, product of Ca and P, IL-6 and TNF-α (P<0.05 for all) (Table 2). When these parameters were entered into the multiple logistic regression analysis, TNF-α, product of Ca and P were independently positively associated with plasma mtDNA, while HGB and serum ALB were independently negatively associated with plasma mtDNA (P<0.05 for all) (Table 3). The logistic regression analyses demonstrated that circulatory mtDNA might be associated to the formation of chronic inflammation state in MHD patients.
Table 2. Univariate logistic regression analysis—variables associated with plasma mtDNA in MHD patients.
| P value | 95% CI | |
| Age | 0.963 | 0.955–1.045 |
| Male gender | 0.910 | 0.329–2.640 |
| BMI | 0.257 | 0.927–1.326 |
| SBP | 0.438 | 0.967–1.015 |
| DBP | 0.626 | 0.961–1.069 |
| HD age | 0.716 | 0.991–1.014 |
| DM | 0.675 | 0.325–5.686 |
| HF | 0.957 | 0.234–4.644 |
| Smoking | 0.851 | 0.299–2.710 |
| HGB | 0.003 | 0.858–0.968 |
| Alb | 0.002 | 0.606–0.891 |
| Ca | 0.161 | 0.455–113.227 |
| Alb adjusted Ca | 0.061 | 0.88–288.857 |
| P | 0.009 | 1.556–20.551 |
| Product of Ca and P | 0.003 | 1.023–1.120 |
| CHOL | 0.234 | 0.676–4.956 |
| LDL-C | 0.576 | 0.523–3.210 |
| Ln-Intact-PTH | 0.233 | 0.532–1.166 |
| CRP | 0.832 | 0.207–3.552 |
| IL-6 | 0.041 | 1.003–1.148 |
| TNF-α | 0.016 | 1.012–1.124 |
Table 3. Multiple logistic regression analysis—variables independently associated with plasma mtDNA in MHD patients.
| P value | 95% CI | |
| TNF-α | 0.045 | 1.002–1.132 |
| IL-6 | 0.131 | 0.981–1.157 |
| HGB | 0.036 | 0.880–0.996 |
| ALB | 0.015 | 0.607–0.949 |
| Phosphate | 0.105 | 0.751–20.795 |
| Product of Ca and P | 0.026 | 1.008–1.137 |
MtDNA induces secretion of inflammatory cytokines in vitro
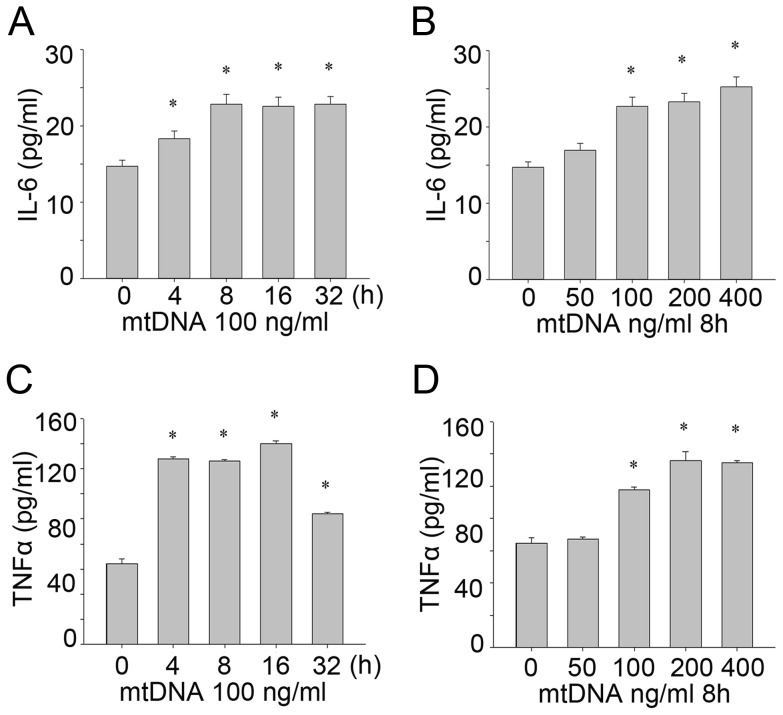
To clarify the relationship between circulatory mtDNA and inflammation, human acute monocytic leukemia cell (THP-1) were treated with mtDNA isolated from human skeletal muscles tissue. For various periods of time at the concentration of 100 ng/ml or different dosage of mtDNA for 8 hours, two typical inflammatory cytokines in the supernatant were measured respectively. The concentration of IL-6 and TNF-α in the supernatant were 14.7±0.8 pg/ml and 64.6±3.4 pg/ml at basal conditions. MtDNA induced IL-6 and TNF-α secretion in a time-dependent manner, starting at a time point as early as 4 hours. The peak values were 22.8±1.3 pg/ml for IL-6 at 8 hours and 159.8±2.6 pg/ml for TNF-α at 16 hours (Fig. 2 A, C). The effect of mtDNA on cytokines secretion was also dose-dependent (Fig. 2 B, D). These results implied that mtDNA could induce secretion of IL-6 and TNF-α from THP-1 cells into the extracellular space, suggesting a potential causal relationship between circulatory mtDNA and inflammation.
Figure 2. mtDNA induces the cultured THP-1 cells to secrete IL-6 and TNF-α.
THP-1 cells (106/ml) were induced to differentiate toward the monocyte/macrophage phenotype by 100 ng/ml of PMA for 36 hours. And then they were incubated with mtDNA for different time and dosage as indicated. (A, B) The effect of mtDNA on release of IL-6 from THP-1. (C, D) The effect of mtDNA on release of TNF-α from THP-1. * P<0.05.
Compared to LF-HD, HF-HD and OL-HDF could partially reduce plasma mtDNA in MHD patients
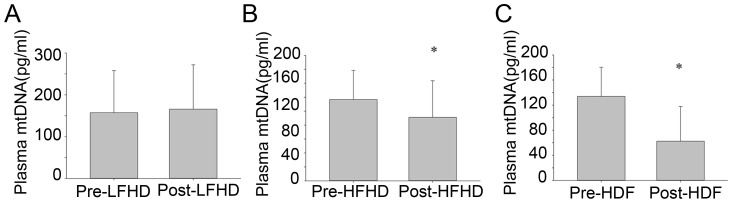
The effect of different dialysis patterns, including LF-HD, HF-HD and OL-HDF on the clearance of circulatory mtDNA was evaluated in MHD patients. The abundance of plasma mtDNA before and after a single dialysis was evaluated. There was no difference of the degree of blood concentration and the change of blood pressure among these three groups. In the patients undergoing LF-HD, plasma mtDNA remained unchanged from 157.1±100.1 ng/ml to 165.7±106.1 ng/ml (P = 0.818) (Fig. 3 A). While in patients undergoing HF-HD, plasma mtDNA was decreased from 108.1±40.0 ng/ml to 90.1±53.7 ng/ml (P = 0.023) (Fig. 3 B). Similar results were observed in patients undergoing OL-HDF, plasma mtDNA decreased from 134.0±46.5 ng/ml to 62.6±52.1 ng/ml (P = 0.000) (Fig. 3 C). These data indicated that HF-HD and OL-HDF but not LF-HD could partially reduce plasma mtDNA, which might ameliorate the chronic inflammation state in MHD patients.
Figure 3. The effects of different dialysis modes on clearance of circulatory mtDNA.
A. The effect of LF-HD on plasma mtDNA. B. The effect of HF-HD on plasma mtDNA. C. The effect of OL-HDF on plasma mtDNA. * P<0.05.
In vitro, HD and HF could fractional remove circulatory mtDNA
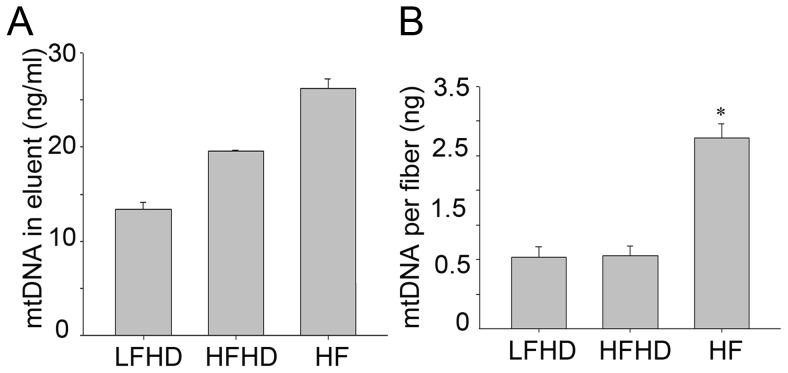
To clarify the mechanism of dialysis modalities on circulatory mtDNA, three simulated dialysis modes were established in vitro. For the LF-HD, combined with the concentration of mtDNA and the final volumes, mtDNA passed through the dialyzer was 5453 ng and remained inside the dialyzer was 2000 ng. For the HF-HD, the amount of mtDNA passed through the dialyzer was 9940 ng and remained inside the dialyzer was 1885 ng. While for the HF, the amount of mtDNA outside the filter was 2002 ng and inside the filter was 3040 ng. Considering the total amount of mtDNA before the treatment was 20000 ng for each dialysis modality, parts of them were absorbed by the dialyzer or filter. mtDNA could be detected in the eluents of the dialyzers and the filter (Fig. 4 A). Following soaking with the eluents, the mtDNA absorbed per fiber could still be detected (Fig. 4 B). It was concluded that mtDNA could partially be removed by LF-HD, HF-HD and HF although quite a number of mtDNA could be absorbed by the dialyzer or filter.
Figure 4. mtDNA absorbed by the dialyzer or filter in the simulated dialysis systems.
A. The concentration of mtDNA in eluent of dialyzers. B. The quantity of mtDNA per fiber after soaked overnight by tissue lysis buffer. * P<0.05.
Discussion
End-stage renal disease (ESRD) is a worldwide public health problem with high morbidity and mortality [20]. Although the long-term survival rate of MHD patients has significantly improved, the mortality of MHD patients was extremely higher than the general population of the same age [21]. Chronic inflammation is highly prevalent in MHD patients, which is a strong independent predictor of all-cause and CVD mortality in those population [1] [22] [23]. It may explain the excess CVD risk in MHD patients, in whom inflammation is common [3]. Certain pro-inflammatory cytokines, such as IL-6 and TNF-α, are considered as early drivers of the inflammatory response. Their elevations in MHD patients are always associated with increased risk of mortality. Common etiologies of inflammation in MHD patients include dialysis-related factors such as the extracorporeal circulation, the quality of the dialysate, the biocompatibility of the dialysis membranes and dialysis catheters as well as non–dialysis-related factors, such as decreased clearance and increased production of cytokines, retention of uremic solutes, increased oxidative stress burden, and high prevalence of co-morbidities associated with inflammation [24].
Circulatory mtDNA is one of the DAMPs reflecting cellular injury, which participating the systemic inflammation in several diseases. Zhang showed that injury released mtDNA into the circulation with functionally important immune consequences. MtDNA could activate human polymorphonuclear neutrophils (PMN) through TLR-9 to induce systemic inflammation [10]. It was also reported recently that mtDNA could act on endothelial cell (EC) and/or PMN via multiple pathways, which could enhance PMN adherence to EC, activate PMN-EC interactions and subsequently increase systemic endothelial permeability [25]. The clinical study implied that plasma mtDNA was an independent indictor with moderate discriminative power to predict the risk of post-traumatic systemic inflammation [26]. Circulatory mtDNA was also elevated in several other diseases such as tumors and sepsis [12], [27], [28]. It has been reported that plasma mtDNA levels could be used as the predictor of outcome in severe sepsis patients in the emergency room [29]. Therefore, circulatory mtDNA might be a pro-inflammatory agent participating systemic inflammation in a variety of diseases.
Given the dependence of renal function on aerobic metabolism, it is not surprising that impairment of normal mitochondrial function—due to insults such as ischaemia, drug toxicity and genetic mitochondrial disease can lead to kidney failure. MtDNA is apt to be exposed to reactive oxygen species (ROS) stress without histone protection. As a result, it is highly susceptible to damage and mutations, with a 10–1,000-fold greater mutation rate than nDNA. Studies have shown significantly increased ROS production, upregulation of COX I and IV expressions, and inactivation of complex IV in peripheral blood mononuclear cells (PBMCs) of patients with phase IV–V chronic kidney disease (CKD), thereby demonstrating the close association between mitochondrial dysfunction and CKD progression [30]. It has been also reported that mtDNA injury was associated with the mortality in MHD patients. The mitochondrial injury markers studied in PBMCs was the mtDNA copy number per cell, and a reduced mitochondrial copy number seems to predict a poor outcome in MHD patients [31]. Although the plasma cf-DNA has been focused recently in MHD patients, the role and mechanisms of circulatory mtDNA in chronic inflammation of MHD patients remains unknown [32].
It was found that circulatory mtDNA was significantly elevated in MHD patients. The mechanism of the elevation of mtDNA in circulation in MHD patients has not been explored. Logistic analysis was done to clarify whether circulatory mtDNA was associated to the chronic inflammatory state in MHD patients. It was demonstrated that plasma mtDNA in MHD patients was positively associated with plasma TNF-α, and was associated with clinical events which have been proven participating in the inflammatory state in MHD patients, such as anemia, hypoproteinemia and hyperphosphatemia [33]. The pro-inflammatory effect of mtDNA was conformed directly in THP-1 cells in vitro. mtDNA could induce the secretion of IL-6 and TNF-α in a time and dose dependent way.
To date, it has been still difficult to ameliorate the chronic inflammatory state in MHD patients. Cytokines such as IL-6 and TNF-α could also not be removed by routine hemodialysis patterns [34]. Considering the pro-inflammatory effect of mtDNA, we further investigated the effect of different routine hemodialysis modes on plasma mtDNA in MHD patients. We found that only HF-HD and OL-HDF but not LF-HD could partially reduce the level of plasma mtDNA. mtDNA in the dialysate could not be detected might due to the large volume of dialysate in single dialysis. Subsequently, in order to further clarify whether mtDNA could go through the dialysis membrane, the simulated dialysis systems in vitro were established. It was demonstrated that mtDNA could be going through the dialysis membrane not only in HF-HD but also in LF-HD. It could also be going through the filer membrane in HF. It was indicated that the form of mtDNA in plasma might be small molecular fragments. This result in vitro was not entirely consistent with the MHD patients undergoing different dialysis modes in vivo. The metabolism of plasma mtDNA might be sophisticated in MHD patients which needs to be explored further.
In conclusion, this is a preliminary clinical study focusing on the circulatory mtDNA on chronic inflammation in MHD patients. It is demonstrated that circulatory mtDNA is elevated in MHD patients. Its level is closely correlated to chronic inflammation in MHD patients. HF-HD and HDF but not LF-HD can partially reduce the circulatory mtDNA. It might be a new marker that provides significant insights into understanding the chronic inflammation of MHD patients and the efficacy of hemodialysis.
Data Availability
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Our data are available upon request because of the ethical restriction, which are related to the privacy of the patients. The readers could contact to the corresponding author Junwei Yang to get the details of the data.
Funding Statement
This work was supported by Jiangsu Province's Science and Technology Programs of Construction and the People's Livelihood (BL2013037) to Junwei Yang. Hongdi Cao was supported by the Yong fellowship from the Nanjing Medical University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hung A, Pupim L, Yu C, Shintani A, Siew E, et al. (2008) Determinants of C-reactive protein in chronic hemodialysis patients: relevance of dialysis catheter utilization. Hemodial Int 12:236–243. [DOI] [PubMed] [Google Scholar]
- 2. Stenvinkel P, Carrero JJ, Axelsson J, Lindholm B, Heimburger O, et al. (2008) Emerging biomarkers for evaluating cardiovascular risk in the chronic kidney disease patient: how do new pieces fit into the uremic puzzle? Clin J Am Soc Nephrol 3:505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kimmel PL, Phillips TM, Simmens SJ, Peterson RA, Weihs KL, et al. (1998) Immunologic function and survival in hemodialysis patients. Kidney Int 54:236–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Malaponte G, Bevelacqua V, Fatuzzo P, Rapisarda F, Emmanuele G, et al. (2002) IL-1beta, TNF-alpha and IL-6 release from monocytes in haemodialysis patients in relation to dialytic age. Nephrol Dial Transplant 17:1964–1970. [DOI] [PubMed] [Google Scholar]
- 5. Morena M, Delbosc S, Dupuy AM, Canaud B, Cristol JP (2005) Overproduction of reactive oxygen species in end-stage renal disease patients: a potential component of hemodialysis-associated inflammation. Hemodial Int 9:37–46. [DOI] [PubMed] [Google Scholar]
- 6. Caglar K, Peng Y, Pupim LB, Flakoll PJ, Levenhagen D, et al. (2002) Inflammatory signals associated with hemodialysis. Kidney Int 62:1408–1416. [DOI] [PubMed] [Google Scholar]
- 7. Arizono K, Nomura K, Motoyama T, Matsushita Y, Matsuoka K, et al. (2004) Use of ultrapure dialysate in reduction of chronic inflammation during hemodialysis. Blood Purif 22 Suppl 2:26–29. [DOI] [PubMed] [Google Scholar]
- 8. Duranton F, Cohen G, De Smet R, Rodriguez M, Jankowski J, et al. (2012) Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol 23:1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Taanman JW (1999) The mitochondrial genome: structure, transcription, translation and replication. Biochim Biophys Acta 1410:103–123. [DOI] [PubMed] [Google Scholar]
- 10. Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, et al. (2010) Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464:104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, et al. (2012) Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature 485:251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Puskarich MA, Shapiro NI, Trzeciak S, Kline JA, Jones AE (2012) Plasma levels of mitochondrial DNA in patients presenting to the emergency department with sepsis. Shock 38:337–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tovbin D, Novack V, Wiessman MP, Abd Elkadir A, Zlotnik M, et al. (2012) Circulating cell-free DNA in hemodialysis patients predicts mortality. Nephrol Dial Transplant 27:3929–3935. [DOI] [PubMed] [Google Scholar]
- 14. Atamaniuk J, Kopecky C, Skoupy S, Saemann MD, Weichhart T (2012) Apoptotic cell-free DNA promotes inflammation in haemodialysis patients. Nephrol Dial Transplant 27:902–905. [DOI] [PubMed] [Google Scholar]
- 15. Tsai NW, Lin TK, Chen SD, Chang WN, Wang HC, et al. (2011) The value of serial plasma nuclear and mitochondrial DNA levels in patients with acute ischemic stroke. Clin Chim Acta 412:476–479. [DOI] [PubMed] [Google Scholar]
- 16. Chiu RW, Chan LY, Lam NY, Tsui NB, Ng EK, et al. (2003) Quantitative analysis of circulating mitochondrial DNA in plasma. Clin Chem 49:719–726. [DOI] [PubMed] [Google Scholar]
- 17. Krauss S, Zhang CY, Scorrano L, Dalgaard LT, St-Pierre J, et al. (2003) Superoxide-mediated activation of uncoupling protein 2 causes pancreatic beta cell dysfunction. J Clin Invest 112:1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang Q, Itagaki K, Hauser CJ (2010) Mitochondrial DNA is released by shock and activates neutrophils via p38 map kinase. Shock 34:55–59. [DOI] [PubMed] [Google Scholar]
- 19. Brand K, Fowler BJ, Edgington TS, Mackman N (1991) Tissue factor mRNA in THP-1 monocytic cells is regulated at both transcriptional and posttranscriptional levels in response to lipopolysaccharide. Mol Cell Biol 11:4732–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Levey AS, Coresh J, Balk E, Kausz AT, Levin A, et al. (2003) National Kidney Foundation practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Ann Intern Med 139:137–147. [DOI] [PubMed] [Google Scholar]
- 21. Nolan CR (2005) Strategies for improving long-term survival in patients with ESRD. J Am Soc Nephrol 16 Suppl 2:S120–127. [DOI] [PubMed] [Google Scholar]
- 22. Stenvinkel P, Wanner C, Metzger T, Heimburger O, Mallamaci F, et al. (2002) Inflammation and outcome in end-stage renal failure: does female gender constitute a survival advantage? Kidney Int 62:1791–1798. [DOI] [PubMed] [Google Scholar]
- 23. Zimmermann J, Herrlinger S, Pruy A, Metzger T, Wanner C (1999) Inflammation enhances cardiovascular risk and mortality in hemodialysis patients. Kidney Int 55:648–658. [DOI] [PubMed] [Google Scholar]
- 24. Hung AM, Ellis CD, Shintani A, Booker C, Ikizler TA (2011) IL-1beta receptor antagonist reduces inflammation in hemodialysis patients. J Am Soc Nephrol 22:437–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun S, Sursal T, Adibnia Y, Zhao C, Zheng Y, et al. (2013) Mitochondrial DAMPs increase endothelial permeability through neutrophil dependent and independent pathways. PLoS One 8:e59989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, et al. (2013) Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med 10:e1001577 discussion e1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang L, Wang F, Wang L, Wang W, Liu B, et al. (2012) Prevalence of chronic kidney disease in China: a cross-sectional survey. Lancet 379:815–822. [DOI] [PubMed] [Google Scholar]
- 28. Sursal T, Stearns-Kurosawa DJ, Itagaki K, Oh SY, Sun S, et al. (2013) Plasma bacterial and mitochondrial DNA distinguish bacterial sepsis from sterile systemic inflammatory response syndrome and quantify inflammatory tissue injury in nonhuman primates. Shock 39:55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kung CT, Hsiao SY, Tsai TC, Su CM, Chang WN, et al. (2012) Plasma nuclear and mitochondrial DNA levels as predictors of outcome in severe sepsis patients in the emergency room. J Transl Med 10:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Che R, Yuan Y, Huang S, Zhang A (2014) Mitochondrial dysfunction in the pathophysiology of renal diseases. Am J Physiol Renal Physiol 306:F367–378. [DOI] [PubMed] [Google Scholar]
- 31. Rao M, Li L, Demello C, Guo D, Jaber BL, et al. (2009) Mitochondrial DNA injury and mortality in hemodialysis patients. J Am Soc Nephrol 20:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Korabecna M, Pazourkova E, Horinek A, Mokrejsova M, Tesar V (2012) Alterations in methylation status of immune response genes promoters in cell-free DNA during a hemodialysis procedure. Expert Opin Biol Ther 12 Suppl 1:S27–33. [DOI] [PubMed] [Google Scholar]
- 33. Cunningham J, Locatelli F, Rodriguez M (2011) Secondary hyperparathyroidism: pathogenesis, disease progression, and therapeutic options. Clin J Am Soc Nephrol 6:913–921. [DOI] [PubMed] [Google Scholar]
- 34. Jourde-Chiche N, Dou L, Cerini C, Dignat-George F, Vanholder R, et al. (2009) Protein-bound toxins–update 2009. Semin Dial 22:334–339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Our data are available upon request because of the ethical restriction, which are related to the privacy of the patients. The readers could contact to the corresponding author Junwei Yang to get the details of the data.