Abstract
Microsatellites occur in all plant genomes and provide useful markers for studies of genetic diversity and structure. Chloroplast microsatellites (cpSSRs) are frequently targeted because they are more easily isolated than nuclear microsatellites. Here, we quantified the frequency and uses of cpSSRs based on a literature review of over 400 studies published 1995–2013. These markers are an important and economical tool for plant biologists and continue to be used alongside modern genomics approaches to study genetic diversity and structure, evolutionary history, and hybridization in native and agricultural species. Studies using species-specific primers reported a greater number of polymorphic loci than those employing universal primers. A major disadvantage to cpSSRs is fragment size homoplasy; therefore, we documented its occurrence at several cpSSR loci within and between species of Acmispon (Fabaceae). Based on our empirical data set, we recommend targeted sequencing of a subset of samples combined with fragment genotyping as a cost-efficient, data-rich approach to the use of cpSSRs and as a test of homoplasy. The availability of genomic resources for plants aids in the development of primers for new study systems, thereby enhancing the utility of cpSSRs across plant biology.
Keywords: chloroplast microsatellite, cpSSR, plastid microsatellite, population genetics, size homoplasy
Chloroplast genomes (cpDNA) have provided a wealth of data for discerning phylogenetic relationships within and among species (e.g., Olmstead and Palmer, 1994; Parks et al., 2009; Moore et al., 2010; Stech and Quandt, 2010; Drew et al., 2014), for evaluating uniparental patterns of gene flow within and among populations (e.g., Ennos et al., 1999; Wallace et al., 2011; Bai et al., 2014), for barcoding efforts to distinguish taxa (e.g., Hollingsworth et al., 2009, 2011; Chen et al., 2010), as well as for other studies. The appeal of using cpDNA markers lies in the fact that universal primers are capable of amplifying homologous regions across diverse taxa (Shaw et al., 2005, 2007), the cpDNA genome often exhibits uniparental inheritance (reviewed in Reboud and Zeyl, 1994; Birky, 1995), and the cpDNA genome is generally nonrecombining (Clegg, 1993; but see Ogihara et al., 1988; Marshall et al., 2001). However, instances of biparental inheritance leading to heteroplasmy are also reported (e.g., Hansen et al., 2007). Much of the cpDNA genome evolves slowly, which reduces its general utility for evolutionary and population genetic studies, especially at lower taxonomic levels. Therefore, many researchers have sought to use noncoding regions, including introns and intergenic spacers, of the cpDNA genome to characterize genetic variation (Shaw et al., 2005, 2007). Like the nuclear genome, these regions have been found to contain tandem repeat regions or simple sequence repeats (SSRs) (Shaw et al., 2005, 2007; Dong et al., 2012), which often show higher levels of allelic variation than single nucleotide polymorphisms (Haasl and Payseur, 2011).
Since their introduction as a readily usable chloroplast marker by Powell et al. (1995), chloroplast SSRs (cpSSRs) have been increasingly used as genetic markers in a broad range of studies, in both basic plant sciences and applied agricultural research (Ebert and Peakall, 2009). They are commonly used in population genetic studies because of their ability to differentiate recently diverged groups (e.g., Echt et al., 1998), to function as a DNA barcode (e.g., Kuang et al., 2011), to allow for the quick identification of a genetic group or species (e.g., Decroocq et al., 2004), and to study hybridization and introgression (e.g., Wallace et al., 2011). Despite their widespread use in the past two decades, cpSSRs are not failsafe. Among the limitations of using cpSSRs are lack of variation in some species, lack of universal primers, size homoplasy (i.e., the occurrence of underlying sequence variation that does not change the length of the region as determined by genotyping), heteroplasmy, and cytoplasmic introgression due to interspecific hybridization. Among these issues, size homoplasy may be the most serious because it can overestimate relatedness among samples and lead to incorrect evolutionary and ecological inferences. Cytoplasmic introgression may also be very serious if undetected because it reflects the history of the donor genome rather than the full history of the focal species.
Given the continued use of cpSSRs in a wide diversity of systems, reviews of published studies employing them are worthwhile for guiding future research and for addressing outstanding problems with their use in plant biology. The foci of previous reviews of cpSSRs include potential problems associated with homoplasy (Navascués and Emerson, 2005) and technical resources available for using cpSSRs (Ebert and Peakall, 2009). This latter study is particularly notable as it called attention to the great potential for cpSSRs to be informative for studies of noneconomic species and highlighted methods that could be used to identify variable loci and develop suitable primers (Ebert and Peakall, 2009). Several papers have called attention to the issue of size homoplasy and the importance of examining underlying sequence variation, but it is not clear that these warnings have been realized among published studies. In this review, we document the continued frequency and nature of published studies employing cpSSRs, and discuss the potential for fragment size homoplasy using an empirical data set of cpSSR loci that spans intraspecific and interspecific taxa of the genus Acmispon (Fabaceae) as well as reports from published papers. In light of the increased use of genomics techniques, we sought to determine whether researchers are still employing single locus approaches involving cpSSRs and whether there is a shift from using chloroplast markers to genome-wide markers. Finally, we use these data to discuss the strengths and weaknesses of cpSSRs for studies of plant biology, and we make recommendations for developing and using cpSSRs in future studies.
MATERIALS AND METHODS
Review of published studies
To understand how cpSSRs have been used in plant biology, we conducted a survey of primary literature sources on the subject. This survey was performed using the search engine Scopus (Elsevier B.V., Amsterdam, The Netherlands). We searched papers published between 1960 and 2013 using the following terms in the abstract, title, or key words: chloroplast SSRs, chloroplastic SSRs, plastid SSRs, cpSSRs, chloroplast MSATs, chloroplastic MSATs, plastid MSATs, cpMSATs, chloroplast microsatellites, chloroplastic microsatellites, plastid microsatellites, chloroplast tandem repeats, chloroplastic tandem repeats, plastid tandem repeats, chloroplast simple sequence repeats, chloroplastic simple sequence repeats, and plastid simple sequence repeats. Only publications with an explicit use or discussion of cpSSR loci were included. Publications that were not available in English-language full-text version were not considered. This review is based on 439 papers (see Appendix S1 (82.6KB, docx) ). From these papers, we recorded the family (or families) of species studied, type of study based on the focal species (agricultural or native), number of loci used, and origin of primers used (e.g., developed de novo or published primer sequences). Studies of agricultural species focused on cultivated and/or economically important species, whereas studies of native species focused on increasing knowledge of species with little economic importance. We further classified by subcategories to develop a better understanding of the types of studies for which cpSSRs have been most useful. Subcategories and their definitions are presented in Table 1.
Table 1.
Subcategories of published papers using cpSSRs reviewed in this study.
| Category | Primary focus |
| Agricultural species | |
| Diversity | Assessment of genetic diversity present within a cultivated crop unit or across a cultivated crop species, breed, or strain |
| History | Assessment of ancestry or origins of a cultivated taxon based on genetic relatedness to wild or cultivated parental taxa |
| Identification | Identification or development of methods to identify crop plant species, breeds, or strains |
| Primer note | Reporting the development and/or testing of cpSSR primers |
| Other | Papers not fitting other categories |
| Native species | |
| Hybridization | Assessment of hybridization and/or documentation of parental taxa or populations of hybrid individuals |
| Methodology | Introducing new methods of data collection or analysis for cpSSR loci |
| Population genetics | Assessment of genetic diversity and structure within and among natural plant populations, including phylogeographic studies |
| Primer note | Reporting the development and/or testing of cpSSR primers |
| Review | Synthesis of published studies using cpSSRs |
| Systematics | Assessment of evolutionary relatedness among distinct taxa, including studies for the purpose of taxonomic revision |
| Other | Papers not fitting other categories |
Quantifying problems with fragment size homoplasy
An assessment of whether appropriate methods were used to determine and/or correct for size homoplasy was made for each publication examined in the literature review. Publications in which microsatellite loci were assayed by genotyping (i.e., only fragment sizes were determined) were considered to be “uncorrected” for issues of size homoplasy. We defined “corrected” publications as those in which the authors stated that size homoplasy was examined after fragment genotyping or when they used a direct sequencing approach.
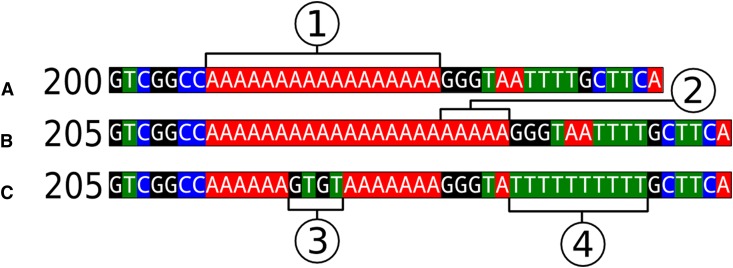
We also evaluated issues of fragment size homoplasy and aberrant mutation motifs (i.e., size changes that deviate from the expected stepwise pattern of microsatellite mutation) in an empirical data set of nine chloroplast microsatellite loci developed for five species of Acmispon Raf. (Fabaceae) (Wheeler et al., 2012). Species included were A. argophyllus (A. Gray) Brouillet, A. dendroideus (Greene) Brouillet, A. micranthus (Torr. & A. Gray) Brouillet, A. glaber (Vogel) Brouillet, and A. heermannii (Durand & Hilg.) Brouillet. Data for four loci (ACcp2, ACcp3, ACcp4, and ACcp5) were derived from a set of direct sequences of 452 Acmispon individuals (i.e., 216 from A. argophyllus, 166 from A. dendroideus, 36 from A. micranthus, 22 from A. glaber, and 12 from A. heermannii). All other loci were sized as fragments, with each unique allele per species also sequenced. Genotyping details are described in Wheeler et al. (2012), and sequencing details are described in Wheeler (2013). Sequences of most alleles were examined in two individuals per species; however, in some cases, particularly when an allele is only present in a single individual of a given species, only one individual was sequenced. As this study does not use sequence data from every individual, nor does it include every species in the genus, the frequencies of motif abnormality and size homoplasy found are likely underestimated. The size of each microsatellite region based on sequence data was compared to size determined by fragment genotyping. Loci were considered to exhibit size homoplasy if any single fragment size corresponded to two or more unique arrangements of sequence gaps (Fig. 1B vs. 1C). A locus was considered to have an abnormal motif if one or more individuals showed a detectable size change that deviated from the expected stepwise pattern of microsatellite expansion or contraction (Fig. 1C3). Single-nucleotide mutations that did not change the length of the sequence were ignored.
Fig. 1.
Three examples of chloroplast sequence regions containing tandem repeats, with indicated size when genotyped as fragments. (A) A segment of genome sequence used to design primers to be amplified in a number of individuals; (1) is the presumed variable region. (B) The sequence of an individual that meets all the assumptions of a chloroplast microsatellite model, with (2) indicating a 5-step expansion of the repeat region. (C) The sequence of an individual that violates these assumptions because there is a size change due to the addition of a nonrepetitive indel, violating the assumed microsatellite motif (3) and expansion of a shorter secondary T-mononucleotide repeat region (4). When genotyped as fragments, (B) and (C) are indistinguishable, showing an example of size homoplasy.
RESULTS
Prevalence and uses of cpSSRs
Of the 439 papers considered in this review, 405 contained original empirical data from cpSSR loci; four of these used minisatellites rather than microsatellites. Seven papers reused cpSSR data from a previous publication. Eight papers were classified as reviews of microsatellites (n = 3) or molecular markers in plants (n = 5). Nineteen papers included whole chloroplast genome sequences or mRNA sequences and identified SSR regions.
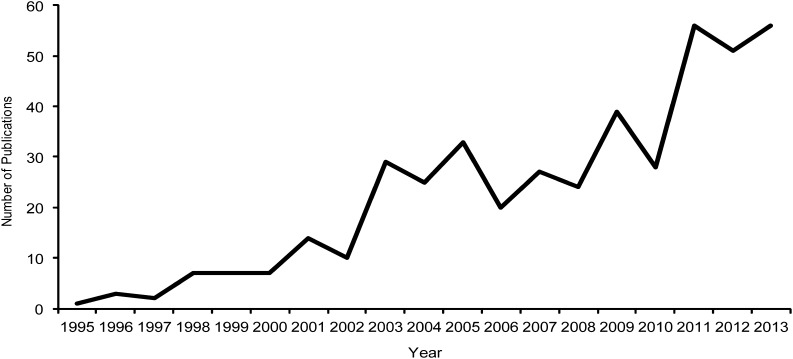
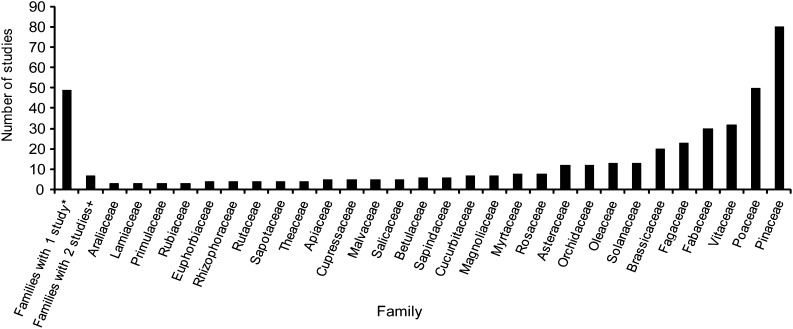
The use of cpSSRs has expanded substantially from 1995, when a single study was published, to 56 publications in 2013 (Fig. 2). cpSSRs have been used as informative genetic characters in a wide range of plant groups, including green algae (n = 2), bryophytes (n = 1), lycophytes (n = 1), gymnosperms (n = 86), magnoliids (n = 9), monocots (n = 79), and eudicots (n = 264). These studies include species from 85 families (Fig. 3). Pinaceae is the most represented family, with 80 studies, followed by several economically important angiosperm families: Poaceae (50 studies), Vitaceae (32 studies), Fabaceae (30 studies), and Brassicaceae (20 studies). However, the majority of families are represented by five or fewer studies employing cpSSRs.
Fig. 2.
Number of publications involving cpSSRs by year. All empirical papers using original or previously published cpSSR data, reviews of microsatellites or molecular markers, and papers involving the identification of cpSSRs in sequenced chloroplast papers were included.
Fig. 3.
Number of cpSSR studies by plant family. Families represented by one or two studies have been grouped together in the first two bars, respectively. Family counts are based on 424 papers, but families were counted separately in studies covering multiple families; review papers and those using previously published data were not included. *Families represented by one published study: Acoraceae, Alliaceae, Amaranthaceae, Aquifoliaceae, Arecaceae, Asparagaceae, Balsaminaceae, Begoniaceae, Bignoniaceae, Calycanthaceae, Cannabaceae, Caricaceae, Caryocaraceae, Caryophyllaceae, Cephalotaxaceae, Chlamydomonaceae, Cistaceae, Clusiaceae, Codiaceae, Combretaceae, Cyperaceae, Dioscoreaceae, Elaeagnaceae, Ericaceae, Gentianaceae, Hydrocharitaceae, Juglandaceae, Liliaceae, Lycopodiaceae, Meliaceae, Moraceae, Musaceae, Myrsinaceae, Nelumbonaceae, Pedaliaceae, Posidoniaceae, Potamogetonaceae, Proteaceae, Ptilidiaceae, Punicaceae, Rhamnaceae, Ruppiaceae, Santalaceae, Tamaricaceae, Trochodendraceae, Verbenaceae, Winteraceae, Xanthorrhoeaceae, Zosteraceae. +Families represented by two published studies: Acanthaceae, Anacardiaceae, Boraginaceae, Bromeliaceae, Convolvulaceae, Ranunculaceae, Saxifragaceae.
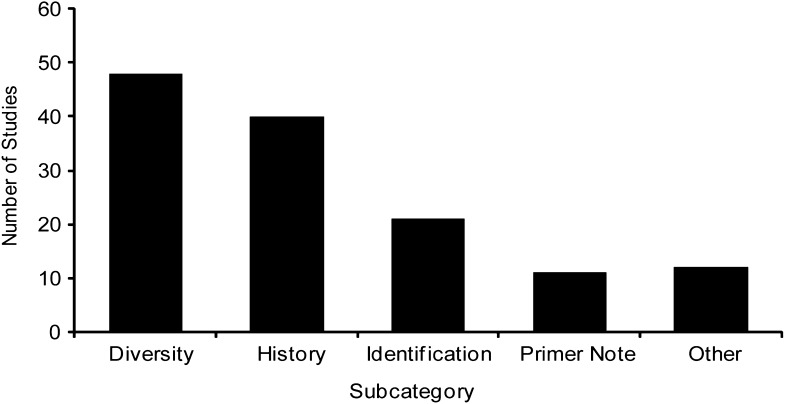
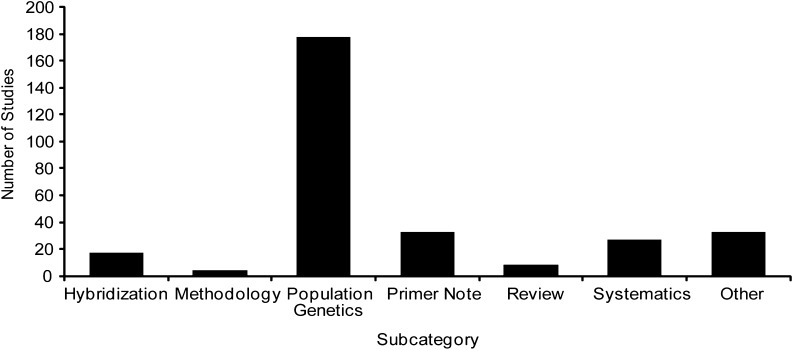
Publications utilizing chloroplast microsatellites in native species (69%) far outweigh those applying cpSSR techniques in agricultural species (31%). In agricultural species, cpSSRs have been used extensively to assess genetic diversity in cultivated taxa (36.4%) and to study the origins or relatedness of cultivars to native species (30.3%; Fig. 4). The other subcategories account for the remaining studies, including identification of cultivars (15.9%), primer notes (8.3%), and various other purposes (9.1%). In native species, cpSSRs have been used most frequently in population genetics studies (59.3%), followed by primer notes or other foci (11.0% each), systematics (9.0%), hybridization (5.7%), reviews (2.7%), and methodology (1.3%; Fig. 5).
Fig. 4.
Publications employing cpSSRs in agricultural species sorted by subcategory.
Fig. 5.
Publications employing cpSSRs in native species sorted by subcategory.
Many different numbers of loci were used across studies, but there are no significant trends in number of loci used and taxonomic category or number of loci used and number of studies published by family. The correlation between number of published studies per family and number of loci used is very weak (r2 = 0.065, P > 0.05). On average, slightly more loci (mean = 8.34) were used in studies of monocot taxa compared to those focused on eudicot taxa (mean = 7.61 loci), although this difference was not significant (t0.05[304] = 0.36, P > 0.05). The other groups did not contain a sufficient number of published studies for comparison. An overwhelming majority of studies used published primers to amplify loci, but more than twice as many loci were used when primers were developed for the taxon of interest (mean = 12.5 loci used) compared to studies using published primers to amplify loci (mean = 5.8 loci used). The most commonly reported published primers that were used across plants are from Weising and Gardner (1999) (131 references), Bryan et al. (1999) (29 references), and Chung and Staub (2003) (24 references). Amplification of primer sets from Weising and Gardner (1999) and Chung and Staub (2003) may have been so highly successful across phylogenetically disparate taxa because these authors considered multiple taxa in the design and testing of their primers. The primers of Bryan et al. (1999) were developed and tested only in species of Solanaceae. These primers have been used primarily in eudicot taxa, although Chaïr et al. (2005) reported polymorphism at these loci in the monocot Dioscorea L. For other taxonomic groups, published primer sets have been used with great success, including those of Vendramin et al. (1996) for gymnosperms (69 references), Deguilloux et al. (2003) for Fagaceae (14 references), Sebastiani et al. (2004) for Fagaceae and Betulaceae (8 references), Ueno et al. (2005) for Magnoliaceae (2 references), and Ishii et al. (2001) for Poaceae (14 references). Authors often reported using primer sets from multiple publications. Many chloroplast genome sequences are now becoming available. We identified 19 studies in this data set in which SSR loci were determined by the authors (Table 2). These may be useful in identifying variable loci for related taxa for which existing published primers have not been highly successful.
Table 2.
Publications in the data set reporting cpSSR loci from whole chloroplast genome sequences.
| Family | Species | No. of SSR loci | Reference |
| Araliaceae | Eleutherococcus senticosus (Rupr. ex Maxim.) Maxim. | 35 | Yi et al. (2012) |
| Araliaceae | Panax schin-seng Nees | 18 | Kim and Lee (2004) |
| Asteraceae | Artemisia frigida Willd. | 41 | Liu et al. (2013) |
| Asteraceae | Jacobaea vulgaris Gaertn. | 34a | Doorduin et al. (2011) |
| Cephalotaxaceae | Cephalotaxus oliveri Mast. | 24 | Yi et al. (2013) |
| Cupressaceae | Cryptomeria japonica (Thunb. ex L. f.) D. Don | 9 | Hirao et al. (2009) |
| Fabaceae | Prosopis alba Griseb. | 36 | Torales et al. (2013) |
| Hydrocharitaceae | Elodea canadensis Michx. | 127 | Huotari and Korpelainen (2012)b |
| Lamiaceae | Origanum vulgare L. | 56 | Lukas and Novak (2013) |
| Lamiaceae | Salvia miltiorrhiza Bunge | 166 | Qian et al. (2013)c |
| Magnoliaceae | Magnolia grandiflora L. | 218 | Li et al. (2013) |
| Magnoliaceae | Magnolia kwangsiensis Figlar & Noot. | 237 | Kuang et al. (2011) |
| Myrtaceae | Eucalyptus L’Hér. spp., Corymbia K. D. Hill & L. A. S. Johnson spp., Angophora Cav. spp., Allosyncarpia ternata S. T. Blake, Stockwellia quadrifida D. J. Carr, S. G. M. Carr & B. Hyland | 248 across all taxa | Bayly et al. (2013) |
| Pedaliaceae | Sesamum indicum L. | 28 | Yi and Kim (2012) |
| Poaceae | Brachypodium distachyon (L.) P. Beauv. | 17 | Sonah et al. (2011) |
| Ranunculaceae | Megaleranthis saniculifolia Ohwi | 54 | Kim et al. (2009) |
| Rosaceae | Pyrus pyrifolia (Burm. f.) Nakai | 67 | Terakami et al. (2012) |
| Solanaceae | Solanum L. spp. | 32 across all taxa | Gargano et al. (2012) |
| Trochodendraceae | Tetracentron sinense Oliv., Trochodendron aralioides Siebold & Zucc. | 154 in both taxa | Sun et al. (2013) |
Ten primer sets were tested in a sample of the focal species.
This study provides a synthesis of cpSSR regions across many monocots for which whole chloroplast genomes have been sequenced.
This study provides a synthesis of cpSSR regions across many species of the Asterid clade.
Size homoplasy
The potential for issues with disagreement between fragment size and underlying sequence has been known for some time (Doyle et al., 1998; Provan et al., 2001; Navascués and Emerson, 2005), but still relatively few papers reported investigating size homoplasy in their data sets. Among the 405 studies reporting empirical cpSSR data, only 135 papers (33%) indicated an approach beyond fragment length genotyping that would enable detection of an abnormal motif or correction for size homoplasy. Within the two categories of papers, the number of studies investigating size homoplasy was similar (32% of agricultural species studies and 34% of native species studies).
In the survey of nine Acmispon loci, two-thirds showed size homoplasy or deviation from a standard microsatellite mutation motif. Three loci exhibited size homoplasy, two loci had deviations from an assumed motif, and one locus contained both problems. Departures from expected patterns were found both within and between species, although more alleles exhibited these problems between species (Table 3).
Table 3.
Deviation from a stepwise mutation motif (i.e., “abnormal motif”) and homoplasy of detected size alleles among cpSSR loci examined in five species of Acmispon.
| Locus | Repeat motif | No. of alleles | Abnormal motif | Within speciesa | Between speciesb |
| ACcp1 | (A)10(T)9(A)10(T)8 | 7 | No | 4 | 5 |
| ACcp2 | (T)13(T)7 | 6 | No | 0 | 0 |
| ACcp3 | (A)14 | 6 | No | 0 | 0 |
| ACcp4 | (T)11 | 6 | Yes | 2 | 0 |
| ACcp5 | (A)12 | 4 | No | 0 | 0 |
| ACcp6 | (T)8A(T)11 | 4 | No | 1 | 2 |
| ACcp7 | (T)12(A)9 | 7 | No | 1 | 3 |
| ACcp8 | (T)9 | 4 | Yes | 0 | 0 |
| ACcp9 | (ATTTATTCTAT)2(AT)3(T)5 | 6 | Yes | 0 | 0 |
Number of alleles showing size homoplasy among conspecific samples and across all species examined.
Number of alleles that exhibit similar sizes yet distinct sequences between species.
DISCUSSION
Use of cpSSRs across plants
Since 1995, cpSSRs have been widely used in plant biology to address basic and applied questions, and they continue to be a useful genetic tool (Fig. 2). The ease of PCR amplification and the polymorphic nature of cpSSRs have made them readily available markers for characterizing genetic variation when few genomic resources exist for a study system. For example, the majority of families in our data set were represented by five or fewer studies, suggesting that markers of this type can be developed and put into use for new groups without much a priori genetic information.
Studies of species in certain families have benefitted more than other families from the use of cpSSRs. Among gymnosperms, studies of taxa in Pinaceae are strongly represented (Fig. 3). Most of these studies were conducted within Pinus L. (56 references), which may reflect greater interest in this genus, which is more speciose than other genera in this family, or perhaps may indicate greater success across the genus using loci designed from P. thunbergii Parl. (Vendramin et al., 1996), the most commonly reported primers used in Pinaceae. Common foci of studies within Pinaceae include understanding the effects of Quaternary glaciation on species range distributions (e.g., Gugerli et al., 2001; Gómez et al., 2005; Bucci et al., 2007; Rodríguez-Banderas et al., 2009; Godbout et al., 2010), characterizing genetic structure (e.g., Vendramin et al., 1999; Dyer and Sork, 2001; Viard et al., 2001; Nasri et al., 2008; Scalfi et al., 2009; Wang et al., 2013), testing for hybridization (e.g., Fady et al., 2003; Epperson et al., 2009), paternity analysis (e.g., Lambeth et al., 2001), and testing taxonomic hypotheses (e.g., Clark et al., 2000; Ledig et al., 2004; Liu et al., 2012).
Within angiosperms, Poaceae, Vitaceae, and Fabaceae are well represented in published studies employing cpSSRs. This finding likely reflects the great economic importance of these families and availability of fully sequenced chloroplast genomes, which enables development of suitable polymorphic loci. Given the size and diversity of Poaceae and Fabaceae, it is not surprising that studies are varied in their foci and include agricultural and natural species. For Poaceae, notable studies exemplifying these varied foci include documenting movement of herbicide resistance from cultivated rice to weedy rice (Busconi et al., 2012); elucidating evolutionary relatedness among polyploids of the genus Cynodon Rich., an important group of grasses used for lawns in temperate areas of the world (Gulsen and Ceylan, 2011); quantifying seed movement in comparison to pollen flow to evaluate the drivers of spatial genetic structure in Anthoxanthum odoratum L. (Freeland et al., 2012); and dating the origin of sympatry among three fire-adapted Triodia R. Br. species with varying life history strategies (Armstrong, 2011). Within Fabaceae, notable studies using cpSSRs include evaluating current anthropogenic causes vs. historical fragmentation as a reason for the genetic structure in the endangered Caesalpinia echinata Lam. (Lira et al., 2003), a comparison of the ethnotaxonomy of Phaseolus L. species among Mexican farmers with genetic differentiation of the species (Soleri et al., 2013), and characterization of genetic diversity in landraces of P. vulgaris L. throughout Europe (Angioi et al., 2010).
The use of cpSSR loci in taxa of other diverse families, such as Asteraceae and Orchidaceae, though, is surprisingly uncommon. In all studies reporting empirical data for species of Asteraceae, we found that published primer sequences were used, and the average number of loci was 6.4. For Orchidaceae, a comparably large family, relatively few studies have employed cpSSRs, and in those, six or fewer loci on average were used. This finding may reflect the lack of widespread genomic resources for these families and the difficulty in transferring primer sequences across taxonomic levels, as reported by Ebert and Peakall (2009). Given the usefulness of next-generation sequencing (NGS) methods for primer design in nonmodel organisms and the increasing accessibility of these methods (Csencsics et al., 2010; Ekblom and Galindo, 2011; Guichoux et al., 2011), a wider range of families may see more representation in the future.
The taxonomic scale of studies employing cpSSRs has ranged from conspecific populations to intergeneric comparisons, although the greatest applicability of cpSSRs may be at the intraspecific level, as suggested by the high number of studies in the category of diversity and history for agricultural species (Fig. 4) and population genetics for native species (Fig. 5). Other common uses of cpSSR data are in understanding uniparental genetic structure (e.g., seed or pollen dispersal) and in studies of hybridization. For example, Richardson et al. (2002) were able to compare rates of seed dispersal via birds using mitochondrial markers with rates of pollen dispersal via wind using cpSSRs in whitebark pine and to determine past patterns of species range expansion in association with habitat and climatic changes. Marsico et al. (2009) also leveraged uniparentally inherited cpSSRs with nuclear data to contrast differing patterns of seed and pollen dispersal in Quercus garryana Douglas ex Hook. Bucci et al. (1998) elegantly demonstrated unidirectional introgression in the Pinus halepensis Mill. complex. Heuertz et al. (2006) identified shared chloroplast haplotypes among Fraxinus L. species in Europe and inferred that they had hybridized historically when coming in contact in common refugia or upon species range expansion. The use of cpSSRs across native and agricultural species as well as in studies ranging from population genetics to systematics demonstrates that they can be used to study many different types of questions in plant biology. The chloroplast genome has the potential to reveal much more about evolutionary history across plants than currently realized, and studies of many other taxa could benefit from the development of cpSSR markers.
Variability of cpSSRs
In many ways, cpSSRs are ideal markers for addressing a variety of genetic questions in plant biology. Genotypic variation is easily assessed using fragment-based methods, and the data are easy to interpret due to uniparental inheritance of the chloroplast genome in most species. However, polymorphism of cpSSR loci is highly variable across taxa, making the testing of primers a necessary precursor to data collection. We found that when primers were adapted from another species, many fewer loci were used (5.8) compared to studies employing primers developed for the study system (12.5). This was due to loci not amplifying in the focal species as well as their discontinued use due to lack of polymorphism. Ebert and Peakall (2009) also found a reduction in amplification success and a decrease in polymorphism among amplified loci with increasing phylogenetic distance.
There is substantial variation in repeat regions of the chloroplast genome across plants. In the extensive review by Ebert and Peakall (2009), the number of cpSSRs of at least eight mononucleotide repeats varied from one in the green alga Nephroselmis olivacea F. Stein to more than 700 in the green alga Chlorella vulgaris Beyerinck [Beijerinck]. Within angiosperms, the number of cpSSRs varied from less than 40 to more than 150 (Ebert and Peakall, 2009). We also found extensive variation in the whole chloroplast genome sequences included in this data set (Table 2). Slightly more loci (mean = 8.34) were used in studies of monocot taxa compared to those focused on eudicot taxa (mean = 7.61). If it is assumed that usable loci were also variable loci, then this finding may indicate a greater number of cpSSR loci in the chloroplast of monocots. However, greater taxonomic coverage of monocot chloroplast genomes is needed to evaluate if this difference is real.
Given that not all chloroplast loci are likely to be equally diverse across plants, it is important to consider a more targeted approach to developing cpSSR loci specific to a study system rather than relying on universal primers. The increasing availability of whole plastome sequences is likely to enhance one’s ability to identify suitable loci in nonmodel species, and is becoming a common pathway to developing variable markers (e.g., Wheeler et al., 2012). The whole chloroplast genome sequences identified in this study as well as those from Ebert and Peakall (2009) could be extremely helpful for the development of primers in novel taxa. Ebert and Peakall (2009) provide an excellent technical review on developing markers utilizing these resources. As Ebert and Peakall (2009) and others have suggested, we also recommend developing markers using sequences from the target species or a closely related species to maximize the number of polymorphic markers available. For taxa lacking genomic resources from a related species, the universal primers of Weising and Gardner (1999), Bryan et al. (1999), and Chung and Staub (2003) may be a good option to finding variable markers.
Fragment size homoplasy
All genetic markers that are assayed using fragment size genotyping are subject to erroneous inferences if the sizes do not reflect similarity of the underlying nucleotide sequence. Size homoplasy is more common in repetitive regions because of the higher mutation rate and complex mutational mechanisms compared to nucleotide substitutions and insertions/deletions involving longer stretches of nucleotides (Provan et al., 2001; Vachon and Freeland, 2011). Previous studies have called attention to the potential for and problems with fragment size homoplasy in cpSSRs (Liepelt et al., 2001; Provan et al., 2001; Ebert and Peakall, 2009; Vachon and Freeland, 2011), and others have studied homoplasy in nuclear SSRs (e.g., Estoup et al., 2002; Curtu et al., 2004; Barthe et al., 2012). Despite these calls in the literature to consider fragment size homoplasy of cpSSRs, two-thirds of the studies we reviewed did not indicate that the authors tested for homoplasy. Given that size homoplasy at certain cpSSR loci is expected to be prominent, we urge researchers employing cpSSRs to incorporate tests for homoplasy into their experimental design and data analyses.
Homoplasy can lead to over-estimation of relatedness among samples with similar fragment sizes when they actually have different evolutionary histories that may be detectable in the underlying sequence. The severity of this problem will vary by taxonomic and geographic scale, being greatest at higher taxonomic levels (e.g., among comparisons of congeneric species; Provan et al., 2001) and in large-scale analyses, such as phylogeographic studies (Provan et al., 2001; Vachon and Freeland, 2011). Repeated evolution of similar-sized alleles has been documented in interspecific studies employing cpSSRs, including sympatric orchid taxa (Soliva and Widmer, 1999) and members of Glycine subg. Glycine J. C. Wendl. (Doyle et al., 1998). Erroneous inferences involving comparisons across taxa may be expected to be more common due to cpSSR homoplasy, but intraspecific comparisons involving populations or subspecific taxa are commonly affected by size homoplasy as well. For example, Ebert and Peakall (2009) found size homoplasy among conspecific individuals sampled from a single population of the orchid Chiloglottis valida D. L. Jones. We found similar issues in Acmispon, both within and across species (Table 3). The greatest differences in underlying sequences when sizes were similar occurred when interspecific comparisons were made.
Undetected size homoplasy can affect estimates of genetic diversity and genetic distance (e.g., Provan et al., 2001; Navascués and Emerson, 2005), give misleading phylogenetic patterns (e.g., Doyle et al., 1998; Hale et al., 2004), and result in ambiguous ancestor-descendant relationships (e.g., Vachon and Freeland, 2011). For example, Navascués et al. (2006) found that homoplasy in cpSSRs affected inference of demographic scenarios, specifically testing for population expansion using the FS test (Fu, 1997), because it is based on genetic distance and haplotype diversity. Underestimation of haplotype diversity due to homoplasy reduced their power to detect population expansion in a simulated data set. The greatest distortions occurred in cases of recent population growth because the errors in estimates of genetic distance and number of haplotypes are more unbalanced in recent expansions (Navascués et al., 2006). The use of cpSSRs in phylogenetic analyses of species has been reported to produce reduced resolution and conflicting results when compared to other data sets. For example, Doyle et al. (1998) compared maximum and minimum estimates of the effects of size homoplasy on a phylogeny of species in Glycine subg. Glycine by mapping the distribution of cpSSR sizes onto a phylogeny based on restriction fragment length polymorphisms (RFLPs; i.e., maximum estimate) and reconstructing the phylogeny using the cpSSR and RFLP data (i.e., minimum estimate). They found that the addition of the cpSSR slightly reduced resolution of the resulting tree and the cpSSR did not reflect patterns of relationship that were suggested by plastome RFLP profiles (Doyle et al., 1998). In a similar study, Hale et al. (2004) compared phylogenies of Clusia L. species based on cpSSRs and chloroplast sequence variation and concluded that the cpSSRs were rich with homoplasy and too variable to be useful in reconstructing evolutionary relationships within the genus. In our data set of Acmispon, we have found conflicting phylogenies that are based on cpSSRs compared to nucleotide polymorphisms or other indels from the chloroplast genome (Wallace, unpublished data). Inference of ancestor-descendant relationships based on haplotype networks of cpSSR loci may also be complicated by homoplasy (Vachon and Freeland, 2011) because they increase the probability of loop structures in the network (Saltonstall, 2002; Jimenez et al., 2004).
The presence of size homoplasy in cpSSR loci is troublesome, but this does not void the usefulness of these markers. If the true underlying sequence of a microsatellite locus can be determined, then deviations from the standard assumptions can be taken into account. For example, Liepelt et al. (2001) noted that polymerase error can occur at a higher rate in regions containing multiple variable repeat regions. They suggested that these loci be split into two (or more, if necessary) loci by designing internal primers to increase DNA polymerase fidelity; this solution would also remove issues with size homoplasy by insuring that all variation is produced by a single variable region. Navascués and Emerson (2005) recommended the use of Nei’s unbiased haplotype diversity (Nei, 1978) over the measure developed by Goldstein et al. (1995), which assumes a pairwise mutation model and is affected more severely by homoplasy. The use of greater numbers of loci can also mitigate the effects of homoplasy, as chloroplast haplotypes that appear identical at a small number of sites are more likely to be distinguishable with greater marker coverage (Navascués and Emerson, 2005). Sequencing all chloroplast microsatellite alleles, regardless of method of discovery, can ensure that genotyped fragments reflect both actual length and nucleotide variation (Hale et al., 2004; Ebert and Peakall, 2009). However, Vachon and Freeland (2011) suggested that repetitive regions, whether genotyped or sequenced, should be excluded from certain types of large-scale analyses, such as haplotype networks, because of their high potential for repeated evolution. Instead, they advocate for the incorporation of repetitive regions into other types of analyses that take into account relative mutation rates or that use comparisons of population allele frequencies.
Given the potential for fragment size homoplasy to occur between and within species, we recommend that all studies making use of cpSSRs perform some sequencing of alleles to verify the mutational patterns of underlying sequences. Our approach to sequence at least two representative samples for each allele in each species of Acmispon provided a reasonable, cost-effective assessment of size homoplasy in this data set. For studies where deep genetic divergence is expected, further testing may be warranted. If one assumes that all individuals with the same sized allele also have the same underlying sequence, then sequencing cpSSRs also provides one with the ability to convert from fragment sizes to nucleotide sequences, thereby opening up additional types of analyses. Analyzing fragment data assuming a stepwise mutation model or complete independence of repeats can cause problems in detected divergence. However, by converting fragments to sequence using representative samples, data can be analyzed more accurately, taking all sequence changes into consideration appropriately. Although the cost of this approach varies based on the allelic diversity of the sample and the number of species (or taxa) included, it should always be more cost- and time-efficient than comparable studies using direct sequencing for each individual.
Sequencing of repeat regions can be difficult using Sanger sequencing because of slipped-strand mispairing leading to disassociation of the DNA polymerase from the template. Mononucleotide repeats composed of eight or more nucleotides are the most problematic because the active site of Taq DNA polymerase is presumed to be approximately the same length (Eom et al., 1996). Given that most cpSSR loci are composed of mononucleotide A/T repeats, generating clean sequences of alleles may be impossible. Proofreading DNA polymerases can improve the accuracy of PCR and the resulting sequences (Fazekas et al., 2010). For example, Guicking et al. (2008) provided a single nucleotide sequence protocol in which a repeat region is sequenced using one nucleotide instead of all four. Finally, regions that have multiple repeat regions can be broken up and sequenced separately (Liepelt et al., 2001).
Next-generation sequencing methodologies are increasingly used in the development of cpSSR loci (e.g., Doorduin et al., 2011; Kuang et al., 2011) and may provide an alternative to Sanger sequencing for genotyping cpSSRs (Guichoux et al., 2011) in nonmodel organisms (McPherson et al., 2013). NGS approaches experience a different set of issues when facing stretches of repetitive nucleotide sequences. NGS data are produced in the form of many short DNA sequence reads that must be assembled using bioinformatics software (Miller et al., 2010). Sequencing platforms employing shorter reads may pose difficulty in the accurate assembly of repetitive regions because it may be impossible to place reads that lack flanking sequences, which should be distinct for a locus, in a single location with 100% certainty (Treangen and Salzberg, 2012). Given that the majority of microsatellites in the chloroplast genome are mononucleotide A/T repeats, divergent flanking regions are especially important for correct assembly. Fortunately, chloroplast microsatellites are often quite short in length, and NGS methods are available that use reads long enough to capture repeat tracts as well as flanking sequences in their entirety. Finally, while polymerase error can potentially occur with NGS methods, computational assembly protocols are designed to handle this issue if read depth is sufficient (Miller et al., 2010; Treangen and Salzberg, 2012).
Cytoplasmic introgression
An additional drawback to consider with the use of cpSSRs is the potential for misleading patterns due to cytoplasmic introgression. As more studies have incorporated nuclear and chloroplast markers into the development of phylogenies, many of these have revealed discordant patterns between data sets, which is attributed to cytoplasmic introgression between samples even when there is little morphological or ecological evidence of hybridization. Such a conclusion is supported by Currat et al. (2008), who demonstrated a high likelihood that cytoplasmic introgression will be retained within a lineage from just a single hybridization event. Because the chloroplast genome is uniparentally inherited and nonrecombining in most plants, all chloroplast regions should exhibit introgression when it occurs. As a result, introgression could mask true evolutionary patterns of divergence. On the other hand, cpSSRs can be very helpful when there is a hypothesis of hybridization between species or populations. In our data set, these markers have been used to identify the direction of hybridization in natural populations and in identifying the parents of cultivated species. Because cpSSRs tend to be used more commonly below than above the species level and multiple samples are typically examined in such studies, cytoplasmic introgression may be readily detected and accounted for in these studies. However, studies employing cpSSRs above the species level and with the use of few samples per species should exercise caution when making inferences about evolutionary history.
Heteroplasmy
Uniparental mode of inheritance is considered a strength of chloroplast markers because the genes involved should not exhibit recombination or heterozygosity. However, some studies have identified biparental inheritance of chloroplasts leading to heteroplasmy (Lee et al., 1988; Johnson and Palmer, 1989; Chat et al., 2002; Frey et al., 2005; Hansen et al., 2007). If heteroplasmic individuals contain multiple variable chloroplast genomes, then genotyping with any chloroplast molecular marker will be problematic. If multiple haplotypes are present in sufficient numbers in the sampled tissue, then heteroplasmy is likely to be detectable in sequence chromatograms via the presence of double peaks or in the presence of more than one allele if genotyping is used. However, if copy number of the individual genomes varies, then it is possible that only the most frequent type will be selected in PCR and sequenced. In this case, a researcher may never know heteroplasmy exists in the study system, and cloning is needed to identify the presence of multiple chloroplast genomes. Given that an estimated one-third of surveyed angiosperms have at least occasional biparental inheritance of plastid DNA (Smith, 1989) and new instances continue to be reported (e.g., Hansen et al., 2007), testing for heteroplasmy prior to widespread use of chloroplast markers may be warranted.
CONCLUSIONS
This review demonstrates the continued widespread use of cpSSRs in plant biology. These markers have been most used for studies of population genetic structure in native species and in characterizing diversity and the origins of cultivated species. Even in the face of genomic approaches commonly available today, cpSSRs continue to be routinely used. This review demonstrates that researchers who have used primers developed in closely related species have had greater success in finding polymorphic loci. Thus, we believe future studies will benefit from a targeted approach using species-specific primers, rather than adapting published universal primers. The abundance of chloroplast genome sequences should facilitate primer design for many taxa. Impediments to the use of cpSSRs include fragment size homoplasy, introgression, and heteroplasmy. Fragment size homoplasy can be overcome by careful use of these markers, such as targeted sequencing of unique alleles to evaluate potential for homoplasy. We find that problems of size homoplasy are likely to be greatest in studies using cpSSRs for comparisons between species. Tests of homoplasy should be a standard component of all studies employing cpSSRs, regardless of the taxonomic level. Introgression and heteroplasmy can be tested via comparisons of alleles with closely related species and between parent and offspring, respectively. Given the greater prevalence of cpSSRs for intraspecific studies reported in the literature and the lower propensity for size homoplasy, their greatest utility is likely to be below the species level.
Supplementary Material
LITERATURE CITED
- Angioi S. A., Rau D., Attene G., Nanni L., Bellucci E., Logozzo G., Negri V., et al. 2010. Beans in Europe: Origin and structure of the European landraces of Phaseolus vulgaris L. Theoretical and Applied Genetics 121: 829–843. [DOI] [PubMed] [Google Scholar]
- Armstrong G. 2011. Evidence for the equal resilience of Triodia spp. (Poaceae), from different functional groups, to frequent fire dating back to the late Pleistocene. Heredity 107: 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai W. N., Wang W. T., Zhang D. Y. 2014. Contrasts between the phylogeographic patterns of chloroplast and nuclear DNA highlight a role for pollen-mediated gene flow in preventing population divergence in an East Asian temperate tree. Molecular Phylogenetics and Evolution 81: 37–48. 10.1016/j.ympev.2014.08.024. [DOI] [PubMed] [Google Scholar]
- Barthe S., Gugerli F., Barkley N. A., Maggia L., Cardi C., Scotti I. 2012. Always look on both sides: Phylogenetic information conveyed by simple sequence repeat allele sequences. PLoS ONE 7: e40699 10.1371/journal.pone.0040699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayly M. J., Rigault P., Spokevicius A., Ladiges P. Y., Ades P. K., Anderson C., Bossinger G., et al. 2013. Chloroplast genome analysis of Australian eucalypts—Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Molecular Phylogenetics and Evolution 69: 704–716. [DOI] [PubMed] [Google Scholar]
- Birky C. W., Jr 1995. Uniparental inheritance of mitochondrial and chloroplast genes: Mechanisms and evolution. Proceedings of the National Academy of Sciences, USA 92: 11331–11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan G. J., McNicoll J., Ramsay G., Meyer R. C., De Jong W. S. 1999. Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theoretical and Applied Genetics 99: 859–867. [Google Scholar]
- Bucci G., Anzedei M., Madaghiele A., Vendramin G. G. 1998. Detection of haplotypic variation and natural hybridization in halepensis-complex pine species using chloroplast simple sequence repeat (SSR) markers. Molecular Ecology 7: 1633–1643. [Google Scholar]
- Bucci G., Gonzalez-Martinez S. C., Provost G. L., Plomion C., Ribeiro M. M., Sebastiani F., Alia R., Vendramin G. G. 2007. Range-wide phylogeography and gene zones in Pinus pinaster Ait. revealed by chloroplast microsatellite markers. Molecular Ecology 16: 2137–2153. [DOI] [PubMed] [Google Scholar]
- Busconi M., Rossi D., Lorenzoni C., Baldi G., Fogher C. 2012. Spread of herbicide-resistant weedy rice (red rice, Oryza sativa L.) after 5 years of clearfield rice cultivation in Italy. Plant Biology 14: 751–759. [DOI] [PubMed] [Google Scholar]
- Chaïr H., Perrier X., Agbangla C., Marchand J. L., Dainou O., Noyer J. L. 2005. Use of cpSSRs for the characterisation of yam phylogeny in Benin. Genome 48: 674–684. [DOI] [PubMed] [Google Scholar]
- Chat J., Decroocq S., Decroocq V., Petit R. J. 2002. A case of chloroplast heteroplasmy in kiwifruit (Actinidia deliciousa) that is not transmitted during sexual reproduction. Journal of Heredity 93: 293–300. [DOI] [PubMed] [Google Scholar]
- Chen S., Yao H., Han J., Liu C., Song J., Shi L., Zhu Y., et al. 2010. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 5: e8613. 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S.-M., Staub J. E. 2003. The development and evaluation of consensus chloroplast primers pairs that possess highly variable sequence regions in a diverse array of plant taxa. Theoretical and Applied Genetics 107: 757–767. [DOI] [PubMed] [Google Scholar]
- Clark C. M., Wentworth T. R., O’Malley D. M. 2000. Genetic discontinuity revealed by chloroplast microsatellites in eastern North American Abies (Pinaceae). American Journal of Botany 87: 774–782. [PubMed] [Google Scholar]
- Clegg M. T. 1993. Chloroplast gene sequences and the study of plant evolution. Proceedings of the National Academy of Sciences, USA 90: 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csencsics D., Brodbeck S., Holderegger R. 2010. Cost-effective, species-specific microsatellite development for the endangered dwarf bulrush (Typha minima) using next-generation sequencing technology. Journal of Heredity 101: 789–793. [DOI] [PubMed] [Google Scholar]
- Currat M., Ruedi M., Petit R. J., Excoffier L. 2008. The hidden side of invasions: Massive introgression by local genes. Evolution 62: 1908–1920. [DOI] [PubMed] [Google Scholar]
- Curtu A.-L., Finkeldey R., Gailing O. 2004. Comparative sequencing of a microsatellite locus reveals size homoplasy within and between European oak species (Quercus spp.). Plant Molecular Biology Reporter 22: 339–346. [Google Scholar]
- Decroocq V., Hagen L. S., Favé M. G., Eyquard J. P., Pierronnet A. 2004. Microsatellite markers in the hexaploid Prunus domestica species and parentage lineage of three European plum cultivars using nuclear and chloroplast simple-sequence repeats. Molecular Breeding 13: 135–142. [Google Scholar]
- Deguilloux M. F., Dumolin-Lapègue S., Gielly L., Grivet D., Petit R. J. 2003. A set of primers for the amplification of chloroplast microsatellites in Quercus. Molecular Ecology Notes 3: 24–27. [Google Scholar]
- Dong W., Liu J., Yu J., Wang L., Zhou S. 2012. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE 7: e35071 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorduin L., Gravendeel B., Lammers Y., Ariyurek Y., Chin-A-Woeng T., Vrieling K. 2011. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Research 18: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Morgante M., Tingey S. V., Powell W. 1998. Size homoplasy in chloroplast microsatellites of wild perennial relatives of soybean (Glycine subgenus Glycine). Molecular Biology and Evolution 15: 215–218. [DOI] [PubMed] [Google Scholar]
- Drew B. T., Ruhfel B. R., Smith S. A., Moore M. J., Briggs B. G., Gitzendanner M. A., Soltis P. S., Soltis D. E. 2014. Another look at the root of the angiosperms reveals a familiar tale. Systematic Biology 63: 368–382. [DOI] [PubMed] [Google Scholar]
- Dyer R. J., Sork V. L. 2001. Pollen pool heterogeneity in shortleaf pine, Pinus echinata Mill. Molecular Ecology 10: 859–866. [DOI] [PubMed] [Google Scholar]
- Ebert D., Peakall R. 2009. Chloroplast simple sequence repeats (cpSSRs): Technical resources and recommendations for expanding cpSSR discovery and applications to a wide array of plant species. Molecular Ecology Resources 9: 673–690. [DOI] [PubMed] [Google Scholar]
- Echt C. S., DeVerno L. L., Anzidei M., Vendramin G. G. 1998. Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Molecular Ecology 7: 307–316. [Google Scholar]
- Ekblom R., Galindo J. 2011. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 107: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennos R. A., Sinclair W. T., Hu X.-X., Langdon A. 1999. Using organelle markers to elucidate the history, ecology, and evolution of plant populations. In P. M. Hollingsworth, R. M. Bateman, and R. J. Gornall [eds.], Molecular systematics and plant evolution, 1–19. Taylor and Francis, London, United Kingdom. [Google Scholar]
- Eom S. H., Wang J., Steitz T. A. 1996. Structure of Taq polymerase with DNA at the polymerase active site. Nature 382: 278–281. [DOI] [PubMed] [Google Scholar]
- Epperson B. K., Telewski F. W., Willyard A. 2009. Chloroplast diversity in a putative hybrid swarm of Ponderosae (Pinaceae). American Journal of Botany 96: 707–712. [DOI] [PubMed] [Google Scholar]
- Estoup A., Jarne P., Cornuet J. M. 2002. Homoplasy and mutation model at microsatellite loci and their consequence for population genetic analysis. Molecular Ecology 11: 1591–1604. [DOI] [PubMed] [Google Scholar]
- Fady B., Lefèvre F., Reynaud M., Vendramin G. G., Bou Dagher-Karrat M., Anzidei M., Pastorelli R., et al. 2003. Gene flow among different taxonomic units: Evidence from nuclear and cytoplasmic markers in Cedrus plantation forests. Theoretical and Applied Genetics 107: 1132–1138. [DOI] [PubMed] [Google Scholar]
- Fazekas A. J., Steeves R., Newmaster S. G. 2010. Improving sequencing quality from PCR products containing long mononucleotide repeats. BioTechniques 48: 277–285. [DOI] [PubMed] [Google Scholar]
- Freeland J. R., Biss P., Silvertown J. 2012. Contrasting patterns of pollen and seed flow influence the spatial genetic structure of sweet vernal grass (Anthoxanthum odoratum) populations. Journal of Heredity 103: 28–35. [DOI] [PubMed] [Google Scholar]
- Frey J., Frey B., Forcioli D. 2005. Quantitative assessment of heteroplasmy levels in Senecio vulgaris chloroplast DNA. Genetica 123: 255–261. [DOI] [PubMed] [Google Scholar]
- Fu Y. X. 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargano D., Scotti N., Vezzi A., Bilardi A., Valle G., Grillo S., Cozzolino S., Cardi T. 2012. Genome-wide analysis of plastome sequence variation and development of plastidial CAPS markers in common potato and related Solanum species. Genetic Resources and Crop Evolution 59: 419–430. [Google Scholar]
- Godbout J., Beaulieu J., Bousquet J. 2010. Phylogeographic structure of jack pine (Pinus banksiana; Pinaceae) supports the existence of a coastal glacial refugium in northeastern North America. American Journal of Botany 97: 1903–1912. [DOI] [PubMed] [Google Scholar]
- Goldstein D. B., Linares A. R., Cavalli-Sforza L. L., Feldman M. W. 1995. An evaluation of genetic distances for use with microsatellite loci. Genetics 139: 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez A., Vendramin G. G., González-Martínez S. C., Alía R. 2005. Genetic diversity and differentiation of two Mediterranean pines (Pinus halepensis Mill. and Pinus pinaster Ait.) along a latitudinal cline using chloroplast microsatellite markers. Diversity and Distributions 11: 257–263. [Google Scholar]
- Gugerli F., Senn J., Anzidei M., Madaghiele A., Büchler U., Sperisen C., Vendramin G. G. 2001. Chloroplast microsatellites and mitochondrial nad1 intron 2 sequences indicate congruent phylogenetic relationships among Swiss stone pine (Pinus cembra), Siberian stone pine (Pinus sibirica), and Siberian dwarf pine (Pinus pumila). Molecular Ecology 10: 1489–1497. [DOI] [PubMed] [Google Scholar]
- Guichoux E., Lagache L., Wagner S., Chaumeil P., Léger P., Lepais O., Lepoittevin C., et al. 2011. Current trends in microsatellite genotyping. Molecular Ecology Resources 11: 591–611. [DOI] [PubMed] [Google Scholar]
- Guicking D., Kröger-Kilian T., Weising K., Blattner F. R. 2008. Single nucleotide sequence analysis: A cost- and time-effective protocol for the analysis of microsatellite- and indel-rich chloroplast DNA regions. Molecular Ecology Resources 8: 62–65. [DOI] [PubMed] [Google Scholar]
- Gulsen O., Ceylan A. 2011. Elucidating polyploidization of bermudagrasses as assessed by organelle and nuclear DNA markers. OMICS: A Journal of Integrative Biology 15: 903–912. [DOI] [PubMed] [Google Scholar]
- Haasl R. J., Payseur B. A. 2011. Multi-locus inference of population structure: A comparison between single nucleotide polymorphisms and microsatellites. Heredity 106: 158–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale M. L., Borland A. M., Gustafsson M. H. G., Wolff K. 2004. Causes of size homoplasy among chloroplast microsatellites in closely related Clusia species. Journal of Molecular Evolution 58: 182–190. [DOI] [PubMed] [Google Scholar]
- Hansen A. K., Escobar L. K., Gilbert L. E., Jansen R. K. 2007. Paternal, maternal, and biparental inheritance of the chloroplast genome in Passiflora (Passifloraceae): Implications for phylogenetic studies. American Journal of Botany 94: 42–46. [DOI] [PubMed] [Google Scholar]
- Heuertz M., Carnevales S., Fineschi S., Sebastiani F., Hausman F., Paule L., Vendramin G. G. 2006. Chloroplast DNA phylogeography of European ashes, Fraxinus sp. (Oleaceae): Roles of hybridization and life history traits. Molecular Ecology 15: 2131–2140. [DOI] [PubMed] [Google Scholar]
- Hirao T., Watanabe A., Kurita M., Kondo T., Takata K. 2009. A frameshift mutation of the chloroplast mat K coding region is associated with chlorophyll deficiency in the Cryptomeria japonica virescent mutant Wogon-Sugi. Current Genetics 55: 311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth P. M., Forrest L. L., Spouge J. L., Hajibabaei M., Ratnasingham S., van der Bank M., Chase M. W., et al. 2009. A DNA barcode for land plants. Proceedings of the National Academy of Sciences, USA 106: 12794–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth P. M., Graham S. W., Little D. P. 2011. Choosing and using a plant DNA barcode. PLoS ONE 6: e19254. 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huotari T., Korpelainen H. 2012. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene 508: 96–105. [DOI] [PubMed] [Google Scholar]
- Ishii T., Mori N., Ogihara Y. 2001. Evaluation of allelic diversity at chloroplast microsatellite loci among common wheat and its ancestral species. Theoretical and Applied Genetics 103: 896–904. [Google Scholar]
- Jimenez P., Lopez de Heredia U., Collada C., Lorenzo L., Gil L. 2004. High variability of chloroplast DNA in three Mediterranean evergreen oaks indicates complex evolutionary history. Heredity 93: 510–515. [DOI] [PubMed] [Google Scholar]
- Johnson L. B., Palmer J. D. 1989. Heteroplasmy of chloroplast DNA in Medicago. Plant Molecular Biology 12: 3–11. [DOI] [PubMed] [Google Scholar]
- Kim K. J., Lee H. L. 2004. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng Nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Research 11: 247–261. [DOI] [PubMed] [Google Scholar]
- Kim Y. K., Park C. W., Kim K. J. 2009. Complete chloroplast DNA sequence from a Korean endemic genus, Megaleranthis saniculifolia, and its evolutionary implications. Molecules and Cells 27: 365–381. [DOI] [PubMed] [Google Scholar]
- Kuang D.-Y., Wu H., Wang Y.-L., Gao L.-M., Zhang S.-Z., Lu L. 2011. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 54: 663–673. [DOI] [PubMed] [Google Scholar]
- Lambeth C., Lee B.-C., O’Malley D., Wheeler N. 2001. Polymix breeding with parental analysis of progeny: An alternative to full-sib breeding and testing. Theoretical and Applied Genetics 103: 930–943. [Google Scholar]
- Ledig F. T., Hodgskiss P. D., Krutovskii K. V., Neale D. B., Eguiluz-Piedra T. 2004. Relationships among the spruces (Picea, Pinaceae) of southwestern North America. Systematic Botany 29: 275–295. [Google Scholar]
- Lee D. J., Blake T. K., Smith S. E. 1988. Biparental inheritance of chloroplast DNA and the existence of heteroplasmic cells in alfalfa. Theoretical and Applied Genetics 76: 545–549. [DOI] [PubMed] [Google Scholar]
- Li X. W., Gao H. H., Wang Y. T., Song J. Y., Henry R., Wu H. Z., Hu Z. G., et al. 2013. Complete chloroplast genome sequence of Magnolia grandiflora and comparative analysis with related species. Science China Life Sciences 56: 189–198. [DOI] [PubMed] [Google Scholar]
- Liepelt S., Kuhlenkamp V., Anzidei M., Vendramin G. G., Ziegenhagen B. 2001. Pitfalls in determining size homoplasy in microsatellite loci. Molecular Ecology Notes 1: 332–335. [Google Scholar]
- Lira C. F., Cardoso S. R. S., Ferreira P. C. G., Cardoso M. A., Provan J. 2003. Long-term population isolation in the endangered tropical tree species Caesalpinia echinata Lam. revealed by chloroplast microsatellites. Molecular Ecology 12: 3219–3225. [DOI] [PubMed] [Google Scholar]
- Liu Y., Huo N., Dong L., Wang Y., Zhang S., Young H. A., Feng X., Gu Y. Q. 2013. Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS ONE 8: e57533. 10.1371/journal.pone.0057533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z.-L., Yang X., Zhang H., Zhang J., Li J.-F. 2012. Systematic position of Pinus henryi (Pinaceae) as revealed by multiple evidence. Nordic Journal of Botany 30: 671–679. [Google Scholar]
- Lukas B., Novak J. 2013. The complete chloroplast genome of Origanum vulgare L. (Lamiaceae). Gene 528: 163–169. [DOI] [PubMed] [Google Scholar]
- Marshall H. D., Newton C., Ritland K. 2001. Sequence-repeat polymorphisms exhibit the signature of recombination in lodgepole pine chloroplast DNA. Molecular Biology and Evolution 18: 2136–2138. [DOI] [PubMed] [Google Scholar]
- Marsico T. D., Hellmann J. J., Romero-Severson J. 2009. Patterns of seed dispersal and pollen flow in Quercus garryana (Fagaceae) following post-glacial climatic changes. Journal of Biogeography 36: 929–941. [Google Scholar]
- McPherson H., van der Merwe M., Delaney S. K., Edwards M. A., Henry R. J., McIntosh E., Rymer P. D., et al. 2013. Capturing chloroplast variation for molecular ecology studies: A simple next generation sequencing approach applied to a rainforest tree. BMC Ecology 13: 8. 10.1186/1472-6785-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J., Koren S., Sutton G. 2010. Assembly algorithms for next-generation sequence data. Genomics 95: 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M. J., Soltis P. S., Bell C. D., Burleigh J. G., Soltis D. E. 2010. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proceedings of the National Academy of Sciences, USA 107: 4623–4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasri N., Bojovic S., Vendramin G. G., Fady B. 2008. Population genetic structure of the relict Serbian spruce, Picea omorika, inferred from plastid DNA. Plant Systematics and Evolution 271: 1–7. [Google Scholar]
- Navascués M., Emerson B. C. 2005. Chloroplast microsatellites: Measures of genetic diversity and the effect of homoplasy. Molecular Ecology 14: 1333–1341. [DOI] [PubMed] [Google Scholar]
- Navascués M., Vaxevanidou Z., González-Martínez S. C., Climent J., Gil L., Emerson B. C. 2006. Chloroplast microsatellites reveal colonization and metapopulation dynamics in the Canary Island pine. Molecular Ecology 15: 2691–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara Y., Terachi T., Sasakuma T. 1988. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proceedings of the National Academy of Sciences, USA 85: 8573–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmstead R. G., Palmer J. D. 1994. Chloroplast DNA systematics: A review of methods and data analysis. American Journal of Botany 81: 1205–1224. [Google Scholar]
- Parks M., Cronn R., Liston A. 2009. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology 7: 84. 10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell W., Morgante M., Andre C., McNicol J. W., Machray G. C., Doyle J. J., Tingey S. V., Rafalski J. A. 1995. Hypervariable microsatellites provide a general source of polymorphic DNA markers for the chloroplast genome. Current Biology 5: 1023–1029. [DOI] [PubMed] [Google Scholar]
- Provan J., Powell W., Hollingsworth P. M. 2001. Chloroplast microsatellites: New tools for studies in plant ecology and evolution. Trends in Ecology & Evolution 16: 142–147. [DOI] [PubMed] [Google Scholar]
- Qian J., Song J., Gao H., Zhu Y., Xu J., Pang X., Yao H., et al. 2013. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE 8: e57607. 10.1371/journal.pone.0057607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboud X., Zeyl C. 1994. Organelle inheritance in plants. Heredity 72: 132–140. [Google Scholar]
- Richardson B. A., Brunsfeld S. J., Klopfenstein N. B. 2002. DNA from bird-dispersed seed and wind-disseminated pollen provides insights into postglacial colonization and population genetic structure of whitebark pine (Pinus albicaulis). Molecular Ecology 11: 215–227. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Banderas A., Vargas-Mendoza C. F., Buonamici A., Vendramin G. G. 2009. Genetic diversity and phylogeographic analysis of Pinus leiophylla: A post-glacial range expansion. Journal of Biogeography 36: 1807–1820. [Google Scholar]
- Saltonstall K. 2002. Cryptic invasion by a non-native genotype of the common reed, Phragmites australis, into North America. Proceedings of the National Academy of Sciences, USA 99: 2445–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalfi M., Piotti A., Rossi M., Piovani P. 2009. Genetic variability of Italian southern Scots pine (Pinus sylvestris L.) populations: The rear edge of the range. European Journal of Forest Research 128: 377–386. [Google Scholar]
- Sebastiani F., Carnevale S., Vendramin G. G. 2004. A new set of mono- and dinucleotide chloroplast microsatellites in Fagaceae. Molecular Ecology Notes 4: 259–261. [Google Scholar]
- Shaw J., Lickey E. B., Beck J. T., Farmer S. B., Liu W., Miller J., Siripun K. C., et al. 2005. The tortoise and the hare II: Relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analyses. American Journal of Botany 92: 142–166. [DOI] [PubMed] [Google Scholar]
- Shaw J., Lickey E. B., Schilling E. E., Small R. L. 2007. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: The tortoise and the hare III. American Journal of Botany 94: 275–288. [DOI] [PubMed] [Google Scholar]
- Smith S. E. 1989. Biparental inheritance of organelles and its implications in crop improvement. Plant Breeding Reviews 6: 361–393. [Google Scholar]
- Soleri D., Worthington M., Aragón-Cuevas F., Smith S. E., Gepts P. 2013. Farmers’ varietal identification in a reference sample of local Phaseolus species in the Sierra Juárez, Oaxaca, Mexico. Economic Botany 67: 283–298. [Google Scholar]
- Soliva M., Widmer A. 1999. Genetic and floral divergence among sympatric populations of Gymnadenia conopsea s.l. (Orchidaceae) with different flowering phenology. International Journal of Plant Sciences 160: 897–905. [DOI] [PubMed] [Google Scholar]
- Sonah H., Deshmukh R. K., Sharma A., Singh V. P., Gupta D. K., Gacche R. N., Rana J. C., et al. 2011. Genome-wide distribution and organization of microsatellites in plants: An insight into marker development in Brachypodium. PLoS ONE 6: e21298. 10.1371/journal.pone.0021298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stech M., Quandt D. 2010. 20,000 species and five key markers: The status of molecular bryophyte phylogenetics. Phytotaxa 9: 196–228. [Google Scholar]
- Sun Y.-X., Moore M. J., Meng A.-P., Soltis P. S., Soltis D. E., Li J.-Q., Wang H.-C. 2013. Complete plastid genome sequencing of Trochodendraceae reveals a significant expansion of the inverted repeat and suggests a Paleogene divergence between the two extant species. PLoS ONE 8: e60429. 10.1371/journal.pone.0060429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terakami S., Matsumura Y., Kurita K., Kanamori H., Katayose Y., Yamamoto T., Katayama H. 2012. Complete sequence of the chloroplast genome from pear (Pyrus pyrifolia): Genome structure and comparative analysis. Tree Genetics & Genomes 8: 841–854. [Google Scholar]
- Torales S. L., Rivarola M., Pomponio M. F., Gonzalez S., Acuna C. V., Fernandez P., Lauenstein D. L., et al. 2013. De novo assembly and characterization of leaf transcriptome for the development of functional molecular markers of the extremophile multipurpose tree species Prosopis alba. BMC Genomics 14: 705. 10.1186/1471-2164-14-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T., Salzberg S. 2012. Repetitive DNA and next-generation sequencing computational challenges and solutions. Nature Reviews Genetics 13: 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno S., Setsuko S., Kawahara T., Yoshimaru H. 2005. Genetic diversity and differentiation of the endangered Japanese endemic tree Magnolia stellata using nuclear and chloroplast microsatellite markers. Conservation Genetics 6: 563–574. [Google Scholar]
- Vachon N., Freeland J. R. 2011. Phylogeographic inferences from chloroplast DNA: Quantifying the effects of mutations in repetitive and non-repetitive sequences. Molecular Ecology Resources 11: 279–285. [DOI] [PubMed] [Google Scholar]
- Vendramin G. G., Lelli L., Rossi P., Morgante M. 1996. A set of primers for the amplification of 20 chloroplast microsatellites in Pinaceae. Molecular Ecology 5: 595–598. [DOI] [PubMed] [Google Scholar]
- Vendramin G. G., Degen B., Petit R. J., Anzidei M., Madaghiele A., Ziegenhagen B. 1999. High level of variation at Abies alba chloroplast microsatellite loci in Europe. Molecular Ecology 8: 1117–1126. [DOI] [PubMed] [Google Scholar]
- Viard F., El-Kassaby Y. A., Ritland K. 2001. Diversity and genetic structure in populations of Pseudotsuga menziesii (Pinaceae) at chloroplast microsatellite loci. Genome 44: 336–344. [DOI] [PubMed] [Google Scholar]
- Wallace L. E., Culley T. M., Weller S. G., Sakai A. K., Kuenzi A., Roy T., Wagner W. L., Nepokroeff M. 2011. Asymmetrical gene flow in a hybrid zone of Hawaiian Schiedea (Caryophyllaceae) species with contrasting mating systems. PLoS ONE 6: e24845. 10.1371/journal.pone.0024845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Mao J. F., Zhao W., Wang X.-R. 2013. Impact of geography and climate on the genetic differentiation of the subtropical pine Pinus yunnanensis. PLoS ONE 8: e67345. 10.1371/journal.pone.0067345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weising K., Gardner R. C. 1999. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42: 9–19. [PubMed] [Google Scholar]
- Wheeler G. L.2013. Population-level genetic structure of Acmispon argophyllus on the Channel Islands of California. M.S. thesis, Mississippi State University, Mississippi State, Mississippi, USA.
- Wheeler G. L., McGlaughlin M. E., Wallace L. E. 2012. Variable length chloroplast markers for population genetic studies in Acmispon (Fabaceae). American Journal of Botany 99: e408–e410. [DOI] [PubMed] [Google Scholar]
- Yi D.-K., Kim K.-J. 2012. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS ONE 7: e35872. 10.1371/journal.pone.0035872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D.-K., Lee H.-L., Sun B.-Y., Chung M. Y., Kim K.-J. 2012. The complete chloroplast DNA sequence of Eleutherococcus senticosus (Araliaceae): Comparative evolutionary analyses with other three asterids. Molecules and Cells 33: 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Gao L., Wang B., Su Y.-J., Wang T. 2013. The complete chloroplast genome sequence of Cephalotaxus oliveri (Cephalotaxaceae): Evolutionary comparison of Cephalotaxus chloroplast DNAs and insights into the loss of inverted repeat copies in gymnosperms. Genome Biology and Evolution 5: 688–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.