Abstract
High-density lipoprotein (HDL), a lipid nanoparticle containing many different low abundance proteins, is an attractive target for clinical proteomics because its compositional heterogeneity is linked to its cardioprotective effects. Selected reaction monitoring (SRM) is currently the method of choice for targeted quantification of proteins in such a complex biological matrix. However, model system studies suggest that parallel reaction monitoring (PRM) is more specific than SRM because many product ions can be used to confirm the identity of a peptide. We therefore compared PRM and SRM for their abilities to quantify proteins in HDL, using 15N-labeled apolipoprotein A-I (HDL’s most abundant protein) as the internal standard. PRM and SRM exhibited comparable linearity, dynamic range, precision, and repeatability for protein quantification of HDL. Moreover, the single internal standard protein performed as well as protein-specific peptide internal standards when quantifying 3 different proteins. Importantly, PRM and SRM yielded virtually identical quantitative results for 26 proteins in HDL isolated from 44 subjects. Because PRM requires less method development than SRM and is potentially more specific, our observations indicate that PRM in concert with a single isotope-labeled protein is a promising new strategy for quantifying HDL proteins in translational studies.
Introduction
One widely used strategy for biomarker discovery uses untargeted mass spectrometry (MS) to search for differentially expressed proteins in samples. To validate candidate proteins, however, it is necessary to use targeted methods for sensitive and specific protein quantification [1, 2]. Selected reaction monitoring (SRM, also termed multiple reaction monitoring [MRM]) of peptides as surrogate markers for precursor proteins is well suited for targeted proteomics, because it is quantitative when used with isotope-labeled peptides or proteins as internal standards.
An SRM experiment is generally performed in a triple quadrupole (QqQ) mass spectrometer. A predefined series of transitions (precursor/product ion pairs) is monitored over time for precise quantification [3]. One drawback is that the intensities of individual fragment product ions derived from a single precursor ion can differ substantially. To obtain a sensitive assay, it is essential to select the most intense product ions, which can be challenging and time-consuming. This is particularly relevant when data from ion trap instruments are used to select transitions, because fragmentation mechanisms in ion trap versus triple quadrupole mass spectrometers are vastly different, resulting in different patterns of product ions and/or product ion intensities. Moreover, the two stages of mass filtering of the QqQ (selecting a precursor at Q1 and its product ions at Q3) do not prevent concomitant detection of interfering ions caused by the quadrupole’s low resolution, especially in complex biological samples [4].
Another disadvantage is that quantifying peptides in candidate proteins in concert with stable isotope-labeled internal standards, though highly reproducible [5], fails to account for variability in the proteolysis step required before MS. Including isotope-labeled full-length proteins as standards overcomes this problem [6], and use of a single labeled protein rather than one labeled peptide or protein per precursor of interest was recently validated for relative quantification in complex mixtures [7].
An alternative method that promises to speed assay development is parallel reaction monitoring (PRM), which can quantify multiple peptides with increased sensitivity and specificity [4, 8–11]. Typically performed on high-resolution hybrid quadrupole-Orbitrap (Q-OT) or time-of-flight instruments, PRM uses targeted tandem MS to simultaneously monitor product ions of a targeted peptide with high resolution and mass accuracy [10, 12]. In brief, the precursor ion of interest is isolated by the quadrupole and fragmented in the high-energy collisional dissociation (HCD) cell. The fragment ions are then analyzed with an Orbitrap mass analyzer [12]. Because of this parallel monitoring, there is no need for prior selection of target peptide transitions. Moreover, PRM offers higher specificity than SRM on QqQ instruments, because it monitors product ions with high resolution and is therefore less likely to be affected by interfering ions. Thus, PRM has the potential to require much less effort than the traditional SRM assay [12].
The differences between PRM and SRM were recently investigated, using model isotope-labeled peptides and tryptic digests of yeast [10]. In that study, both methods exhibited the same linearity. However, PRM yielded quantitative data over a wider dynamic range, likely because of its higher selectivity, while SRM produced more precise measurements, possibly due to the higher sampling rate. Another study used 35 isotopically labeled peptides for target proteins, with urine as the biological matrix [4]. PRM obtained high resolution by separating ions of interest from interferences, increasing selectivity. Although this improved quantification, in some cases lower limits of quantification were achieved by SRM, especially for transitions without interference due to SRM’s higher intrinsic sensitivity.
Selected ion monitoring (SIM) on a Q-OT instrument is another strategy for quantifying complex samples. It uses accurate mass measurements of the precursor ions in a narrow mass range [12]. Recent studies have compared standard-free SIM and PRM approaches with SRM, using labeled peptides in model systems [4, 10, 12]. However, it is unclear whether these high resolution methods are comparable to SRM for analyzing complex clinical samples or whether they can be used with a single isotopically labeled standard. We therefore determined whether SIM and PRM perform as accurately as SRM when analyzing high density lipoprotein (HDL), a complex mixture of lipids and proteins.
We selected HDL because it is a particularly challenging biological matrix to analyze by MS. Defined by its flotation on ultracentrifugation in the density range 1.063 to 1.210 g/mL [13], HDL is a heterogeneous collection of particles that differ greatly in size, lipid composition, and contain a wide range of proteins [14, 15]. Its major constituents, apolipoprotein A-I (APOA1) and apolipoprotein A-II (APOA2), comprise ~90% of its protein mass; therefore, many of the proteins of biological interest are much less abundant (<1% total protein). It is difficult to separate these proteins from the matrix without complex biochemical approaches that alter relative abundances.
HDL is of clinical interest because its concentration in plasma correlates inversely with the incidence and severity of cardiovascular disease (CVD), the leading cause of myocardial infarction and death in the industrialized world [16]. Low levels of HDL cholesterol (HDL-C) associate robustly with increased CVD risk, likely because HDL promotes the removal of excess cholesterol from macrophages in the artery wall [17]. HDL is typically quantified in clinical practice by its cholesterol content, but recent clinical studies indicate that drug treatments that boost HDL-C levels fail to confer clinical benefit in statin-treated humans with established atherosclerosis [18, 19]. It is therefore critical to develop new metrics for quantifying HDL’s cardioprotective proteins.
The combination of SRM with isotopically labeled peptides has emerged as the standard approach in quantitative MS-based proteomics because of its sensitivity, specificity, and precision with complex biological samples [5, 20, 21]. This targeted approach enables the development of multiplexed assays, using tryptic peptides as surrogates for the protein of interest. However, representative peptide candidates must be selected for each protein, and an isotope-labeled internal standard for each selected peptide has to be synthesized [22]. Moreover, assay optimization requires peptides that are reproducibly generated during sample preparation, give a linear response in terms of protein concentration, and correlate with other candidate peptides for the same protein (not affected by biological variation, genetic mutations, post-translational modifications, etc.)[23]. It is therefore critical to establish generalizable approaches that can efficiently identify and prioritize a subset of potential biomarker candidates for use in the development of targeted analyses.
In the current studies, we determined whether PRM in concert with a single labeled protein—15N-labeled apolipoprotein A-I ([15N]APOA1)—as an internal standard could successfully provide a relative quantification of proteins in HDL and compared this with SRM and SIM. We first investigated the linearity of peptide quantification of [15N]APOA1 in a matrix of HDL over a wide range of concentrations. Using clinical samples, we then compared the analytical performance of PRM with that of SRM. Finally, we compared the use of a single isotope-labeled protein with the use of isotopically labeled peptides and an immunoassay. Our observations indicate that, for quantifying HDL proteins, PRM with a single labeled protein is as precise and accurate as SRM.
Materials and Methods
Materials and Reagents
Unless otherwise specified, all reagents were obtained from Sigma Aldrich (St. Louis, MO). Water and acetonitrile for MS analyses were Optima LC/MS grade (Fischer Scientific, Pittsburg, PA). Formic acid was purchased from EMD Millipore (Billerica, MA). The 15N-labeled apolipoprotein A-I ([15N]APOA1,15N enrichment 99+%) was produced by using a bacterial expression system as described [24]. The purity of labeled peptides (>95% purity; Pierce Biotechnology) was confirmed by MS.
Sample collection
Plasma samples from healthy subjects were acquired from the CLEAR (Carotid Lesion Epidemiology and Risk) study. The CLEAR study was designed to compare subjects with severe carotid disease (>80% stenosis) with healthy controls. However, for the current work, only a subset of the control population (44 samples, randomly selected from more than 1000 healthy volunteers) was utilized. Control subjects were recruited using criteria that excluded anyone with atherosclerosis-related diagnoses. These subjects presented less than 10% carotid stenosis bilaterally in the carotid ultrasound. All subjects completed an extensive medical history questionnaire and a complete physical exam. We used previously collected samples that are anonymous and contain no information that might allow for the identification of individuals. More detailed demographics are given elsewhere [25]. Fasting blood was collected in EDTA-treated tubes and centrifuged at 4 °C for 10 min at 1600 g to generate plasma. Plasma was pipetted into cryovials and immediately frozen and stored at −80 °C. The Human Studies Committee at the University of Washington approved all studies involving human material.
HDL isolation and proteolytic digestion
Plasma was quickly thawed at 37 °C, and 335 μL were used to isolate HDL (density 1.063–1.210 g/mL) by sequential ultracentrifugation as described previously [26]. Total protein concentration in HDL was measured using the Bradford assay with albumin as the standard. HDL (10 μg protein) was solubilized with 0.2% RapiGest (Waters, Milford, MA) in 100 mM ammonium bicarbonate, reduced with dithiothreitol, alkylated with iodoacetamide, and digested with trypsin (1:20, w/w HDL protein; Promega, Madison, WI) for 4 h at 37 °C. A second aliquot of trypsin (1:20, w/w HDL protein) was added and samples were incubated overnight at 37 °C [7]. After acidic hydrolysis of RapiGest with 0.5% trifluoroacetic acid, samples were dried, and stored at −20 °C until MS analysis. [15N]APOA1 was added to the sample before the digestion (10:1 w/w HDL/[15N]APOA1). For data-dependent experiments, after the HDL isolation procedure, aliquots containing 3.3 μg of protein (from different healthy subjects from the CLEAR study) were randomly taken. These aliquots were randomly combined into 10 pools containing 9.9 μg HDL protein (3.3 μg of HDL protein from each subject). The HDL was digested, using the protocol described above, though internal standard was not added. The results were analyzed by data-dependent shotgun analysis.
Dilution series of [15N]APOA1 in pooled HDL
A pool of control HDL was digested as described above, but without the addition of [15N]APOA1. After that, increasing concentrations of digested [15N]APOA1 were added to the pooled HDL, generating a standard curve ranging from 0 (blank) to 10 pmols of [15N]APOA1 per 0.25 μg of pooled HDL as the biological matrix. In this set of experiments, we chose to add the [15N]APOA1 after digestion rather than before in order to generate uniform samples because our aim was to compare different mass spectrometric methods. For quantification, [15N]APOA1 peptide peak areas were normalized to the corresponding unlabeled APOA1 peptide peak area. To remove the N-terminal affinity tag used in [15N]apoA-I isolation, a Glu2Asp mutation was introduced into the apoA-I cDNA to generate the N-terminal linker sequence NH2–1DDPPQS5–. For this reason, it was not possible to quantify the N-terminal peptide [24].
Data-dependent shotgun analysis
A nanoACQUITY UPLC (Waters, Milford, MA) was used for the separation with a linear gradient of solvents A and B (solvent A −0.1% formic acid in water; solvent B −0.1% formic acid in acetonitrile). Pooled digested HDL (0.25 μg) was loaded onto a trap column (Magic C18 AQ 200 A, 5 μM, 0.1 × 30 mm, Michrom Bioresources, Inc.) and washed for 5 minutes with a flow rate of 4 μL/min with an isocratic gradient of 2% solvent B. After this period, the trapped peptides were eluted onto an in-house packed C18 column (Magic C18 AQ 10 A, 5 μM, 0.1 × 300 mm, Michrom Bioresources, Inc.) with an integrated electrospray emitter pulled using a laser micropipette puller (Sutter Instrument, Novato, CA). Peptides were eluted at a flow rate of 0.35 μL/min, using with a linear gradient of 2% to 65% B in 180 minutes. Finally, the column was washed for 5 minutes with 80% B, followed by re-equilibration of the system with 2% B for 23 minutes. Data were acquired in a Q-Exactive mass spectrometer (Q-OT, Thermo Scientific, Bremen, Germany) using a full scan followed by data dependent MS2 scans. Precursor ions selected for MS2 were excluded for subsequent MS2 scans for 30 seconds. The resolution for the full scan mode was set as 35,000 (at m/z 200) the AGC target at 1×106 monitoring the m/z range 300 to 2000. Full scan was followed by a data dependent MS2 acquisition with resolution of 17,500 (at m/z 200), maximum ion fill time 100 ms, isolation window of 2 Th and normalized collision energy of 25.
Protein identification
MS/MS spectra were searched against the human International Protein Index database (September 2011, version 3.87, 91464 entries), using the Sequest search engine (version 2.7) with fixed Cys carbamidomethylation and variable Met oxidation of peptides [27]. The mass tolerance for precursor ions was 50 ppm and Sequest default tolerance was used for product ions. Trypsin was selected as the enzyme, and semi-tryptic specificity and one missed cleavage were allowed. Sequest results were further processed with PeptideProphet [28] and ProteinProphet [29] using an adjusted probability of ≥ 0.90 for peptides and ≥ 0.95 for proteins to give estimated FDR 5%.
Liquid chromatography for targeted MS analyses
Identical chromatographic conditions were used to allow direct comparison of the three MS methods. Following desalting on a C18 trap (Waters XBridge BEH C18, 5 μm, 0.075 × 40 mm), peptides were separated using an in house packed C18 column (Waters XBridge BEH C18, 3.5 μm, 0.075 × 100 mm) with an integrated electrospray emitter pulled using a laser micropipette puller (Sutter Instrument, Novato, CA). The column was kept at 50 °C. A nanoACQUITY UPLC (Waters, Milford, MA) was used for the separation with a linear gradient of solvents A and B (0.1% formic acid in water; solvent B −0.1% formic acid in acetonitrile). Peptide digest (0.25 μg) were injected onto the trap column at a flow of 3 μL/min of 99% solvent A. After 6 minutes the valve was switched, and the peptides were eluted from the trap column onto the analytical column at a flow rate of 0.6 μL/min. A multi-step gradient was applied as follows: linear 1 to 7% solvent B in 3 minutes, a linear gradient increasing solvent B from 7 to 25% in 16 minutes, followed by an increase from 25 to 35% B in 3 minutes. The column was subsequently washed for 3 minutes at 80% B and re-equilibrated at 99% A for 11 minutes.
SRM analyses
SRM experiments were performed on a TSQ Vantage Triple Stage Quadrupole Mass Spectrometer (QqQ, Thermo Scientific, San Jose, CA). Resolution for Q1 and Q3 were set to 0.7 Da (full width at half-maximum). Each transition had a dwell time of 15 ms. Skyline software (see below) was used for collision energy and retention time optimization and to generate the transition list [30]. The collision gas (argon) pressure of Q2 was set to 1.5 mTorr.
PRM and SIM analyses
Experiments were performed using a Q-Exactive mass spectrometer (Q-OT, Thermo Scientific, Bremen, Germany). For both methods, the resolution was set at 17,500 (at m/z 200) the AGC target at 5×104, maximum fill time 30 ms, and the individual isolation window of 2 Th window. Normalized collision energy of 25 was employed for fragmentation.
Linearity of [15N]APOA1 in HDL
A standard curve was prepared by serial dilution of [15N]APOA1 into HDL. Each HDL sample was supplemented with increasing amounts of digested [15N]APOA1, spanning a 10,000-fold range. Triplicate injections of each concentration were performed by SRM, PRM and SIM. Linear regression of all calibration curves for all methods was performed using a 1/x2 weighting, due to the wide dynamic range [31]. SRM experiments were performed using a scheduled (3-minute window) transition list generated in Skyline, which contained each precursor/product transition pair along with the collision energy and retention time (see Supplemental Table 1 for transitions and scheduling). Three different transitions for each peptide were monitored for SRM quantification. The quantification was performed using the sum of peak areas obtained for each transition. A scheduled (3-minute window) inclusion list for PRM analyses was generated using Skyline software [30]. The inclusion list consisted of m/z of precursor peptides of interest and corresponding retention times (see Supplemental Table 2 for isolation list, products monitored and scheduling). For quantification, the sum of peak areas of 10 most intense product ions was considered. Skyline software was used for integration and any product ion signal showing interferences was excluded. We excluded ions that did not match the retention time of the other monitored ions, or that gave intense signal in other regions of the chromatogram. SIM method was also optimized using Skyline software, with a list of precursors scheduled in a 3-minute window. The precursor isotopes M, M+1 and M+2 were selected for SIM quantifications (see Supplemental Table 3 for precursors monitored and scheduling).
Lower Limit of quantification (LLOQ)
The LLOQ was defined as the lowest concentration where the coefficient of variation (CV) of triplicate injections was less than 20% and had an average accuracy within 80 – 120%. All of the standard points for any level above the LLOQ had to fall within 75 – 125% of accuracy, otherwise the lower limit level would be removed and the linear regression equation recalculated.
Comparison of [15N]APOA1 and labeled peptides as internal standard
Human HDL was diluted into mouse HDL (10–75% human HDL), keeping the total amount of protein constant (5 μg). We used this strategy to maintain the same matrix for sample processing while varying the concentration of the human HDL proteins. Importantly, we confirmed that the sequences of the human peptides monitored differed from those of the mouse peptides. Each point of the standard curve was digested in quadruplicate, in the presence of a constant amount (0. 5 μg) of [15N]APOA1. After digestion, labeled peptides for the proteins phospholipid transfer protein (PLTP, peptide FLEQELETITIPDLR [R(13C6;15N4)], 18 pmols/ug HDL), alpha-1-antitrypsin (SERPINA1, peptide LSITGTYDLK [K(13C6;15N2)], 3 pmols/ug HDL), and serum paraoxonase/lactonase 3 (PON3, peptide LLNYNPEDPPGSEVLR [R(13C6;15N4)], 0.75 pmols/ug HDL) were added to the samples. The results obtained dividing each peptide by its corresponding labeled standard were compared to those obtained dividing the peptide by [15N]APOA1 peptides, using PRM and SRM methods. The labeled peptides were selected empirically, based on their relative abundances in spectral libraries built from initial shotgun proteomic analysis of human HDL and their correlations (r>0.85) with other peptides from the same protein. Supplemental Table 4 presents a list of peptides and transitions selected for both SRM and PRM methods.
Selection of HDL peptides and their transitions for targeted quantification
To compare targeted methods of quantification, 26 proteins present in widely different amounts (ranging from<1% to ~70% of total protein) in HDL were chosen. At least two peptides for each protein were selected, based on the observed frequency in the pooled analyses of HDL by shotgun proteomics and previous results [27]. Supplemental Table 5 shows a list of proteins, peptides and transitions selected for both SRM and PRM methods.
Relative quantification of HDL proteins in biological samples
For the relative quantification of HDL proteins in biological samples, 0.25 μg of digested HDL protein mixture containing [15N]APOA1 were analyzed by SRM and PRM using the transitions and inclusion list presented in Supplemental Table 5.
Immunoassay
Alpha-1-antitrypsin (SERPINA1) in HDL was quantified using a sandwich enzyme immunoassay (USBiological, Swampscott, Massachusetts). The reported intra-assay and inter-assay CVs are <10% and <12%, respectively. The results are expressed in ng/μg protein.
Data processing
Data analyses were performed using Skyline daily (version 2.5.1.6094), an open source software tool application for quantitative data processing and proteomic analysis [30]. All integrated peaks were manually inspected to ensure correct peak detection and integration.
Statistical analyses
Pearson’s correlations (r) were performed with STATA software version 12 (Stata Corp, College Park, TX).
Results
Study rationale and experimental approach
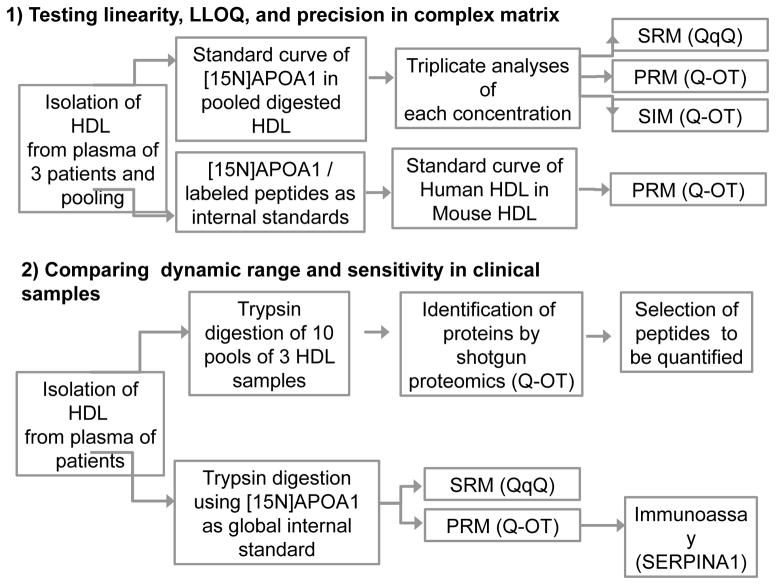
Our studies were divided into two parts (Fig. 1). First, we used pooled HDL as a matrix for generating a calibration curve for [15N]APOA1 to determine the linearity, LLOQ, and precision of SRM, PRM, and SIM. Each concentration of the standard curve was determined in triplicate, using a QqQ (SRM mode) and a Q-OT (PRM and SIM modes). We also compared [15N]APOA1 and labeled peptides as internal standards by serially diluting human HDL into mouse HDL.
Figure 1.
Schematic representation of study design and sample workflow.
Second, we investigated the potential use of PRM in translational studies of HDL. We initially identified candidate proteins by shotgun analyses of pooled HDL. Then, we performed a relative quantification of peptides present in HDL isolated from plasma of apparently healthy control subjects, using labeled [15N]APOA1 as a global internal standard. In this stage, we compared the quantification capabilities of PRM and SRM for 26 different proteins in HDL ranging widely in size (<10 kDa to >40 kDa) and concentration (<1% to ~70% of total HDL protein). Finally, we validated the relative quantification of a different protein, SERPINA1, by comparing the MS results with those obtained by an immunoassay.
Evaluation of PRM, SRM, and SIM with [15N]APOA1 in HDL
Our initial studies centered on APOA1 (28 kDa), HDL’s major structural protein. It contains 243 amino acids, including 21 lysine and 16 arginine residues. Based on sequence analysis, we anticipated generating 17 tryptic peptides that were at least 7 amino acids long, assuming no missed cleavage sites. In our analyses, we detected all 17 anticipated tryptic peptides. Because the N-terminal peptide was mutated to facilitate [15N]APOA1 purification (Materials and Methods), we did not monitor that peptide in its labeled form. Five of the 17 peptides did not give satisfactory results, because their poor retention by the C18 beads in the trap column caused variable loss. We were able to quantify the other 12 peptides over a 10,000-fold range of labeled to unlabeled APOA1.
We first determined how well PRM, SRM, and SIM quantified levels of [15N]APOA1 in the presence of a biological matrix of HDL (Fig. 1, workflow). Our criteria were linearity, lower limits of quantification (LLOQ), and precision. The relative responses of increasing concentrations of [15N]APOA1 peptides were determined by normalizing the sum of the peak areas for each transition of a given labeled peptide to that of the corresponding native peptide (effectively using the native APOA1 and its tryptic peptides as an internal standard for the 15N[APOA1]). Due to the wide dynamic range, a 1/x2 weighted regression was used [31]. The presence of a constant matrix prevented the overestimation of LLOQ and linear range that can occur when analyzing dilution series of pure proteins [32].
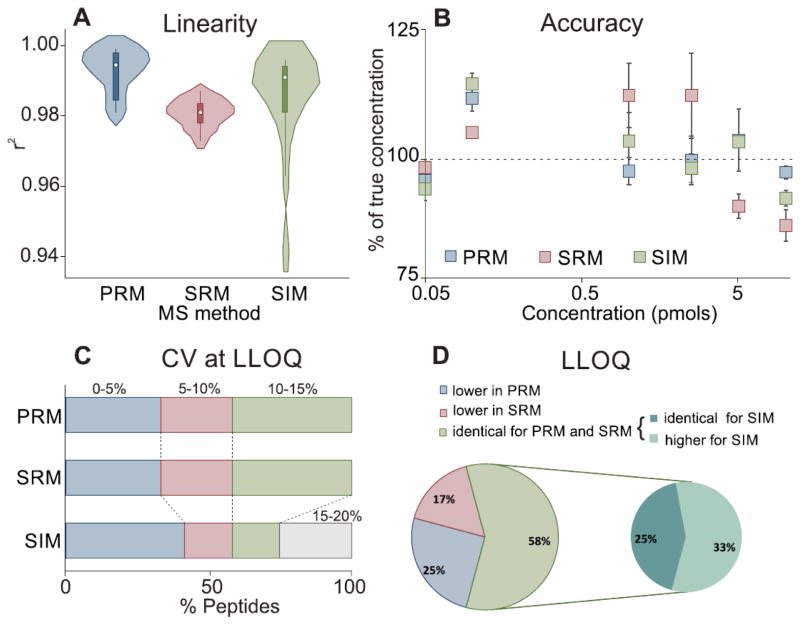
We evaluated linearity by the coefficient of determination (Pearson’s r2) of the standard curves of [15N]APOA1 peptides. Using r2 to assess the fit of the data to a straight line assumes that the data are homoscedastic (have constant variance over the whole range of concentrations). If so, the larger the squared correlation coefficient, the better the curve fit. However, peptide quantification does not necessarily exhibit homogeneity of variance at all concentrations. In such cases, the value of r2 can be misleading, especially over large dynamic ranges of relative peptide concentration. Nonetheless, it is still very informative to compare r2 across the different methods, provided one is aware of the limitations. Fig. 2A provides the distribution of r2 for the 12 measured peptides of [15N]APOA1. The r2 for PRM was ≥ 0.98 for all 12. With SRM, five of the peptides gave r2 of 0.97–0.98, while seven gave r2 of ≥0.98. The lowest r2 were obtained with SIM: 0.94–0.96 for two peptides, though for all the remaining peptides r2 was ≥0.98 (See Supplemental Table 6).
Figure 2.
Comparison of the performances of PRM, SRM, and SIM for quantifying a standard curve of 12 [15N]APOA1 peptides in HDL matrix. (A) Distribution of r2 values obtained for PRM, SRM, and SIM presented as a violin plot. The white dot in the center of each solid rectangular box is the median of the data, the upper and lower values of the rectangular box indicate the 75th and 25th percentiles, and the spikes of the rectangles are the range of the data. The width of the plot outside the modified box plot is the density of values (i.e., the fraction of Y values within each interval of Y values). (B) Residuals (difference between the measured concentration and expected value) obtained by PRM, SRM, and SIM for the heavy peptide DYVSQFEGSALGK. The scale of the X axis is logarithmic. (C) Percentage distribution of CVs at LLOQ for PRM, SRM, and SIM. (D) Comparison of the LLOQ values for PRM, SRM, and SIM for each quantified peptide.
A good way to test curve fit is to consider the accuracy of each measurement (the residuals) together with coefficients of determination. We therefore determined the accuracy of each measurement for all concentrations of APOA1 peptides for all calibration curves determined by PRM, SRM, and SIM. Representative residuals for the three methods are shown in Fig. 2B for the peptide DYVSQFEGSALGK. Taking together, linearity results show that the three methods exhibit a good curve fit, with the great majority of r2 above 0.98 and with values for each standard point with no more than 25% deviation of the “true” concentration.
The LLOQ of [15N]APOA1 and the precision of the measurements were evaluated by determining the CVs of triplicate injection for each standard curve point. Almost 60% (7 of 12) of the peptides quantified by the three methods yielded CVs at LLOQ <10%. For PRM and SRM, all of the remaining peptides (41%, 5 of 12) had CVs at LLOQ between 10% and 15%. In contrast, 2 peptides quantified by SIM had CVs at LLOQ between 10% and 15%, and the remaining 3 (25%) of [15N]APOA1 peptides had CVs between 15% and 20% (Fig. 2C).
Different APOA1 peptides in the same protein digest had different LLOQs, even though we used the same quantification method. A key component of protein quantification is the use of signature peptides that exhibit favorable characteristics for MS. In particular, peptides that are susceptible to ex vivo modification (e.g., methionine and tryptophan) or that contain amino acid sequences with high potential for miscleavage should be avoided when possible to obtain optimal results. However, because we were testing the performance of the three methods, we quantified all of the readily detectable peptides in the protein in all analyses. The values obtained for LLOQ were very comparable between PRM and SRM. For many peptides, SIM provided similar results to PRM and SRM. Overall, however, SIM provided inferior performance, with lower precision and accuracy increasing its LLOQs (Fig. 2D). Of the 12 peptides analyzed, 58% showed identical LLOQ for PRM and SRM. Of this 58%, only 25% also had similar LLOQ when measured by SIM, while the remaining 33% had lower performance. PRM gave the best LLOQ for 25% of the peptides and SRM for 17%. A detailed comparison of LLOQ, analytical response (slope), r2, and % CV at LLOQ for doubly charged [15N]APOA1 peptides is provided in Supplemental Table 6. The same criteria were evaluated for triply charged peptides (Supplemental Table 7), which generally yielded inferior performance compared with their doubly charged counterparts when quantified by PRM or SRM. For the majority of the peptides, the intensity of the signal for the triply charged state was much lower than that for doubly charged peptides, likely explaining the lower performance. Triply charged peptides performed so poorly with SIM that their linear regression curves were not acceptable by our criteria.
Taken together, the systematic comparison of assay criteria (in terms of linearity, accuracy, precision at LLOQ, and LLOQ) extracted from the curves of 12 APOA1 peptides suggests that PRM and SRM are quantitatively comparable. For both methods, the accuracy and precision of peptide quantification were within 20% of the true concentration with <15% CVs (Fig. 2B,C). In contrast, SIM performed worse, with only 4 of peptides matching the LLOQ obtained for PRM and SRM methods.
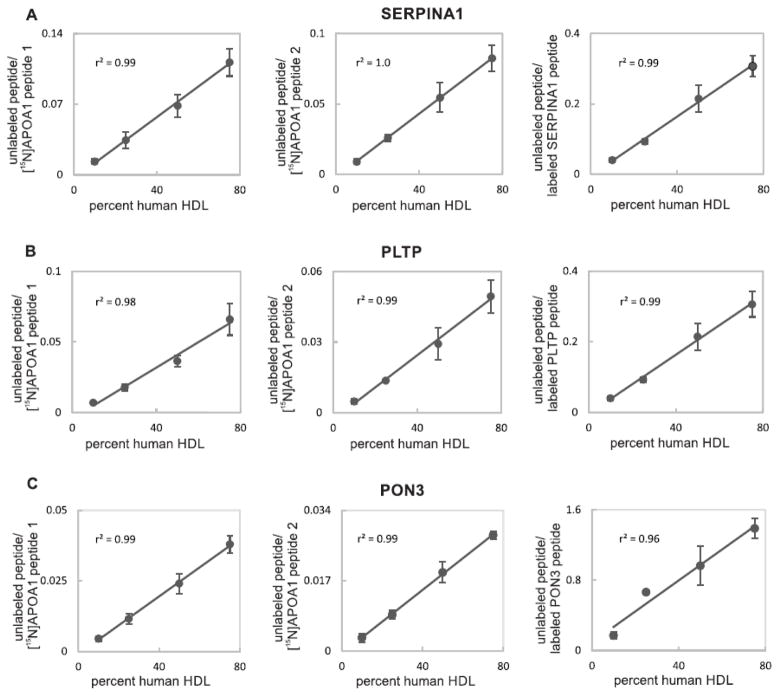
We next determined if a single labeled protein, [15N]APOA1, could be used as a single, global internal standard for APOA1 peptides as well as for peptides derived from other proteins (Fig. 1, workflow). The strategy involved a series dilution of human HDL into mouse HDL, addition of a constant amount of [15N]APOA1, and then tryptic digestion. To compare this approach with that of using synthetic peptides as internal standards, we also added constant amounts of three heavy-labeled peptides, FLEQELETITIPDLR, LLNYNPEDPPGSEVLR, and LSITGTYDLK. These were respectively derived by proteolysis of PLTP (phospholipid transfer protein), PON3 (serum paraoxonase/lactonase 3), and SERPINA1 (alpha-1-antitrypsin). Two [15N]APOA1 peptides, [15N]APOA1 peptide 1 ([15N]DYVSQFEGSALGK) and [15N]APOA1 peptide 2 ([15N]VQPYLDDFQK) were used as global internal standards. We did not investigate the single labeled protein standard approach with label-free quantification because the latter method was shown to perform poorly when compared with methods utilizing either protein or peptides as internal standards [7]. The standard curves with different internal standards were monitored by PRM. Fig. 3A, B, and C show the respective results obtained for SERPINA1, PLTP, and PON3. The left, central, and right panels, respectively display the ratios obtained by dividing the peak area of the endogenous peptide by the peak area of [15N]APOA1 peptide 1, [15N]APOA1 peptide 2, or the corresponding labeled synthetic peptide. Importantly, using the three different internal standards as means of normalization gave linear results for the three proteins tested, with r2 ≥ 0.96. These results indicate that the performance of the [15N]APOA1 peptides was equivalent to that of the corresponding synthetic label peptides for each of the three proteins tested.
Figure 3.
Quantification of a dilution series of human HDL peptides in mouse HDL background using different internal standards. (A) PRM quantification of SERPINA1 peptide (LSITGTYDLK). (B) PRM quantification of PLTP peptide (FLEQELETITIPDLR). (C) PRM quantification of PON3 peptide (LLNYNPEDPPGSEVLR). Left, central and right panels respectively display quantification results based on the ratios obtained by dividing the peak area of endogenous peptide by the peak area of [15N]APOA1 peptide 1 ([15N]DYVSQFEGSALGK), [15N]APOA1 peptide 2 ([15N]VQPYLDDFQK), or the corresponding synthetic peptide for each protein. For some points, SDs are smaller than the dots. Coefficients of determination (r2) are indicated.
Relative quantification of proteins in human HDL
Because PRM quantified APOA1 peptides in HDL as well as SRM, we next tested whether the two methods performed comparably when quantifying multiple proteins in HDL isolated from the plasma of apparently healthy subjects (Fig. 1, workflow). Details of the clinical population are provided in Materials and Methods. We chose proteins, peptides, and transitions based on the shotgun proteomic experiments and a previous study [27]. A list of the proteins and peptides is provided in Supplemental Table 5.
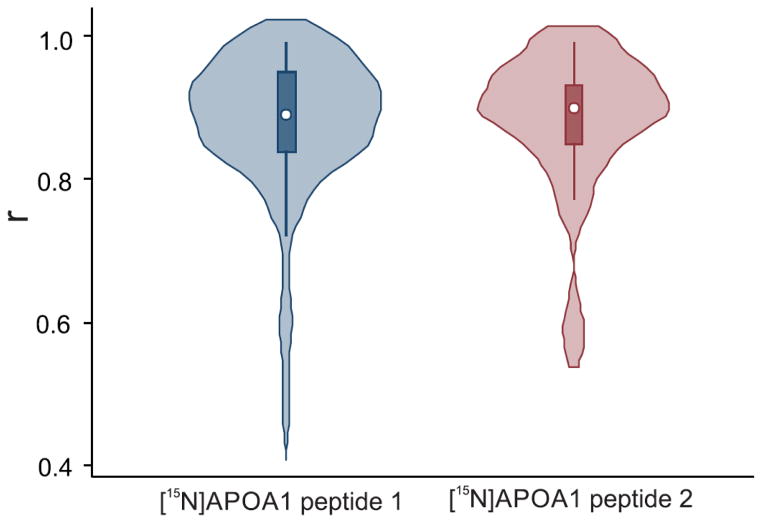
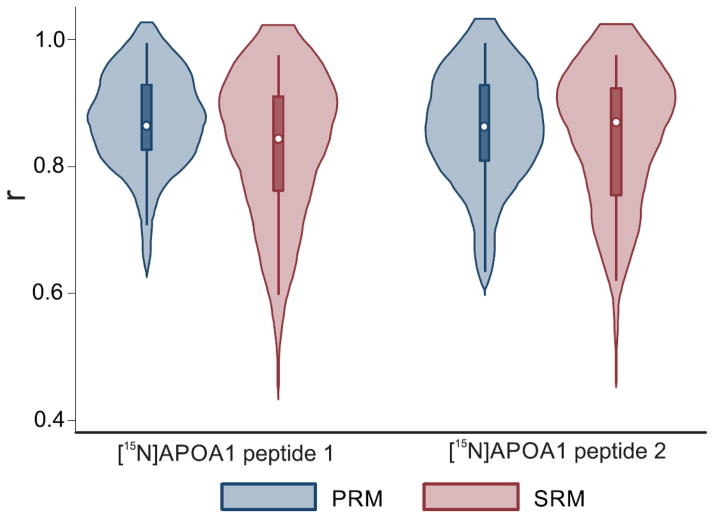
We quantified 26 different proteins in HDL, using at least 2 peptides per protein. To determine if the two methods gave consistent results, we calculated Pearson’s correlation between SRM and PRM for each peptide, using ratios obtained by dividing the integrated peak area of each specific peptide by the peak area of two internal standards for [15N]APOA1—[15N]APOA1 peptide 1, DYVSQFEGSALGK, and [15N]APOA1 peptide 2, VQPYLDDFQK (Fig. 4, Supplemental Table 8). The two methods yielded correlations ≥0.90 for 38% of the peptides when peptide 1 was used as an internal standard and for 50% of the peptides when peptide 2 was used. For >80% of the peptides monitored, correlations between SRM and PRM were >0.80, regardless of the internal standard used. Only 5% of the correlations were <0.6. These results point to a high degree of agreement between PRM and SRM and confirm previous data showing that [15N]APOA1 can serve as a global internal standard for quantifying proteins in HDL [7].
Figure 4.
Relationship between SRM and PRM methods for peptides in HDL of human subjects. Pearson correlations were performed using the ratios obtained by dividing the integrated area of a specific peptide by the areas of [15N]APOA1 peptide 1 ([15N]DYVSQFEGSALGK) or [15N]APOA1 peptide 2 ([15N]VQPYLDDFQK). The white dot marker shows the median of the data, while the box indicates the interquartile range, and spikes extend to the upper- and lower-adjacent values. Overlaid with this modified box plot is the estimated density.
When using peptides as surrogates for protein abundance, it is necessary to avoid peptides that are digested inefficiently or have unknown modifications that might affect quantification. Thus in the early stages of method development, many peptides should be tested to find the right surrogates for a protein of interest. Ideally, the chosen peptides should correlate tightly across clinical samples (e.g., be insensitive to biological variations, genetic mutations, etc.). We therefore determined how the peptides of a specific protein correlated with each other across the 44 clinical samples and whether they provided similar correlations with PRM and SRM. Pearson correlations for each peptide from 26 HDL proteins normalized using ([15N]APOA1 peptide 1 (DYVSQFEGSALGK) and ([15N]APOA1 peptide 2 ([VQPYLDDFQK) and quantified by PRM and SRM are shown in Fig. 5. The r for each peptide pair is reported in Supplemental Table 9.
Figure 5.
Correlations for peptide pairs of proteins in HDL obtained by SRM and PRM. Pearson correlations were performed using the ratios obtained by dividing the integrated natural peptide area by the areas of [15N]APOA1 peptide 1 ([15N]DYVSQFEGSALGK) or [15N]APOA1 peptide 2 ([15N]VQPYLDDFQK). The white dot marker shows the median of the data, while the box indicates the interquartile range, and spikes extend to the upper- and lower-adjacent values. Overlaid with this modified box plot is the estimated density.
When peptide 1 was the internal standard, comparable fractions of the peptide pairs had correlations ≥0.90 for PRM and SRM (41% and 36%, respectively). Similar results were obtained with peptide 2 (correlation ≥0.90 for 38% of the peptide pairs) for both PRM and SRM. Moreover, when the peptide peak area was normalized to peptide 1, 95% and 82% of the peptide correlations were ≥0.7 for PRM and SRM, respectively. Similar results were obtained with peptide 2 as internal standard, with 90% and 87% of the peptide pair correlations ≥0.7. Measurements of different peptides in the same protein agreed when performed by either PRM or SRM. Using two different internal standards provided similar results, confirming that a single labeled protein can be successfully used as a global internal standard.
Validation of the PRM/single labeled protein approach by immunoassay
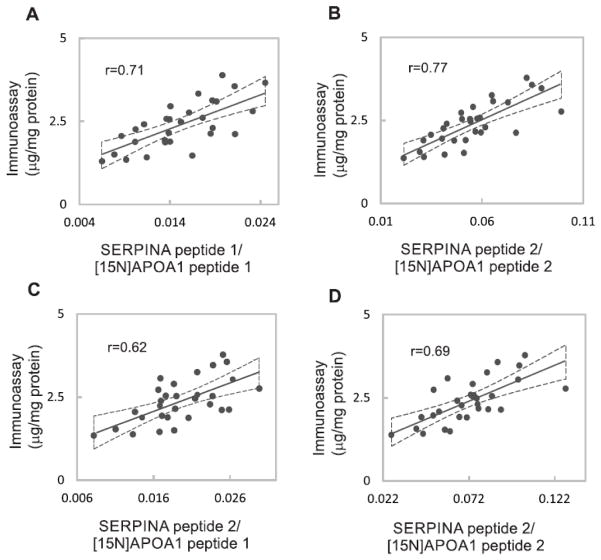
We further tested our method by comparing the results obtained by PRM and [15N]APOA1 peptides with an immunoassay for the protein SERPINA1 in 30 clinical samples (Fig. 1, workflow). Two different SERPINA1 peptides (SERPINA peptide 1, SVLGQLGITK, and SERPINA peptide 2, LSITGTYDLK) were employed as surrogates of the protein, and normalization was accomplished by using the two [15N]APOA1 peptides as described above. Fig. 6 shows the correlation between the immunoassay (μg/mg protein) and the following measurements of SERPINA1 obtained by PRM: ratio between the areas of SERPINA peptide 1 and [15N]APOA1 peptide 1(Fig. 6A); ratio between the areas of SERPINA peptide 1 and [15N]APOA1 peptide 2 (Fig. 6B); ratio between the areas of SERPINA peptide 2 and [15N]APOA1 peptide 1 (Fig. 6C); ratio between the areas of SERPINA peptide 2 and [15N]APOA1 peptide 2 (Fig. 6D). All the PRM measurements of the protein correlated relatively well with the results obtained by the immunoassay (r = 0.62–0.77), providing further evidence of the validity of our approach for quantifying proteins in HDL.
Figure 6.
Comparison of SERPINA1 quantification using immunoassay or PRM/[15N]APOA1 peptides. (A) Comparison of immunoassay (μg/mg protein) and PRM quantification using the ratio between the areas of SERPINA peptide 1 and [15N]APOA1 peptide 1 ([15N]DYVSQFEGSALGK). (B) Comparison of immunoassay (μg/mg protein) and quantification by the areas ratio of SERPINA peptide 1 and [15N]APOA1 peptide 2 ([15N]VQPYLDDFQK). (C) Comparison of immunoassay (μg/mg protein) and quantification by the areas ratio of SERPINA peptide 2 and [15N]APOA1 peptide 1([15N]DYVSQFEGSALGK). (D) Comparison of immunoassay (μg/mg protein) and quantification by the areas ratio of SERPINA peptide 2 and [15N]APOA1 peptide 2 ([15N]VQPYLDDFQK). The Pearson correlation coefficients (r) are shown. Dashed lines represent the 95% confidence interval of the measurements.
Discussion
This study aimed to test the accuracy and sensitivity of PRM coupled to a single internal standard as an efficient approach for relative quantification of HDL proteins. Ideally, adopting PRM should improve throughput without compromising sensitivity and precision, which are necessary for quantitative studies. To this end, we systematically compared both SIM and PRM with SRM. We first created a standard curve for [15N]APOA1, using HDL as a biological matrix, as other proteins in the HDL proteome are potential confounders for quantifying labeled APOA1 peptides. For all of the 12 APOA1 peptides we quantified, SRM and PRM exhibited comparable linearity, limits of quantification, and precision. As assessed by these metrics, SIM was inferior to the other two methods, likely due to the superior selectivity conferred on SRM and PRM by a second stage of MS [12]. SIM relies on single-stage MS, in which precursor ions with a limited range of m/z are isolated in the quadrupole and analyzed with high resolution in an Orbitrap mass analyzer. Actual quantification is performed on peak areas of precursor ions, and no fragmentation is involved [12]. Although straightforward, the SIM mode is more likely to have interferences, especially when complex matrices are involved, because it lacks the second-stage filter when it monitors precursor/product ion pairs.
In contrast to SIM, both PRM and SRM fragment the quadrupole-selected precursor peptide ion to generate product ions that are used to confirm the peptide’s identity and provide an additional level of specificity. In SRM, a predefined and experimentally validated set of precursor/product pairs is acquired [33]. Similar to SRM, PRM requires selection of a set of target peptides, but unlike for SRM all products of a selected precursor ion are monitored in parallel to confirm and quantify the peptide. This eliminates the substantial time and effort required to optimize the best transitions and collision energy for each peptide. These steps are especially challenging when starting without a standard protein or when transferring discovery analyses from an ion trap instrument to a triple quadrupole instrument [10, 12]. Moreover, the low resolution of the QqQ can result in co-isolation of interfering ions, especially from complex biological samples.
Measuring several transitions for each peptide improves selectivity, though the number of predefined transitions measured during one acquisition cycle is limited to maintain a practical dwell time [4]. While mass selection on the Q-OT instrument also has low resolution, the high resolution and mass accuracy of the Orbitrap mass analyzer (< 3 ppm) used to analyze product ions significantly increases selectivity during data analysis [10, 12]. Importantly, our observations clearly demonstrate that PRM and SRM exhibit similar linearity, accuracy, and sensitivity when quantifying standard curves of [15N]APOA1 in the complex HDL matrix. These observations suggest that PRM can indeed rapidly quantify candidate proteins in a complex biological system.
We also determined whether other proteins could be quantified as precisely if PRM used a single labeled protein rather than individual synthetic peptides for each peptide. A previous study validated this approach for HDL, using [15N]APOA1 as a global internal standard to correct for digestion efficiency and run-to-run variability in the MS analyses [7]. Importantly, that study showed that [15N]APOA1 peptides performed as well as or better than the corresponding labeled peptide for relative quantification of 6 different HDL proteins (APOA1, APOB, APOC2, APOC3, APOE, and APOJ) [7]. In the current study, we used PRM to quantify three other proteins: a serpin peptidase inhibitor (SERPINA1), phospholipid transfer protein (PLTP), and a paraoxonase (PON3). For all three, [15N]APOA1 peptides corrected for variability as well as the corresponding labeled peptides, again suggesting that a single protein (stable isotope-labeled or exogenous to the sample analyzed) can serve as an internal standard for relative quantification of proteins in complex systems.
To compare the performances of PRM and SRM with clinical samples, we determined how well the two methods quantify multiple HDL proteins that vary widely in abundance. Across 44 subjects, PRM and SRM yielded highly similar quantitative results, regardless of the [15N]APOA1 peptide used as internal standard. We also obtained good correlations when we compared an immunoassay of SERPINA1 in HDL with quantification by PRM and [15N]APOA1 peptides. These results validate our method for quantifying proteins in HDL.
We note several potential limitations of our work. First, because the study aimed to compare PRM and SRM, we did not evaluate batch-to-batch reproducibility using different trypsin batches, nor did we examine CVs for multiple nonconsecutive days of analysis. To implement routine quantification of HDL proteins for clinical investigations, in future studies it will be important to address these issues. Second, we did not obtain absolute quantification of HDL proteins. Our studies with relative quantification of SERPINA1 by PRM and immunoassay showed that the two methods are well correlated: however, neither method provides absolute quantification [34]. We are aware correlation measures the strength of a relation between two variables, not the agreement between them [34]. Nonetheless, the differences in units of measurement precluded an agreement analyses. As an alternative, the 95 % confidence intervals of the regression between the two measurements were provided (Fig. 6).
One motivation for our study was our long-term goal of devising metrics that better capture HDL’s cardioprotective effects than its cholesterol content, the currently used metric. Recent studies demonstrate that HDL carries a wide range of proteins in addition to APOA1 and APOA2, many of which have biological effects that could play significant roles in HDL’s biology [14, 27, 35, 36]. Importantly, those proteins are much less abundant in HDL than APOA1 and APOA2, which makes their quantification more challenging. The availability of a method that increases sample throughput without compromising the reproducibility, sensitivity, and accuracy could therefore accelerate biomarker discovery. In this study, the remarkable concordance between PRM and SRM strongly supports the proposal that PRM, in concert with a single labeled protein as internal standard, might offer an alternative approach to prioritizing and verifying candidate protein biomarkers in HDL, a complex biological matrix implicated in cardioprotection.
Supplementary Material
Biological significance.
HDL, a complex matrix composed of lipids and proteins, is implicated in cardioprotection. Its cholesterol content correlates inversely with cardiovascular disease and it is the current metric to assess cardiovascular risk. However, the cholesterol content does not capture HDL’s complexity and heterogeneity. Devising metrics that better capture HDL’s cardioprotective effects, we developed an optimized method for quantification of HDL proteome, using PRM in concert with a single labeled protein as internal standard. The availability of a method that increases sample throughput without compromising the reproducibility, sensitivity, and accuracy could therefore point to better risk assessment for CVD or other diseases.
Highlights.
Development of an efficient approach to analyze HDL proteins in clinical samples
PRM showed to be as effective as SRM in quantifying HDL proteins
A single protein as internal standard performed as well as protein-specific peptides
Acknowledgments
Mass spectrometry experiments were performed in the University of Washington’s Proteomics Resource (UWPR95794) and the Quantitative and the Functional Proteomics Core of the Diabetes Research Center, University of Washington. This work was supported by grants from the American Heart Association (14POST18620020) and the National Institutes of Health (HL086798, HL091055, HL092969, HL108897, HL112625). None of the sponsors had any role in study design, data analysis, or reporting of the results.
Abbreviations
- [15N]APOA1
15N-labeled Apolipoprotein A-I
- APOA1
Apolipoprotein A-I
- APOA2
Apolipoprotein A-II
- CVD
cardiovascular disease
- HCD
high-energy collisional dissociation
- HDL
high density lipoprotein
- LLOQ
lower limit of quantification
- MRM
multiple reaction monitoring
- PRM
parallel reaction monitoring
- Q-OT
quadrupole Orbitrap
- QqQ
triple quadrupole
- SIM
selected ion monitoring
- SRM
selected reaction monitoring
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jaffe JD, Keshishian H, Chang B, Addona TA, Gillette MA, Carr SA. Accurate inclusion mass screening: a bridge from unbiased discovery to targeted assay development for biomarker verification. Mol Cell Proteomics. 2008;7:1952–62. doi: 10.1074/mcp.M800218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith RD. Mass spectrometry in biomarker applications: from untargeted discovery to targeted verification, and implications for platform convergence and clinical application. Clin Chem. 2012;58:528–30. doi: 10.1373/clinchem.2011.180596. [DOI] [PubMed] [Google Scholar]
- 3.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular Syst Biol. 2008;4:1–14. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gallien S, Duriez E, Demeure K, Domon B. Selectivity of LC-MS/MS analysis: Implication for proteomics experiments. J Proteomics. 2013;81:148–58. doi: 10.1016/j.jprot.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat Biotechnol. 2009;27:633–41. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun V, Dupuis A, Adrait A, Marcellin M, Thomas D, Court M, et al. Isotope-labeled protein standards: toward absolute quantitative proteomics. Mol Cell Proteomics. 2007;6:2139–49. doi: 10.1074/mcp.M700163-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Hoofnagle AN, Becker JO, Oda MN, Cavigiolio G, Mayer P, Vaisar T. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clin Chem. 2012;58:777–81. doi: 10.1373/clinchem.2011.173856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallien S, Bourmaud A, Kim SY, Domon B. Technical considerations for large-scale parallel reaction monitoring analysis. J Proteomics. 2013;4:00544–7. doi: 10.1016/j.jprot.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 9.Law KP, Lim YP. Recent advances in mass spectrometry: data independent analysis and hyper reaction monitoring. Expert Rev Proteomics. 2013;10:551–66. doi: 10.1586/14789450.2013.858022. [DOI] [PubMed] [Google Scholar]
- 10.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11:1475–88. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schiffmann C, Hansen R, Baumann S, Kublik A, Nielsen PH, Adrian L, et al. Comparison of targeted peptide quantification assays for reductive dehalogenases by selective reaction monitoring (SRM) and precursor reaction monitoring (PRM) Anal Bioanal Chem. 2014;406:283–91. doi: 10.1007/s00216-013-7451-7. [DOI] [PubMed] [Google Scholar]
- 12.Gallien S, Duriez E, Crone C, Kellmann M, Moehring T, Domon B. Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol Cell Proteomics. 2012;11:1709–23. doi: 10.1074/mcp.O112.019802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Havel RJ, Eder HA, Bragdon JH. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J Clin Invest. 1955;34:1345–53. doi: 10.1172/JCI103182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davidson WS, Silva RA, Chantepie S, Lagor WR, Chapman MJ, Kontush A. Proteomic analysis of defined HDL subpopulations reveals particle-specific protein clusters: relevance to antioxidative function. Arterioscler Thromb Vasc Biol. 2009;29:870–6. doi: 10.1161/ATVBAHA.109.186031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah AS, Tan L, Long JL, Davidson WS. Proteomic diversity of high density lipoproteins: our emerging understanding of its importance in lipid transport and beyond. J Lipid Res. 2013;54:2575–85. doi: 10.1194/jlr.R035725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics--2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khera AV, Cuchel M, de la Llera-Moya M, Rodrigues A, Burke MF, Jafri K, et al. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. New Engl J Med. 2011;364:127–35. doi: 10.1056/NEJMoa1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rader DJ, Tall AR. The not-so-simple HDL story: Is it time to revise the HDL cholesterol hypothesis? Nat Med. 2012;18:1344–6. doi: 10.1038/nm.2937. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz GG, Olsson AG, Abt M, Ballantyne CM, Barter PJ, Brumm J, et al. Effects of dalcetrapib in patients with a recent acute coronary syndrome. New Engl J Med. 2012;367:2089–99. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 20.Gillette MA, Carr SA. Quantitative analysis of peptides and proteins in biomedicine by targeted mass spectrometry. Nat Methods. 2013;10:28–34. doi: 10.1038/nmeth.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–66. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 22.Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, et al. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat Methods. 2010;7:43–6. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 23.Mohammed Y, Domański D, Jackson AM, Smith DS, Deelder AM, Palmblad M, et al. PeptidePicker: A scientific workflow with web interface for selecting appropriate peptides for targeted proteomics experiments. J Proteomics. 2014;106:151–61. doi: 10.1016/j.jprot.2014.04.018. [DOI] [PubMed] [Google Scholar]
- 24.Ryan RO, Forte TM, Oda MN. Optimized bacterial expression of human apolipoprotein A-I. Protein Expr Purif. 2003;27:98–103. doi: 10.1016/s1046-5928(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 25.Jarvik GP, Hatsukami TS, Carlson C, Richter RJ, Jampsa R, Brophy VH, et al. Paraoxonase activity, but not haplotype utilizing the linkage disequilibrium structure, predicts vascular disease. Arterioscler Thromb Vasc Biol. 2003;23:1465–71. doi: 10.1161/01.ATV.0000081635.96290.D3. [DOI] [PubMed] [Google Scholar]
- 26.Mendez A, Oram J, Bierman E. Protein kinase C as a mediator of high density lipoprotein receptor- dependent efflux of intracellular cholesterol. J Biol Chem. 1991;266:10104–11. [PubMed] [Google Scholar]
- 27.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117:746–56. doi: 10.1172/JCI26206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 29.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 30.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–8. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almeida AM, Castel-Branco MM, Falcão AC. Linear regression for calibration lines revisited: weighting schemes for bioanalytical methods. J Chromatogr B. 2002;774:215–22. doi: 10.1016/s1570-0232(02)00244-1. [DOI] [PubMed] [Google Scholar]
- 32.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol Cell Proteomics. 2007;6:2212–29. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domon B. Considerations on selected reaction monitoring experiments: Implications for the selectivity and accuracy of measurements. Proteomics – Clin Appl. 2012;6:609–14. doi: 10.1002/prca.201200111. [DOI] [PubMed] [Google Scholar]
- 34.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–10. [PubMed] [Google Scholar]
- 35.Alwaili K, Bailey D, Awan Z, Bailey SD, Ruel I, Hafiane A, et al. The HDL proteome in acute coronary syndromes shifts to an inflammatory profile. Biochim Biophys Acta. 2012;3:405–15. doi: 10.1016/j.bbalip.2011.07.013. [DOI] [PubMed] [Google Scholar]
- 36.Yassine HN, Jackson AM, Borges CR, Billheimer D, Koh H, Smith D, et al. The application of multiple reaction monitoring and multi-analyte profiling to HDL proteins. Lipids Health Dis. 2014;13:13–8. doi: 10.1186/1476-511X-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.