Abstract
Proteins involved in iron regulation are modifiers of cancer risk and progression. Of these, the HFE protein (high iron gene and its protein product) is of particular interest because of its interaction with both iron handling and immune function and the high rate of genetic polymorphisms resulting in a mutant protein. Clinical studies suggest that HFE polymorphisms increase the risk of certain cancers, but the inconsistent outcomes suggest a more nuanced effect, possibly interacting with other genetic or environmental factors. Some basic science research has been conducted to begin to understand the implications of variant HFE genotype on cancer, but the story is far from complete. In particular, putative mechanisms exist for HFE to affect tumor progression through its role in iron handling and its major histocompatibility complex class I structural features. In this review, the current understanding of the role of HFE in cancer is described and models for future directions are identified.
Keywords: iron, HFE, hemochromatosis, overload, tumor progression
Introduction
Cancer is one of the leading causes of mortality in the United States.1 Tumors require iron and have increased ferritin and transferrin receptor 1 (TfR1) expression compared to normal tissue and stroma.2 Intracellularly, iron regulates several key cell cycle proteins including p53, p27kip1, cyclin D1, cdk1, and p21CIP1/WAF1.3–5 Proteins involved in iron regulation are modifiers of cancer risk and progression. For example, in clinical specimens, overexpression of iron import proteins was associated with increased progression of colorectal cancer6 and malignant progression of esophageal adenocarcinoma and risk of metastasis.7 In breast cancer, cell lines with aggressive, mesenchymal phenotypes expressed more ferritin, transferrin, TfR, and iron regulatory proteins compared to less aggressive lines.8 We have reported that elevated ferritin in serum is associated with poor prognosis in breast cancer and that ferritin may have a direct impact on the tumor.9 In addition, ferroportin levels are linked to tumor progression in breast cancer such that impaired iron export is linked to a more aggressive phenotype.10 Another study in MAL-12 murine hepatocytes showed that transforming growth factor β1, a known promoter of tumor aggression, induces epithelial to mesenchymal transition (EMT) through inhibition of ferritin heavy chain, increasing the labile iron pool and generating reactive oxygen species (ROS).11 EMT is a widely studied phenomenon believed to be associated with the metastatic progression of tumor cells as well as certain developmental migratory processes. We have recently reported that decreased expression of H-ferritin in glioblastoma multiforme increases the vulnerability of these cells to therapeutic strategies.12 In the field of metastasis, the protein n-myc–downregulated gene 1, the iron-related metastasis suppressor, links iron metabolism to cancer progression.13 The intent of the present review is to describe the relationship between cancer progression and the most common gene variant in Caucasians: HFE (high iron protein). The HFE protein is involved in iron regulation and immune system function and thus sits at the intersection of two major physiological processes in cancer biology. HFE polymorphisms are extremely common in the United States, with prevalence of 5.4% for C282Y and 13.5% for H63D and higher numbers in Caucasians (6.2% and 15.1%, respectively).14 Because of the prevalence of these variants, focusing on HFE provides the possibility of understanding the special risks and treatment considerations that may apply to cancer patients who are carriers of variant HFE alleles.
The HFE protein
The HFE protein is an atypical class I major histocompatibility complex (MHC) molecule that acts on iron homeostasis through its interaction with β2 microglobulin (β2 m) and the TfRs (1 and 2). Normal HFE function limits iron intake into the cell, preventing iron overload (Fig. 1). The mutant form of this protein has long been thought to be related to hereditary hemochromatosis (HH) in humans,15 although the penetrance of HH relative to the gene variant is very low.16 The H63D polymorphism translocates to the cell surface but fails to participate in the interactions with the TfR1, and the C282Y polymorphism cannot interact with β2 m, preventing its surface translocation.17 An illustration of the normal function of HFE in a typical parenchymal cell is given in Figure 2. Furthermore, HFE polymorphisms have been shown to directly affect immune function. For example, peripheral blood mononuclear cells from patients with variant HFE expression have lower levels of surface MHC I molecule expression, which are important presentation molecules affecting leukocyte activation in cancer.18–20
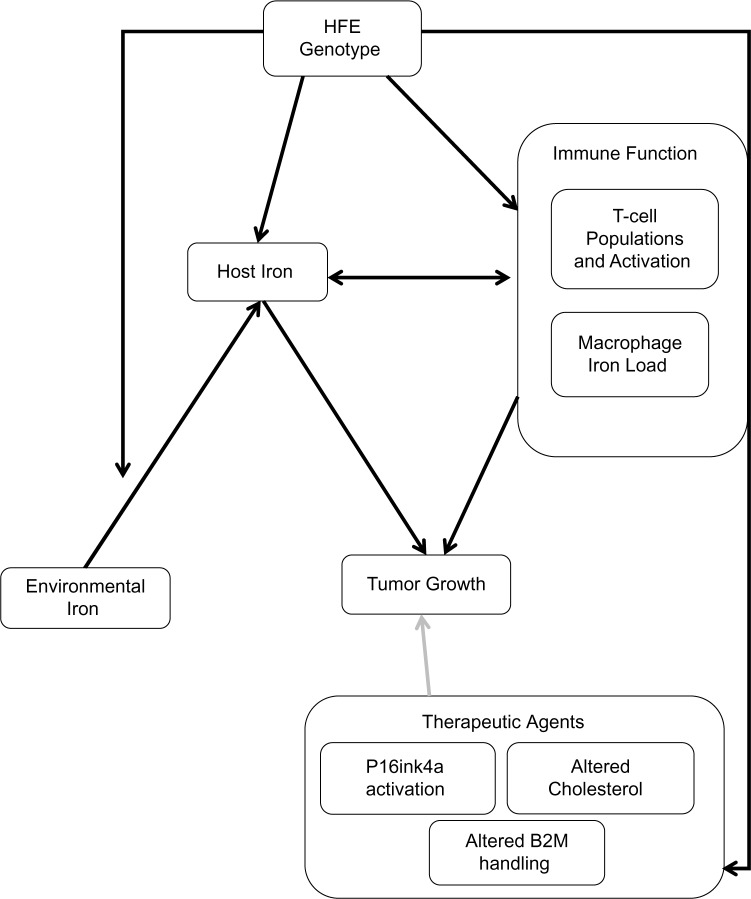
Figure 1.
Conceptual framework of HFE as it relates to cancer. HFE genotype has been shown to affect both iron load and immune function. Although the relative importance of each cell type remains to be tested, some combination of these factors appears to affect tumor growth across a variety of cancer types. In addition, it is expected that HFE interacts with environmental factors, particularly dietary iron, to yield the observed effect on cancer risk and tumor progression. Lastly, HFE variants have been shown to affect the handling of β2m, the response to cholesterol-altering medications, and the levels of P16ink4a. These findings strongly suggest that HFE mutations will affect the efficacy of therapeutic interventions. Therefore, taking HFE genotype into account may one day provide valuable information on prognosis and could lead to more efficient application of pharmacologic interventions.
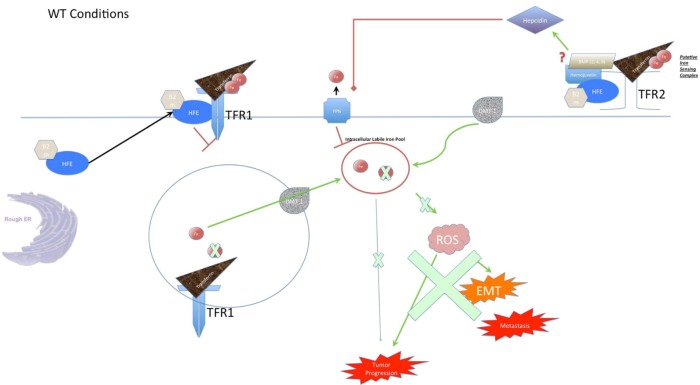
Figure 2.
Function of the HFE protein in iron metabolism. When HFE is functioning normally, it associates with β2m and transits to the membrane, where it complexes with TfRs. At TfR1, it competes with transferrin to limit the rate of iron uptake into the cell, promoting a homeostatic level of iron load. Iron is taken into the cell when the entire TfR1 complex is endocytosed, and the pH of the compartment is acidified to the point where the iron dissociates from transferrin. From here, iron can pass through divalent metal transporter 1 (DMT-1), where it enters the cytoplasm. From here, the iron storage protein ferritin normally sequesters the free iron to limit its unwanted reactivity (not shown). For completeness, the diagram also shows the normal role of HFE in forming a complex with TfR2, which is thought to perform whole-body iron sensing in hepatocytes. In this way, host HFE is partially responsible for the proper secretion of Hepcidin, which suppresses the iron export protein ferroportin and encourages cells to retain iron intracellularly. Importantly, the regulation of iron load by HFE prevents an excess of ROS (generated via the Fenton reaction), which in turn are linked to EMT and metastasis. Furthermore, the limitation of iron supply prevents the cell from properly undergoing the cell division and metabolism that a progressing tumor would demand. Disruption of HFE function in the case of variant H63D genotype leads to iron overload because the HFE is unable to perform its normal function at TfR1. This leads to iron overload in the cell, which can lead to tumor progression and metastasis through increased stress on the endoplasmic reticulum, generation of ROS, and the increased availability of iron for cell metabolism and division. As the diagram indicates, this excess of ROS causes cellular damage and promotes EMT, leading to greater metastatic propensity. In addition, the decrease in interaction with TfR1 is thought to promote an interaction with TfR2 in hepatocytes, causing a systemic increase in hepcidin and suppression of ferroportin. Disruption of HFE function in the case of variant C282Y has similar effects to H63D in many ways. The key difference is that the mutation prevents the association of HFE with β2m, which prevents it from localizing to the cell surface at all. This would be expected to have similar effects on TfR1, since there is a loss of function in both cases. However, several lines of evidence show that the two mutations have divergent effects, so there must be a more nuanced effect on the cell than a simple loss of TfR1 blockade.
An exploration of the relationship between HFE genotype and cancer must, of course, include a consideration of the role of iron in cancer. The relationship is considered complex and is incompletely understood, yet iron metabolism and cancer progression are clearly linked.21 While a detailed examination of the relationship between iron and cancer is beyond the scope of this review, the authors refer the reader to previous reviews on the subject and supply here a cursory overview.22,23 The favored mechanism for linking iron and cancer is through the generation of ROS, which in turn induce a more aggressive tumor phenotype.24,25 Supporting this mechanism, it has been shown that HFE knockout mice are at greater risk for oxidative damage, and the intake of iron exacerbates this phenomenon.26 However, we will explore this concept in greater detail and provide evidence that the mutant HFE protein itself may impose altered cellular phenotype that alters cancer biology.
The concept that HFE mutant protein may directly impact cancer phenotype would not exclude a relationship with iron. In clinical studies, iron loading has shown a modest association with increased overall cancer risk, and specifically, lung cancer risk.8,27 In addition, patients with HH, who experience severe iron overload, showed a risk of liver cancer over two hundred times greater than matched controls.28 Conversely, regular blood donors, who have diminished iron load, have decreased overall rates of cancer.29 Studies of spontaneous tumorigenesis in rats found that iron loading led to increased rates of tumor formation.30–32 By the same token, feeding mice an iron-deficient diet results in smaller tumors in a xenograft model using three different cancer cell lines.33 Experimentally, the iron chelator deferoxamine blocks the proliferation of glioma and neuroblastoma cells, supporting the manipulation of iron balance as a therapeutic approach in some cancers.34,35 In cell culture, iron loading in Caco-2 and SW480 cells led to increased proliferation and loss of E-cadherin expression, suggesting EMT.6 More recently, a 2011 study in several cell culture models found that suppression of HFE or β2m, which binds to HFE and facilitates trafficking, caused a reversal of EMT.36
Taken together, these studies highlight how the proteins of iron metabolism can modify tumor aggression and are thus potential targets of interest in the development of therapeutics. Although both H63D and C282Y fail to produce a properly functioning protein and are associated with increased iron uptake, cell culture studies have shown that these variants can impact cancer phenotype in dramatically different ways,37 supporting the proposition in this review that the HFE impact on cancer biology is not limited to its impact on iron homeostasis.
While much of the focus on HFE and cancer has been on its connection to iron metabolism, as an MHC molecule, it is not surprising that HFE interacts with the immune system. For example, patients with C282Y homozygous genotype have been shown to have significantly lower total lymphocytes and CD4+ lymphocytes specifically compared to control subjects.38 Also, HFE knockout mice have been shown to induce expression of lipocalin 2 (LCN2), an immune-signaling molecule.39 Given that LCN2 has been implicated in tumorigenesis of leukemias and both tumorigenesis and metastasis in breast tumors, this is one possible mechanism for HFE to modulate tumor aggression.40–42 The interaction of HFE with β2m may also have immunological consequences.43,44 Given that β2m in the cerebrospinal fluid is associated with leptomeningeal spread of cancers, a relationship between central nervous system metastasis and HFE is feasible.45 Lastly, analysis of patient plasma samples using a biochip array analyzer revealed decreased L-selectin, increased E-selectin, and increased intracellular adhesion molecule 1 in HH patients, suggesting that immune response is affected in the context of HFE mutation by way of altered adhesion molecules.46 This is important to the progression of cancer in two ways: First, any disruption in the migration of leukocytes to the tumor would be expected to affect the tumor microenvironment and limit the ability of these cells to influence tumor growth. Second, at the metastatic stage, circulating tumor cells have been shown to mimic leukocyte behavior and utilize the selectin-mediated leukocyte adhesion cascade to infiltrate tissues.47–49
In addition to the impairment of protein function, HFE mutations may also alter cell biology due to the stress of producing and breaking down large quantities of extraneous protein. In our cell culture and animal models, the C282Y and H63D mutations have both been shown to create ER stress and activate the unfolded protein response (UPR).50,51 In addition, C282Y HFE has been shown to create intracellular aggregates, which could affect cell signaling and survival through the binding sites on these aggregates.50 UPR is known to affect cellular apoptosis and may contribute to promoting the survival of tumor cells.52 Another mechanism by which HFE mutation–induced UPR could affect tumor spread is through effects on MHC I expression at the cell surface, altering the immune response to the cancer cells.53
Specifically, HFE C282Y mutant patients had peripheral blood mononuclear cells with reduced surface expression of β2m-assembled MHC class I molecules and increased free MHC class I molecules. Within the cell, the C282Y mutants had fewer free class I MHCs and more β2m-assembled class I molecules.18
In support of the ability of HFE to affect the immune system, it has been found that hepcidin response to inflammation depends on HFE and the TfR2.54 Hepcidin normally sequesters iron in response to inflammation, which limits access to iron by microbes and should alter iron availability for neoplasms. In mice with knocked down HFE, TfR2, or both, the levels of hepcidin and the downstream iron sequestration were hindered.
Although this review focuses on HFE polymorphisms because of their high prevalence in the general population, there is also evidence that the HFE gene undergoes copy number alterations in many cancers.55,56 If these copy number alterations affect mRNA levels of Wild Type (WT) or polymorphic HFE, the evidence suggests that they will affect cellular iron handling and immune presentation. Specifically, increased levels of WT HFE would be expected to limit iron uptake in cancer cells and decreased levels would lead to iron overload. This question has not been specifically studied, and future investigation would provide insight into the ability of HFE to affect tumor progression.
The effects of HFE on tumor progression have the potential to affect clinical practice not only as a risk factor but also HFE appears to modulate the response of cancers to standard therapeutic regimens. The population studies we will review in the rest of this document will focus on evaluating if HFE genotype is a risk factor for various cancers, only because there is a considerable knowledge gap on the impact of HFE genotype on therapeutic response. There is enough evidence to suggest that HFE is capable of affecting therapeutic response. For example, a 2006 study showed that expression of HFE reduces the effect of doxorubicin on proliferation in breast cancer cells.57 In addition, a 2011 study conducted in our laboratory revealed that the C282Y mutation in HFE induced an elevation in p16INK4A, which was associated with treatment resistance in both neuroblastoma and astrocytoma cell lines.58 One potential mechanism to explore further is the interaction between β2m and HFE, because β2m manipulation has been shown to affect therapeutic response.59 Another mechanism by which HFE is likely to affect tumor progression and therapeutic response is by its effects on cholesterol and sphingolipid metabolism.60 In particular, it is noteworthy that neuroblastoma cells with the C282Y variant of HFE exhibit improved survival in the presence of sphingosine kinase inhibitors, as this mechanism is already recognized as a chemotherapeutic strategy. Given the prevalence of HFE polymorphisms in the population, in the emerging era of personalized medicine, considering HFE variants has a strong likelihood to improve patient care.
Clinical Studies
The data on whether HFE mutations are clinically important in cancer progression are quite variable, suggesting that non-genetic factors are affecting the penetrance of HFE genotype. HFE has shown to affect response to environmental stimuli in other contexts, so there is reason to believe that its effects would interact with the environment in tumor progression (n = 518).61 Intuitively, dietary habits and iron content of drinking water could impact iron availability. In support of this idea, women have onset of iron overload about two years later than men in cases of hemochromatosis, and phlebotomy appears to be protective against the development of cancer, suggesting that removal of iron in blood is a significant modifier of these processes.29,62 In breast cancer, Abraham and colleagues found no effect of H63D or C282Y on incidence, although a trend was seen toward more C282Y alleles in aggressive cases (n = 688 cancer patients, 724 matched controls).63 In cervical cancer, H63D has been found to be associated with a lower risk (n = 346 samples, 201 with cervical cancer).64 In contrast to these findings, H63D has been found to be associated with an increased risk of gastric cancer and with an increased risk of hepatocellular carcinoma (HCC; n = 365 gastric adenocarcinoma with 1284 matched controls, n = 100 cirrhosis patients, 100 HCC patients, and 100 controls).65,66 A study comparing 100 consecutive oncology patients in central Alabama with 318 healthy controls found no significant differences in HFE variant allele frequency between the two populations.67 This study argues against a large effect across several cancer types, but it was not sufficiently powered to detect more subtle differences in specific cancers. As the studies below will show, the literature shows both positive and negative results, either an absence of effect or an exacerbation of cancer risk or severity with HFE variant genotype. Thus, these data suggest there is a relationship between HFE genotype and cancer. The clinical studies evaluating the relationship between H63D and cancer have been summarized in Table 1, and the studies evaluating C282Y and cancer have been summarized in Table 2.
Table 1.
Clinical studies of H63D and cancer have found, in general, either no effect or an increased risk of cancer for patients with H63D mutations. The finding that H63D appears to be protective in cervical cancer stands out and suggests a need for further study.
| CANCER | REGION | H63D FINDING | SOURCE |
|---|---|---|---|
| Adult acute leukemia | Italy | No differences in frequency compared to general population | Veneri et al., 2005 |
| ALL | United Kingdom | Increased Risk of childhood ALL in females | Dorak et al., 2009 |
| ALL (Childhood) | Spain | No association between HFE variant Alleles and Childhood ALL | Rodriguez-Lopez et al., 2013 |
| AML | Spain | No differences in frequency compared to blood donor controls | Gimferrer et al., 1999 |
| AML | Finland | No significant differences in HFE allele frequency compared to general population | Hannuksela et al., 2002 |
| Breast Cancer | Germany | No differences in frequency compared to general population | Abraham et al., 2005 |
| Breast Cancer | Turkey | Significantly increased frequency of H63D in breast cancer patients compared to controls | Gunzel-Ozcan et al., 2006 |
| Cervical Cancer | Portugal | Decreased Risk, later onset of cervical lesions | Cardoso et al., 2006 |
| Colorectal Cancer | USA | No increased risk for colorectal adenoma in women | Chan et al., 2005 |
| Colorectal cancer | Spain | No significant difference in HFE allele frequency compared to blood donor controls | Altes et al., 1999 |
| Colorectal cancer | Poland and Australia | H63D homozygosity associated with significantly increased risk of colorectal cancer and earlier age of onset in patients with mismatch repair gene mutations | Shi et al., 2009 |
| Colorectal cancer | USA | Significant increase in the risk of canger in patients with either HFE allele after adjusting for race, iron intake, red meat consumption, and NSAID use. | Shaheen et al., 2003 |
| Colorectal cancer, breast cancer in women, prostate cancer | Australia | No increased risk noted | Osborne et al., 2010 |
| Gastric Cancer | Europe (10 countries) | Increased Risk (Particularly Non-cardia site, Intestinal Histologic Subtype) | Agudo et al., 2013 |
| Glioma | Italy | Significantly higher rate of H63D in high-grade gliomas compared to controls | Martinez di Montemuros et al., 2001 |
| Hepatocellular Carcinoma | Italy | No increase in HFE mutations in HCC | Racchi et al., 2002 |
| Hepatocellular Carcinoma | Sweden | 20 fold increase in rates* | Elmberg et al., 2003 |
| Hepatocellular Carcinoma | Spain | All H63D homozygotes did not have cirrhosis | Lauret et al., 2002 |
| Hepatocellular Carcinoma | Egypt | Increased frequency of H63D in HCC cases compared to the general population in both alcoholic cirrhosis and hepatis C cohorts | Gharib et al., 2011 |
| Hepatocellular Carcinoma | France | No significant diffrences in allele frequency between patients with cirrhosis and HCC and patients with cirrhosis without HCC | Boige et al., 2003 |
| Hepatocellular Carcinoma | Spain | H63D was associated with risk of HCC in cirrhotic patients | Ropero et al., 2007 |
| Male Breast, Prostate | Finland | No significant differences in H63D frequency compared to general population | Syrjakoski et al., 2006 |
| Non-small-cell lung cancer | USA (Cooperative Human Tissue Network) | No evidence that NSCLCs select for HFE mutation | Muller et al., 2005 |
| Several Types | Alabama, USA | No significant differences in HFE variant allele frequency compared to healthy controls | Barton et al., 2004 |
Note:
Elmberg et al94 only examined patients with HH and did not assess which HFE polymorphisms were present.
Table 2.
Clinical studies of C282Y and cancer similarly show either no effect or increased risk of cancer for patients with C282Y mutations. Going forward, controlled studies in disease models will help separate HFE effects from other genetic and environmental influences.
| CANCER | REGION | C282Y FINDING | SOURCE |
|---|---|---|---|
| Adult acute leukemia | Italy | No differences in frequency compared to general population | Veneri et al., 2005 |
| ALL | United Kingdom | Increased Risk of childhood ALL in females | Dorak et al., 2009 |
| ALL (Childhood) | United Kingdom | Male-specific association between C282Y and ALL | Dorak et al., 1999 |
| ALL (Childhood) | Spain | No association between HFE variant alleles and Childhood ALL | Rodriguez-Lopez et al., 2013 |
| ALL (Childhood) | Poland | Examination of polymorphisms in the chromosomal region near HFE considerably weakened the power of the C282Y association with Childhood ALL. Further studies are needed | Sikorska et al., 2011 |
| AML | Spain | No differences in frequency compared to blood donor controls | Gimferrer et al., 1999 |
| AML | Finland | No significant differences in HFE allele frequency compared to general population | Hannuksela et al., 2002 |
| Breast Cancer | Germany | No differences in frequency compared to general population, greater frequency in patients with lymph node involvement. | Abraham et al., 2005 |
| Breast Cancer | Tenessee | Increased prevalence of C282Y mutations in breast cancer cases compared to healthy controls | Kallianpur et al., 2004 |
| Cervical Cancer | Portugal | Not associated (no increase or decrease) | Cardoso et al., 2006 |
| Colorectal and Primary Liver Cancer | Norway | Increased risk of colorectal cancer and primary liver cancer in C282Y homozygotes. | Asberg et al., 2013 |
| Colorectal Cancer | Australia | Heterozygosity not associated with risk, site, or tumor stage | Macdonald et al., 1999 |
| Colorectal Cancer | USA | No increased risk for colorectal adenoma in women | Chan et al., 2005 |
| Colorectal cancer | Spain | No significant difference in HFE allele frequency compared to blood donor controls | Altes et al., 1999 |
| Colorectal Cancer | Several (Meta-analysis) | C282Y mutation significantly associated with colorectal cancer in caucasians | Chen et al., 2013 |
| Colorectal cancer | USA | Significant increase in the risk of cancer in patients with either HFE allele after adjusting for race, iron intake, red meat consumption, and NSAID use. | Shaheen et al., 2003 |
| Colorectal cancer, breast cancer in women, prostate cancer | Australia | C282Y homozygotes were significantly more likely to contract colorectal cancer and (in females) breast cancer compared to WT individuals (no increased risk for prostate cancer) | Osborne et al., 2010 |
| Hepatocellular Carcinoma | Italy | No increase in HFE mutations in HCC | Racchi et al., 2002 |
| Hepatocellular Carcinoma | Europe | C282Y is associated with hepatocellular carcinoma | Jin et al., 2010 |
| Hepatocellular Carcinoma | Sweden | 20 fold increase in rates* | Elmberg et al., 2003 |
| Hepatocellular Carcinoma | Spain | C282Y mutations were significantly more common in HCC + alcoholic cirrhosis than in alcoholic cirrhosis without HCC | Lauret et al., 2002 |
| Hepatocellular Carcinoma | France | No significant diffrences in allele frequency between patients with cirrhosis and HCC and patients with cirrhosis without HCC | Boige et al., 2003 |
| Hepatocellular Carcinoma | Italy | Increased prevalence of C282Y in HCC patients compared to healthy controls, trends toward an interaction with alcohol exposure and hepatitis virus markers. | Fargion et al., 2001 |
| Hepatocellular Carcinoma | Germany | C282Y heterozygosity was significantly higher in HCC cases than control individuals | Hellerbrand et al., 2003 |
| Hepatocellular Carcinoma | France | C282Y and iron load were associated with HCC risk in alcoholic cirrhotic patients but not HCV-related cirrhotic patients | Nahon et al., 2008 |
| Hepatocellular Carcinoma | Spain | C282Y was not associated with increased risk of HCC in cirrhotic patients | Ropero et al., 2007 |
| Hepatocellular Carcinoma | United Kingdom | C282Y was associated with an increased risk of HCC | Willis et al., 2000 |
| Male Breast, Prostate | Finland | No significant differences in C282Y frequency compared to general population | Syrjakoski et al., 2006 |
| Non-small-cell lung cancer | USA (Cooperative Human Tissue Network) | No evidence that NSCLCs select for HFE mutation | Muller et al., 2005 |
| Ovarian | Canada | Increased both risk and aggressiveness | Gannon et al., 2010 |
| Several Types | Alabama, USA | No significant differences in HFE variant allele frequency compared to healthy controls | Barton et al., 2004 |
Note:
Elmberg et al94 only examined patients with HH and did not assess which HFE polymorphisms were present.
Leukemia
The available evidence suggests a possible effect of HFE genotype on acute leukemias, particularly in childhood. However, more recent studies have found that this interpretation may be influenced by nearby genes that segregate with HFE.
Initially, a 1999 case-control study by Dorak and colleagues in the United Kingdom found a male-specific association between HFE C282Y and childhood acute lymphocytic leukemia (ALL) (n(ALL) = 117, n(control) = 135).68 In contrast, a small study of 36 acute myelocytic leukemia (AML) patients and 108 blood donor controls did not find an increased HFE variant allele frequency in AML patients.69 A later Finnish study found no significant differences in HFE allele frequency in AML, ET, and AML in their populations, although they may have used too few subjects to detect subtle differences (232 total patients).70 Similarly, a 2005study did not find differences in HFE gene variant allele frequencies in adult acute leukemia cases compared with the general population (n = 82).71 However, HFE genotype (H63D and C282Y) has been found to be associated with increased risk of childhood ALL in females (n = 163 ALL, 995 controls).72 It must be noted that the childhood ALL risk finding for C282Y was considerably weakened when it was examined in a more detailed study of polymorphisms in that chromosomal region. This argues for a contribution of nearby HLA genes and may suggest a smaller or noncontributory role for HFE in leukemia, but the animal and basic science data specific to HFE manipulation suggest that it has some effect. A follow-up study suggests that more statistical power is needed to determine if the effect is meaningful (n = 117 ALL, 414 newborn controls).73 Finally, a 2013 study conducted in Spain did not find an association between HFE variant alleles and childhood ALL, which disagreed with earlier findings (see above) (n = 475 patients, 179 controls).74 In toto, the evidence does not strongly support a relationship between HFE genotype and acute leukemias, especially in adults. The childhood effect warrants further investigation, and confounding factors will need to be considered going forward.
Breast Cancer
Breast cancer is a well-studied cancer that has already been linked to iron metabolism and inflammation.9,75 Consistent with this is strong evidence for an effect of HFE variant genotype on breast cancer risk and prognosis. A 2004 study conducted on 168 patients and 169 matched controls in Tennessee showed an increased prevalence for C282Y polymorphisms in breast cancers than in healthy controls.63,76 A 2005 study did not find differences in HFE gene variant allele frequencies in breast cancer cases compared to the general population, but did find that C282Y was associated with greater frequency in patients with greater lymph node involvement (n = 688 patients, 724 matched controls).63 In 2006, a Turkish study compared 176 breast cancer patients to 200 healthy volunteers, finding no cases of C282Y in any of the studied patients and reporting a significantly increased frequency of H63D in breast cancer patients (39/176 versus 28/200, P = 0.02).77 This is a notable example of regional effects on such studies, because an absence of C282Y would be quite rare in a European or North American population but is not surprising in a Middle Eastern population. Although a 2011 study conducted in Brazil found that H63D was associated with p53 mutations in breast cancer, it did not find an increased prevalence of HFE polymorphisms in breast cancer patients or a direct diagnostic or prognostic application for HFE genotype in this context (n = 68 patients, 85 controls).78 A 2013 Norwegian study comparing 292 C282Y homozygotes with 62,568 controls from the HUNT 2 population screening study found that C282Y homozygosity was not associated with an increased risk of breast cancer, although it found increased risks of other cancer types.79 Therefore, the evidence is suggestive of a risk but not totally conclusive, and there may be ethnic differences or gene–environment interactions that explain the discrepancy in results among different studies. In particular, it would be useful to fully understand the effects of dietary iron intake and iron loss through blood donation and menstruation.
Cervical Cancer
Although the research on HFE polymorphism in cervical cancer is limited, it is particularly interesting because it is a rare (perhaps unique) example of HFE variant genotype conferring a protective effect against cancer. Specifically, H63D carriers were at lower risk of developing cervical neoplasia, and they had a later onset of cervical lesions compared to controls. In this study, C282Y was not associated with cervical cancer progression (n = 346 total, 201 with cervical cancer).64 This result suggests that HFE polymorphisms may be able to both antagonize and promote tumor progression depending on the carcinogenic mechanism of cancer. In this case, the strong association between human papilloma virus and cervical cancer may explain the divergent effect of HFE. If HFE H63D is disrupting some aspect of the viral process, it may be acting against the onset of cervical cancer before neoplastic changes ever set in.
Colorectal Cancer
Probably because of the known effects of HFE on intestinal iron absorption, the relationship between HFE genotype and colorectal cancer has been well studied.80 In an Australian case-control study, HFE C282Y heterozygosity was not found in higher frequency in colorectal cancer cases (relative risk = 0.90). There was no association between heterozygosity of C282Y and tumor site or stage, either (n = 229 patients, 228 matched controls).81 The same year, a study comparing 116 colorectal cancer cases to 108 blood donor controls found no significant difference in allelic frequency between the two groups.82 However, an American study examining the relationship between colon cancer and HFE genotype found a significant increase in risk of colon cancer in patients with either H63D or C282Y alleles (odds ratio [OR] 1.4, 95% confidence interval [CI] 1.07–1.87) after controlling for several confounding variables including race, iron intake, red meat consumption, and nonsteroidal anti-inflammatory drug use (n = 475 patients, 833 controls).83
A study in women showed no increased risk for colorectal adenoma with HFE gene polymorphisms (n = 527 cases, 527 matched controls).84 A 2009 study of Polish and Australian cohorts featuring 362 patients with mismatch repair gene mutations found that HFE H63D homozygosity was associated with a significantly increased risk of colorectal cancer (HR = 2.93, P = 0.007) and that H63D homozygous individuals had an earlier age of onset (44 years versus 50 years of age, P < 0.05).85 A large prospective study conducted in Australia derived from the Melbourne Collaborative Cohort Study found that C282Y homozygotes were about twice as likely to contract colorectal cancer (HR 2.28, 95% CI 1.22–4.25, P = 0.01) and, in females, breast cancer (HR = 2.39, 95% CI 1.24–4.61, P = 0.01) compared to individuals with WT HFE. They found no evidence for increased risk of prostate cancer, nor did they see an increased risk of cancers in compound heterozygotes (with H63D and C282Y alleles) (n = 28,509).86
The previously mentioned 2013 Norwegian study found that C282Y homozygosity was associated with an increased risk of colorectal cancer and primary liver cancer in men (colorectal cancer hazard ratio 3.03, hazard ratio 3.03, 95% CI 1.17–7.82, P = 0.022; C282Y liver cancer hazard ratio 54.0, 95% CI 2.68–1089, P = 0.009; n = 292 homozygotes, 62,568 other subjects).79 Most recently, a 2013 meta-analysis showed that the C282Y polymorphism was significantly associated with colorectal cancer in Caucasians (OR 2.00, 95% CI 1.32–3.04) (pooled n = 7588 colorectal cancer cases, 81,571 controls).87 Taken together, the available studies favor the conclusion that HFE polymorphisms are associated with increased risk of colorectal cancer, but this association may be limited to specific populations and thus possibly dietary habits.
Gastric Cancer
Although the evidence is limited, there is reason to believe that the HFE polymorphism H63D is a risk factor for gastric cancer. Specifically, a study conducted within the European Prospective Investigation into Cancer and Nutrition showed an increased risk of gastric cancer, particularly the noncardia site and intestinal histological type, in patients with H63D (n = 365 patients, 1284 controls).65
Glioma
As with gastric cancer, studies examining the link between HFE and glioma are limited. A 2001 study conducted on patients with low- and high-grade gliomas found a significantly higher rate of H63D allele (mostly in the form of heterozygotes) in high-grade gliomas compared to controls (n = 203).88 This study offers the suggestion that further work is warranted.
Hepatocellular Carcinoma
Because liver dysfunction is readily seen in hemochromatosis, HCC was the first cancer to be linked to HFE polymorphism. Therefore, the literature is extensive compared to other cancers and strongly suggests that the C282Y variant increases the risk for HCC. An early study found that C282Y homozygosity was more common than expected in HCC, although the numbers were low and the penetrance of HCC as a phenotype related to C282Y homozygous genotype was low (n = 3600).89 Later, a study conducted in Italy on 12 HCC patients and 130 controls did not find an increased prevalence of HFE polymorphisms in HCC, although 12 patients is too few to detect the effects that other groups reported.90 A study of liver cancer and cirrhosis patients showed that C282Y homozygous genotype occurred significantly more often in both conditions compared to healthy controls. They did not see any differences in the frequency of HFE variant heterozygous genotypes (n = 34 liver cancer, 190 cirrhosis).91 An Italian study conducted in 2001 found an increase in C282Y in HCC patients compared to healthy controls (P < 0.03) and found trends toward an association between C282Y and hepatitis virus markers and alcohol exposure, suggesting that C282Y could be interacting with these environmental factors to increase HCC risk (n = 81).92 A 2002 study of patients with alcoholic and viral cirrhosis showed that C282Y polymorphisms were significantly more common in alcoholic cirrhosis patients with HCC compared to alcoholic cirrhosis patients without HCC. This effect was not seen in viral cirrhosis. In addition, all the H63D homozygotes in their study had cirrhosis, which points to a protective effect for H63D heterozygosity in alcoholic cirrhosis patients (n = 179 alcoholic cirrhosis patients, 98 viral cirrhosis patients, and 159 blood donor controls).93 In a Swedish study, patients with HH had a 20-fold increase in liver cancer rates compared to expected population prevalence, but no other increase in GI cancers. The same study also saw nothing of note in first- degree relatives, but they did not perform genotyping and their study did not delve directly into iron levels (n = 1847 patients, 5973 first-degree relative controls).94 A large multicenter study conducted in 2003 found no differences in HFE-variant allele frequency between patients with cirrhosis and HCC and patients with cirrhosis without HCC (n = 133 patients, 100 cirrhosis controls).95 Another 2003 study found that C282Y heterozygosity was significantly more common in HCC cases than in control individuals and that C282Y heterozygosity in HCC was associated with higher levels of hepatic iron, serum ferritin, and transferrin saturation compared to WT HCC patients (n = 137 HCC, 107 cirrhosis, and 126 healthy).96 This study suggests for some level of penetrance at the heterozygote genotype. A 2005 British study found that C282Y was associated with an increased risk of HCC (OR 14, 95% CI 5–37). They did not examine H63D (n = 144).97 A 2007 Spanish study found that H63D was associated with risk of HCC in cirrhotic patients, while no significant difference in risk was seen for C282Y. For H63D, the OR of carriers versus wild type was 2.03 (95% CI 1.25–3.28; n = 196, 181 controls).98 A 2008 study of cirrhotic patients showed that C282Y polymorphism and iron load were associated with a risk of HCC in alcoholic cirrhotic patients but not in HCV-related cirrhotic patients (n = 301).99 A meta-analysis showed that, at least in cases with alcoholic liver cirrhosis as controls, the C282Y polymorphism was associated with HCC (n = 1102 HCC cases, 3766 controls).100 In Egyptian HCC cases, both heterozygotes and homozygotes for H63D were more common in an HCC group than in matched controls, suggesting an association between the H63D allele and HCC in the context of cirrhosis and hepatitis C infection (n = 200 patients, 100 controls).66 A 2011 study of Polish cirrhosis patients and controls did not find a significant association between HFE variant alleles and HCC. However, their study was fairly small (61 cirrhosis patients and 42 controls, with only 36 of the patients biopsied for liver iron stores), so it is possible that they were not sufficiently powered to detect subtler effects.101 Although the evidence of effect is not universal, and the specific interactions with alcohol and hepatitis are not completely clear, there is sufficient evidence at present to safely claim that HFE variants are a risk factor for HCC. At present, it is not known whether these risk factors are due to iron overload alone or an off-target effect of HFE, and this question is best answered outside of the clinical population because it is ethically imperative to manage iron overload in known hemochromatosis patients. Furthermore, if untreated or late-treated populations of hemochromatosis patients could be identified, it would be useful to compare their oncologic epidemiologic data to a more aggressively treated set.
Prostate Cancer
The only available study on prostate cancer and HFE found no statistically significant difference in frequency of H63D or C282Y in 116 male breast cancer and 843 prostate cancer cases compared to 480 blood donor controls.102 This study took place in Finland and used blood donors as controls, so more general population–based studies and a more representative control group are required before drawing a conclusion on prostate cancer.
Lung Cancer
As with several of the extrahepatic cancers, only one study has been published examining the association between lung cancer and HFE. A 2005 study of NSCLC found no evidence that these cancers select for HFE polymorphisms. The few cases of C282Y they found were present in both the tumor tissue and the normal surrounding lung (n = 36).103 This piece of evidence alone says little about the effect of a host HFE polymorphism on cancer, because it was designed to address the phenomenon of clonal selection by the tumor itself. Thus, at present, it is difficult to say whether lung cancer is among the cancers influenced by HFE status.
Ovarian Cancer
The only study exploring ovarian cancer and HFE variants found that C282Y polymorphism increased both the risk and aggressiveness of ovarian cancer in clinical populations (n = 677).104 This finding demands follow-up, and more work is necessary before the repeatability and generalizability of these results are evident.
Conclusion
There exists a considerable body of evidence studying HFE in human populations, and the results are mostly inconsistent between studies. Given the prevalence of the HFE gene variants in the population and the ranges in dietary iron among different populations, the exact nature of the relationship of HFE polymorphisms to cancer is tremendously important but will be difficult to establish in population-based studies. Perhaps the most apparent shortcoming in the present literature is the overwhelming focus on risk. To fully understand the effect of HFE on tumor progression, we must also investigate the extent to which HFE modifies treatment effects and prognosis in the various cancers. In addition, we must consider whether variant HFE interacts with premalignant phenomena such as viral infection to affect tumor initiation. The findings in cervical cancer and virally mediated HCC support this hypothesis. The cell culture data suggest the existence of such effects, and how these findings translate to a clinically relevant effect requires further study.
In addition, to better understand the mechanistic significance of HFE polymorphisms in tumor progression, more controllable experimental systems are needed. One important factor that is sure to influence clinical studies from region to region is the association of HFE with ethnicity. This association introduces potentially confounding variables due to the links between ethnicity, socioeconomic status, gender (including menstruating versus nonmenstruating women), and various culturally influenced environmental exposures that need to be controlled in order to derive meaningful conclusions from this work. Importantly, an animal model would be free from these considerations, offering a means to directly interrogate mechanistic questions.
Glossary
List of Abbreviations
- ALL
Acute lymphocytic leukemia
- AML
Acute myelocytic leukemia
- β2m
β2 microglobulin
- C282Y
Cysteine-282-Tyrosine
- Caco-2
A human colonic adenocarcinoma cell line
- CD4
Cluster of differentiation 4 – Designates a protein on T-cells
- CD8
Cluster of differentiation 8 – Designates a protein on T-cells
- CDK1
Cyclin-dependent kinase 1
- CNS
Central nervous system
- CSF
Cerebrospinal fluid
- DFO
Deferoxamine
- EMT
Epithelial to mesenchymal transition
- EPIC
European Prospective Investigation into Cancer and Nutrition
- H63D
Histidine-63-aspartic acid
- HCC
Hepatocellular carcinoma
- HFE
high iron gene and its protein product
- HH
Hereditary hemochromatosis
- HPV
Human papilloma virus
- ICAM-1
Intracellular adhesion molecule one
- LCN2
Lipocalin 2
- MAL-12
Murine hepatocyte cell line
- MHC1
Major histocompatibility complex 1
- N-Myc
DNA-binding transcription factor
- NDRG1
N-myc downregulated gene 1
- NSAID
Nonsteroidal anti-inflammatory drug
- OR
Odds ratio
- P16INK4A
Cyclin-dependent kinase inhibitor 2A
- P21CIP1/WAF1
Cyclin-dependent kinase inhibitor 1
- P27KIP1
A cyclin-dependent kinase inhibitor
- P53
Tumor suppressor gene
- ROS
Reactive oxygen species
- RR
Relative risk
- SW480
A human adenocarcinoma cell line
- TFR1
Transferrin receptor 1
- TGF-β1
Transforming growth factor β 1
- UPR
Unfolded protein response
Footnotes
Author Contributions
Conceived the concepts: CLW, JRC. Analyzed the data: CLW. Wrote the first draft of the manuscript: CLW. Contributed to the writing of the manuscript: CLW, JRC. Agree with manuscript results and conclusions: CLW, JRC. Jointly developed the structure and arguments for the paper: CLW, JRC. Made critical revisions and approved final version: CLW, JRC. Both authors reviewed and approved of the final manuscript.
ACADEMIC EDITOR: Todd W. Miller, Editor in Chief
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
REFERENCES
- 1.Miniño AM Murphy SL. Death in the United States, 2010. Hyattsville, MD: National Center for Health Statistics; 2012. (NCHS data brief, no 99). http://www.cdc.gov/nchs/data/databriefs/db99.pdf. [PubMed] [Google Scholar]
- 2.Kukulj S, Jaganjac M, Boranic M, Krizanac S, Santic Z, Poljak-Blazi M. Altered iron metabolism, inflammation, transferrin receptors, and ferritin expression in non-small-cell lung cancer. Med Oncol. 2010;27(2):268–77. doi: 10.1007/s12032-009-9203-2. [DOI] [PubMed] [Google Scholar]
- 3.Yu Y, Kovacevic Z, Richardson DR. Tuning cell cycle regulation with an iron key. Cell Cycle. 2007;6(16):1982–94. doi: 10.4161/cc.6.16.4603. [DOI] [PubMed] [Google Scholar]
- 4.Cazzola M, Bergamaschi G, Dezza L, Arosio P. Manipulations of cellular iron metabolism for modulating normal and malignant cell proliferation: achievements and prospects. Blood. 1990;75(10):1903–19. [PubMed] [Google Scholar]
- 5.Saletta F, Suryo Rahmanto Y, Siafakas AR, Richardson DR. Cellular iron depletion and the mechanisms involved in the iron-dependent regulation of the growth arrest and DNA damage family of genes. J Biol Chem. 2011;286(41):35396–406. doi: 10.1074/jbc.M111.273060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brookes MJ, Hughes S, Turner FE, et al. Modulation of iron transport proteins in human colorectal carcinogenesis. Gut. 2006;55(10):1449–60. doi: 10.1136/gut.2006.094060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boult J, Roberts K, Brookes MJ, et al. Overexpression of cellular iron import proteins is associated with malignant progression of esophageal adenocarcinoma. Clin Cancer Res. 2008;14(2):379–87. doi: 10.1158/1078-0432.CCR-07-1054. [DOI] [PubMed] [Google Scholar]
- 8.Mainous AG, III, Gill JM, Everett CJ. Transferrin saturation, dietary iron intake, and risk of cancer. Ann Fam Med. 2005;3(2):131–7. doi: 10.1370/afm.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alkhateeb AA, Han B, Connor JR. Ferritin stimulates breast cancer cells through an iron-independent mechanism and is localized within tumor-associated macrophages. Breast Cancer Res Treat. 2013;137(3):733–44. doi: 10.1007/s10549-012-2405-x. [DOI] [PubMed] [Google Scholar]
- 10.Pinnix ZK, Miller LD, Wang W, et al. Ferroportin and iron regulation in breast cancer progression and prognosis. Sci Transl Med. 2010;2(43):ra456–63. doi: 10.1126/scisignal.3001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang KH, Tian HY, Gao X, et al. Ferritin heavy chain-mediated iron homeostasis and subsequent increased reactive oxygen species production are essential for epithelial-mesenchymal transition. Cancer Res. 2009;69(13):5340–8. doi: 10.1158/0008-5472.CAN-09-0112. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Madhankumar AB, Slagle-Webb B, Sheehan JM, Surguladze N, Connor JR. Heavy chain ferritin siRNA delivered by cationic liposomes increases sensitivity of cancer cells to chemotherapeutic agents. Cancer Res. 2011;71(6):2240–9. doi: 10.1158/0008-5472.CAN-10-1375. [DOI] [PubMed] [Google Scholar]
- 13.Kovacevic Z, Chikhani S, Lui GY, Sivagurunathan S, Richardson DR. The iron-regulated metastasis suppressor NDRG1 targets NEDD4 L, PTEN, and SMAD4 and inhibits the PI3K and Ras signaling pathways. Antioxid Redox Signal. 2012;18(8):874–87. doi: 10.1089/ars.2011.4273. [DOI] [PubMed] [Google Scholar]
- 14.Steinberg KK, Cogswell ME, Chang JC, et al. Prevalence of C282Y and H63D mutations in the hemochromatosis (HFE) gene in the United States. JAMA. 2001;285(17):2216–22. doi: 10.1001/jama.285.17.2216. [DOI] [PubMed] [Google Scholar]
- 15.Feder JN, Gnirke A, Thomas W, Tsuchihashi Z. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13(4):399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 16.Beutler E, Felitti VJ, Koziol JA, Ho NJ, Gelbart T. Penetrance of 845G--> A (C282Y) HFE hereditary haemochromatosis mutation in the USA. Lancet. 2002;359(9302):211–8. doi: 10.1016/S0140-6736(02)07447-0. [DOI] [PubMed] [Google Scholar]
- 17.Connor JR, Lee SY. HFE mutations and Alzheimer’s disease. J Alzheimer Dis. 2006;10(2–3):267–76. doi: 10.3233/jad-2006-102-311. [DOI] [PubMed] [Google Scholar]
- 18.de Almeida SF, Carvalho IF, Cardoso CS, et al. HFE cross-talks with the MHC class I antigen presentation pathway. Blood. 2005;106(3):971–7. doi: 10.1182/blood-2004-12-4640. [DOI] [PubMed] [Google Scholar]
- 19.Korkolopoulou P, Kaklamanis L, Pezzella F, Harris AL, Gatter KC. Loss of antigen-presenting molecules (MHC class I and TAP-1) in lung cancer. Br J Cancer. 1996;73(2):148–53. doi: 10.1038/bjc.1996.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vetter CS, Groh V, Thor Straten P, Spies T, Bröcker EB, Becker JC. Expression of stress-induced MHC class I related chain molecules on human melanoma. J Invest Dermatol. 2002;118(4):600–5. doi: 10.1046/j.1523-1747.2002.01700.x. [DOI] [PubMed] [Google Scholar]
- 21.Connor JR, Lee SY. Bioactive Compounds and Cancer. In: Milner JA, Donato F, editors. Iron and cancer. New York: Humana Press; 2010. p. 882. [Google Scholar]
- 22.Kwok JC, Richardson DR. The iron metabolism of neoplastic cells: alterations that facilitate proliferation? Crit Rev Oncol Hematol. 2002;42(1):65–78. doi: 10.1016/s1040-8428(01)00213-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen J, Chloupkova M. Abnormal iron uptake and liver cancer. Cancer Biol Ther. 2009;8(18):1699–708. doi: 10.4161/cbt.8.18.9146. [DOI] [PubMed] [Google Scholar]
- 24.Smith MA, Harris PL, Sayre LM, Perry G. Iron accumulation in Alzheimer disease is a source of redox-generated free radicals. Proc Natl Acad Sci USA. 1997;94(18):9866–8. doi: 10.1073/pnas.94.18.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luanpitpong S, Talbott SJ, Rojanasakul Y, et al. Regulation of lung cancer cell migration and invasion by reactive oxygen species and caveolin−1. J Biol Chem. 2010;285(50):38832–40. doi: 10.1074/jbc.M110.124958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stevens RG, Morris JE, Cordis GA, Anderson LE, Rosenberg DW, Sasser LB. Oxidative damage in colon and mammary tissue of the HFE-knockout mouse. Free Radic Biol Med. 2003;34(9):1212–6. doi: 10.1016/s0891-5849(03)00072-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhou W, Park S, Liu G, et al. Dietary iron, zinc, and calcium and the risk of lung cancer. Epidemiology. 2005;16(6):772–9. doi: 10.1097/01.ede.0000181311.11585.59. [DOI] [PubMed] [Google Scholar]
- 28.Niederau C, Fischer R, Sonnenberg A, Stremmel W, Trampisch HJ, Strohmeyer G. Survival and causes of death in cirrhotic and in noncirrhotic patients with primary hemochromatosis. N Engl J Med. 1985;313(20):1256–62. doi: 10.1056/NEJM198511143132004. [DOI] [PubMed] [Google Scholar]
- 29.MERK K, Mattsson B, Mattsson A, Holm G, Gullbring B, Björkholm M. The incidence of cancer among blood donors. Int J Epidemiol. 1990;19(3):505–9. doi: 10.1093/ije/19.3.505. [DOI] [PubMed] [Google Scholar]
- 30.Diwan BA, Kasprzak KS, Anderson LM. Promotion of dimethylbenz[a]anthracene-initiated mammary carcinogenesis by iron in female Sprague-Dawley rats. Carcinogenesis. 1997;18(9):1757–62. doi: 10.1093/carcin/18.9.1757. [DOI] [PubMed] [Google Scholar]
- 31.Singh M, Lu J, Briggs SP, McGinley JN, Haegele AD, Thompson HJ. Effect of excess dietary iron on the promotion stage of 1-methyl-1-nitrosourea-induced mammary carcinogenesis: pathogenetic characteristics and distribution of iron. Carcinogenesis. 1994;15(8):1567–70. doi: 10.1093/carcin/15.8.1567. [DOI] [PubMed] [Google Scholar]
- 32.Thompson HJ, Kennedy K, Witt M, Juzefyk J. Effect of dietary iron deficiency or excess on the induction of mammary carcinogenesis by 1-methyl-1-nitrosourea. Carcinogenesis. 1991;12(1):111–4. doi: 10.1093/carcin/12.1.111. [DOI] [PubMed] [Google Scholar]
- 33.Hann HW, Stahlhut MW, Blumberg BS. Iron nutrition and tumor growth: decreased tumor growth in iron-deficient mice. Cancer Res. 1988;48(15):4168–70. [PubMed] [Google Scholar]
- 34.Renton FJ, Jeitner TM. Cell cycle-dependent inhibition of the proliferation of human neural tumor cell lines by iron chelators. Biochem Pharmacol. 1996;51(11):1553–61. doi: 10.1016/0006-2952(96)00099-8. [DOI] [PubMed] [Google Scholar]
- 35.Brodie C, Siriwardana G, Lucas J, et al. Neuroblastoma sensitivity to growth inhibition by deferrioxamine: evidence for a block in G1 phase of the cell cycle. Cancer Res. 1993;53(17):3968–75. [PubMed] [Google Scholar]
- 36.Josson S, Nomura T, Lin JT, et al. Beta2-microglobulin induces epithelial to mesenchymal transition and confers cancer lethality and bone metastasis in human cancer cells. Cancer Res. 2011;71(7):2600–10. doi: 10.1158/0008-5472.CAN-10-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SY, Patton SM, Henderson RJ, Connor JR. Consequences of expressing mutants of the hemochromatosis gene (HFE) into a human neuronal cell line lacking endogenous HFE. FASEB J. 2007;21(2):564–76. doi: 10.1096/fj.06-6397com. [DOI] [PubMed] [Google Scholar]
- 38.Fabio G, Zarantonello M, Mocellin C, et al. Peripheral lymphocytes and intracellular cytokines in C282Y homozygous hemochromatosis patients. J Hepatol. 2002;37(6):753–61. doi: 10.1016/s0168-8278(02)00276-3. [DOI] [PubMed] [Google Scholar]
- 39.Nairz M, Theurl I, Schroll A, et al. Absence of functional Hfe protects mice from invasive Salmonella enterica serovar Typhimurium infection via induction of lipocalin−2. Blood. 2009;114(17):3642–51. doi: 10.1182/blood-2009-05-223354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leng X, Lin H, Ding T, et al. Lipocalin 2 is required for BCR-ABL-induced tumorigenesis. Oncogene. 2008;27(47):6110–9. doi: 10.1038/onc.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leng X, Ding T, Lin H, et al. Inhibition of lipocalin 2 impairs breast tumorigenesis and metastasis. Cancer Res. 2009;69(22):8579–84. doi: 10.1158/0008-5472.CAN-09-1934. [DOI] [PubMed] [Google Scholar]
- 42.Yang J, Bielenberg DR, Rodig SJ, et al. Lipocalin 2 promotes breast cancer progression. Proc Natl Acad Sci USA. 2009;106(10):3913–8. doi: 10.1073/pnas.0810617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fergelot P, Orhant M, Thénié A, et al. Over-expression of wild-type and mutant HFE in a human melanocytic cell line reveals an intracellular bridge between MHC class I pathway and transferrin iron uptake. Biol Cell. 2003;95(5):243–55. doi: 10.1016/s0248-4900(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 44.Nomura T, Huang WC, Zhau HE, et al. Beta2-microglobulin promotes the growth of human renal cell carcinoma through the activation of the protein kinase A, cyclic AMP-responsive element-binding protein, and vascular endothelial growth factor axis. Clin Cancer Res. 2006;12(24):7294–305. doi: 10.1158/1078-0432.CCR-06-2060. [DOI] [PubMed] [Google Scholar]
- 45.Hansen PB, Kjeldsen L, Dalhoff K, Olesen B. Cerebrospinal fluid beta-2-microglobulin in adult patients with acute leukemia or lymphoma: a useful marker in early diagnosis and monitoring of CNS-involvement. Acta Neurol Scand. 1992;85(3):224–7. doi: 10.1111/j.1600-0404.1992.tb04033.x. [DOI] [PubMed] [Google Scholar]
- 46.Norris S, White M, Mankan AK, Lawless MW. Highly sensitivity adhesion molecules detection in hereditary haemochromatosis patients reveals altered expression. Int J Immunogenet. 2010;37(2):125–33. doi: 10.1111/j.1744-313X.2010.00904.x. [DOI] [PubMed] [Google Scholar]
- 47.Ley K. The role of selectins in inflammation and disease. Trends Mol Med. 2003;9(6):263–8. doi: 10.1016/s1471-4914(03)00071-6. [DOI] [PubMed] [Google Scholar]
- 48.Witz I. The Link between Inflammation and Cancer. In: Dalgleish A, Haefner B, editors. Tumor-microenvironment interactions. US: Springer; 2006. pp. 125–40. [Google Scholar]
- 49.Borsig L, Wong R, Hynes RO, Varki NM, Varki A. Synergistic effects of L-and P-selectin in facilitating tumor metastasis can involve non-mucin ligands and implicate leukocytes as enhancers of metastasis. Proc Natl Acad Sci USA. 2002;99(4):2193–8. doi: 10.1073/pnas.261704098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Almeida SF, Picarote G, Fleming JV, Carmo-Fonseca M, Azevedo JE, de Sousa M. Chemical chaperones reduce endoplasmic reticulum stress and prevent mutant HFE aggregate formation. J Biol Chem. 2007;282(38):27905–12. doi: 10.1074/jbc.M702672200. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Lee SY, Neely E, et al. Mutant HFE H63D protein is associated with prolonged endoplasmic reticulum stress and increased neuronal vulnerability. J Biol Chem. 2011;286(15):13161–70. doi: 10.1074/jbc.M110.170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim R, Emi M, Tanabe K, Murakami S. Role of the unfolded protein response in cell death. Apoptosis. 2006;11(1):5–13. doi: 10.1007/s10495-005-3088-0. [DOI] [PubMed] [Google Scholar]
- 53.de Almeida SF, Fleming JV, Azevedo JE, Carmo-Fonseca M, de Sousa M. Stimulation of an unfolded protein response impairs MHC class I expression. J Immunol. 2007;178(6):3612–9. doi: 10.4049/jimmunol.178.6.3612. [DOI] [PubMed] [Google Scholar]
- 54.Wallace DF, McDonald CJ, Ostini L, Subramaniam VN. Blunted hepcidin response to inflammation in the absence of Hfe and transferrin receptor 2. Blood. 2011;117(10):2960–6. doi: 10.1182/blood-2010-08-303859. [DOI] [PubMed] [Google Scholar]
- 55.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chitambar CR, Kotamraju S, Wereley JP. Expression of the hemochromatosis gene modulates the cytotoxicity of doxorubicin in breast cancer cells. Int J Cancer. 2006;119(9):2200–4. doi: 10.1002/ijc.22079. [DOI] [PubMed] [Google Scholar]
- 58.Lee SY, Liu S, Mitchell RM, et al. HFE polymorphisms influence the response to chemotherapeutic agents via induction of p16INK4A. Int J Cancer. 2011;129(9):2104–14. doi: 10.1002/ijc.25888. [DOI] [PubMed] [Google Scholar]
- 59.Josson S, Matsuoka Y, Gururajan M, et al. Inhibition of beta2-microglobulin/hemochromatosis enhances radiation sensitivity by induction of iron overload in prostate cancer cells. PLoS One. 2013;8(7):e68366. doi: 10.1371/journal.pone.0068366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ali-Rahmani F, Huang MA, Schengrund CL, Connor JR, Lee SY. C282Y-HFE gene variant affects cholesterol metabolism in human neuroblastoma cells. PLoS One. 2014;9(2):e88724. doi: 10.1371/journal.pone.0088724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park SK, O’Neill MS, Wright RO, et al. HFE genotype, particulate air pollution, and heart rate variability: a gene-environment interaction. Circulation. 2006;114(25):2798–805. doi: 10.1161/CIRCULATIONAHA.106.643197. [DOI] [PubMed] [Google Scholar]
- 62.Moirand R, Adams PC, Bicheler V, Brissot P, Deugnier Y. Clinical features of genetic hemochromatosis in women compared with men. Ann Intern Med. 1997;127(2):105–10. doi: 10.7326/0003-4819-127-2-199707150-00002. [DOI] [PubMed] [Google Scholar]
- 63.Abraham BK, Justenhoven C, Pesch B, et al. Investigation of genetic variants of genes of the hemochromatosis pathway and their role in breast cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1102–7. doi: 10.1158/1055-9965.EPI-05-0013. [DOI] [PubMed] [Google Scholar]
- 64.Cardoso CS, Araújo HC, Cruz E, et al. Haemochromatosis gene (HFE) mutations in viral-associated neoplasia: linkage to cervical cancer. Biochem Biophys Res Commun. 2006;341(1):232–8. doi: 10.1016/j.bbrc.2005.12.174. [DOI] [PubMed] [Google Scholar]
- 65.Agudo A, Bonet C, Sala N, et al. Hemochromatosis (HFE) gene mutations and risk of gastric cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Carcinogenesis. 2013;34(6):1244–50. doi: 10.1093/carcin/bgt045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gharib AF, Karam RA, Pasha HF, Radwan MI, Elsawy WH. Polymorphisms of hemochromatosis, and alpha-1 antitrypsin genes in Egyptian HCV patients with and without hepatocellular carcinoma. Gene. 2011;489(2):98–102. doi: 10.1016/j.gene.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 67.Barton JC, Bertoli LF, Acton RT. HFE C282Y and H63D in adults with malignancies in a community medical oncology practice. BMC Cancer. 2004;4:6. doi: 10.1186/1471-2407-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dorak MT, Burnett AK, Worwood M, Sproul AM, Gibson BE. The C282Y mutation of HFE is another male-specific risk factor for childhood acute lymphoblastic leukemia. Blood. 1999;94(11):3957. [PubMed] [Google Scholar]
- 69.Gimferrer E, Nomdedeu J, Gich I, Barceló MJ, Baiget M. Prevalence of hemochromatosis related HFE gene mutations in patients with acute myeloid leukemia. Leuk Res. 1999;23(6):597–8. doi: 10.1016/s0145-2126(99)00043-0. [DOI] [PubMed] [Google Scholar]
- 70.Hannuksela J, Savolainen ER, Koistinen P, Parkkila S. Prevalence of HFE genotypes, C282Y and H63D, in patients with hematologic disorders. Haematologica. 2002;87(2):131–5. [PubMed] [Google Scholar]
- 71.Veneri D, Franchini M, Krampera M, de Matteis G, Solero P, Pizzolo G. Analysis of HFE and TFR2 gene mutations in patients with acute leukemia. Leuk Res. 2005;29(6):661–4. doi: 10.1016/j.leukres.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 72.Dorak MT, Mackay RK, Relton CL, Worwood M, Parker L, Hall AG. Hereditary hemochromatosis gene (HFE) variants are associated with birth weight and childhood leukemia risk. Pediatr Blood Cancer. 2009;53(7):1242–8. doi: 10.1002/pbc.22236. [DOI] [PubMed] [Google Scholar]
- 73.Davis CF, Dorak MT. An extensive analysis of the hereditary hemochromatosis gene HFE and neighboring histone genes: associations with childhood leukemia. Ann Hematol. 2010;89(4):375–84. doi: 10.1007/s00277-009-0839-y. [DOI] [PubMed] [Google Scholar]
- 74.Rodríguez-López R, Donoso M, Fernández-Cavada M, et al. Diagnostic utility of HFE variants in Spanish patients: association with HLA alleles and role in susceptibility to acute lymphoblastic leukemia. Gene. 2013;514(1):31–5. doi: 10.1016/j.gene.2012.10.090. [DOI] [PubMed] [Google Scholar]
- 75.Miller LD, Ferrari P, Fallahi P, Antonelli A. An iron regulatory gene signature predicts outcome in breast cancer. Cancer Res. 2011;71(21):6728–37. doi: 10.1158/0008-5472.CAN-11-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kallianpur AR. Increased prevalence of the HFE C282Y hemochromatosis allele in women with breast cancer. Cancer Epidemiol Biomarkers Prev. 2004;13(2):205–12. doi: 10.1158/1055-9965.epi-03-0188. [DOI] [PubMed] [Google Scholar]
- 77.Gunel-Ozcan A, Alyilmaz-Bekmez S, Guler EN, Guc D. HFE H63D mutation frequency shows an increase in Turkish women with breast cancer. BMC Cancer. 2006;6:37. doi: 10.1186/1471-2407-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Batschauer AP, Cruz NG, Oliveira VC, et al. HFE, MTHFR, and FGFR4 genes polymorphisms and breast cancer in Brazilian women. Mol Cell Biochem. 2011;357(1–2):247–53. doi: 10.1007/s11010-011-0895-1. [DOI] [PubMed] [Google Scholar]
- 79.Asberg A, Thorstensen K, Irgens WØ, Romundstad PR, Hveem K. Cancer risk in HFE C282Y homozygotes: results from the HUNT 2 study. Scand J Gastroenterol. 2013;48(2):189–95. doi: 10.3109/00365521.2012.752028. [DOI] [PubMed] [Google Scholar]
- 80.Flanagan PR, Hajdu A, Adams PC. Iron-responsive element-binding protein in hemochromatosis liver and intestine. Hepatology. 1995;22(3):828–32. [PubMed] [Google Scholar]
- 81.Macdonald GA, Tarish J, Whitehall VJ, et al. No evidence of increased risk of colorectal cancer in individuals heterozygous for the Cys282Tyr haemochromatosis mutation. J Gastroenterol Hepatol. 1999;14(12):1188–91. doi: 10.1046/j.1440-1746.1999.02027.x. [DOI] [PubMed] [Google Scholar]
- 82.Altes A, Gimferrer E, Capella G, Barcelo MJ, Baiget M. Colorectal cancer and HFE gene mutations. Haematologica. 1999;84(5):479–80. [PubMed] [Google Scholar]
- 83.Shaheen NJ, Silverman LM, Keku T, et al. Association between hemochromatosis (HFE) gene mutation carrier status and the risk of colon cancer. J Natl Cancer Inst. 2003;95(2):154–9. doi: 10.1093/jnci/95.2.154. [DOI] [PubMed] [Google Scholar]
- 84.Chan AT, Ma J, Tranah GJ, et al. Hemochromatosis gene mutations, body iron stores, dietary iron, and risk of colorectal adenoma in women. J Natl Cancer Inst. 2005;97(12):917–26. doi: 10.1093/jnci/dji165. [DOI] [PubMed] [Google Scholar]
- 85.Shi Z, Johnstone D, Talseth-Palmer BA, et al. Haemochromatosis HFE gene polymorphisms as potential modifiers of hereditary nonpolyposis colorectal cancer risk and onset age. Int J Cancer. 2009;125(1):78–83. doi: 10.1002/ijc.24304. [DOI] [PubMed] [Google Scholar]
- 86.Osborne NJ, Gurrin LC, Allen KJ, et al. HFE C282Y homozygotes are at increased risk of breast and colorectal cancer. Hepatology. 2010;51(4):1311–8. doi: 10.1002/hep.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen W, Zhao H, Li T, Yao H. HFE gene C282Y variant is associated with colorectal cancer in Caucasians: a meta-analysis. Tumour Biol. 2013;34(4):2255–9. doi: 10.1007/s13277-013-0766-3. [DOI] [PubMed] [Google Scholar]
- 88.Martinez di Montemuros F, Tavazzi D, Salsano E, et al. High frequency of the H63D mutation of the hemochromatosis gene (HFE) in malignant gliomas. Neurology. 2001;57(7):1342. doi: 10.1212/wnl.57.7.1342. [DOI] [PubMed] [Google Scholar]
- 89.Willis G, Wimperis JZ, Lonsdale R, Jennings BA. Haemochromatosis gene mutation in hepatocellular cancer. Lancet. 1997;350(9077):565–6. doi: 10.1016/s0140-6736(05)63143-1. [DOI] [PubMed] [Google Scholar]
- 90.Racchi O, Mangerini R, Rapezzi D, et al. Mutations of the HFE gene and the risk of hepatocellular carcinoma. Blood Cells Mol Dis. 1999;25(5–6):350–3. doi: 10.1006/bcmd.1999.0263. [DOI] [PubMed] [Google Scholar]
- 91.Willis G, Wimperis JZ, Lonsdale R, et al. Incidence of liver disease in people with HFE mutations. Gut. 2000;46(3):401–4. doi: 10.1136/gut.46.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fargion S, Stazi MA, Fracanzani AL, et al. Mutations in the HFE gene and their interaction with exogenous risk factors in hepatocellular carcinoma. Blood Cells Mol Dis. 2001;27(2):505–11. doi: 10.1006/bcmd.2001.0411. [DOI] [PubMed] [Google Scholar]
- 93.Lauret E, Rodríguez M, González S, et al. HFE gene mutations in alcoholic and virus-related cirrhotic patients with hepatocellular carcinoma. Am J Gastroenterol. 2002;97(4):1016–21. doi: 10.1111/j.1572-0241.2002.05553.x. [DOI] [PubMed] [Google Scholar]
- 94.Elmberg M, Hultcrantz R, Ekbom A, et al. Cancer risk in patients with hereditary hemochromatosis and in their first-degree relatives. Gastroenterology. 2003;125(6):1733–41. doi: 10.1053/j.gastro.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 95.Boige V, Castéra L, de Roux N, et al. Lack of association between HFE gene mutations and hepatocellular carcinoma in patients with cirrhosis. Gut. 2003;52(8):1178–81. doi: 10.1136/gut.52.8.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hellerbrand C, Pöppl A, Hartmann A, Schölmerich J, Lock G. HFE C282Y heterozygosity in hepatocellular carcinoma: evidence for an increased prevalence. Clin Gastroenterol Hepatol. 2003;1(4):279–84. doi: 10.1016/s1542-3565(03)00132-0. [DOI] [PubMed] [Google Scholar]
- 97.Willis G, Bardsley V, Fellows IW, Lonsdale R, Wimperis JZ, Jennings BA. Hepatocellular carcinoma and the penetrance of HFE C282Y mutations: a cross sectional study. BMC Gastroenterol. 2005;5:17. doi: 10.1186/1471-230X-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ropero P, Briceño O, López-Alonso G, et al. The H63D mutation in the HFE gene is related to the risk of hepatocellular carcinoma. Rev Esp Enferm Dig. 2007;99(7):376–81. doi: 10.4321/s1130-01082007000700002. [DOI] [PubMed] [Google Scholar]
- 99.Nahon P, Sutton A, Rufat P, et al. Liver iron, HFE gene mutations, and hepatocellular carcinoma occurrence in patients with cirrhosis. Gastroenterology. 2008;134(1):102–10. doi: 10.1053/j.gastro.2007.10.038. [DOI] [PubMed] [Google Scholar]
- 100.Jin F, Qu LS, Shen XZ. Association between C282Y and H63D mutations of the HFE gene with hepatocellular carcinoma in European populations: a meta-analysis. J Exp Clin Cancer Res. 2010;29:18. doi: 10.1186/1756-9966-29-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sikorska K, Romanowski T, Stalke P, Iżycka-Świeszewska E, Bielawski KP. Iron overload and HFE gene mutations in Polish patients with liver cirrhosis. Hepatobiliary Pancreat Dis Int. 2011;10(3):270–5. doi: 10.1016/s1499-3872(11)60045-3. [DOI] [PubMed] [Google Scholar]
- 102.Syrjäkoski K, Fredriksson H, Ikonen T, et al. Hemochromatosis gene mutations among Finnish male breast and prostate cancer patients. Int J Cancer. 2006;118(2):518–20. doi: 10.1002/ijc.21331. [DOI] [PubMed] [Google Scholar]
- 103.Müller CI, Miller CW, Kawabata H, McKenna RJ, Jr, Marchevsky AM, Koeffler HP. Do cancer cells selectively mutate HFE to increase their intracellular iron? Oncol Rep. 2005;14(2):299–303. [PubMed] [Google Scholar]
- 104.Gannon PO, Medelci S, Le Page C, et al. Impact of hemochromatosis gene (HFE) mutations on epithelial ovarian cancer risk and prognosis. Int J Cancer. 2010;128(10):2326–34. doi: 10.1002/ijc.25577. [DOI] [PMC free article] [PubMed] [Google Scholar]