Abstract
Background: Apoptosis is an important mechanism that is responsible for the physiological deletion of harmful, damaged, or unwanted cells. Changed expression of apoptosis-related genes may lead to abnormal cell proliferation and finally to tumorigenesis. Our aims were to analyze the promoter methylation and gene expression profiles of FADD and FAS genes in risk of OSCC. Material and Methods: we analyze the promoter methylation status of FADD and FAS genes using Methylation - Specific PCR (MSP) in 86 OSCC tissues were kept in paraffin and 68 normal oral tissues applied as control. Also, FADD and FAS genes expression were analyzed in 19 cases and 20 normal specimens by Real-Time Reverse-Transcripts PCR. Results: Aberrant promoter methylation of FADD and FAS genes were detected in 12.79 % (11 of 86) and 60.46 % (52 of 86) of the OSCC cases, respectively, with a significant difference between cases and healthy controls for both FADD and FAS genes (P<0.001). The gene expression analysis showed statistically significant difference between cases and healthy controls for both FADD (p<0.02) and FAS (p<0.007) genes. Conclusions: To the best our knowledge, the data of this study are the first report regarding, the effect of promoter hypermethylation of the FADD and FAS genes in development of OSCC. To confirm the data, it is recommended doing further study in large sample sizes in various genetic populations.
Key words:OSCC, FADD, FAS, DNA methylation, gene expression.
Introduction
Head and neck cancer holds the sixth place in the cancer incidence ranking worldwide, influencing almost 650,000 people and causing nearly 350,000 cancer deaths each year (1,2). Among all of the head and neck cancers, Oral Squamous Cell Carcinoma (OSCC) is the most prevalent malignant epithelial neoplasm influencing the oral cavity (3).
The incidence and development of OSCC are multi stage processes, which arises through a collection of genetic and epigenetic variations (4). DNA methylation, as a key epigenetic variation is necessary for normal differentiation and development. The aberrant DNA promoter methylation that influences gene expression is a common feature of many human cancers (5). With respect to OSCC development, recent works demonstrated that hypermethylation of CpG islands of genes that are implicated in apoptosis, DNA-repair, cell-cell adhesion, and cell cycle regulation plays a vital role in cancer progression (6-9).
Apoptosis or programmed cell death deals with a significant task in the maintenance of cellular homeostasis. The inactivation of apoptosis related genes may lead to unusual cell proliferation and tumorgenesis (10,11). Generally, apoptosis is regulated by two major pathways: the receptor-mediated and the intrinsic (mitochondrial) pathways (12). Fas (CD95/Apo1) are a cell surface receptor that belongs to tumor necrosis factor receptor (TNF-R) family. Physiologically, it is expressed in various tissues such as lymph nodes, spleen and on mature hematologic cells (13,14).
Fas Ligand (FasL) is a homotrimeric protein act as a ligand for Fas receptor and causes its oligomerization. This process vast through the death domain (DD) and Fasassociated death domain (FADD). The N-terminal region of FADD which comprises DED (Death Effector Domain) motif binds to a homologous motif in Procaspase-8. Caspase-8 activates caspase-3 and -7 that mediate cell death. Furthermore, it cleaves Bid to generate truncated Bid (tBid) which translocates to the mitochondria and triggers the mitochondrial apoptotic pathway (15,16). Aberrant promoter methylation of FAS and FADD gene were exposed in different types of human cancers (17). Further, Fas is expressed in high quantities in lower stage of OSCC and a high incidence of FADD expression was significantly correlated with lymph node metastasis of SCCs (18,19). The data have been reported rarely, regarding to the status of the methylation and expression profile of FADD and FAS genes in OSCC tissues. Hence, the present study is trying to highlight the expression and methylation profile of FADD and FAS genes in patients with OSCC.
Material and Methods
-Samples and DNA preparation
This study involves 86 tumor specimens of OSCC (mean age 54.37 ±14) that had been fixed in Paraffin and 68 oral mucosa biopsies as controls (mean age 41±14) were collected during surgical resections of oral region squamous cell of patient’s (gingival aria) without a history of OSCC who were referred to Periodontics Department, after explanation of study purpose and signing of consent form.
Clinic pathological data of the patients and the controls such as age, sex, and clinical stage are shown in Table 1 and Table 2. Genomic DNA was isolated from tumor and healthy tissue samples using QIAamp DNA extraction kit (Cat. No. 56404, Qiagen) according to the manufacturer’s instructions and then its quality was estimated by Spectrophotometer.
Table 1. Association between Fas gene promoter methylation and clinicocopathological parameters in patients with OSCC and health controls.
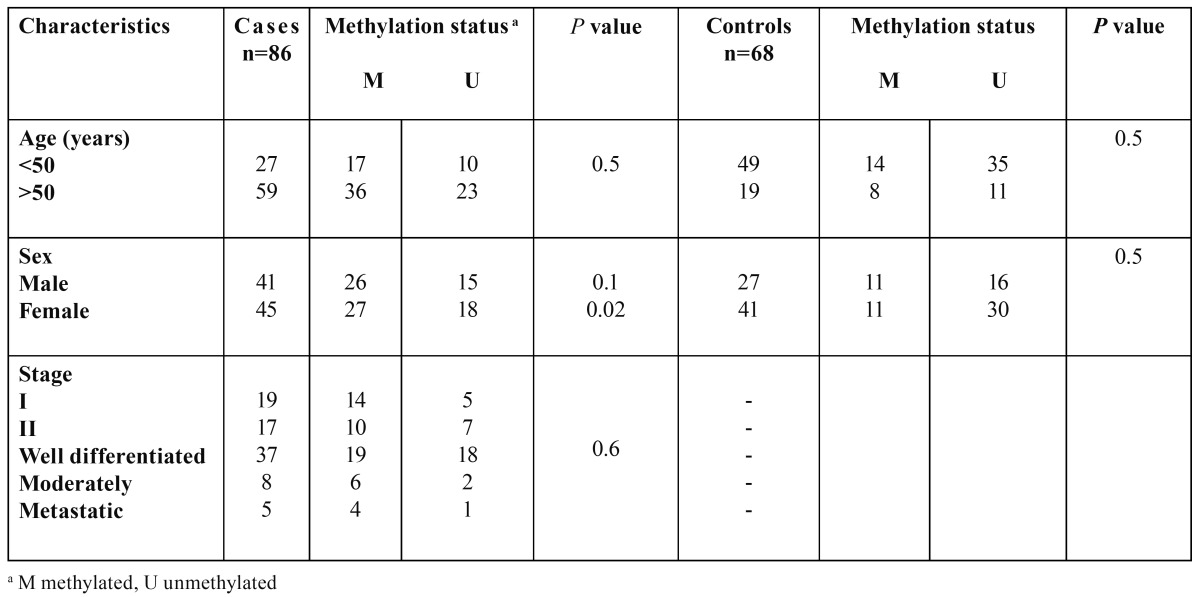
Table 2. Association between FADD gene promoter methylation and clinicocopathological parameters in patients with OSCC and health controls.
-Methylation-Specific PCR (MSP)
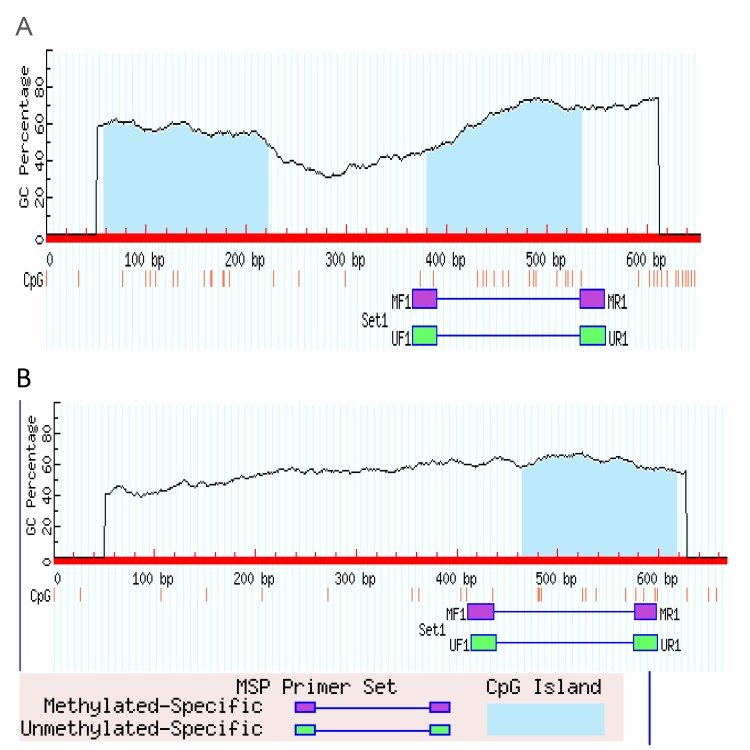
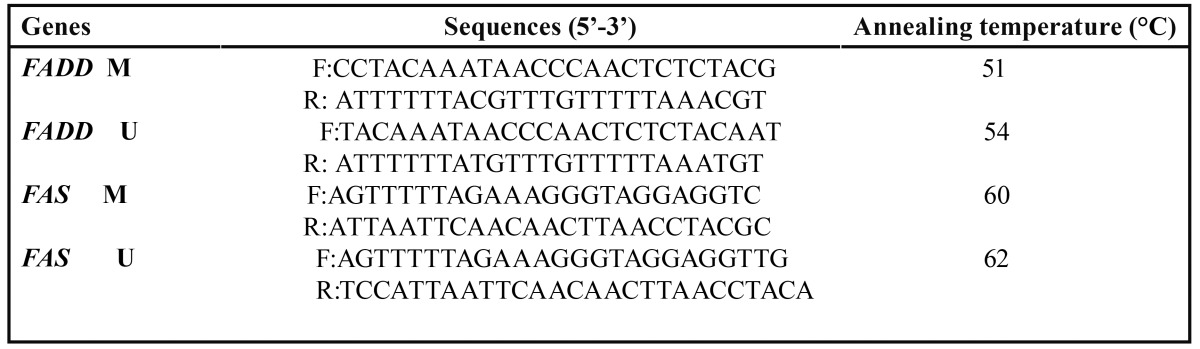
The process of bisulfit modification of DNA samples was performed as previously described (20). Methylation status of the promoter regions of FADD and FAS was determined by Methylation-Specific PCR (MSP) using methylated specific and unmethylated specific primers was designed at CpG sites of the promoter region using MatPrime online software (Fig. 1, Table 3).
Figure 1.
Selcted CPG island for MSP amplification, near to transcription start point A: FADD-001 ENST00000301838 and B; FAS-003 ENST00000460510.
Table 3. Primer sequences and annealing temperatures.
The PCR reaction mixture included 2 μL of modified template DNA, 0.2 µL of HotStarTaq®, 1 µL of dNTP mix (10 µmol/L), 16.3 µL of RNase free double distilled water, 2.5 µL of 10× buffer, 0.5 µL of each primer (10 µmol/L), and 2 µL of Mg2+ (25 µmol/L) in total (a) volume of 25 µL.
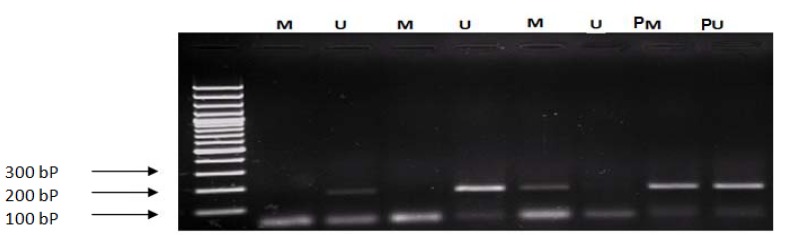
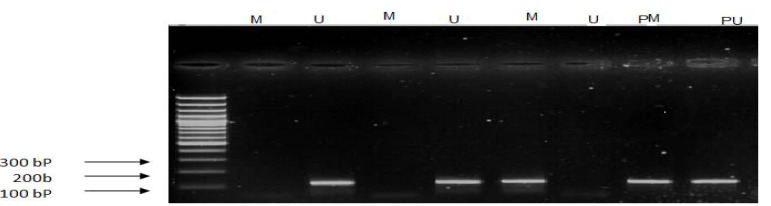
MSP amplification was performed as follows: 94°C for 10 min; and then 40 cycles consisting of (40 s at 94°C, 30 s at 51°C for FADD (M), 54°C (U), and for FAS 60°C (M), 62°C (U), 1 min at 72), and a final extension at 72°C for 10 min. PCR products were loaded onto 4% agarose gel and stained with ethidium bromide (Figs. 2,3).
Figure 2.
Methylation analysis of FADD gene: M: amplified product regoconizse by methylated primer (194 bp) U: amplified product regoconizse by unmethylated primer (192bp); L: ladder 100 bp: PM and PU (Human HCT116 DKO Methylated and Unmethylation DNA, D5014-1,2: Zymo Research California, USA).
Figure 3.
Methylation analysis of FAS gen, M: amplified product regoconizse by methylated primer( 160bp), U: amplified product regoconizse by unmethylated primer( 165bp), L: ladder 100 bp.
-Gene expression analysis
Total RNA was extracted from OSCC fixed paraffin embedded tissue sections and fresh normal samples that had been taken during various surgical operations in oral region, except of cancerous cases. It were used (Using) the High Pure FFPE RNA Micro Kit) Cat No: 04823125001 (and Cinna Pure RNA Purification Kit) Cat No: PR891620 (respectively, according to the manufacturer’s instructions.
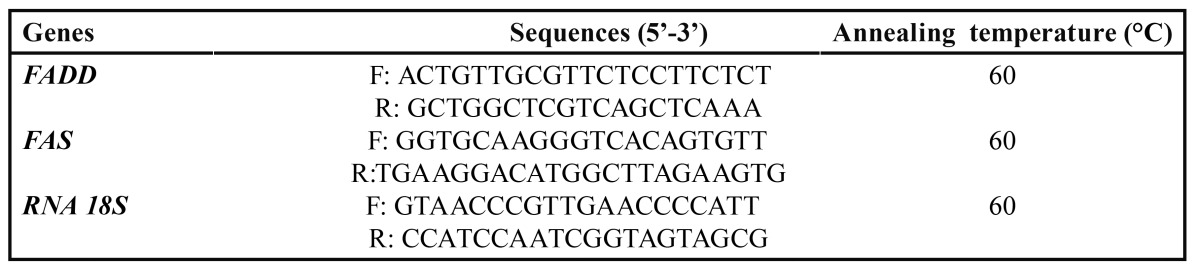
The cDNA Synthesis Kit (Fermentas, Cat No: K1621) was used to reverse-transcribe 1 μg of RNA in a final volume of 20 μl. As an internal standard, RNA18S was used. Real time-PCR of Fadd and Fas were performed using the primers and annealing temperatures in Table 4. Cycle threshold (CT) at which the fluorescence for the reaction well crosses was recognized for each gene in all samples and then, normalized CT (CT target gene/CT housekeeping gene) was used for comparison of genes expression between groups.
Table 4. Real-time primer sequences and annealing temperatures.
-Statistical analysis
Data were analyzed using SPSS software. The chi-square test was used for categorical variables. The effect of the methylation of FADD and FAS genes on the risk of OSCC was detected by estimating odds ratios (OR) and 95% confidence intervals (95% CI) using the binary logistic regression test. Analysis of relative gene expression between patients and controls was done by mann-whitney test. The significance level was set at p≤0.05 for all the tests.
Results
-Promoter methylation of FADD and FAS
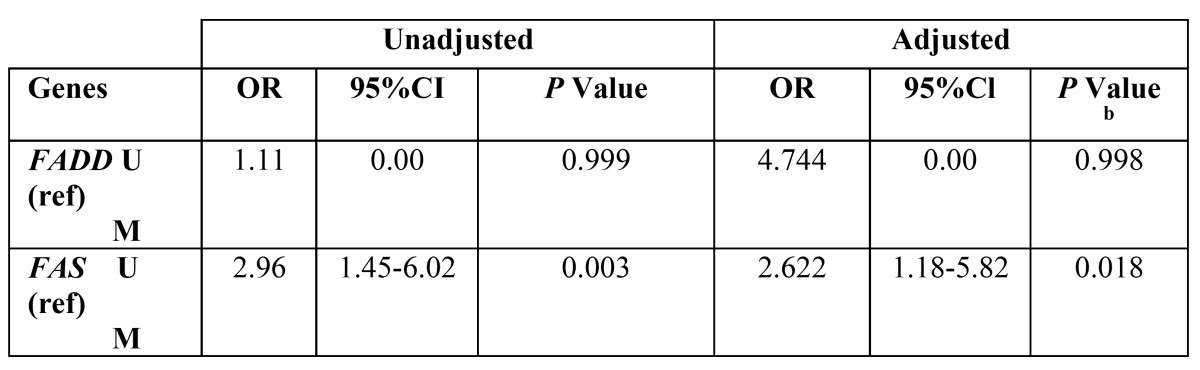
Promoter methylation status of the FADD and FAS genes in patients and healthy individuals and their relationship with risk of OSCC is indicated in Table 5 and Table 6. As shown, the frequency of methylation status for FADD gene was 12.79 % in tumor tissues (11 of 86 cases) and zero for normal mucosa (68). So (On the other side), it was not appeared a significant association between methylation status of FADD gene and risk of OSCC. Regarding the FAS gene, the amount of methylation was 60.46 % for cases (52 of 86) and 39.54% (22 of 68) for controls that this difference statistically, was significant between groups (P< 0.001). In addition, it was appeared a significant association between methylation status of FAS gene and increased risk of OSCC (OR=2.622, 95% CI; 1.18-5.82, P< 0.018).
Table 5. Promoter methylation frequency of FADD and FAS genes in patients with OSCC and healthy controls.
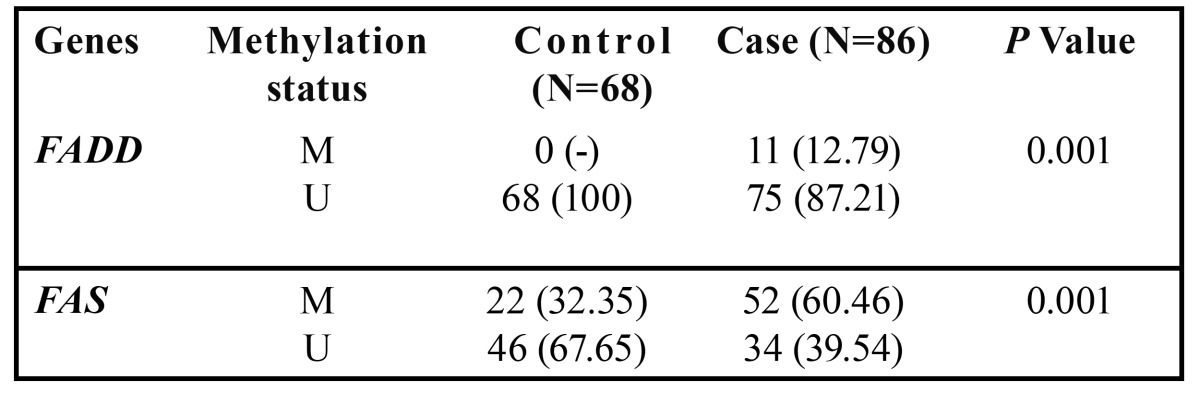
Table 6. Risk of OSCC based on gene promoter methylation.
-FADD and FAS mRNA levels
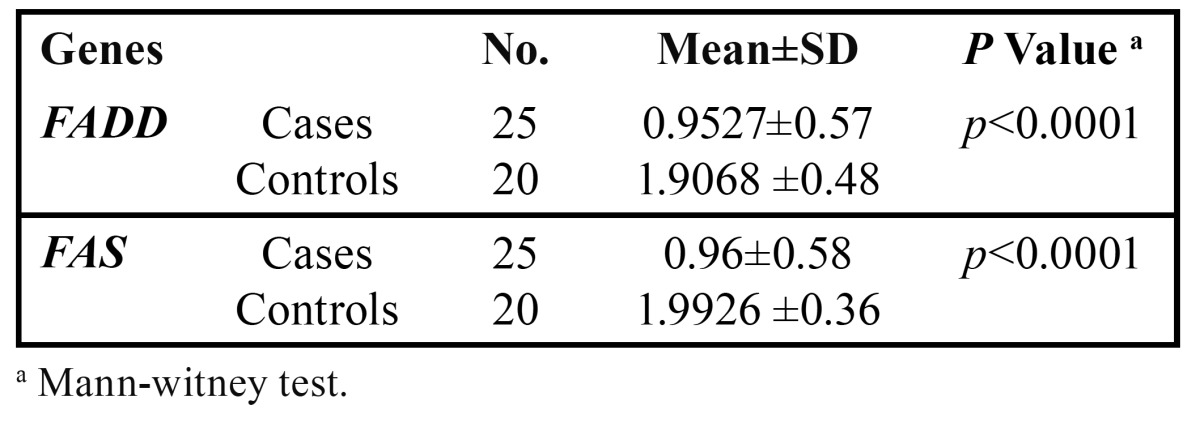
Assessment of relative gene expression was done according to dividing CT target gene to CT housekeeping gene for FADD and FAS between groups. As shown in Table 7, the mean of relative expression for FADD was 0.9527±0.57 in cases (n=25) and 1.9068 ±0.48 in controls (n=19). The FAS outcomes were 0.96±0.58 for cases (n=25) and 1.9926 ±0.36 for controls (n=20). The differences of relative gene expression between patients and healthy individuals were statistically significant for both of them (P< 0.0001).
Table 7. Comparison of relative gene expression for FADD and FAS genes between patients with OSCC and healthy controls.
Discussion
Epigenetics is a study of heritable variations that interferes in gene function without modifying the DNA sequences (21). It is responsible for the stable maintenance of a particular gene expression pattern through the cell cycle. The realizing of epigenetic mechanisms, including DNA methylation and chromatin remodeling, have shown a rapid progress in diagnosis and treatment of various diseases (22). These variations may induce gene silencing, imprinting and RNA interference that may lead to unusual modification as tumorigenesis (23). The results of methylation analysis in this study showed statistically, significant difference in amount of promoter methylation status between cases and healthy controls.
In line our results, Li W et al., (2011) shown that the rate of FAS promoter methylation in bladder urothelial carcinoma samples is higher than normal samples (p<0.01) (24). In addition, a vast literatures have been highlighted the aberrant promoter methylation of FAS gene in different types of cancers including Lymphomas, CXCA, melanoma, Colon, Prostatic and Lung (25-28).
To play a significant role in down regulation of FAS and FADD expression in early stage of tumorogenesis. The outcomes of the present gene expression analysis exposed a higher ratio of expression for FADD and FAS in patients with OSCC than healthy controls.
FAS and FADD are the most important elements of the apoptotic pathway with role of removing of harmful, damaged, or unwanted cells (29). Some studies have suggested that impairment of FAS gene expression links with development of various tumors such as; stomach, esophagus and liver (30-32). Muraki et al., (2000) have reported the increased expression of FAS gene in lower stage of SCC, it might operate as a controlling factor to promote apoptosis at the first step of disease but, in advanced stage of the disease has been detected to be down-regulated (33,34). The expression of the FADD has been found to be linked with non-small cell lung cancer and poor survival in laryngeal carcinoma (35,36). Accordance to our study, Lo Mozio et al., (2008) reported that there is a significant difference for expression of FADD gene between OSCC patients and healthy controls (37,38). In summary, this study tried to demonstrate the patterns of FAS and FADD genes methylation and expression profile in OSCC within a Southeastern Iranian population. The different expression of these genes between ill and normal groups is highlighting their significant role in development of OSCC. Ultimately. It should be mentioned that mathylation could be one of the reasons of gene expression changes. Therefore, we would like to suggest further studies to identify exact molecular process of the disease using advanced molecular techniques such as Micro Array and Meth Light in various and larger genetic populations.
Acknowledgments
We thank the Zahedan University of Medical Sciences, and University of Sistan and Baluchestan, for supporting this project financially.
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet. 2008;371:1695–709. doi: 10.1016/S0140-6736(08)60728-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goot-Heah K, Kwai-Lin T, Froemming GR, Abraham MT, Nik Mohd Rosdy NM, Zain RB. Human papilloma virus 18 detection in oral squamous cell carcinoma and potentially malignant lesions using saliva samples. Asian Pac J Cancer Prev. 2012;13:6109–13. doi: 10.7314/apjcp.2012.13.12.6109. [DOI] [PubMed] [Google Scholar]
- 4.Rigi-Ladiz MA, Kordi-Tamandani DM, Torkamanzehi A. Analysis of hypermethylation and expression profiles of APC and ATM genes in patients with oral squamous cell carcinoma. Clin Epigenetics. 2011;3:6. doi: 10.1186/1868-7083-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasegawa M, Nelson HH, Peters E, Ringstrom E, Posner M, Kelsey KT. Patterns of gene promoter methylation in squamous cell cancer of the head and neck. Oncogene. 2002;21:4231–6. doi: 10.1038/sj.onc.1205528. [DOI] [PubMed] [Google Scholar]
- 6.Shaw R. The epigenetics of oral cancer. Int J Oral Maxillofac Surg. 2006;35:101–8. doi: 10.1016/j.ijom.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Shaw RJ, Hall GL, Woolgar JA, Lowe D, Rogers SN, Field JK. Quantitative methylation analysis of resection margins and lymph nodes in oral squamous cell carcinoma. Br J Oral Maxillofac Surg. 2007;45:617–22. doi: 10.1016/j.bjoms.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 8.Supić G, Kozomara R, Branković-Magić M, Jović N, Magić Z. Gene hypermethylation in tumor tissue of advanced oral squamous cell carcinoma patients. Oral Oncol. 2009;45:1051–7. doi: 10.1016/j.oraloncology.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Supic G, Jovic N, Kozomara R, Zeljic K, Magic Z. Interaction between the MTHFR C677T polymorphism and alcohol--impact on oral cancer risk and multiple DNA methylation of tumor-related genes. J Dent Res. 2011;90:65–70. doi: 10.1177/0022034510385243. [DOI] [PubMed] [Google Scholar]
- 10.Gopisetty G, Ramachandran K, Singal R. DNA methylation and apoptosis. Mol Immunol. 2006;43:1729–40. doi: 10.1016/j.molimm.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Li C, Wang L, Su J, Zhang R, Fu L, Zhou Y. mRNA expression and hypermethylation of tumor suppressor genes apoptosis protease activating factor-1 and death-associated protein kinase in oral squamous cell carcinoma. Oncol Lett. 2013;6:280–286. doi: 10.3892/ol.2013.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zacks DN, Zheng QD, Han Y, Bakhru R, Miller JW. FAS-mediated apoptosis and its relation to intrinsic pathway activation in an experimental model of retinal detachment. Invest Ophthalmol Vis Sci. 2004;45:4563–9. doi: 10.1167/iovs.04-0598. [DOI] [PubMed] [Google Scholar]
- 13.Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet. 2005;14:1465–74. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- 14.Maurillo L, Del Poeta G, Venditti A, Buccisano F, Battaglia A, Santinelli S. Quantitative analysis of Fas and bcl-2 expression in hematopoietic precursors. Haematologica. 2001;86:237–43. [PubMed] [Google Scholar]
- 15.Nagata S. Apoptosis by death factor. Cell. 1997;88:355–65. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 16.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 17.Asuthkar S, Velpula KK, Chetty C, Gorantla B, Rao JS. Epigenetic regulation of miRNA-211 by MMP-9 governs glioma cell apoptosis, chemosensitivity and radiosensitivity. Oncotarget. 2012;3:1439–54. doi: 10.18632/oncotarget.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prapinjumrune C, Morita K, Kuribayashi Y, Hanabata Y, Shi Q, Nakajima Y. DNA amplification and expression of FADD in oral squamous cell carcinoma. J Oral Pathol Med. 2010;39:525–32. doi: 10.1111/j.1600-0714.2009.00847.x. [DOI] [PubMed] [Google Scholar]
- 19.Guler N, Uckan S, Celik I, Oznurlu Y, Uckan D. Expression of Fas and Fas-ligand and analysis of argyrophilic nucleolar organizer regions in squamous cell carcinoma: relationships with tumor stage and grade, and apoptosis. Int J Oral Maxillofac Surg. 2005;34:900–6. doi: 10.1016/j.ijom.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Kordi-Tamandani DM, Ladies MA, Hashemi M, Moazeni-Roodi AK, Krishna S, Torkamanzehi A. Analysis of p15INK4b and p16INK4a gene methylation in patients with oral squamous cell carcinoma. Biochem Genet. 2012;50:448–53. doi: 10.1007/s10528-011-9489-6. [DOI] [PubMed] [Google Scholar]
- 21.Conerly M, Grady WM. Insights into the role of DNA methylation in disease through the use of mouse models. Dis Model Mech. 2010;3:290–7. doi: 10.1242/dmm.004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitsiades CS, Anderson KC. Epigenetic modulation in hematologic malignancies: challenges and progress. J Natl Compr Canc Netw. 2009;7 Suppl 8:S1–12. doi: 10.6004/jnccn.2009.0081. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Serra L, Esteller M. Proteins that bind methylated DNA and human cancer: reading the wrong words. Br J Cancer. 2008;98:1881–5. doi: 10.1038/sj.bjc.6604374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li W, Xia D, Wang Y, Li Y, Xue Y, Wu X. Relationship between aberrant methylation of FAS promoter and biological behavior of bladder urothelial carcinoma. J Huazhong Univ Sci Technolog Med Sci. 2011;31:794–8. doi: 10.1007/s11596-011-0679-6. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Wood GS. Reduction of Fas/CD95 promoter methylation, upregulation of Fas protein, and enhancement of sensitivity to apoptosis in cutaneous T-cell lymphoma. Arch Dermatol. 2011;147:443–9. doi: 10.1001/archdermatol.2010.376. [DOI] [PubMed] [Google Scholar]
- 26.Chaopatchayakul P, Jearanaikoon P, Yuenyao P, Limpaiboon T. Aberrant DNA methylation of apoptotic signaling genes in patients responsive and nonresponsive to therapy for cervical carcinoma. Am J Obstet Gynecol. 2010;202:281–9. doi: 10.1016/j.ajog.2009.11.037. [DOI] [PubMed] [Google Scholar]
- 27.Petak I, Danam RP, Tillman DM, Vernes R, Howell SR, Berczi L. Hypermethylation of the gene promoter and enhancer region can regulate Fas expression and sensitivity in colon carcinoma. Cell Death Differ. 2003;10:211–7. doi: 10.1038/sj.cdd.4401132. [DOI] [PubMed] [Google Scholar]
- 28.Hopkins-Donaldson S, Ziegler A, Kurtz S, Bigosch C, Kandioler D, Ludwig C. Silencing of death receptor and caspase-8 expression in small cell lung carcinoma cell lines and tumors by DNA methylation. Cell Death Differ. 2003;10:356–64. doi: 10.1038/sj.cdd.4401157. [DOI] [PubMed] [Google Scholar]
- 29.Alenzi FQ. Links between apoptosis, proliferation and the cell cycle. Br J Biomed Sci. 2004;61:99–102. doi: 10.1080/09674845.2004.11732652. [DOI] [PubMed] [Google Scholar]
- 30.Ohno S, Tachibana M, Shibakita M, Dhar DK, Yoshimura H, Kinugasa S. Prognostic significance of Fas and Fas ligand system-associated apoptosis in gastric cancer. Ann Surg Oncol. 2000;7:750–7. doi: 10.1007/s10434-000-0750-1. [DOI] [PubMed] [Google Scholar]
- 31.Gratas C, Tohma Y, Barnas C, Taniere P, Hainaut P, Ohgaki H. Up-regulation of Fas (APO-1/CD95) ligand and down-regulation of Fas expression in human esophageal cancer. Cancer Res. 1998;58:2057–62. [PubMed] [Google Scholar]
- 32.Nagao M, Nakajima Y, Hisanaga M, Kayagaki N, Kanehiro H, Aomatsu Y. The alteration of Fas receptor and ligand system in hepatocellular carcinomas: how do hepatoma cells escape from the host immune surveillance in vivo? Hepatology. 1999;30:413–21. doi: 10.1002/hep.510300237. [DOI] [PubMed] [Google Scholar]
- 33.Muraki Y, Tateishi A, Seta C, Fukuda J, Haneji T, Oya R. Fas antigen expression and outcome of oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2000;29:360–5. [PubMed] [Google Scholar]
- 34.Chen Q, Samaranayake LP, Zhen X, Luo G, Nie M, Li B. Up-regulation of Fas ligand and down-regulation of Fas expression in oral carcinogenesis. Oral Oncol. 1999;35:548–53. doi: 10.1016/s1368-8375(99)00029-9. [DOI] [PubMed] [Google Scholar]
- 35.Shin MS, Kim HS, Lee SH, Lee JW, Song YH, Kim YS. Alterations of Fas-pathway genes associated with nodal metastasis in non-small cell lung cancer. Oncogene. 2002;21:4129–36. doi: 10.1038/sj.onc.1205527. [DOI] [PubMed] [Google Scholar]
- 36.Gibcus JH, Menkema L, Mastik MF, Hermsen MA, de Bock GH, van Velthuysen ML. Amplicon mapping and expression profiling identify the Fas-associated death domain gene as a new driver in the 11q13.3 amplicon in laryngeal/pharyngeal cancer. Clin Cancer Res. 2007;13:6257–66. doi: 10.1158/1078-0432.CCR-07-1247. [DOI] [PubMed] [Google Scholar]
- 37.Lo Muzio L, Santarelli A, Emanuelli M, Pierella F, Sartini D, Staibano S. Genetic analysis of oral squamous cell carcinoma by cDNA microarrays focused apoptotic pathway. Int J Immunopathol Pharmacol. 2006;19:675–82. doi: 10.1177/039463200601900323. [DOI] [PubMed] [Google Scholar]
- 38.Lo Muzio L, Sartini D, Santarelli A, Rocchetti R, Morganti S, Pozzi V. Expression and prognostic significance of apoptotic genes in oral squamous cell carcinoma. Mol Carcinog. 2014;53:264–71. doi: 10.1002/mc.21960. [DOI] [PubMed] [Google Scholar]