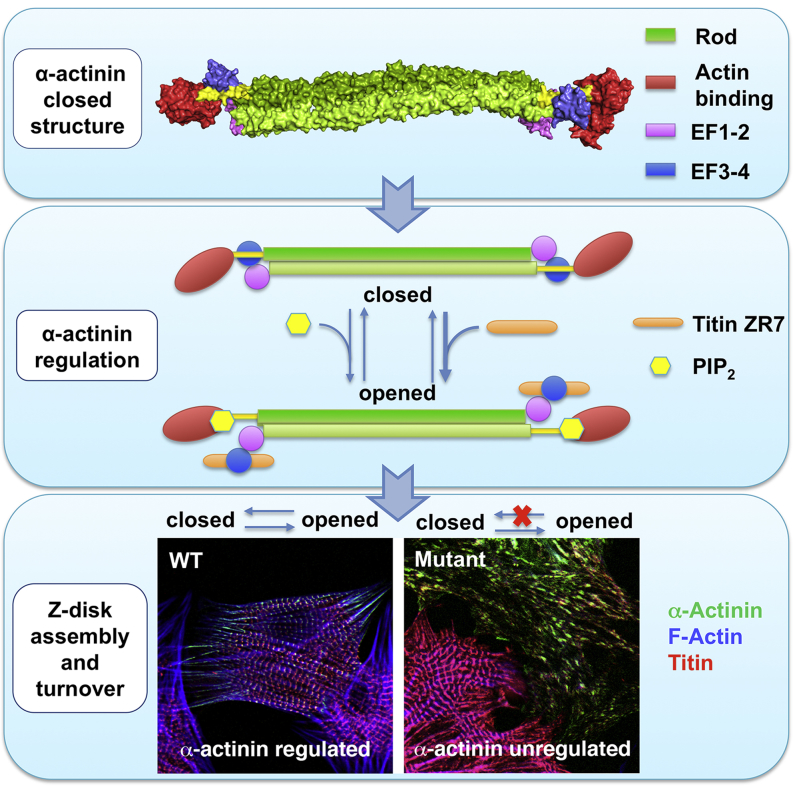
Summary
The spectrin superfamily of proteins plays key roles in assembling the actin cytoskeleton in various cell types, crosslinks actin filaments, and acts as scaffolds for the assembly of large protein complexes involved in structural integrity and mechanosensation, as well as cell signaling. α-actinins in particular are the major actin crosslinkers in muscle Z-disks, focal adhesions, and actin stress fibers. We report a complete high-resolution structure of the 200 kDa α-actinin-2 dimer from striated muscle and explore its functional implications on the biochemical and cellular level. The structure provides insight into the phosphoinositide-based mechanism controlling its interaction with sarcomeric proteins such as titin, lays a foundation for studying the impact of pathogenic mutations at molecular resolution, and is likely to be broadly relevant for the regulation of spectrin-like proteins.
Graphical Abstract
Highlights
-
•
Structure of human α-actinin-2 in an autoinhibited closed conformation
-
•
Facilitation of PIP2-induced allosteric modulation for opening and titin binding
-
•
Essentiality of structural flexibility for crosslinking antiparallel F-actin
-
•
Relevance for the intramolecular pseudoligand regulation mechanism of the spectrin family
The structure of human α-actinin, the major component of the basic contractile units of muscles, reveals a unique phosphoinositide-based mechanism of its regulation, as well as assembly.
Introduction
Mobility is essential to all living organisms, from organelle transport to movement of entire organisms. In many motile systems, actin and myosin filaments assume ordered arrays organized by specific actin or myosin ligands. In higher animals, movement is performed by striated muscle, defined by highly regular arrangements of visible striations. The minimal contractile unit of striated muscle is the sarcomere, which is anchored and stabilized by transverse crosslinking structures at the two lateral Z-disk boundaries, the A-band and the central M-band (Gautel, 2011, Tskhovrebova and Trinick, 2010). In vertebrates, the giant protein titin (connectin) spans Z-disks to M-bands and may act as a blueprint for sarcomere assembly (Gautel, 2011, Tskhovrebova and Trinick, 2010). Within the vertebrate Z-disk, a complicated network of protein-protein interactions anchors and stabilizes the actin and the elastic titin filaments (Luther, 2009).
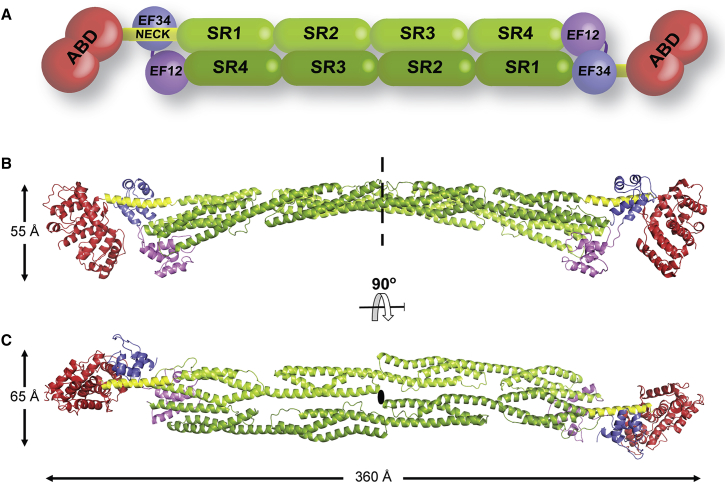
α-actinin was originally described as an actin-crosslinking Z-disk protein in muscle (Masaki et al., 1967), but its four closely related isogenes (ACTN1–4) fulfil similar functions in all cell types (Foley and Young, 2014, Sjöblom et al., 2008). α-actinin, in particular isoform 2 (encoded by ACTN2), is the major Z-disk component, where it plays a central role crosslinking actin and titin filaments. α-actinin is an antiparallel homodimer of more than 200 kDa, comprising an N-terminal actin-binding domain (ABD), a central domain of four spectrin-like repeats (SRs), and a C-terminal calmodulin-like domain (CAMD) with two pairs of EF hand motifs (EFs) (Figure 1A). Because the SR region appears to have a cylindrical shape, it is also called the rod domain.
Figure 1.
Complete Structure of α-Actinin-2 in Closed Conformation
(A) Domain composition of the α-actinin dimer. Color code, as in all the following figures: ABD, red; neck, yellow; SR1–SR4, green; EF1-2, violet; EF3-4, blue.
(B) The dimeric structure of α-actinin-2 assembled from two halves of the α-actinin-2 protomer (ABD-SR1-SR2/SR3-SR4-CaM) through a crystallographic 2-fold axis (dashed line; ellipse in C). Overall dimensions are indicated.
(C) Same as in (B), rotated 90° around the horizontal axis.
See also Table S1.
The elementary structure of the Z-disk is that of a tetragonal array of antiparallel actin filaments spaced 240 Å apart and crosslinked by successive layers of filaments at intervals of ≈190 Å rotated by 90° between each layer along the myofibril axis (Goldstein et al., 1979). These filaments correlate with α-actinin crosslinks, but the molecular layout of α-actinin that allows strict alternating crosslinks between actin filaments remains elusive (reviewed in Luther, 2009).
In striated muscle, α-actinin also binds differentially spliced titin Z-repeats, possibly regulating the number of crosslinking α-actinin molecules (Gautel et al., 1996). These titin Z-repeats contain a short, hydrophobic α-actinin-binding motif, which interacts with the CAMD (Atkinson et al., 2001, Sorimachi et al., 1997, Young et al., 1998). Additionally, α-actinin binds a plethora of cytoplasmic and membrane proteins in striated muscle and nonmuscle tissues (Djinovic-Carugo et al., 2002, Foley and Young, 2014). To achieve ordered cytoskeletal assemblies, the binding properties of α-actinin must be spatiotemporally regulated. Actin binding of nonmuscle isoforms is regulated by binding of Ca2+ ions to the CAMD (Foley and Young, 2014). In contrast, muscle α-actinin is calcium insensitive, and its F-actin- and titin-binding properties are likely regulated by phospholipids (most notably phosphatidylinositol bisphosphate; PIP2) (Fukami et al., 1992, Young et al., 1998, Young and Gautel, 2000). Despite being a major integrator of titin and actin in one of the stiffest structures of the sarcomere, muscle α-actinin shows surprisingly dynamic association with the Z-disk actin cytoskeleton (Sanger and Sanger, 2008), suggesting that its actin and titin binding activity must be dynamically regulated.
Biochemical analysis led us to propose previously that the α-actinin-titin interaction is regulated by an intramolecular mechanism where a short sequence in α-actinin between the ABD and the rod interacts with the CAMD in a pseudoligand mechanism (Young and Gautel, 2000). A similar mode of interaction has been found for the α-actinin ligand palladin (Beck et al., 2011).
Here, we report the crystal structure of human α-actinin-2 at 3.5 Å resolution. It is a complete high-resolution α-actinin-2 structure, revealing insight into the mechanism that promotes the molecular assembly of the Z-disk and the intramolecular contacts that regulate these interactions. Furthermore, the structure provides a template for the α-actinin and spectrin superfamily and insight into the impact of disease-associated genetic variants in ACTN genes.
Results
Closed Structure of α-Actinin-2
Overall Architecture
The structure of α-actinin-2 was solved and refined to 3.5 Å resolution to an Rwork/Rfree of 20.5%/25.8% (Table S1 available online). The α-actinin-2 dimer reveals a cylindrical shape ∼360 Å long and ∼60 Å wide (Figures 1B and 1C). Each protomer comprises an N-terminal ABD followed by an α-helical linker (neck), four spectrin-like repeats (SR1–4), and a C-terminal CAMD of two pairs of EF hands (EF1-2 and EF3-4). The first 34 and last 2 residues are missing from our model. Two antiparallel SR1–4s that assemble the core of the extended structure form the central portion of the dimer (rod). The two ABDs and two CAMDs flank the elongated assembly at its ends.
As expected, in the absence of actin, the ABD is in a closed conformation, in which the two calponin homology domains (CH1 and CH2) are in extensive contact, similar to the arrangement found in α-actinin-3, plectin, and fimbrin ABD domains (Franzot et al., 2005, Klein et al., 2004).
The ABD is linked to the first spectrin-like repeat (SR1) of the rod through the six-turn α-helical neck that is flanked by two hinges, the first on residue G258 linking the ABD to the neck and the second on residue M283 linking the neck to SR1 (Figure 2A).
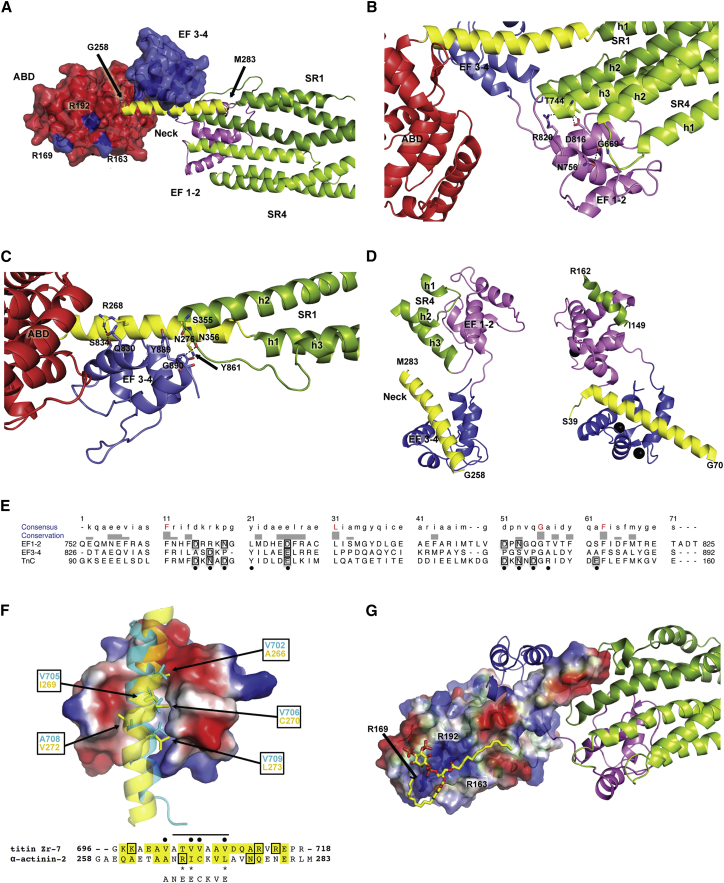
Figure 2.
Close-Up of the Functional Domain Interactions
(A) PIP2 binding site on α-actinin-2 and the EF3-4-neck interaction. ABD and EF3-4 are presented with their solvent-accessible surface areas. The R residues responsible for PIP2 binding are highlighted in blue on the ABD surface.
(B) Detail of EF1-2 interactions with SR4.
(C) Detail of EF3-4 interactions with the neck region and SR1.
(D) Comparison of α-actinin-2 CAMD with TnC bound to TnI, aligned on EF3-4 and the C-terminal lobe of TnC. Left: cartoon representation of α-actinin EF1-2 and EF3-4 with the interacting portion of the neck from the juxtaposed subunit (yellow) and a part of SR4 from the same subunit. Right: cartoon representation of TnC bound to TnI. N-terminal lobe, violet, as in EF1-2, and the C-terminal lobe, blue, as in EF3-4. Bound N-terminal TnI fragment, yellow; C-terminal TnI helix, green. Calcium ions are shown as black spheres (on TnC). The C-terminal lobe of TnC is aligned to EF3-4. To show the direction of bound helices, residues defining the neck domain of actinin and TnI fragments are indicated.
(E) Sequence alignment of EF1-2 and EF3-4 and the C-terminal lobe of TnC. The corresponding calcium-binding positions in Ca2+/CaM are indicated by black dots. Charged residues involved in calcium binding are boxed. Fully conserved residues are highlighted in red.
(F) Comparison of interactions between Zr-7 and the neck α helices with EF3-4. Electrostatic surface representation of the EF hands and cartoon representation for Zr-7 (cyan) and the α-actinin-2 neck (yellow). Side chains of key hydrophobic residues are shown as sticks; sequence numbers are boxed. Structural sequence alignment between titin Zr-7 and the neck. Residues involved in the interface with EF3-4 are highlighted in yellow. Residues involved in H bonding are boxed. Asterisks denote the mutations in the NEECK mutant. Black dots denote the CaM-binding motif 1-4-5-8. Underneath is shown the sequence of the NEEK mutant.
(G) α-actinin-2 with docked PIP2 (the overall top-scoring pose is shown as a yellow stick model) together with the EF3-4-neck interaction. The top-scoring pose with two PIP2 tails in contact with the neck region is presented in Figure S2F. ABD and the neck region are presented with their solvent-accessible surface areas colored by electrostatic potential and the rest by cartoon representation and color coded as in Figure 1. The three R residues responsible for PIP2 binding are indicated.
See also Table S2.
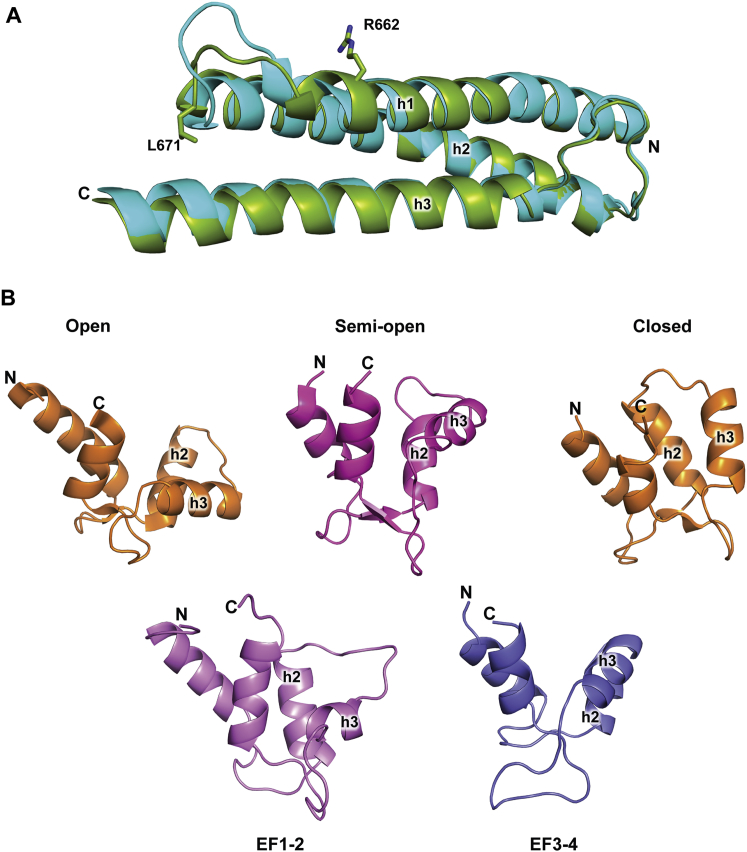
Comparison of the spectrin-like repeats in the full-length α-actinin-2 structure with the previously determined dimeric rod domain (Ylänne et al., 2001) (Protein Data Bank [PDB] ID code 1HCI) reveals high similarity between SR2, SR3, and SR4 and minor differences for SR1 (Table S2). However, although the α helices of the two SR4 domains are well aligned, there are large deviations between α helices h1 and h2 due to contacts of EF1-2 positioned over this loop in the full-length structure (Figures 1 and 2B; Figure S1A). The antiparallel α-actinin-2 dimer assembles predominately via the rod domain. The two ABDs are flanked at both termini along the long axis of the rod and are stabilized in their position by a few polar interactions with the neck region and EF3-4.
Figure S1.
Conformational Variability and Substructures of α-Actinin-2, Related to Figure 2
(A) Loop connecting α helices 1 and 2 in SR4 undergoes a conformational change upon EF1-2 binding. Ribbon representation of superimposed SR4 from the full-length structure of α-actinin-2 (green) and previously determined rod domain structure (light blue) (PDB entry 1HCI) (Ylänne et al., 2001).
(B) Cartoon representation of EF-hand lobes in different conformations. On top from left to right: the open conformation of the N-terminal lobe from calmodulin (PDB code 1CDM) (Meador et al., 1993), the semi-open conformation from the C-terminal lobe of the myosin essential light chain (PDB code 1SCM) (Xie et al., 1994) and the closed conformation from the calcium free C-terminal lobe of calmodulin (PDB code 1DMO) (Zhang et al., 1995). On bottom the EF1-2 and EF3-4 from the α-actinin-2 CAMD. All structures are aligned on the N-terminal α-helix. The N-terminal and C-terminal amino acids as also the middle helices h2 and h3 are indicated on the structures. The h2 and h3 α helices on both EF1-2 and EF3-4 are tilted compared to the positions on the closed lobe conformation, similarly to the semi-open conformation.
The C-terminal CAMD is divided into structurally distinct N- and C-terminal lobes: EF hands 1-2 and 3-4 connected by a short linker. The overall structure of each lobe is well defined through the main-chain atoms on the α helices of the EF hands. As expected, no density conforming to bound calcium was detected, and the architecture of the CAMD resembles calcium-free conformations of calmodulin (see below).
Calmodulin-like Domain Conformation and Interactions
The EF3-4 accommodates the α helix of the neck from the juxtaposed α-actinin-2 protomer through its hydrophobic cleft. This classifies the EF3-4-neck interaction as 1:1 or canonical (Hoeflich and Ikura, 2002), involving the cavity formed by the first helix of the EF3-4, the linker between the EF3-4 helices, and the C-terminal helix of the lobe (Figures 2C and 2D). The EF3-4-neck interface is mainly hydrophobic, supported with a few H-bonds (Figure 2C). The neck region displays the known hydrophobic Ca2+/calmodulin (CaM)-binding motif, termed 1-4-5-8 (Bayley et al., 1996), with hydrophobic A266, I269, C270, and L273 at these positions (Figure 2F). Additionally, EF3-4 interact with the connecting loop of α helices 2 and 3 from SR1 via an H-bond and a π-stacking interaction, supporting previous reports that SR1 stabilizes the interaction with EF3-4 (Young and Gautel, 2000). The overall binding interface of EF3-4 is ∼500 Å2, representing 11% of its total surface area. The root-mean-square deviation (rmsd) with the C-terminal lobe in complex with Zr-7 of titin (Atkinson et al., 2001) (PDB ID code 1H8B) is 1.1 Å for 64 equivalent Cα atoms, implying no significant conformational difference.
EF1-2 are less well defined in the electron density compared to the C-terminal lobe, and similar to EF3-4 show a binding interface of ∼500 Å2 with SR4. The interactions occur between the N-terminal α helix of EF1-2, the loop connecting EF1 and EF2 intercalates between α helices 2 and 3 of SR4, whereas the C-terminal α helix of EF1-2 lies in a parallel orientation on the third α helix 3 of SR4 (Figure 2B). In essence, only the N- and C-terminal helices of EF1-2 are involved in the binding, and this mode has no resemblance to any known classification of calmodulin-like domains but rather is similar to a bound-free lobe (Hoeflich and Ikura, 2002).
Interestingly, even though EF1-2 and EF3-4 have different interaction interfaces, they both adopt the same semiopen conformation (Chin and Means, 2000, Swindells and Ikura, 1996) (Figure S1B). The interaction of EF3-4 with the neck resembles that with titin Zr-7, both in terms of domain structure and ligand binding (Atkinson et al., 2001) (Figure 2F). EF1-2 and EF3-4 are mostly similar to the C-terminal lobe of human cardiac troponin C (TnC) (rmsd 3.1 and 3.2 Å for EF1-2 and EF3-4, respectively, with the C-terminal TnC lobe; PDB ID code 1J1E) (Takeda et al., 2003), confirming a previous observation that side-chain clusters in the EF hands are not related to the semiopen conformation (Atkinson et al., 2001). Further details on CAMD conformation can be found in Supplemental Information.
Sequence alignment of the EF hand pairs from α-actinin-2 and TnC reveals that in α-actinin-2, several calcium ligands in TnC, typically D, N, and E, are replaced by bulkier charged or smaller residues (Figure 2E). In essence, an R inhibits calcium binding in EF1 and a longer Q in EF2, where also a smaller S occupies the last position. In EF3, two positions typically occupied by a negatively charged D host an A and a P, whereas no residue promoting calcium binding is found in EF4.
Molecular Determinants of EF3-4 Ligand Specificity
Young and Gautel (2000) showed that CAMD binds to the neck region with lower affinity than to Zr-7 (Kd of 0.57 μM and 0.19 μM, respectively). In order to understand the higher affinity of CAMD for Zr-7, we compared the interaction interfaces of CAMD with Zr-7 (Atkinson et al., 2001) and the neck region.
Structural superposition of EF3-4 domains in complex with the ligands shows a similar layout between the α helices of the neck and Zr-7 in complex with CAMD (Figure 2F). Comparison of the 1-4-5-8 motifs shows that titin Zr-7 hosts a bulkier hydrophobic residue at position 1 (V702/A266), whereas position 5 in the neck is a less bulky Cys residue at position 270 (cf V706 in Zr-7) (Carugo, 2014), resulting in reduced stabilization of the interaction. This is reflected in the increase of the probability measure PΔG,IF for the interface derived from the gain in solvation energy upon complexation, where PΔG,IF > 0.5 points to hydrophilic/unspecific and PΔG,IF < 0.5 points to hydrophobic/specific interfaces using PISA (Krissinel and Henrick, 2007). Calculated for the ligand, these range from 0.31 in the EF3-4-Zr-7 complex to 0.49 for the EF3-4-neck complex, indicating a reduced specificity with a concomitant increased hydrophilic nature of the interaction.
PIP2 Binding Site
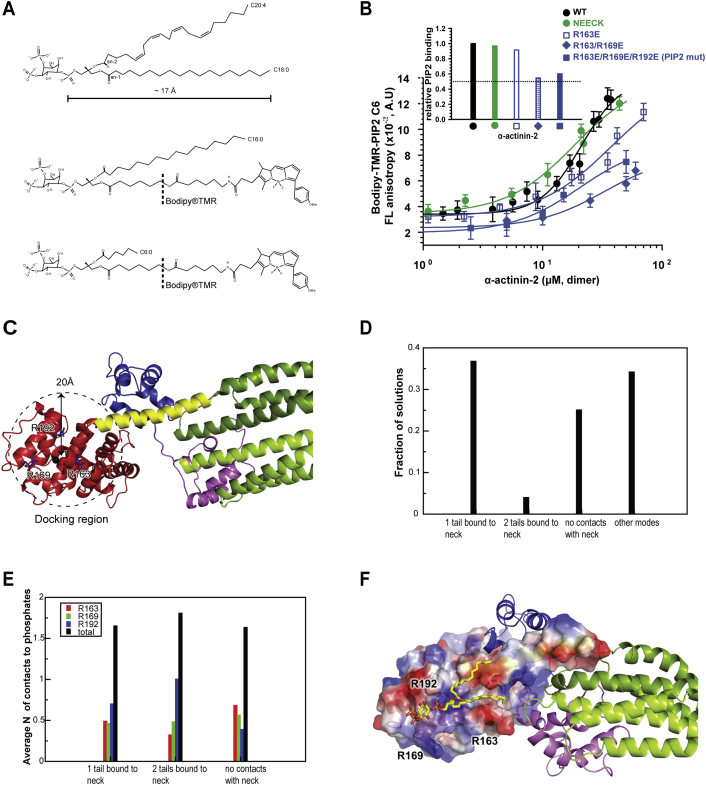
The model of α-actinin-2 and -3 activation by PIP2 hypothesizes that PIP2 docks with the polar head group on the CH2 domain, whereas its aliphatic chain reaches the CAMD binding site on the neck, perturbing this interaction (Franzot et al., 2005). This regulatory mechanism requires spatial proximity of the PIP2 binding site and neck region. The PIP2 binding site was originally mapped to residues 165–181 (Fraley et al., 2003, Fukami et al., 1996) on a loop connecting the first and second α helix of the CH2 domain. Structural analysis of the ABD of α-actinin-3 suggested a triplet of positively charged residues, which form a platform for PIP2 binding (α-actinin-2 residues R163, R169, and R192) (Franzot et al., 2005).
We used molecular dynamics (MD) simulations and flexible ligand docking to place PIP2 into the structure of α-actinin-2. Although the results suggest that the PIP2 binding site is likely not rigid, a large fraction of docked poses exhibits important commonalities. Namely, in about 40% of 10,000 generated models of the complex, the polar PIP2 head directly interacts with the above-mentioned arginine platform. This was supported by fluorescence anisotropy using PIP2 binding site mutants (Figures S2A and S2B). At the same time, one (∼35% of models; Figure 2G) or both (∼4% of models; Figure S2F) PIP2 aliphatic chains, which span some 17 Å, lean on the partially hydrophobic surface of the ABD and extend toward the 1-4-5-8 motif in the neck region (Figure 2G). A similar binding mode, where not only the polar head is involved but also the aliphatic chain, has been observed in the matrix domain of HIV-1 (Saad et al., 2006). This suggests that the architecture of the α-actinin-2 wild-type (WT) provides a suitable spatial orientation of both PIP2 and CAMD binding sites.
Figure S2.
Molecular Structures of L-α-Phosphatidylinositol 4,5-Diphosphate Used Mapping of PIP2 Binding Sites and Docking Experiments, Related to Figures 2 and 3
(A) Top: Native PIP2 containing two long-chain fatty acid C20:4 and C18:0 (PubChem database compound ID: 53480324 - National Center for Biotechnology Information, http://pubchem.ncbi.nlm.nih.gov). The maximum length of long-chain fatty acid C18:0 accounted from phosphate at the sn-1 position is indicated (Å). Middle, bottom: PIP2 analogs labeled on sn-1 with the 26-carbon Bodipy®TMR label attached via a 6-carbon linker. Either palmitic acid C16:0 or caproic C6:0 was esterified on the sn-2 position for the bodipy®TMR-PIP2-C16 (PIP2-C16∗, middle) and bodipy®TMR-PIP2-C6 (PIP2-C6∗, bottom), respectively. Chemical structures of PIP2 were edited using Accelrys Draw 4.1 (Accelrys Software Inc).
(B) PIP2 binding to α-actinin-2 variants measured by fluorescence anisotropy. Unlabelled α-actinin-2 was titrated into a fixed concentration of fluorescent PIP2-C6∗ (4 μM). Increases in fluorescence anisotropy signal indicate α-actinin-2 is binding to PIP2-C6∗. NEECK mutant showed similar PIP2 binding, while single mutant R163E, double mutant R163E/R169E, and triple R163E/R169E/R192E mutants showed decreased PIP2 binding (insert: relative PIP2 binding analysis). PIP2-C6∗ analog was used for the screening of the phosphoinositide head-group binding to arginine residues in order to minimize the interference of a longer chain fatty acid, such as PIP2-C16∗, in the binding assay. Based on this assay, triple PIP2 mutant, which displayed about 50% reduction of the anisotropy signal, was used as reference to investigate the relative effect of PIP2 binding inhibition on Zr-7 binding regulation compared to WT (Figures 3D–3F). Average and error bars (SDs) of three experimental replicas are plotted.
(C) Definition of the search region for flexible docking of PIP2 using Gold
(D) Fraction of docked solutions belonging to each of the 4 types as defined in Experimental Procedures.
(E) The average number of contacts to PIP2 phosphates by different R residues in the Arginine platform among docked solutions.
(F) α-actinin-2 structure with docked PIP2 (top scoring pose with two PIP2 tails in contact with the neck region are represented as yellow stick model) together with the EF3-4:neck interaction. ABD and the neck region are presented with their solvent accessible surface areas colored by electrostatic potential, the rest by cartoon representation and color coded as in Figure 1. The three R residues responsible for PIP2 binding are highlighted. The PIP2 molecule used in the flexible docking experiments containing palmitic acid (C16:0) esterified on the sn-1 and sn-2 (i.e., without bodipy-TMR fluorescence tag).
Structure of Activated α-Actinin
The biochemical model suggests that α-actinin is activated by PIP2 binding to ABD, resulting in a release of EF3-4 from the neck, thus facilitating its interaction with titin (Young and Gautel, 2000).
Structure-guided mutants were designed to disrupt key contacts between the neck segment of α-actinin-2 and EF3-4, producing a constitutively open variant. In particular, one positively charged residue (R268) and two hydrophobic residues (I269, L273) were replaced by negatively charged glutamates (R268E/I269E/L273E), hereafter termed NEECK (Figure 2F). NEECK was used to validate the closed conformation observed in the crystal structure of WT α-actinin and to probe in cellula the biological relevance of the opening/closing mechanism.
Conformational Switch of α-Actinin Is Modulated by PIP2 and Titin Zr-7
We explored whether the molecular architecture observed in the crystal structure of α-actinin-2 is maintained in solution and how this is altered in NEECK as a model of the open state.
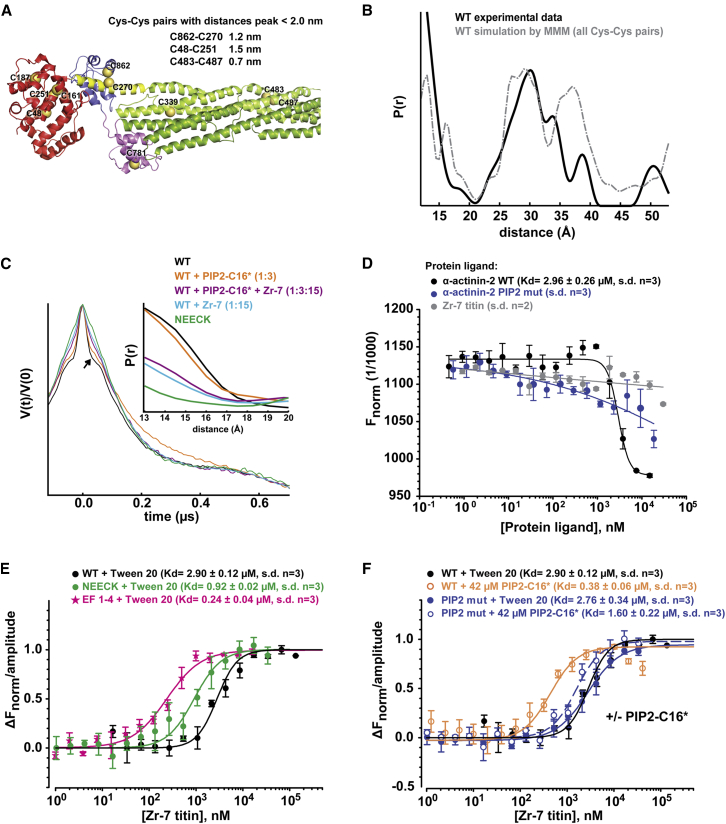
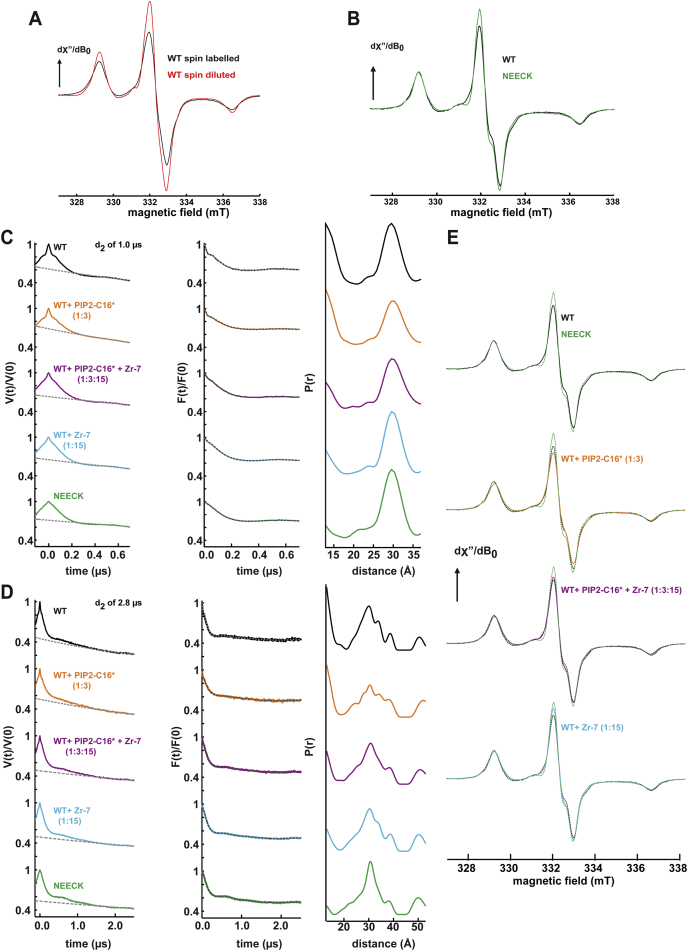
We used site-directed spin labeling and electron paramagnetic resonance (EPR) spectroscopy to obtain structural information on WT and NEECK α-actinin. All possible pairwise distances between labeled cysteines were computed (Figure 3A) and compared to those obtained by Q band double electron-electron resonance (DEER) experiments, showing a bimodal distribution peaking at 30 Å and below 20 Å (Figure 3B; Table S3). The experimental and computed distributions fit well in the distance range up to 35 Å (Figure 3B), confirming that the structures of α-actinin-2 in the crystal and solution are comparable.
Figure 3.
Structural Plasticity and Regulation of α-Actinin-2 Assessed by EPR and MST
(A) Cluster of the ten cysteine residues (Cα in dark yellow spheres) of α-actinin-2 used for the computed DEER distance distribution shown in (B). The inset shows pairs with interspin distances <20 Å.
(B) Experimental distance distribution of spin-labeled α-actinin WT (black) and simulation of the distance distribution (gray) based on spin-labeled cysteine residues from the crystal structure using the program MMM (see Extended Experimental Procedures and Table S3).
(C) DEER traces (for better comparison, adjusted by the modulation depth) and distance distributions from Q band DEER experiments using a DEER dipolar evolution time of 1 μs at 50 K of α-actinin-2 WT (black), WT plus PIP2-C16∗ (orange), WT plus PIP2-C16∗ plus Zr-7 (magenta), WT plus Zr-7 (light blue), and the NEECK mutant (green). The arrow indicates the change in the time domain trace, which is reflected in the variation of the fraction of distances <20 Å (inset).
(D) PIP2-C16∗ binding to α-actinin-2 measured by MST.
(E) CAMD, α-actinin-2 WT, and NEECK variant binding to Zr-7 measured by MST. The affinity determined for α-actinin-2 (+PIP2-C16∗) binding to Zr-7 is in a similar range of affinity for α-actinin-2 CAMD and is implicated in Zr-7 interaction, as well as for the NEECK variant.
(F) α-actinin-2 variant binding to Zr-7 with and without PIP2-C16∗ measured by MST. Unlabeled Zr-7 was titrated into a fixed concentration of fluorescently labeled α-actinin-2 (50 nM).
Average and error bars (SDs) of three MST experimental replicas are plotted. Mean and SD of Kd values were calculated from these plots.
We focused on the distance range below and around 20 Å, because the distance between C270 in the neck and C862 in EF3-4 is 12 ± 0.2 Å (Figure 3A). This spin label pair could sense open and closed conformations of α-actinin-2. Other spin label sites within a 20 Å distance are buried or located on the rigid rod domain (Table S3).
An open conformation of NEECK was inferred from a decreased fraction of short distances between C270 and C862 in the DEER distribution compared with WT (Figure 3C). This change in distances was validated with low-temperature continuous-wave (cw) EPR (Figures S3A and S3B).
Figure S3.
Interspin Distance Measurements by Continuous-Wave and Pulsed EPR, Related to Figure 3
(A) Spin-normalized cw EPR spectra (in H2O) detected at 160 K of α-actinin-2 WT spin labeled with 1:10-fold cysteine:MTSL stoichiometric ratios (black) and with 1:1:6 ratio of cysteine:MTSL(paramagnetic):MTSL(diamagnetic) (red). The increase in the spectral intensity in the spin diluted sample is indicative of decreased dipolar broadening arising from interspin distances < 2 nm.
(B) Spin-normalized cw EPR spectra (in D2O) at 160 K of α-actinin-2 WT (black) and NEECK (green). The increased spectral intensity in the NEECK mutation indicates decreased dipolar broadening (disappearance of short interspin distances).
(C) Q-band DEER primary traces (V(t)/V(0)), form factors (F(t)/F(0)) and distance distributions (P(r)). A dipolar evolution time of 1 μs was used to obtain a good S/N ratio focusing on the distance features < 1.8 nm (borderline region of the DEER sensitivity).
(D) Longer DEER traces with a dipolar evolution time of 2.8 μs (V(t)/V(0)), form factors (F(t)/F(0)) and distance distributions (P(r)). Samples and color codes are the same as in (C).
(E) Low temperature CW EPR spectra (spin normalized) of the corresponding sample used for DEER (full line), compared with the WT and NEECK mutant (dashed lines). The comparison corroborates the DEER results. Color codes are the same as in (C).
The analysis of the crystal structure using MD/docking suggests that the PIP2 binding site on ABD maps at a suitable position and distance from the CAMD-neck interaction to sense the hydrophobic tail of PIP2 (Figures 2A and 2G; Figures S2C–S2F). Can PIP2 alone induce opening of α-actinin-2? DEER measurements were carried out on WT α-actinin-2 using the more hydrophilic PIP2 analog Bodipy-TMR-PIP2-C16 (PIP2-C16∗; Figure 3C; Figures S2 and S3). No significant changes were detectable in the short distance range (Figure 3C; Figure S3).
We tested whether titin Zr-7 could act on the conformational equilibrium of α-actinin-2. We addressed this question structurally by EPR spectroscopy and quantitatively by microscale thermophoresis (MST). Addition of Zr-7 plus PIP2-C16∗ to α-actinin-2 significantly reduced the short distance peaks in the DEER distance distribution, indicating conversion to an open conformation. A similar effect was observed after addition of 15-fold molar excess of Zr-7 to α-actinin alone in the absence of PIP2-C16∗ (Figure 3C; Figures S3C and S3D). This decrease was validated by cw EPR at low temperature (Figure S3E).
The effect of PIP2-C16∗ on α-actinin affinity to Zr-7 was quantified by MST (Figures 3D–3F; Table S4). PIP2-C16∗ bound to WT α-actinin with Kd 2.96 ± 0.26 μM (Figure 3D). The results showed a significantly higher affinity of titin Zr-7 for the PIP2-C16∗-α-actinin complex (Figure 3F; Kd 0.38 ± 0.06 μM) compared to α-actinin alone (Kd 2.90 ± 0.12 μM). The nanomolar Zr-7 binding affinity for the PIP2-C16∗-α-actinin complex is comparable to that observed for the isolated CAMD (EF1–4) and Zr-7 (Figure 3E; Kd 0.24 ± 0.04 μM), whereas NEECK shows an intermediate affinity (Figure 3E; Kd 0.92 ± 0.02 μM) in the absence of PIP2-C16∗. The results for CAMD agree with earlier binding studies (Table S4). MST thus confirms that PIP2-C16∗ increases Zr-7 affinity for α-actinin-2 ∼10-fold, whereas the PIP2 mutant—having lower PIP2-C16∗ affinity (Figure 3D)—showed an insignificant increase of Zr-7 binding by PIP2-C16∗ (Figure 3F; Table S4).
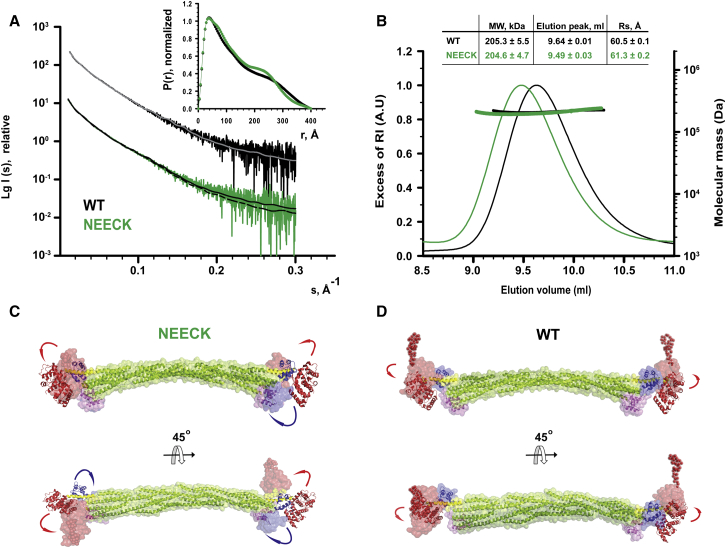
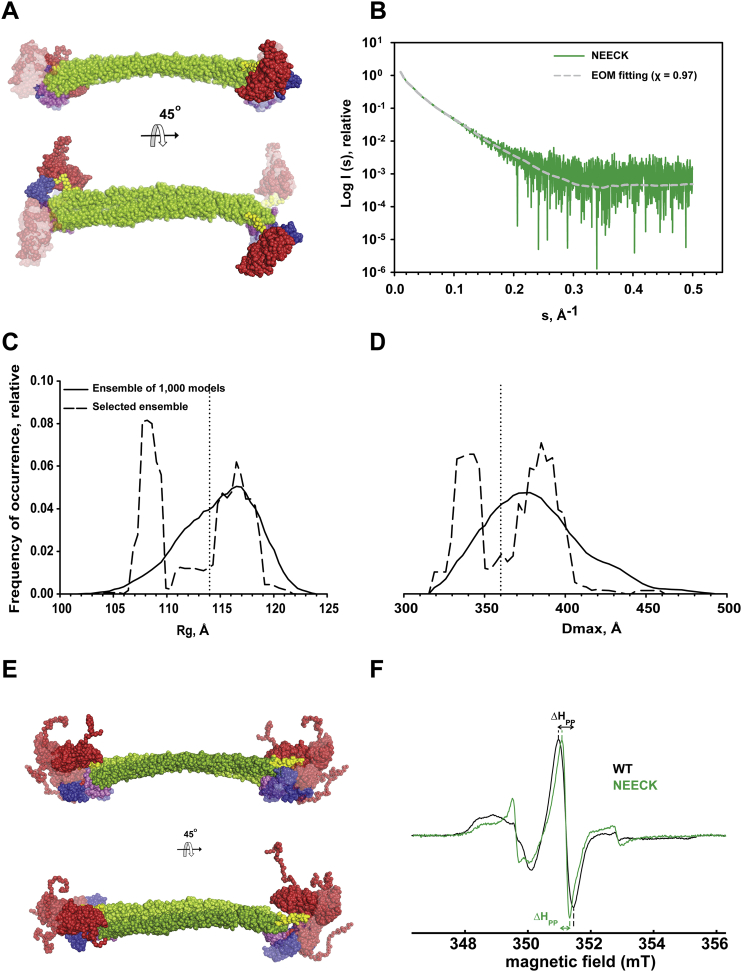
We used small-angle X-ray scattering (SAXS), multiangle static laser light scattering (MALLS), and size-exclusion chromatography (SEC) on WT and NEECK α-actinin to understand the structural differences between closed and open conformations. The derived molecular parameters are given in Figures 4A and 4B and Table S5. SEC-MALLS shows that both α-actinin samples display the same molecular mass but that NEECK has an increased Stokes radius. In order to model the NEECK variant, we first addressed the structural consequences for the α-helical neck upon release of EF3-4 by performing NMR of the WT free neck peptide (amino acids A259–Y286). Analysis of 2D nuclear Overhauser effect spectroscopy (NOESY) showed no evidence of stable secondary structure (Figure S4). NEECK was thus modeled with a flexible neck region (Figure 4C), whereas the neck for WT was modeled as a rigid-body α helix, as in the X-ray structure (Figure 4D). The best WT model fits with the crystal structure (χ = 1.25). In NEECK, the ABDs deviate from the linear alignment with the rod and EF3-4 are in open conformation (Figure 4C), giving a fit of χ = 1.14. This agrees with increased mobility of NEECK and unfolding of the neck. In addition, we found a change in spin label mobility between WT and NEECK α-actinin using cw EPR room temperature measurements (Figure S5F). The narrower lines in the EPR spectrum of NEECK (Figure S5F) indicated increased mobility compared to WT, in line with an open conformation of NEECK. Thus, the solution structure of NEECK can best be modeled by an ensemble of conformations (Bernadó et al., 2007) where ABD and CAMD adopt various orientations (Figures S5A and S5E).
Figure 4.
Solution Structure of α-Actinin-2 and the NEECK Mutant Derived from SAXS
(A) Experimental SAXS data of WT (black) and the NEECK mutant (green) of α-actinin-2. SAXS curves are computed from a rigid-body (RB) model for WT (gray) and NEECK (black). The logarithm of scattering intensity (I) is plotted as a function of the momentum transfers (s, Å−1). Successive curves are displaced by one logarithmic unit for better visualization. Distance distribution functions (inset) P(r) for WT and NEECK assume slightly different shapes. RB modeling fits the experimental WT data with χ 1.25 (gray line) and experimental NEECK data with χ 1.14 (dashed black line). The fit discrepancy for NEECK increased to 1.32, assuming a helical neck (solid black line).
(B) Characterization of hydrodynamic properties of α-actinin WT and the NEECK mutant by SEC-MALLS. The lines across the protein elution volume show the molecular masses (MWs) of proteins. SEC-MALLS shows that NEECK has the same molecular weight as WT α-actinin-2 but a higher Stokes radius Rs (inset; data are represented as mean ± SD of three experiments), corroborating the open conformation for NEECK suggested by SAXS (C). AU, arbitrary units.
(C) RB model of NEECK in solvent-accessible surface representation. The neck region was modeled as a flexible linker between the rigid bodies ABD and rod, with no contact restraint. Only one RB model is shown for clarity out of three independent BUNCH runs (Figure S5A).
(D) The best RB model of WT α-actinin-2 in solvent-accessible surface representation superimposed on the crystal structure. For WT RB modeling, only ABD was allowed a variable position, whereas EF hands 3-4 were fixed in contact with the neck.
In all models, N-terminal residues missing from the crystal structure were modeled as dummy atoms. Arrows highlight the movement of ABD and EF hands 3-4 relative to the superimposed crystal structure. See also Table S5.
Figure S4.
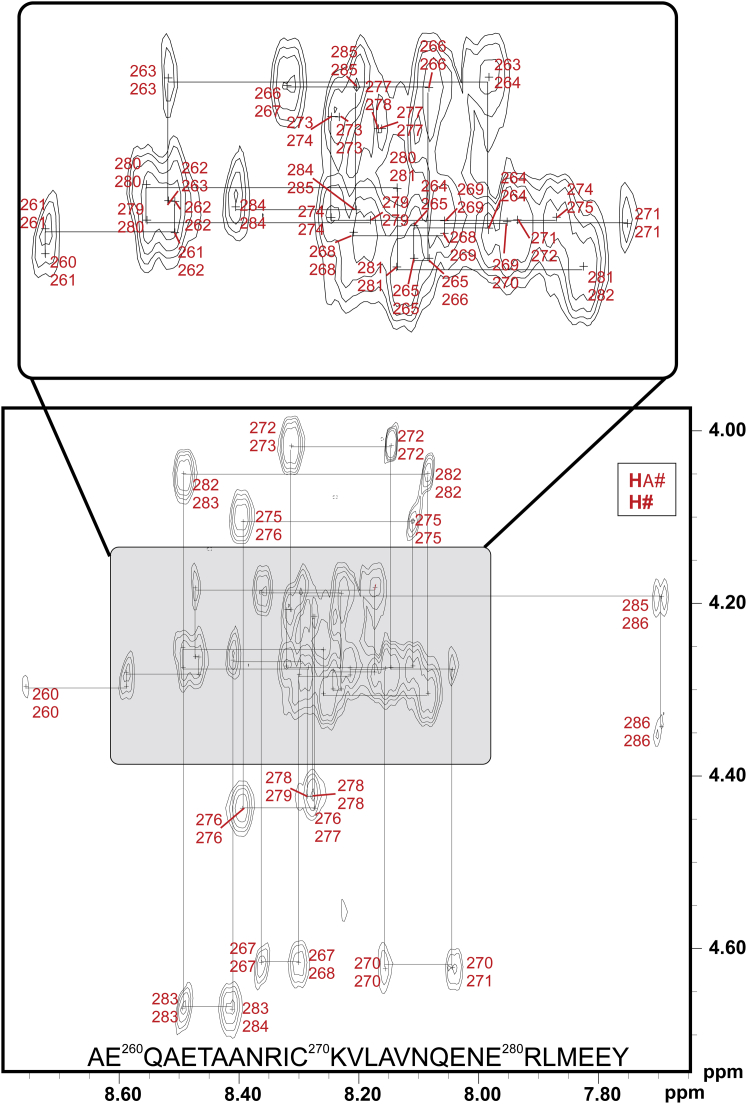
Neck Peptide of Wild-Type α-Actinin-2 Is Unstructured when Not Bound to the CaM-like Domain, Related to Figure 4
Fingerprint area of the 2D NOESY spectrum of the WT neck-peptide in 90:10 H2O:D2O with neck-peptide in 25mM phosphate pH 6.5, 25 mM NaCl, 3 mM DTT and 0.5 mM EDTA recorded at 298K. Hαi-HNi and Hαi-HNi+1 cross peaks are shown in red and the sequential walk of the peptide is indicated with horizontal and vertical lines. No additional NOE cross peaks were found that indicated an existence of any kind of stable α-helical structural in the fingerprint area.
Figure S5.
Structural Plasticity of NEECK as Assessed by SAXS and Room Temperature Continuous-Wave EPR, Related to Figure 4
(A) BUNCH runs starting from random initial structures provided models of NEECK mutant with varying positions of ABD and EF3-4 assuming nonhelical neck. Three representative BUNCH models (distinguished by degree of transparency) are superimposed, showing that ABDs deviate from the linear alignment of the rod domains and that EF3-4 are in open conformations.
(B–E) The flexibility of the ABD domains and EF3-4 is also supported by EOM analysis of NEECK. The central rod part (green) was fixed as in the crystal structure whereas ABD (red) and EF3-4 (blue) domains position were optimized together with the N-termini residues.
(B) The low discrepancy between experimental data (green plot) and those computed from EOM models (dashed gray plot) of NEECK mutant is defined by χ value of around 1.0. Size and shape descriptor distributions: Rg (C) and Dmax (D).
(D) The distributions correspond to the pool of 1,000 structures (solid lines), and the optimized ensemble (dashed lines) calculated using chromosomes with N = 20 structures; dotted lines are references for Rg and Dmax values calculated from X-ray structure of WT α-actinin-2. The overall Rg and Dmax analysis of the select optimized ensemble show bimodal distribution, which indicates distinct population of conformers (compacted and extended). The peak of compacted models is slightly shifted to smaller distances, due to the larger deviation of ABD domains from the linear alignment of the rod domains, compared to the peak of extended models.
(E) Selected ensemble of models were superimposed and distinguished by degree of transparency. In the lower panel models are rotated 45° around the long axis of the dimer.
(F) Cw EPR spectra (normalized by peak intensity) of spin labeled α-actinin-2 WT (black) and α-actinin-2 NEECK mutant (green), detected at room temperature. ΔHpp indicates the peak-to-peak line-width of the central peak. The narrower lines in the room temperature EPR spectrum of the NEECK mutant spectrum (green) with ΔHpp of 0.26 mT indicates an increase in mobility of at least one part of the spin-labeled structure compared to the WT (ΔHpp = 0.46 mT).
In conclusion, NEECK adopts a constitutively open conformation, and although it has to be seen as a “hyperactive” state, it is likely to approximate the Zr-7-bound structure of α-actinin in the Z-disk.
Impaired α-Actinin Regulation Disrupts Coordinated Z-Disk Assembly
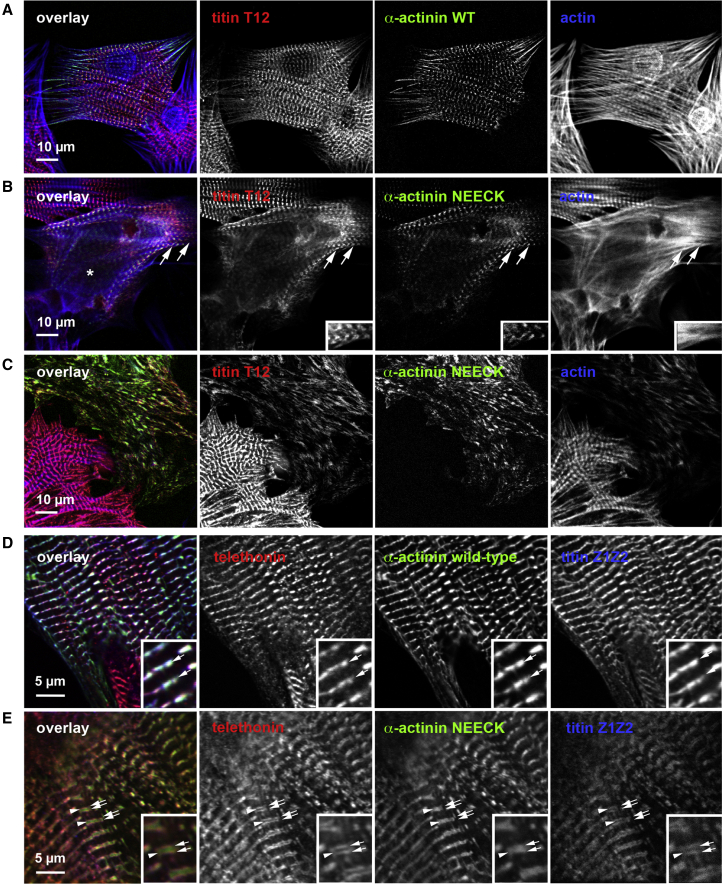
To directly test the role of α-actinin regulation in Z-disk assembly, we performed live-cell imaging. We hypothesized that disrupted coordination of α-actinin-2-ligand interactions in NEECK, which binds titin constitutively, and the PIP2 mutant, where activation by PIP2 is blunted, should reduce dynamic exchange of α-actinin at Z-disks.
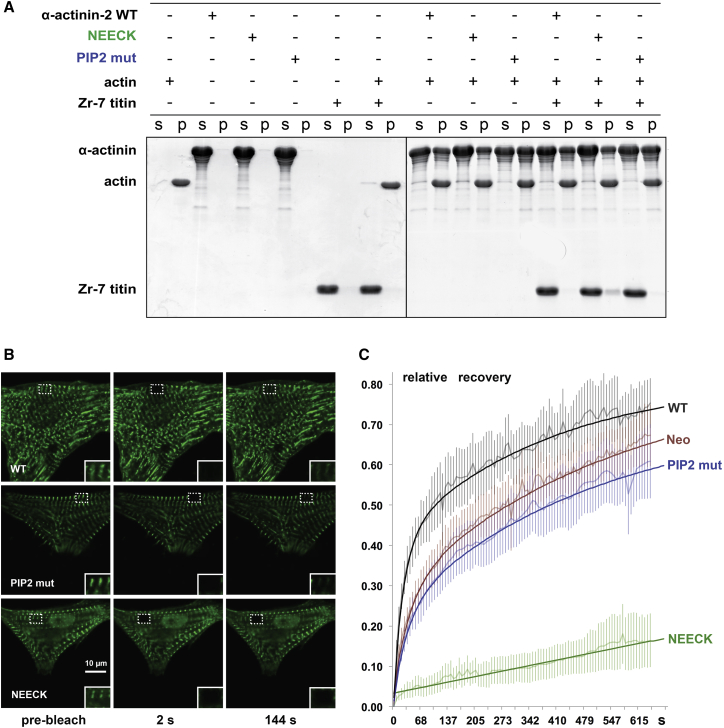
We assessed whether the NEECK and PIP2 mutants can still play their basic roles—binding to F-actin and titin Zr-7—by actin cosedimentation assays. F-actin binding was unaffected in both mutants (Figure 5A), in contrast to PIP2-binding mutants in nonmuscle α-actinin (Fraley et al., 2003). No cosedimentation of Zr-7 was seen with WT or PIP2 mutant, in contrast to NEECK. Together, these results suggest that the PIP2 or NEECK mutations do not impair F-actin binding, and that NEECK induces PIP2-independent Zr-7 binding.
Figure 5.
Mutations Affecting Regulation of α-Actinin-2 with PIP2 Do Not Influence F-Actin Binding but Impact α-Actinin-2 Z-Disk Dynamics
(A) Binding of α-actinin-2 variants to F-actin and titin Zr-7. α-actinin-2 WT, NEECK, and PIP2 mutants (PIP2 mut) were cosedimented with actin, and equal amounts of supernatant (s) and pellet (p) fractions were subjected to SDS-PAGE and visualized by Coomassie blue.
(B and C) FRAP measurements of α-actinin-2 dynamics in live NRCs expressing GFP-labeled α-actinin-2 variants (WT, PIP2 mutants, and NEECK).
(B) Snapshots at prebleach and two time points postbleach; the bleached region of interest (ROI) is highlighted by a dotted box. Note that NEECK fluorescence does not recover within the 144 s time course shown here, whereas rapid recovery is observed for WT α-actinin. Insets: ROIs enlarged 2-fold.
(C) Quantification of fluorescence intensity recovery. Note that the slowed fluorescence recovery of the PIP2 mutant is mirrored by treatment with 500 μM neomycin (Neo). Bold lines, exponential fits; shaded lines, average values. Error bars indicate SD.
We carried out fluorescence recovery after photobleaching (FRAP) studies in neonatal rat cardiomyocytes (NRCs) expressing the GFP-labeled α-actinin-2 mutants to assess the impact of α-actinin-2 regulation on spatiotemporal dynamics in sarcomeres. The exchange of WT α-actinin-2 at Z-disks was rapid, with a fast component t1/2 25 ± 2 s (Figures 5B and 5C). In contrast, the NEECK mutant was dramatically slower, with t1/2 >6,134 s. The PIP2 mutant showed reduced dynamics, with t1/2 35 ± 4 s (Figure 5B). The slower dynamics of α-actinin-2 mutants compared to WT observed in cellula seems not to be due to F-actin binding activity, because both mutants bind F-actin (Figure 5A). Analysis of FRAP kinetics revealed standard fast and slow components in the case of WT and the PIP2 mutant, whereas for NEECK only a slow component could be discriminated. The slow, single-exponential exchange of NEECK agrees with a dominant, high-affinity interaction. Because the NEECK interaction with titin is constitutive, the slow cellular dynamics likely reflect the high affinity of the EF3-4 interaction. Exchange of the PIP2 mutant was also slower, in agreement with reduced phospholipid regulation. To probe the role of PIP2 in regulating α-actinin dynamics independently, we used the aminoglycoside neomycin, an inhibitor of PIP2 signaling (Li and Russell, 2013, Schacht, 1976). Neomycin resulted in slower FRAP recovery of WT α-actinin-2 (≈40 ± 4 s), similar to the PIP2 mutant (Figure 5C), supporting the notion that α-actinin-2 Z-disk dynamics are strongly dependent on PIP2 regulation, in agreement with the Z-disk localization of PIP2 (Figure S6A).
Figure S6.
PIP2 and Vinculin in Relation to the Z-Disk and Temporal Increase of the Titin T12 Epitope in α-Actinin WT- and NEECK-Transfected Cardiomyocytes, Related to Figures 5 and 6
(A) PIP2 is localized to the cardiac Z-disk. Immunofluorescence in untransfected NRC using PIP2 antibody shows a striated pattern that coincides with the Z-disk titin epitope T12 (arrows). Note the noncardiomyocytes above the striated cell, in which PIP2 is localized in a vesicular pattern.
(B) The α-actinin-2 NEECK mutant does not colocalize in abnormal Z-disks. GFP-labeled NEECK α-actinin was transiently expressed in NRC for 18-48 hr. The NEECK mutant leads to widening of the Z-disk (upper row) and ultimately formation of rod-like structures (lower row). Note that vinculin shows no appreciable colocalization with α-actinin-2 but remains predominantly restricted to focal adhesions. Red: monoclonal anti-vinculin stain, green: α-actinin-2 NEECK-GFP, Blue: actin (Alexa643-phalloidine).
(C) The optically resolvable T12 distance was measured in confocal immunofluorescence images 2 and 3 days after transfection of the GFP-labeled constructs. Note the expansion to an average width of over 600 nm at day 2 and over 900 nm at day 3, while the optical T12 distance in WT α-actinin transfected cells does not exceed 300 nm. Data from 9 independent cells each. Dashed lines: means.
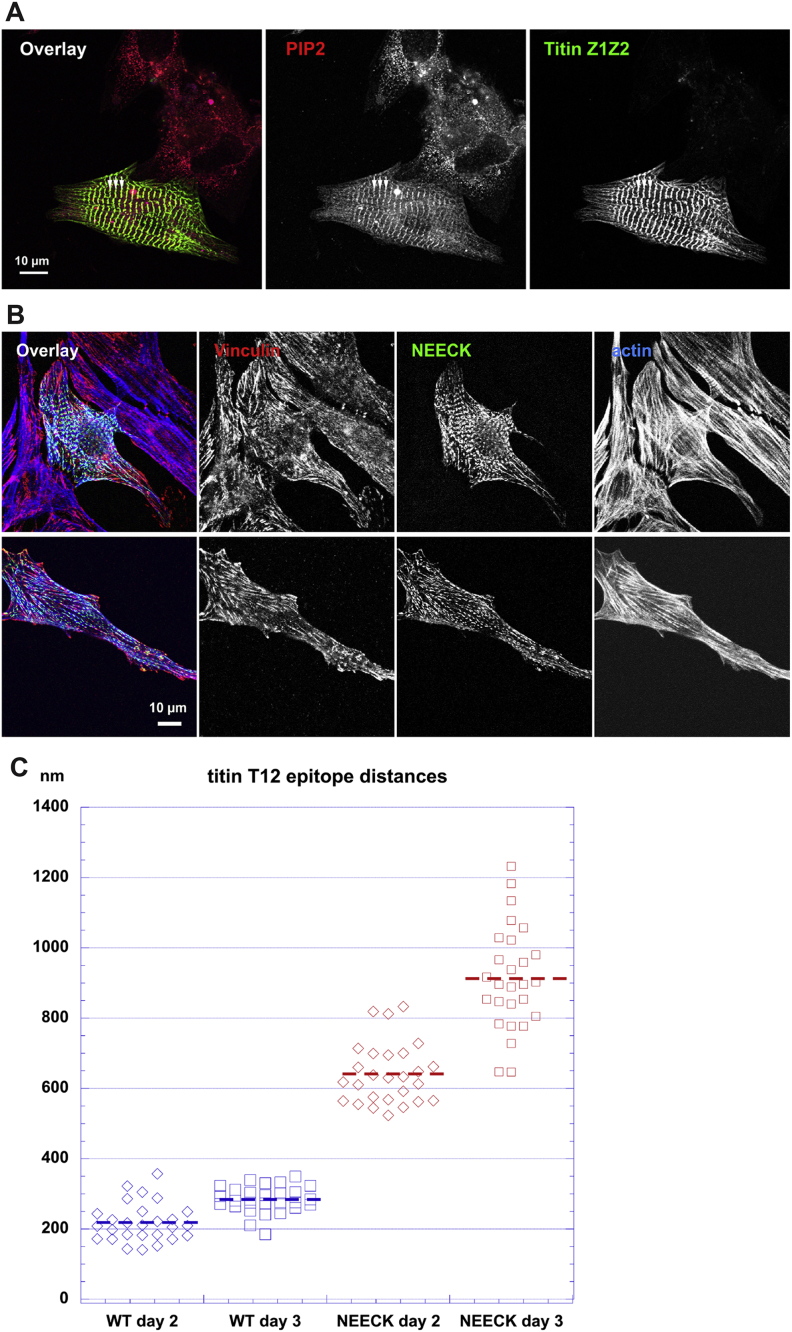
Z-disk morphology in α-actinin-transfected NRCs showed a striking phenotype for the NEECK mutant but not WT or the PIP2 mutant. Cells expressing NEECK showed gradual appearance of sarcomeres with wide α-actinin labeling, where titin epitopes peripheral (T12 antibody) and more central (Z1Z2) to the Z-disk (Young et al., 1998) were resolved as doublets flanking the edge of the α-actinin-labeled central Z-disk. This resulted in formation of actin/α-actinin bundles resembling nemaline rods but containing diffusely localized Z-disk titin (Figures 6A–6C) and ultimately complete disruption of sarcomeres (Figure 6C). Vinculin localization in NEECK-transfected cardiomyocytes was unaffected (Figure S6B).
Figure 6.
Constitutively Activated α-Actinin-2 Disrupts Z-Disks and Leads to Myofibril Disassembly
GFP-labeled WT and NEECK α-actinin-2 were transiently expressed in NRCs for 18–48 hr.
(A) WT α-actinin shows normal Z-disk localization, and the titin T12 epitope is resolved as a single line in standard confocal microscopy.
(B) In contrast, NEECK leads to widening of the Z-disk and splitting of the T12 epitope after ∼18 hr (asterisk); doublet T12 lines are highlighted by arrows.
(C) After 48 hr, Z-disks are completely disrupted and Z-disk titin, actin, and mutant α-actinin are localized in rod-like structures.
(D and E) Superresolution microscopy reveals that epitopes of N-terminal Z1Z2 of titin and their ligand telethonin are unresolvable in WT-transfected cells; NEECK causes widening of Z-disks. Doublet Z1Z2/telethonin lines are highlighted by arrows and the central α-actinin region is indicated by arrowheads. Insets show 2-fold enlargement. Z-disk titin (T12 epitope) or telethonin, red; mutant α-actinin-GFP, green; actin (Alexa 688-phalloidin) or titin Z1Z2, blue.
Whereas the optically resolvable distance between T12 epitopes in WT-transfected cells is ∼200 nm, in NEECK-transfected cells this was >600 nm, and >800 nm after 3 days (Figure S6C). Similar splitting to >200 nm was also seen for Z1Z2, normally at the limit of optical resolution with a separation of ∼100 nm (Young et al., 1998). This suggests that the ordered integration of titin and α-actinin is severely disrupted by NEECK, raising the question of whether the spatiotemporal integration of other titin Z-disk ligands is affected. Current models of titin layout in the Z-disk predict that Z-disk widening could only be achieved by relative slipping of the overlapping N termini of titin molecules entering the Z-disk from two antiparallel sarcomere halves (Gautel, 2011). Titin molecules are crosslinked in an antiparallel palindromic complex of domains Z1-Z2 and the small Z-disk protein telethonin (Zou et al., 2006). Both telethonin and titin-Z1Z2 epitopes strictly colocalize at the Z-disk periphery (Figure 6D) but also in the wide NEECK Z-disks (Figure 6E). These results show that intramolecular autoregulation of α-actinin-ligand interactions is crucial for sarcomere integrity regulating the integration of titin, actin, and α-actinin in Z-disks of controlled width, without affecting interaction of titin with Z-disk proteins such as telethonin.
Discussion
The structure of α-actinin-2 shows a modular architecture, yet is more than just “the sum of its parts”: important intra- and intermolecular contacts lock the molecule in a closed conformation that is crucial for dynamic regulation.
Pseudoligand Model Validation
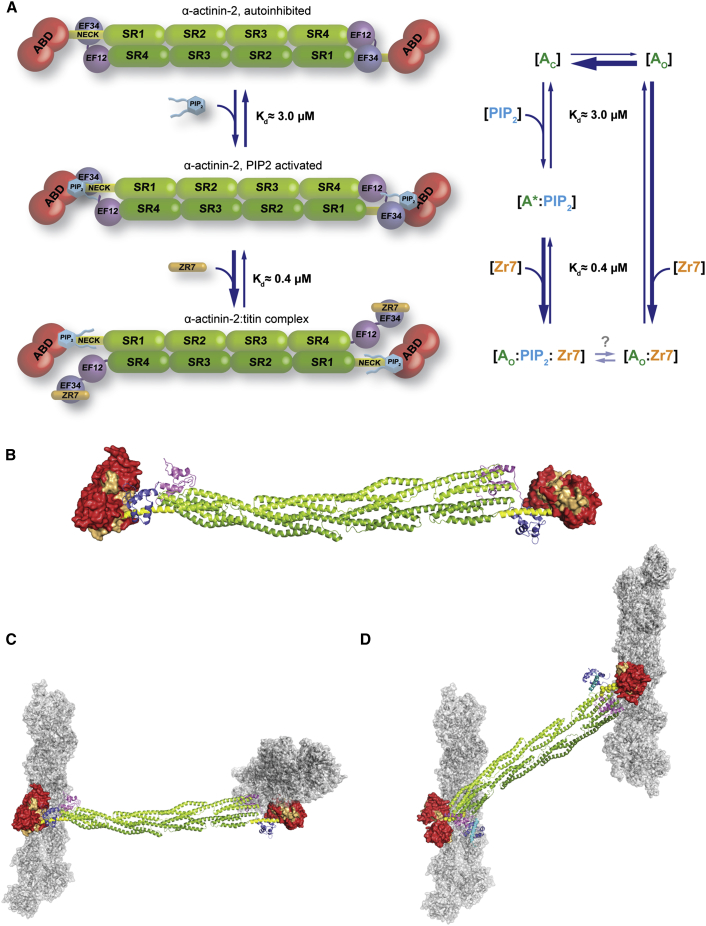
The structure of α-actinin-2 displays a closed, autoinhibited conformation, as suggested by Young and Gautel (2000) (Figure 1B). The closed structure of α-actinin-2 shows, furthermore, that the PIP2 binding site on the ABD (Franzot et al., 2005) maps at a suitable position and distance from the CAMD-neck interaction for sensing the hydrophobic tail of PIP2 (Figure 2A), as supported by MD/docking simulations (Figure 2G; Figures S2C–S2F).
The closed α-actinin-2 conformation shows that a number of interactions between SR1–4 stabilize the formation of antiparallel dimers, providing its structural rigidity and stability. Although addition of PIP2-C16∗ alone does not promote complete opening of α-actinin-2, at least as measurable by DEER, it promotes binding of Zr-7 with nanomolar affinity (Figure 3F), suggesting a positive allosteric modulation for opening and ligand binding. Furthermore, local structural changes in the CH2 domain, as observed upon PIP2 binding to nonmuscle α-actinin (Full et al., 2007), cannot be excluded.
Additional mechanisms might act in cells that cooperate with PIP2 or offer alternative regulation, including posttranslational modifications or protein cofactors. However, we could not identify any such plausible sites conserved between muscle α-actinin-2 and -3 in proteomic databases. No protein cofactors regulating α-actinin-titin interactions have been identified to date.
Structural comparison of interactions between EF3-4, Zr-7, and the neck reveals the basis for the higher affinity of α-actinin-2 for titin versus the pseudoligand neck (Young and Gautel, 2000) (Figure 2F). Our results suggest a model for PIP2 regulation of α-actinin, relying on structural plasticity and conformational dynamics of α-actinin-2 (Figure 7A): in the absence of PIP2, α-actinin-2 exists in two conformational states, a highly populated closed [AC] and a low populated open and active state [AO]. Addition of PIP2 triggers an activated state [A∗:PIP2] with lower activation energy for opening. Binding of Zr-7 recruits α-actinin-2 in the open conformation [AO:Zr-7], which is enriched, because α-actinin-2 binds Zr-7 with higher affinity than it does to the pseudoligand neck (Figure 2F; Table S4).
Figure 7.
Molecular Mechanism of α-Actinin-2 Phosphoinositide-Based Activation and Model for F-Actin/α-Actinin Crosslinking in the Z-Disk
(A) Reaction mechanism depicting α-actinin activation by PIP2. α-actinin in the absence of PIP2 is in equilibrium between highly populated closed [AC] and low populated open states [AO]. Addition of PIP2 generates an activated state [A∗:PIP2], with lower activation energy for opening. Binding of Zr-7 recruits α-actinin to the open conformation, leading to an increase of [AO:Zr-7] due to higher α-actinin affinity for Zr-7 compared to the neck.
(B) Actin binding sites 1–3 (orange) mapped onto the molecular surface of ABD in α-actinin-2 in closed conformation. Color coding of domains is as in Figure 1.
(C) α-actinin-2 crystal structure superimposed on F-actin decorated by the CH1 domain of α-actinin-2 (PDB ID code 3LUE).
(D) Model of α-actinin-2 crosslinking antiparallel actin filaments. α-actinin-2 in open conformation (NEECK) was modeled assuming structural plasticity in the flexible neck, which allows for suitable orientation of ABDs. Titin Zr-7 is in cartoon presentation (cyan).
See also Movie S1.
Implications for Binding F-Actin Crosslinking and Z-Disk Structure
In the Z-disk, antiparallel actin filaments are crosslinked by α-actinin in a paracrystalline tetragonal lattice (Goldstein et al., 1988). To analyze the structural determinants of α-actinin’s principal crosslinking function, we mapped the known actin binding sites (ABSs) to the structure of the α-actinin dimer. ABSs in ABDs of several actin-binding proteins have been located on the first and the last α helix of the CH1 domain and on the first α helix of the CH2 domain (Sjöblom et al., 2008). ABSs in α-actinin-2 are exposed, and not blocked by interdomain interactions (Figure 7B).
To generate a 3D model of F-actin/α-actinin, we superimposed the structure of α-actinin-2 on that of the F-actin-bound CH1 domain (PDB ID code 3LUE) (Galkin et al., 2010). Notably, using the α-actinin-2 structure leads to a model with perpendicular actin filaments (Figure 7C), in strong disagreement with the established antiparallel actin architecture in the Z-disk but agreeing with the α-actinin-2 dimer architecture. Due to the internal twist of about 90° of the central rod (Ylänne et al., 2001), the ABDs in the dimer are rotated by 90°. We next used the structure of the NEECK mutant as the “open” structure in the Z-disk. Here the unbound neck is unstructured (Figure S4), allowing the ABD to explore different orientations, adopting those compatible with interaction with antiparallel actin filaments. Considering the angular distribution of F-actin and α-actinin, which centers at 60° and 120°, respectively (Hampton et al., 2007), we generated a model of two actin filaments crosslinked by an α-actinin-2 dimer (Figure 7D; Movie S1). In this model, the distance between filaments is ∼230 Å, which is in excellent agreement with the observed interfilament distances in the tetragonal Z-disk lattice (240 Å; Goldstein et al., 1979).
The structural plasticity of α-actinin-2 has implications for not only its regulation but also for actin filament binding both in muscle and nonmuscle isoforms, where actin filaments are randomly oriented, requiring ABDs to adopt variable orientations. Interestingly, muscle α-actinin-2 was found to crosslink antiparallel as well as parallel actin filaments in α-actinin-F-actin rafts (Hampton et al., 2007). Assuming the absence of PIP2 in these assays, the flexibility of the ABD likely resides in the hinge region between the ABD and the neck (residue G258) (Figure 2A), similar to the solution structure of closed α-actinin-2 (Figure 4D). Other electron microscopy studies showed that ABDs in smooth muscle α-actinin can attain different orientations through movement in the flexible neck (Taylor and Taylor, 1993, Winkler et al., 1997), crosslinking both antiparallel and parallel actin filaments in vitro and in vivo (Meyer and Aebi, 1990, Tang et al., 2001).
Although alternative paths of titin in the Z-disk are conceivable, our results with NEECK show that even in strongly split Z-disks the Z1Z2 and telethonin epitopes remain in strict colocalization, implying that the two titin molecules crosslinked in the titin-telethonin complex must come from the same half-sarcomere, as suggested previously (Zou et al., 2006). Intriguingly, these findings also suggest that titin capping (via telethonin) and barbed-end actin filament capping by CapZ are not directly correlated, despite the close association of the titin Z1Z2-telethonin complex with the actin barbed end (Zou et al., 2006). However, actin capping by CapZ and crosslinking by α-actinin may crosstalk, as indeed α-actinin was reported to interact with CapZ via a binding site on the rod (Papa et al., 1999) and both proteins are PIP2 regulated (Figure S6A).
Impact of Pathogenic Mutations
Genetic variants in α-actinin genes are associated with several inherited diseases. Missense variants in nonmuscle actinin 1 (ACTN1 gene) cause autosomal-dominant congenital macrothrombocytopenia (Guéguen et al., 2013, Kunishima et al., 2013), and approximately 4% of autosomal-dominant familial focal segmental glomerulosclerosis has been linked to nonmuscle ACTN4 mutations (Kaplan et al., 2000). Missense variants in muscle ACTN2 have been reported in sporadic cases and a few families with dilated or hypertrophic cardiomyopathy (Chiu et al., 2010, Mohapatra et al., 2003, Theis et al., 2006). Our structure now provides a platform for the analysis of mutational impact on structure, ligand binding, and regulation of α-actinin in inherited human diseases (see Supplemental Information and Figure S7).
Figure S7.
Amino Acid Sequence Conservation and Genetic Variants Mapping on Solvent-Accessible Areas of α-Actinin-2, Related to “Impact of Pathogenic Mutations” in Discussion
(A) Representative vertebrate sequences of all 4 α-actinin isoforms were aligned using Clustal Omega (Sievers et al., 2011), and sequence conservation (variable red, average white, conserved blue) mapped on the solvent accessible surface using Consruf server (Celniker et al., 2013). Species included are Homo sapiens, Bos taurus, Mus musculus, Macaca mulatta, Canis familiaris, Orcinus orca, Danio rerio, Gallus gallus, Falco peregrinus, Oryctolagus cuniculus, Lepisosteus oculatus, Equus caballus.
(B) Genetic variants reported in literature (red; see Discussion) were mapped on the solvent accessible surface of α-actinin-2 (white).
Structural Analysis of Selected Genetic Variants
The genetic variants are spread over all domains of α-actinin. Several variants are conservative and would not lead to major structural perturbations, in particular mutations in the CAMD and the rod domain. Changes of rod surface properties might, however, abrogate interactions with ligands, because the rod domain is recognized as the prominent protein interaction platform of α-actinin (Djinovic-Carugo et al., 2002). Interestingly, the mutations on the rod domain map on the less conserved, acidic side of the convex surface (Ylänne et al., 2001) (Figure S7). However, four mutations in the ACTN1 and ACTN4 genes have predicted disruptive potential: E225K (ACTN1 gene, ABD) leads to a loss of a salt bridge and mutation R738W (ACTN1 gene, CAMD) would disrupt the structure of the CAMD, whereas the W59R (ACTN4 gene, ABD) and S262F (ACTN4 gene, ABD) mutations destabilize the domain structure due to introduction of charged or bulky hydrophobic residues to the core of the ABD. Most hypertrophic cardiomyopathy and dilated cardiomyopathy variants were classified as structurally neutral.
Implication for Regulation of the α-Actinin Family in General
Muscle α-actinin interacts with many proteins via multiple binding sites. The CAMD EF3-4 site interacts with helical motifs in the actin- and α-actinin-binding proteins myopalladin, palladin, and myotilin, highly similar to the α-actinin-titin complex and the intramolecular neck complex detailed here (Beck et al., 2011). Dynamic regulation of α-actinin interactions with these proteins is therefore likely governed by the same principles as the one with titin. Additionally, the α-actinin-associated LIM protein (ALP) and ZASP/Cypher bind α-actinin at both the CAMD (via its PDZ domain) and the SR (Faulkner et al., 1999, Klaavuniemi et al., 2004). Although the binding sites for titin Zr-7 and the PDZ domain on CAMD do not coincide, an open structure might be required to accommodate both binding partners and prevent steric hindrance by the spatially close α-actinin domains.
Furthermore, interactions of CAMDs of the structurally related cytoskeletal actin-binding proteins dystrophin, utrophin, and spectrin may play important roles in regulating cytoskeletal interactions near the plasma membrane (Bennett and Healy, 2008), as suggested by recent studies on spectrin-ankyrin, actin, and protein 4.2 interactions (Korsgren and Lux, 2010, Korsgren et al., 2010). Although these EF hand domains retain aspects of calcium regulation (only the N-terminal EF hand binds calcium), the general mode of regulation seems highly similar to α-actinin, namely the CH2-R1 linker region of α/β-spectrin also binds to the CAMD EF3-4 hands, and this regulates protein interactions.
The mechanism we have detailed here is therefore likely to be of general relevance for regulating spectrin-like proteins via intramolecular pseudoligand interactions.
Experimental Procedures
Purification and Crystallization
Proteins were expressed as His fusions in Escherichia coli and purified via Ni-NTA agarose and size-exclusion chromatography. Protein was lysine methylated and crystallized in a precipitant containing 0.2 M Mg formate, 5% PEG smear, and 10 mM EDTA by hanging-drop vapor diffusion at 14°C.
Structure Determination
A 3.5 Å data set was collected at beamline ID23-2 (European Synchrotron Radiation Facility [ESRF]). The phase problem was solved by molecular replacement using structures of the rod domain (PDB ID code 1HCI), the ABD from α-actinin-3 (PDB ID code 1WKU), and the NMR structure of EF3-4 (PDB ID code 1H8B) as search models.
Residues 34–892 were assigned in the final model. Details on data collection, processing, structure determination, and refinement are described in Extended Experimental Procedures and Table S1.
Extended Experimental Procedures.
DNA Constructs, Mutagenesis, and Cloning of α-Actinin-2
For crystallographic purposes, a construct of human α-actinin-2 (Swissprot entry P35609) encompassing the residues 19-894 was cloned into a pETM11 vector (EMBL-Heidelberg) carrying an N-terminal His6 tag followed by a TEV cleavage site. The N-terminal residues (1-18) were removed based on sequence comparison with ABD domain from α-actinin-3 (Franzot et al., 2005) where they are unstructured. To enhance the crystallizability of the protein we designed surface entropy reduction (SER) mutants suggested by the server http://nihserver.mbi.ucla.edu/SER (Goldsmith et al., 1997). Based on the results we mutated the cluster E310A/K311A located on SR1.
Structure-informed mutants were designed to specifically disrupt key contacts between the neck segment of α-actinin-2 and EF3-4. In particular, one positively charged residue (R268), and two hydrophobic residues (I269, L273) were replaced by negatively charged acid glutamic residues (R268E/I269E/L273E), hereafter termed the NEECK mutant.
To explore the regulation of α-actinin-2 by PIP2 binding, we generated mutants where one (R163E), two (R163E/R169E) or three (R163E/R169E/R192E, hereafter the PIP2 mutant) of the predicted main phosphoinositide-binding arginine residues (Fraley et al., 2005, Franzot et al., 2005, Fukami et al., 1992) were exchanged to glutamic acid residues.
All site-directed mutagenesis (SDM) were made following the QuickChange site-directed mutagenesis instructions (Stratagene). Specific oligonucleotides for the SDM reaction were purchased from SIGMA Mutations sites were validated by DNA sequencing.
Protein Expression and Purification
Protein Expression and Purification for Crystallography
The α-actinin-2 Δ1-18, E310A/K311A (α-actinin-2) was expressed in E. coli Rosetta (DE3) pLysS strain (Novagen). The bacterial cultures were grown at 37°C to an optical density (OD) of about 0.6. Then, the culture was induced for four hours with 0.5 mM IPTG. The harvested cells were resuspended in lysis buffer (20 mM Tris-HCl pH 7.6, 150 mM NaCl, 10 mM imidazole) and lysed by sonication. The lysate was cleared from debris by centrifugation and then was loaded onto a Ni-affinity resin. After washing with lysis buffer, bound protein was eluted with elution buffer (20 mM Tris-HCl, 150 mM NaCl, 300 mM imidazole, pH 8.0) and then set for overnight dialysis at 4°C in a buffer containing 50 mM Tris-HCl pH 7.5, 50 mM NaCl, 1 mM EDTA, 2 mM β-mercaptoethanol and TEV protease for the cleavage of the His6-tag. The protein was further purified by ion exchange chromatography and then submitted for reductive methylation of the lysine residues (Walter et al., 2006). In the final purification step α-actinin-2 was passed through a size exclusion chromatography column Superdex S200 column (GE Healthcare) equilibrated in a buffer containing 20 mM HEPES pH 8.0, 150 mM NaCl. SDS-PAGE confirmed the purity of the final protein and dynamic light scattering (DLS) measurements resulted in monomodal distribution with a polydispersity of 15%, suggesting that the protein solution was homogeneous.
Protein Expression and Purification for Biochemical Studies
Full-Length α-Actinin-2 Constructs. α-actinin-2 constructs (wild-type, R268E/I269E/L273E, R163E, R163E/R169E, and R163E/R169E/R192E) were expressed using a recombinant plasmid pET3d containing the full-length gene with a His6-Tag. The plasmids were transfected into suitable E. coli host strains for high-level expression that were defined based on protein expression optimization screened for each constructs. The large-scale expression experiments were carried out in auto-induction media ZYP5052, according the protocol described by Studier et al. (Studier, 2005). Cells growth and protein expression steps were carried out at 30 °C.
After induction, cells were harvested by centrifugation and the cell pellet from one liter of high-density shaking cultures (OD600∼5 - 8 A.U.) were re-suspended in 50 ml of lysis buffer 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5mM EDTA, 0.1% Triton X-100 supplemented with antiproteases (Complete™ Protease Inhibitor Cocktail Tablets, Roche). Cells were disrupted by French Press® and extracts were centrifuged at 18,000xg during 30 min at 4°C.
The supernatant was loaded on a Ni-affinity resin column (2xHis trap FF 5 mL), pre-equilibrated in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 20 mM imidazole, 2 mM β-mercaptoethanol (Buffer A), followed by 3 column volumes of washing using Buffer A. All α-actinin-2 protein constructs were eluted in a two step gradient of imidazole, first with 250 mM eluting the large fraction, followed by 500 mM eluting the remaining protein fraction.
For the anion-exchange purification step, the protein samples were diluted in a low salt buffer, containing 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, 0.1 mM PMSF, 2 mM β-mercaptoethanol, decreasing the final buffer salt concentration by approximately 5-fold. Samples were subsequently loaded on a Resource Q (6 ml column, GE Healthcare) at constant flow rate of 6 ml/min, previously equilibrated in 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, 0.1 mM PMSF, 2 mM β-mercaptoethanol (Buffer B). The α-actinin-2 variants were eluted by increasing salt concentration with 50 mM Tris-HCl (pH 7.5), 2 M NaCl, 2 mM ßME, 5.0 mM EDTA, 1.0 mM PMSF, pH 7.5 (Buffer C). All α-actinin-2 constructs were eluted with 10%–40% of Buffer C. Remaining contaminants/aggregates were removed on a size exclusion chromatography (SEC) column (HiLoad 26/60 Superdex 200 prep grade, GE Healthcare) pre-equilibrated in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM EDTA, 2mM β-mercaptoethanol, 0.1 mM PMSF. SEC separations were performed at a flow rate of 2.5 ml/min at 4°C.
The quality of the protein sample preparation after each purification step was assessed by SDS-PAGE (Laemmli, 1970). Protein concentration was measured by absorbance at 280 nm (extinction coefficient of 127,365 M-1.cm-1). The average protein yield was 10-16 mg of protein/liter of high-density shaking cultures (OD600 = 5-8 A.U.). A sample preparation with ratio protein/nucleic acid (A280/A260) > 1.8 was judged free of nucleic acid contamination (Ribeiro and Ramos, 2004). The proteins were concentrated, by centrifugation using a Centriplus YM-50 (cut-off 50kDa), up to 4.5 mg/ml, flash frozen in liquid nitrogen and stored at −80°C until the spectroscopy characterization.
Titin Zr-7 Construct. cDNA of titin Zr-7 construct (amino acids Lys 692 - Arg 740) was cloned into pETM11 (construct supplied by Dr. Matthias Wilmanns, EMBL Hamburg). The plasmids were transformed in to E. coli BL21(DE3) host strains for high-level expression. For the protein expression, the bacterial cells were grown at 37°C in LB media supplemented with kanamycin at 50 μg/ml to an optical density (OD600) of about 0.8 A.U. The culture was then induced for 16 hr at 30°C with 0.5 mM IPTG. The cells were harvested by centrifugation and the pellet from one liter of cell culture was resuspended in lysis buffer 20 mM Tris-HCl 200 mM NaCl, 10 mM Imidazole, 0.5 mM β-mercaptoethanol (Buffer D), supplemented with DNase I and Lysozyme, and completely lysed by cell sonication. The lysate was centrifuged at 20,000 rpm at 4°C for 30 min. The soluble fractions were loaded onto Ni-affinity resin column, pre-equilibrated with Buffer D. The Ni-affinity resin was washed successively with 3 bed volumes of Buffer D and eluted using 20 mM Tris HCl and 500 mM imidazole at pH 7.2 and loaded onto a SEC column (Superdex 75 HiLoad 16/60 (GE Healthcare) pre-equilibrated with 20 mM Tris HCl 200 mM NaCl 0.5 mM β-mercaptoethanol pH 7.2. The separation was performed at a flow rate of 1.5 ml/min. The quality of the protein sample preparation after each purification step was assessed by SDS-PAGE (Laemmli, 1970). Protein concentration was measured by absorbance at 280 nm. The average protein yield was 1.0-1.5 mg of protein/liter of culture (OD600 = 0.8 U.A.). Absorbance ratio A280/A260 was used to judge nucleic acid contamination (Ribeiro and Ramos, 2004). The proteins were concentrated up to 4.0 mg/ml by centrifugation using a Vivaspin-6 (cut-off 3 kDa, Sartorius Biotech S.A), flashed frozen in liquid nitrogen and stored at −80°C until the spectroscopy characterization.
Analytical Size-Exclusion Chromatography and Multiangle Laser Static Light Scattering
SEC was performed with a Superdex S200 10/300 GL column (GE Healthcare) equilibrated in a buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 1 mM β-mercaptoethanol and 5 mM EDTA. Separations were performed at 20°C with a flow rate of 0.5 ml/min by HPLC (Agilent Technologies 1260 infinity). The samples (50 μl) were injected at a concentration of 2.5 mg/ml. On-line MALLS detection was performed with a miniDawn Treos detector (Wyatt Technology Corp., Santa Barbara, CA) using a laser emitting at 690 nm and by refractive index measurement using a Shodex RI-101 (Shodex). Data analyses were performed as described previously (Gerard et al., 2007). The S200 Superdex column was calibrated with proteins of known Stokes’ radii (RS), according to the method described by (Uversky, 1993), in order to extrapolate the relative RS of WT and NEECK mutant.
Dynamic Laser Light Scattering
DLS was measured at a scattering angle of 90° on a DynaProNanoStar (Wyatt Technology) using a 10 μl microcuvette and a laser emitting at λ = 662 nm. Samples with protein concentration ranging from 1 to 4 mg/ml were centrifuged for 30 min at 13,000 g to eliminate large aggregates. DLS measurements were conducted with laser intensity in autoregulation mode, using 5 acquisition frames of 10 s/acquisition at 25°C. The data were analyzed using online Wyatt software to determine the sample PdI, where a sample with an average PdI ≤ 15% was considered as monodisperse and suitable for structural characterization. The apparent translational diffusion coefficient, D, calculated from the intensity autocorrelation functions fits, was converted into Stokes radii (Rs) according to the Stokes–Einstein equation (Equation 1):
| (Equation 1) |
with R being the gas constant, T the absolute temperature, NA Avogadro’s number and n0 the solvent viscosity. Control experiments carried out with lysozyme yielded the expected apparent translational diffusion coefficient.
Protein Crystallization
Prior to crystallization the protein was lysine methylated following essentially the protocol by (Walter et al., 2006). Pure lysine methylated α-actinin-2 Δ1-18 sample was concentrated to about 5 mg/ml for crystallization experiments. The protein was crystallized in a precipitant containing 0.2 M Mg formate 5% PEG smear and 10 mM EDTA by the hanging drop vapor diffusion method at 14°C. The ratio precipitant:protein was 1:2 (v/v) in hanging drops of final volume 3 μl. All crystals were cryoprotected in the reservoir solution modified with 25% PEG smear. Diffraction data were collected at beamline ID23-2 (ESRF, Grenoble, France).
X-Ray Data Collection, Structure Determination, and Refinement
Crystals belonged to space group P 2 21 21, with unit cell dimensions of a = 73.3 Å, b = 102.6 Å, c = 183.0 Å and one α-actinin-2 molecule in the asymmetric unit. All collected data sets were integrated using XDS (Kabsch, 2010) and two of them showed significantly better statistics (I/σ(I) and CC1/2). These data sets were merged and scaled together using aimless (Evans et al., 2003) up to the maximum resolution of 3.5 Å (Table S1).
The crystal structure comprises one half of α-actinin-2 dimer composed of two halves of the α-actinin-2 protomer (ABD-SR1-SR2/SR3-SR4-CaM) per asymmetric unit and forms the biologically relevant antiparallel homodimer through a crystallographic two-fold axis. The structure was solved by molecular replacement (MR) as implemented in the software PHASER (McCoy et al., 2007). Initially the actin binding domain (ABD) was placed, using as a search model the chain A of the ABD domain from the α-actinin isoform 3 (PDB entry 1WKU). Then, using the placed ABD as a partial solution we positioned the previously determined four spectrin-like repeats of α-actinin in two steps; first we positioned the two C-terminal spectrin-like repeats and then the two N-terminal (PDB entry 1HCI). We finally positioned the two C-terminal EF-hands 3-4, previously determined by NMR (PDB entry 1H8B). The remainder of the structure (linker between ABD domain and spectrin-like domain 1 as well as the EF-hands 1-2) were built manually in coot (Emsley et al., 2010)
The model was refined against native data extending to 3.5 Å by iterations of maximum likelihood and TLS refinement as implemented in the PHENIX suite (Adams et al., 2010) and manual building in coot. To maintain the proper dihedral restraints of the low resolution structure a reference model was used by combining the previously determined high resolution models. Residues 34-892 were assigned in the final model, however, several side chains were not placed due to the weak electron density, specifically in the EF-hands 1-2. Refinement statistics are summarized in Table S1.
Structure Analysis
Analysis of protein-protein interactions and interfaces was performed by PISA (Krissinel and Henrick, 2007). Structural alignments were performed by superpose or PDBeFold (SSM) (Krissinel and Henrick, 2004) using secondary structure matching. Figures of the crystal structure were generated by pymol (http://www.pymol.org). Representation of the structural alignment was generated by Chimera (Pettersen et al., 2004). Sequence conservation analysis was done by Clustal Omega (Sievers et al., 2011) and the ConSruf server (Celniker et al., 2013).
Electron Paramagnetic Resonance
The protein samples for spin labeling experiments were dialyzed in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 0.5 mM EDTA to remove β-mercaptoethanol. The protein concentration was adjusted to 2 μM, and spin labeled with 10 times equivalents of the spin label (SL) (1-Oxyl-2, 2, 5, 5- tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfone, MTSL (Toronto Research Chemicals) to protein which has 10 spin label sites per protomer. For the spin diluted samples the diamagnetic analog (1-Acetyl-2, 5, 5 tetramethyl-Δ3-pyrroline-3-methyl) methanethiosulfone (Toronto Research Chemicals) and MTSL were mixed at the ratio 6:1 prior to addition to α-actinin-2 samples. The mixture of protein and SL (or spin diluted SL) was incubated overnight (∼16 h) under stirring at 4 °C. The free SL was removed by analytical size-exclusion chromatography (Superdex S200 10/300 GL column, GE Healthcare) using a flow rate of 0.5 ml/min. The α-actinin-SL samples were concentrated by centrifugation using CentriplusYM-50 (cutoff 50 kDa) up to 40-60 μM and supplemented with glycerol to a final concentration of 20% (v/v). The spin label efficiency was between 30 and 50% as calculated by the ratio of bound SL versus Cys concentration in the protein where the protein concentration was calculated by absorbance at 280 nm (extinction coefficient of 127,365 M-1.cm-1), and the SL concentration was determined by double integration of the continuous wave EPR (cw EPR) spectrum at room temperature, using the spin probe TEMPOL (4-hydroxy-2,2,6,6- tetramethyl-1-piperidinyl-1-oxyl) as standard. The 10 SL sites were characterized using an open-source software multiscale modeling of macromolecular systems (MMM, (Polyhach et al., 2011) based on the crystal structure data of α-actinin-2. MMM analysis delivers the distances between labeled Cys-residues and estimates the number of rotamers of the SLs, which is dependent on the environment of the labeled Cys. The C270 (neck region), C487 (SR2), C781 (EF1-2) had the highest SL rotamers values (Table S3). We focused on the interspin distance range below and around 20 Å, since the simulated distance between C270 in the neck and the C862 in the EF3-4 is about 12 Å (Table S3). This is the spin label pair that can sense the open and closed conformations of α-actinin-2 (Figure 3A). The other two spin label pairs within 20 Å distance are located on the rod domain (C483-C487) and on the ABD-domain (C48-C251). The first pair is located far from the NEECK mutation sites, and the simulated interspin distance is too short (<10 Å) to be detected by DEER. Additionally the two sites are too close to allow efficient labeling at both sites simultaneously. The ABD-domain pair (C48-C251) instead contains one buried spin label site (C48) therefore the labeling efficiency is predicted to be low (Table S3). These considerations, together with the observed effects on the short distance peak induced in the NEECK mutant (below) strongly suggest that the short distance peak mainly arises from the C270-C862 labeled pair.
After spin labeling, the oligomerization state of the samples was assessed by SEC-MALLS and their quality before and after EPR measurements checked by SDS-PAGE (12%).
Continuous-wave (cw) EPR experiments were performed at 160 K on a Bruker EMX spectrometer operating at 9 GHz equipped with a high sensitivity resonator and a variable temperature unit. All measurements were carried out with 0.2 mW microwave power (MWP), 100 kHz modulation frequency, 0.25 mT modulation amplitude (MA), 41 ms conversion time and 41 ms time constant. Room temperature spectra were detected using 1 mW MWP and 0.1 mT MA.
Q-band DEER experiments were performed at 50 K on a power upgraded Bruker ELEXSYS E580 spectrometer operating at 34 GHz equipped with a 200 W TWT amplifier (Appl. Systems Engineering) and a home-made resonator allowing the insertion of tubes with 3 mm outer diameter. The four-pulse DEER sequence (Pannier et al., 2000) was chosen with π/2(νobs)-τ1-π(νobs)-t’- π(νpump)-(τ1+τ2-t’)-π(νobs)-τ2-echo, where all pulses were set to 12 ns. The pump pulse was placed at the maximum of the spectral intensity and the observer pulses 100 MHz lower in frequency (Polyhach et al., 2012). The DEER spectra were analyzed using the program DeerAnalysis2010 (Jeschke et al., 2006). The background was corrected by a homology 3-dimensional fit. Simulations were checked for stability according to the DEERAnalysis2010 manual.
We used the most common PIP2 found in the plasma membrane of mammalian cells that contains stearoyl (C18:0) and arachidonoyl (C20:4) fatty acid residues (Figure S2A). In vitro, it can assemble micelles that could compromise the homogeneity of the sample preparation and further experimental interpretation. To estimate micelles formation of native PIP2 under the assay conditions used here, the critical micelle concentration (CMC, the amphiphile concentration at which the surface tension of the aqueous phase reaches its minimum) was measured. Stock solution of native PIP2 (Sigma-Aldrich, P9763) was typically prepared at 1-5 mg/ml (1-5 mM) in water (MilliQ) and a series of concentrations was prepared using the same buffer as for EPR experiments (20 mM Tris-HCl 150 mM NaCl 1 mM EDTA pH 7.5 and 20% v/v glycerol). Native PIP2 solutions were sonicated for 5 min at room temperature immediately before dynamic light scattering measurements. DLS analysis showed a CMC of 4-8 μM for native PIP2, which is consistent with the value (10-56 μM) obtained by other groups using slightly different buffer conditions (Moens and Bagatolli, 2007, Walsh et al., 1995). Native PIP2 at concentrations above the CMC induced aggregation in α-actinin-2 monitored by SEC-MALLS-RI; an effect probably due to the association of protein to micelles. In order to reduce sample heterogeneity we subsequently used the water soluble analog of PIP2 labeled in the sn-1 position with the 26-carbon BODIPY-TMR label attached via a 6-carbon linker (Figure S2A). A palmitic acid (C16:0) named here bodipy®TMR-PIP2-C16 (PIP2-C16∗) was esterified on the sn-2 position, purchased from Echelon (Salt Lake City, Utah). Because these PIP2 analogs have shorter alkyl chains and BODIPY®TMR moiety attached on them, they are more water-soluble compared to native PIP2. Previous studies performed by (Moens and Bagatolli, 2007) showed PIP2-C6∗ (Figure S2A) exhibited a CMC of about 500 μM, while PIP2-C16∗ (Figure S2A) a CMC of 100 μM (our experimental conditions), thus > 10-fold higher than the CMC of native PIP2.
In the cw and pulsed EPR experiments, the mean concentration of α-actinin-2 dimer was kept at 15 μM, whereas PIP2-C16∗ was 90 μM, below its critical micelle concentration CMC, and Zr-7 was 450 μM.
Microscale Thermophoresis
PIP2-C16∗ Binding Assay
Specific binding between PIP2-C16∗ and α-actinin-2 was measured by MST method (Duhr and Braun, 2006, Seidel et al., 2013). Unlabelled α-actinin-2 WT or PIP2 mutant was titrated into a fixed concentration of fluorescent bodipy®TMR-PIP2-C16 (PIP2-C16∗, 250 nM). Titration of unlabelled Zr-7 was used as negative control of PIP2-C16∗ binding. Binding reactions were carried out in buffer containing 10 mM HEPES pH 7.4, 150 mM NaCl. Samples were loaded into NT.115 standard treated capillaries (Nanotemper Technologies) immediately after preparation to avoid unspecific adsorption. Before MST measurement the reaction was incubated at room temperature for 10 min mounted in the Monolith NT.115 apparatus (Nanotemper Technologies). The data for microscale thermophoresis analysis were recorded at 25°C using the red LED at 15% (GREEN filter; excitation 515-525 nm, emission 560-585 nm) and IR-Laser power at 40%. Data analyses were performed with NTAnalysis and Sigmaplot software.
CAMD (EF1–4), NEECK Mutant, α-Actinin-2 WT (+/− PIP2-C16∗), and PIP2 Mutant (+/− PIP2-C16∗) Binding to Zr-7
Unlabelled Zr-7 was titrated into a fixed concentration of NT-647 fluorescent cystein labeled α-actinin-2 (EF1-4 at 100 nM, and α-actinin-2 variant at 50 nM), without or with a fixed concentration of PIP2-C16∗ (42 μM). For α-Actinin-2 alone the binding reactions were carried out in buffer containing 10 mM HEPES pH 7.4, 150 mM NaCl plus 0.05% (v/v) Tween 20 (MST buffer), while samples treated with PIP2-C16∗ were in MST buffer without Tween 20 to avoid PIP2-C16∗ incorporation into Tween 20 micelles. Control experiments of NT-647 fluorescent cystein labeled α-actinin-2 WT (50 nM) plus Tween 20 with a fixed concentration of PIP2-C16∗ (80 μM) were made for the direct comparison with the MST results of samples + Tween 20. Protein NT-647-maleimide fluorescent labeling was made using Monolith NT™ protein labeling Kit following manufactures instructions. Labeling efficiencies were in the range 50%–80% and were assessed by measuring the ratio absorbance at 280 nm (α-actinin-2 extinction coefficient of 127,365 M-1.cm-1) and 650 nm (NT-647 extinction coefficient of 250,000 M-1.cm-1). Increasing amounts of unlabeled Zr-7 are indicated in Figures 3E and 3F. Samples of α-actinin-2 WT and NEECK mutant were loaded into NT.115 hydrophilic, while EF1-2 was loaded into hydrophobic capillaries (Nanotemper Technologies). The reactions were incubated at room temperature in dark for 30 min before measurement. The data for microscale thermophoresis analysis were recorded at 25°C using the RED LED at typically 30% and IR-Laser at 40% using the Monolith NT.115 (Nanotemper Technologies). Up to 80% of LED power (RED filter - excitation 605-645 nm, emission 680-685 nm), there was no interference of bodipy®TMR fluorophore (PIP2-C16∗, green spectra) fluorescence intensity on samples containing NT-647 fluorescent cysteines labeled α-actinin-2 in the mixture. Data analysis was performed with software NTAnalysis. The isotherms derived from the raw data were fitted with Hill equation for cooperativity to estimate an apparent dissociation constant (Kd). Average Kd values and standard deviations were calculated from 3 independent measurements (Table S4) and curves fitting were calculated using SigmaPlot 10.0 software (Systat software). Sigmaplot software was used for the transformation of data into ΔFnorm/amplitude (Seidel et al., 2013) allowing the comparison of MST data of samples with different amplitude signal and starting level (Figures 3E and 3F). MST control experiments were carried out using α-actinin-2 EF1-4 and Zr-7 yielded the expected Kd value (Table S4) already characterized by isothermal titration calorimetry (Franzot et al., 2005, Young and Gautel, 2000), and circular dichroism (Joseph et al., 2001).
MST data are represented as means ± standard deviation calculated from three experimental replicas (see also Table S4).
NMR
Samples for NMR studies contained 1.5-2.0 mg of α-actinin-2 wild-type neck-peptide in 25 mM phosphate pH 6.5, 25 mM NaCl, 3 mM DTT and 0.5 mM EDTA in 90% H2O/10% D2O.
All homo- and heteronuclear NMR experiments were recorded on a Bruker Avance 600 MHz spectrometer (Bruker BioSpin AG, Fällanden, Switzerland) equipped with a 5 mm cryogenic CP-TCI z-gradient probe, at 25°C. Proton chemical shift was referenced to the water signal. For chemical shift assignment the following spectra were recorded: 1D proton, 2D double quantum filter correlated spectroscopy (DQF-COSY), 2D In-phase correlation spectroscopy (IP-COSY) (Xia et al., 2005), 2D total correlation spectroscopy (TOCSY) 70 ms mixing time, 2D 1H,1H nuclear Overhauser Enhancement spectroscopy (NOESY), and, 2D 1H,15N heteronuclear single quantum coherence (HSQC). NMR data were processed using BRUKER XWinNMR version 3.5 and TopSpin version 2.1/3.0. NMR spectral analysis was performed using CARA version 1.4.1/1.5.3 (Keller, 2005).
Small-Angle X-Ray Scattering
SAXS data were collected on the EMBL beamline X33 (DESY, Hamburg) (Blanchet et al., 2012) using a photon counting Pilatus 1M detector (DECTRIS, Switzerland). Samples were measured at protein concentrations of 0.8, 1.6 and 3.2 mg/ml in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM β-mercaptoethanol, 0.1 mM PMSF. The scattering intensity I in the range of momentum transfer 0.01 < s < 0.45 Å-1 was recorded (s = 4πsinθ/λ, where λ = 1.5 Å is the X-ray wavelength and 2θ is the scattering angle) at a sample-detector distance of 2.7 m. Radiation damage, monitored by repetitive 15 s exposures, was negligible. Background scattering was subtracted and data reduced, normalized and extrapolated to infinite dilution using PRIMUS (Konarev et al., 2003). The forward scattering I(0) and the radius of gyration Rg were evaluated using the Guinier approximation (Guinier, 1939). These parameters were also computed from the entire scattering patterns using the indirect transform package GNOM (Svergun, 1992), providing also the pair distribution function of the particle P(r) and the maximum size Dmax. The molecular mass (MM) was estimated from the forward scattering by normalization against reference solutions of bovine serum albumin indicating that α-actinin-2 is dimeric in solution in this concentration range.
The scattering pattern of the crystallographic coordinates of wild-type α-actinin-2 was calculated using CRYSOL (Svergun et al., 1995). Given the atomic coordinates, the program fits the experimental intensity by adjusting the excluded volume of the particle and the contrast of the hydration layer to minimize the discrepancy, defined by χ2 given by the equation described by (Svergun et al., 1995).
BUNCH (Petoukhov and Svergun, 2005) was employed to calculate a model of α-actinin-2 (for the wild-type and NEECK mutant) by combined rigid body and ab initio refinement of individual domain components against SAXS data. For this, the crystal structure was fragmented into the dimeric central rod domain and single ABD and EF3-4 hands. The positions and orientations of ABD and EF3-4 were optimized and the missing 49 N-termini residues were restored by BUNCH using P2 symmetry. The low resolution SAXS models obtained by the program BUNCH yielded significantly improved fits to the experimental data (χ = 1.25 and χ = 1.32 for wild-type and NEECK variant, respectively). It should be noted that different BUNCH runs starting from random initial approximations provided models with varying positions of ABD and EF3-4 within similar elongated overall shapes. Five typical rigid body models of NEECK mutant fit the data well with χ values between 1.32 and 1.44. For the wild-type sample the position of EF3-4 was fixed as in the crystal structure, using distance restraint between C862 residues (EF3-4 hands) and C270 (neck region) to be 15 Å, while no such distance restraint was imposed in the BUNCH runs for NEECK mutant, as it was found by EPR experiments (Figure 3C) that the neck region is no longer in contact with the EF3-4 hands in NEECK mutant. NMR analysis showed that the neck-peptide alone does not form a stable α-helical structure (Figure S4). To take into account this information, the neck region in NEECK mutant was subsequently modeled as a flexible link using dummy atoms, which improved SAXS modeling fitting the experimental data to a χ of 1.14. Further flexibility analysis of NEECK mutant was performed using Ensemble Optimization Methods (EOM) analysis (Bernadó et al., 2007), employing P2 symmetry and assuming the linkers between ABD and the rod, and between EF1-2 and EF3-4 are flexible (in other words ABD and EF3-4 could be moved/rotated), improving the fitting to χ of 1.00 (Figure S5).
Molecular Dynamics Simulations of α-Actinin-2 and Docking of PIP2
We have used atomistic molecular dynamics (MD) simulations in order to realistically capture the dynamic behavior of the α-actinin-2 dimer as preparation for subsequent docking of PIP2. For this purpose, the full-length X-ray structure of the dimer was truncated such that the starting conformation for all simulations consisted of two α-actinin-2 fragments: the N-terminal part (residues 34-368) of one monomer (including the ABD domain, the α-helical neck linker and one spectrin-like repeat) and the C-terminal part (residues 633-892) of another monomer (including CAMD with two pairs of EF hands i.e., EF1-2 & EF3-4, and one spectrin-like repeat). This conformation (ABD/CAMD) was placed into a rectangular simulation box (13 × 13 × 13 nm3) and solvated in explicit water molecules (SPC model (Berendsen et al., 1981), 70000 molecules) and 8 sodium counterions that were added in order to keep the net charge neutral. After energy minimization, the system underwent a 5-ns equilibration (including heating up to 300 K) with position restraints applied to all protein atoms. The subsequent production simulation was run without any position restraints for 100 ns with a 1-ps resolution between system snapshots. The simulation was performed using the GROMACS 4.0.7 package (Hess et al., 2008) and the GROMOS96 force field (45a3 parameter set (Schuler et al., 2001)) and was carried out with an integration time step of 2 fs and 3D periodic boundary conditions in the isothermal−isobaric (NPT) ensemble with an isotropic pressure of 1 bar and a constant temperature of 300 K. The pressure and the temperature were controlled using the Berendsen thermostat and barostat (Berendsen et al., 1984) with relaxation times of 1.0 and 0.1 ps, respectively, and a compressibility of 4.5 Å x 10−5 bar−1 for the barostat. A twin-range (10/12 Å) spherical cutoff function was used to truncate van-der-Waals interactions. Electrostatic interactions were treated using the particle-mesh Ewald summation (real space cutoff of 10 Å and 1.2 Å grid with fourth-order spline interpolation).
The thus obtained 100-ns MD trajectory was used for docking of the PIP2 molecule (PIP2-C16:0C16:0) with 100 ABD/CAMD conformations, sampled in 1-ns time steps from the trajectory, as input. The docking was performed using Gold software for flexible ligand docking (version 5.2.2) (Jones et al., 1997). The docking area was specified as a 20 Å sphere centered in the middle of the ABD Arg platform (comprised of Arg163, Arg169, Arg192; Figure S2C). For each protein conformation, 100 different docking solutions were collected, resulting in a total of 10000 PIP2 + ABD/CAMD complex structures over the whole MD trajectory. The obtained solutions were ranked using a scoring function defined as the product of the Gold docking score and a measure of complementarity between hydrophobic/hydrophilic properties of protein and ligand surfaces as calculated by PLATINUM software (Pyrkov et al., 2009). For classification, the docking solutions were grouped in 4 general binding modes: 1) at least one Arg in the Arg platform is in contact with at least one of PIP2 phosphates and one of PIP2 tails is in contact with the ABD neck; 2) at least one Arg in the Arg platform is in contact with at least one of PIP2 phosphates and both PIP2 tails are in contact with the ABD neck; 3) at least one Arg in the Arg platform is in contact with at least one of PIP2 phosphates and none of PIP2 tails are in direct contact with the ABD neck, and 4) other conformations, where PIP2 is in contact with neither the Arg platform nor the ABD neck. Here, a contact between an arginine and a phosphate was defined if the distance between the central carbon in arginine’s guanidinium group and the phosphorus in the phosphate group was < 7 Å, while a contact between a PIP2 tail and the ABD neck was defined if the distance between any atom in one group and any atom in the other group was < 3.5 Å.
PIP2 Binding Assays by Fluorescence Anisotropy
To explore the regulation of α-actinin-2 by PIP2 binding assay using fluorescence anisotropy experiment, we generated mutants on the ABD where one (R163E), two (R163E/R169E) or three (R163E/R169E/R192E, “PIP2 mutant”) of the predicted main phosphoinositide head-group binding arginine residues (see chapter “PIP2 binding site” main text).
Fluorescence anisotropy of Bodipy-TMR-PIP2-C6 (Echelon Bioscience Inc.), PIP2-C6∗ (Figure S2B), at final concentration of 4 μM was measured in the presence of increasing concentrations of α-actinin-2 protein.
Data were recorded in a Perkin Elmer LS 50 B luminescence spectrometer setting an excitation wavelength of 542 nm (slit width of 2.5 nm) and emission of 574 nm (slit width of 5 nm). The calculated grating factor (GF) values were 0.99-1.03 using Bodipy-TMR-PIP2-C6 as reference. The mixture PIP2-C6∗ and protein in 20 mM Tris-HCl (pH 7.5) 150 mM NaCl, 0.5 mM EDTA, 1 mM β-mercaptoethanol were incubated for 10 min at 25°C prior measurement. A very low anisotropy value (0.03 ± 0.01) was observed for PIP2-C6∗ alone, indicating negligible micelle formation as previously reported (Moens and Bagatolli, 2007). The binding assays were carried out at 25°̊C, where an increase of fluorescence anisotropy value indicates PIP2-C6∗-protein interaction. Before all anisotropy measurements, the emission spectra of PIP2-C6∗ (550 to 650 nm) were inspected monitoring signal/noise ratio quality and eventual spectral changes over protein titration. The data were fitted using nonlinear regression with SigmaPlot software. It was not possible to reach the binding saturation curve due to a propensity of α-actinin-2 WT to form other oligomeric species at very high concentration (>40 μM). Although we have not quantified binding affinities constant for this experiment they still deliver information on relative binding affinities (Figure S2B).
F-Actin Cosedimentation Assays
The binding of various α-actinin-2 fragments to F-actin in the presence or absence of titin Zr-7 was examined using the cosedimentation. Actin was purified from rabbit skeletal muscle as described (Spudich and Watt, 1971). Proteins were used at fixed final concentrations i.e., actin at 7 μM, α-actinin-2 variants at 12 μM, and titin Zr-7 at 50 μM in a final total volume of 40 μl. Following 15 min incubation with or without titin Zr-7, α-actinin-2 constructs were mixed with purified rabbit skeletal G-actin. Polymerization of actin was initiated by addition of 1/10 volume of 10x buffer F (50 mM Tris, 0.5 M KCl, 20 mM MgCl2, 20 mM ATP, pH 8). After a 30 min incubation period at room temperature, actin filaments and proteins bound were sedimented by centrifugation for 30 min at 100,000 g at 20°C (Optima Ultracentrifuge, TLA 100 rotor). Supernatants were decanted carefully and added to 5x protein sample buffer. Pellet fractions were dissolved in an equivalent volume of 1x protein sample buffer. Supernatant and pellet fractions were size-separated on a 15% SDS-PAGE and stained with Coomassie Brilliant Blue.
In Vivo Studies
Molecular Cloning for Transient Transfection in Mammalian Cells
The vector pEGFP-N3 containing full-length wild-type human α-actinin-2 cloned between EcoRI and SalI restriction sites was kindly provided by Dr E. Ehler. This was used for the mutagenesis of α-actinin2 R268E/I269E/L273E (NEECK mutant) and R163E/R169E/R192E (PIP2 mutant), using the same oligonucleotides used for mutagenesis in pET3d vector. Between the α-actinin-2 C terminus and GFP N terminus, an additional linker of 12 residues was inserted (VDGATAGPGSIAT). All constructs were validated by DNA sequencing.
Cell Culture and Immunofluorescence
Neonatal rat cardiomyocytes (NRC) were isolated from neonatal rat ventricles and maintained as previously described (Lange et al., 2002). Cells were transfected with 1 μg of plasmid DNA using 3 μl Escort III transfection reagent (Sigma, USA), and fixed with 4%-paraformaldehyde (PFA)/PBS at different time points (24, 48 and 72h). Samples were permeabilized with 0.2% Triton X-100 in PBS for 5 min. Primary and secondary antibodies were diluted in Gold Buffer (20 mM Tris; 155 mM NaCl; 2 mM EGTA; 2 mM MgCl2 pH 7.5) containing 5% BSA. Cells were incubated in primary antibodies for 1 hr at room temperature or overnight at 4°C, then in secondary antibodies for 1 hr at room temperature (primary antibodies: mouse monoclonal anti-telethonin (clone G-11, SantaCruz), anti-vinculin (clone h-VIN1; Sigma), anti-PIP2 (clone 8C2, MBL; fixation in cold methanol and blocking for 30 min in goat serum at room temperature), titin T12 (Fürst et al., 1988) and polyclonal rabbit Z1Z2 antibodies (Zou et al., 2006). Primary antibodies were visualized with species-specific secondary antibodies (Invitrogen) and actin stained with Alexa-633-labeled phalloidin (Invitrogen A22284). Specimens were mounted in anti-fade medium and imaged on a Zeiss LSM510 Meta confocal microscope using a 63X oil immersion objective. Super-resolution microscopy was performed on a Leica TCS STED microscope with a 100X, 1.4 NA STED-grade objective and standard settings for emission and depletion, using atto-647N-labeled goat anti-rabbit secondary antibody (Sigma Aldrich 40839) to visualize titin.
Fluorescence Recovery after Photobleaching
Neonatal rat cardiomyocytes expressing α-actinin-2-GFP variants were imaged 30 h after transfection with a 63 × objective and analyzed with the FRAP module of a Zeiss LSM510 Meta microscope. Regions Of Interest (ROI) were photo-bleached with the laser at 100% intensity and postbleach images were followed with 2 to 4% laser intensity for 100 frames (1 frame every 6 s). FRAP data were analyzed as reported (Wang et al., 2005) and curves fitted to the two-exponential recovery equation (Equation 2):
| (Equation 2) |
where I is the relative intensity compared to the prebleach value, k1 and k2 represent the association rates, and t1 and t2 are the half time recovery of the first and second phase expressed in seconds.
For experiments in the presence of Neomycin, 500 μM of fresh neomycin (Sigma Aldrich) was dissolved in the culture medium and cells equilibrated for 6 hr before imaging, following (Li and Russell, 2013).
Electron Paramagnetic Resonance
Site-directed spin labeling (SDSL) was performed on native cysteine residues of WT and NEECK α-actinin-2. X band cw EPR experiments were carried out at 298 K or 160 K on a Bruker EMX spectrometer. Pulsed EPR measurements were carried out at 50 K on a Q band power upgraded Bruker ELEXSYS E580 spectrometer. Details are given in Extended Experimental Procedures.
SAXS Measurements and Modeling
Small-angle X-ray scattering data were collected at beamline X33 at European Molecular Biology Laboratory (EMBL) Hamburg for WT, NEECK, and PIP2 mutants at three different concentrations and analyzed following standard procedures. Molecular dynamics simulations were carried out using the GROMACS 4.0.7 package (Hess et al., 2008), whereas flexible docking was performed using GOLD version 5.2.2 (Jones et al., 1997). Further details are described in Extended Experimental Procedures.
Cell Biophysics
Experiments in neonatal rat cardiomyocytes were performed using published methods and antibodies using live-cell imaging on a Zeiss LSM510 confocal microscope and superresolution on a Leica TCS STED instrument (see Extended Experimental Procedures).
Author Contributions
Experiments were designed by M.G., K.D.-C., and E.d.A.R. in consultation with K.F.P., E.B., and D.I.S.; all structural work was performed by E.d.A.R., N.P., A.S., B.S., K.F.P., and P.V.K.; E.d.A.R., A.G., and J.K. performed biochemical work; A.G. and M.R.H. performed cell biophysics, and F.L.A. performed the NMR work; C.S. and E.A.G. assisted with protein purification and crystallization; A.A.P. and B.Ž. performed MD; and M.G., K.D.-C., E.d.A.R., and N.P. wrote the manuscript, with all authors contributing to editing the manuscript and supporting the conclusions.
Acknowledgments
We are greatly indebted to Ay Lin Kho for NRC preparations. Thanks also to Dusan Turk (Institute Jozef Stefan, Ljubljana) for initial help with refinement and Oliviero Carugo (University of Vienna and University of Pavia) and Bettina Hartlieb (Baxter Innovations GmbH) for critical reading of the manuscript. We thank the staff of the MX beamlines at the ESRF in Grenoble, SAXS beamline X33 (Deutsches Elektronen-Synchrotron, Hamburg), and SWING beamline (Soleil, Saint-Aubin) for their excellent support. We thank the staff of Campus Science Support Facilities GmbH (Campus Vienna Biocenter) for technical assistance. The K.D.-C. group was supported by Austrian Science Fund (FWF) Projects I525, I1593, P22276, and P19060, by the Federal Ministry of Economy, Family and Youth through the initiative “Laura Bassi Centres of Expertise” funding the Center of Optimized Structural Studies (253275), by the Marie Curie Initial Training Network: MUZIC (238423), and by the University of Vienna. This research was also funded by the European Community Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (283570). F.L.A. thanks BioTek2021 Project 217708/O10 and the Research Council of Norway for financial support. B.Ž. and A.A.P. were supported by the European Research Council (279408). A.A.P. was supported by the Russian Scientific Foundation (14-24-00118). M.G. and A.G. were supported by the Leducq Foundation Transatlantic Network of Excellence: Proteotoxicity (11 CVD 04) and the Medical Research Council of Great Britain (MR/J010456/1). A.G. and E.d.A.R. were also supported by the Marie Curie Initial Training Network: MUZIC (238423). M.G. holds the British Heart Foundation Chair of Molecular Cardiology (CH/08/001). D.I.S. acknowledges support from the Human Frontier Science Program (RGP0017/2012).
Published: November 26, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Extended Experimental Procedures, seven figures, five tables, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2014.10.056.
Contributor Information
Mathias Gautel, Email: mathias.gautel@kcl.ac.uk.
Kristina Djinović-Carugo, Email: kristina.djinovic@univie.ac.at.
Accession Numbers
The coordinates and structure factors of the α-actinin-2 structure have been deposited in the Protein Data Bank under ID code 4D1E.
Supplemental Information
References
- Atkinson R.A., Joseph C., Kelly G., Muskett F.W., Frenkiel T.A., Nietlispach D., Pastore A. Ca2+-independent binding of an EF-hand domain to a novel motif in the α-actinin-titin complex. Nat. Struct. Biol. 2001;8:853–857. doi: 10.1038/nsb1001-853. [DOI] [PubMed] [Google Scholar]
- Bayley P.M., Findlay W.A., Martin S.R. Target recognition by calmodulin: dissecting the kinetics and affinity of interaction using short peptide sequences. Protein Sci. 1996;5:1215–1228. doi: 10.1002/pro.5560050701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M.R., Otey C.A., Campbell S.L. Structural characterization of the interactions between palladin and α-actinin. J. Mol. Biol. 2011;413:712–725. doi: 10.1016/j.jmb.2011.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V., Healy J. Organizing the fluid membrane bilayer: diseases linked to spectrin and ankyrin. Trends Mol. Med. 2008;14:28–36. doi: 10.1016/j.molmed.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Bernadó P., Mylonas E., Petoukhov M.V., Blackledge M., Svergun D.I. Structural characterization of flexible proteins using small-angle X-ray scattering. J. Am. Chem. Soc. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- Carugo, O. (2014). Wolumes - an algorithm to compute the volume of atoms and residues in proteins. arXiv, http://arxiv.org/pdf/1406.3242.pdf.
- Chin D., Means A.R. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- Chiu C., Bagnall R.D., Ingles J., Yeates L., Kennerson M., Donald J.A., Jormakka M., Lind J.M., Semsarian C. Mutations in α-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J. Am. Coll. Cardiol. 2010;55:1127–1135. doi: 10.1016/j.jacc.2009.11.016. [DOI] [PubMed] [Google Scholar]
- Djinovic-Carugo K., Gautel M., Ylänne J., Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- Faulkner G., Pallavicini A., Formentin E., Comelli A., Ievolella C., Trevisan S., Bortoletto G., Scannapieco P., Salamon M., Mouly V. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J. Cell Biol. 1999;146:465–475. doi: 10.1083/jcb.146.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley K.S., Young P.W. The non-muscle functions of actinins: an update. Biochem. J. 2014;459:1–13. doi: 10.1042/BJ20131511. [DOI] [PubMed] [Google Scholar]
- Fraley T.S., Tran T.C., Corgan A.M., Nash C.A., Hao J., Critchley D.R., Greenwood J.A. Phosphoinositide binding inhibits α-actinin bundling activity. J. Biol. Chem. 2003;278:24039–24045. doi: 10.1074/jbc.M213288200. [DOI] [PubMed] [Google Scholar]
- Franzot G., Sjöblom B., Gautel M., Djinović Carugo K. The crystal structure of the actin binding domain from α-actinin in its closed conformation: structural insight into phospholipid regulation of α-actinin. J. Mol. Biol. 2005;348:151–165. doi: 10.1016/j.jmb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Fukami K., Furuhashi K., Inagaki M., Endo T., Hatano S., Takenawa T. Requirement of phosphatidylinositol 4,5-bisphosphate for α-actinin function. Nature. 1992;359:150–152. doi: 10.1038/359150a0. [DOI] [PubMed] [Google Scholar]
- Fukami K., Sawada N., Endo T., Takenawa T. Identification of a phosphatidylinositol 4,5-bisphosphate-binding site in chicken skeletal muscle α-actinin. J. Biol. Chem. 1996;271:2646–2650. doi: 10.1074/jbc.271.5.2646. [DOI] [PubMed] [Google Scholar]
- Full S.J., Deinzer M.L., Ho P.S., Greenwood J.A. Phosphoinositide binding regulates α-actinin CH2 domain structure: analysis by hydrogen/deuterium exchange mass spectrometry. Protein Sci. 2007;16:2597–2604. doi: 10.1110/ps.073146807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin V.E., Orlova A., Salmazo A., Djinović-Carugo K., Egelman E.H. Opening of tandem calponin homology domains regulates their affinity for F-actin. Nat. Struct. Mol. Biol. 2010;17:614–616. doi: 10.1038/nsmb.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautel M. The sarcomeric cytoskeleton: who picks up the strain? Curr. Opin. Cell Biol. 2011;23:39–46. doi: 10.1016/j.ceb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- Gautel M., Goulding D., Bullard B., Weber K., Fürst D.O. The central Z-disk region of titin is assembled from a novel repeat in variable copy numbers. J. Cell Sci. 1996;109:2747–2754. doi: 10.1242/jcs.109.11.2747. [DOI] [PubMed] [Google Scholar]
- Goldstein M.A., Schroeter J.P., Sass R.L. The Z lattice in canine cardiac muscle. J. Cell Biol. 1979;83:187–204. doi: 10.1083/jcb.83.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein M.A., Michael L.H., Schroeter J.P., Sass R.L. Structural states in the Z band of skeletal muscle correlate with states of active and passive tension. J. Gen. Physiol. 1988;92:113–119. doi: 10.1085/jgp.92.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guéguen P., Rouault K., Chen J.M., Raguénès O., Fichou Y., Hardy E., Gobin E., Pan-Petesch B., Kerbiriou M., Trouvé P. A missense mutation in the α-actinin 1 gene (ACTN1) is the cause of autosomal dominant macrothrombocytopenia in a large French family. PLoS ONE. 2013;8:e74728. doi: 10.1371/journal.pone.0074728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton C.M., Taylor D.W., Taylor K.A. Novel structures for α-actinin:F-actin interactions and their implications for actin-membrane attachment and tension sensing in the cytoskeleton. J. Mol. Biol. 2007;368:92–104. doi: 10.1016/j.jmb.2007.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hoeflich K.P., Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108:739–742. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- Jones G., Willett P., Glen R.C., Leach A.R., Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Kaplan J.M., Kim S.H., North K.N., Rennke H., Correia L.A., Tong H.Q., Mathis B.J., Rodríguez-Pérez J.C., Allen P.G., Beggs A.H., Pollak M.R. Mutations in ACTN4, encoding α-actinin-4, cause familial focal segmental glomerulosclerosis. Nat. Genet. 2000;24:251–256. doi: 10.1038/73456. [DOI] [PubMed] [Google Scholar]
- Klaavuniemi T., Kelloniemi A., Ylänne J. The ZASP-like motif in actinin-associated LIM protein is required for interaction with the α-actinin rod and for targeting to the muscle Z-line. J. Biol. Chem. 2004;279:26402–26410. doi: 10.1074/jbc.M401871200. [DOI] [PubMed] [Google Scholar]
- Klein M.G., Shi W., Ramagopal U., Tseng Y., Wirtz D., Kovar D.R., Staiger C.J., Almo S.C. Structure of the actin crosslinking core of fimbrin. Structure. 2004;12:999–1013. doi: 10.1016/j.str.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Korsgren C., Lux S.E. The carboxyterminal EF domain of erythroid α-spectrin is necessary for optimal spectrin-actin binding. Blood. 2010;116:2600–2607. doi: 10.1182/blood-2009-12-260612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsgren C., Peters L.L., Lux S.E. Protein 4.2 binds to the carboxyl-terminal EF-hands of erythroid α-spectrin in a calcium- and calmodulin-dependent manner. J. Biol. Chem. 2010;285:4757–4770. doi: 10.1074/jbc.M109.056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kunishima S., Okuno Y., Yoshida K., Shiraishi Y., Sanada M., Muramatsu H., Chiba K., Tanaka H., Miyazaki K., Sakai M. ACTN1 mutations cause congenital macrothrombocytopenia. Am. J. Hum. Genet. 2013;92:431–438. doi: 10.1016/j.ajhg.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Russell B. Phosphatidylinositol 4,5-bisphosphate regulates CapZβ1 and actin dynamics in response to mechanical strain. Am. J. Physiol. Heart Circ. Physiol. 2013;305:H1614–H1623. doi: 10.1152/ajpheart.00477.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther P.K. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J. Muscle Res. Cell Motil. 2009;30:171–185. doi: 10.1007/s10974-009-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T., Endo M., Ebashi S. Localization of 6S component of α-actinin at Z-band. J. Biochem. 1967;62:630–632. doi: 10.1093/oxfordjournals.jbchem.a128717. [DOI] [PubMed] [Google Scholar]
- Meyer R.K., Aebi U. Bundling of actin filaments by α-actinin depends on its molecular length. J. Cell Biol. 1990;110:2013–2024. doi: 10.1083/jcb.110.6.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra B., Jimenez S., Lin J.H., Bowles K.R., Coveler K.J., Marx J.G., Chrisco M.A., Murphy R.T., Lurie P.R., Schwartz R.J. Mutations in the muscle LIM protein and α-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol. Genet. Metab. 2003;80:207–215. doi: 10.1016/s1096-7192(03)00142-2. [DOI] [PubMed] [Google Scholar]
- Papa I., Astier C., Kwiatek O., Raynaud F., Bonnal C., Lebart M.C., Roustan C., Benyamin Y. αactinin-CapZ, an anchoring complex for thin filaments in Z-line. J. Muscle Res. Cell Motil. 1999;20:187–197. doi: 10.1023/a:1005489319058. [DOI] [PubMed] [Google Scholar]
- Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., Summers M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger J.M., Sanger J.W. The dynamic Z bands of striated muscle cells. Sci. Signal. 2008;1:pe37. doi: 10.1126/scisignal.132pe37. [DOI] [PubMed] [Google Scholar]
- Schacht J. Inhibition by neomycin of polyphosphoinositide turnover in subcellular fractions of guinea-pig cerebral cortex in vitro. J. Neurochem. 1976;27:1119–1124. doi: 10.1111/j.1471-4159.1976.tb00318.x. [DOI] [PubMed] [Google Scholar]
- Sjöblom B., Salmazo A., Djinović-Carugo K. α-actinin structure and regulation. Cell. Mol. Life Sci. 2008;65:2688–2701. doi: 10.1007/s00018-008-8080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi H., Freiburg A., Kolmerer B., Ishiura S., Stier G., Gregorio C.C., Labeit D., Linke W.A., Suzuki K., Labeit S. Tissue-specific expression and α-actinin binding properties of the Z-disc titin: implications for the nature of vertebrate Z-discs. J. Mol. Biol. 1997;270:688–695. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- Swindells M.B., Ikura M. Pre-formation of the semi-open conformation by the apo-calmodulin C-terminal domain and implications for binding IQ-motifs. Nat. Struct. Biol. 1996;3:501–504. doi: 10.1038/nsb0696-501. [DOI] [PubMed] [Google Scholar]
- Takeda S., Yamashita A., Maeda K., Maéda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- Tang J., Taylor D.W., Taylor K.A. The three-dimensional structure of α-actinin obtained by cryoelectron microscopy suggests a model for Ca(2+)-dependent actin binding. J. Mol. Biol. 2001;310:845–858. doi: 10.1006/jmbi.2001.4789. [DOI] [PubMed] [Google Scholar]
- Taylor K.A., Taylor D.W. Projection image of smooth muscle α-actinin from two-dimensional crystals formed on positively charged lipid layers. J. Mol. Biol. 1993;230:196–205. doi: 10.1006/jmbi.1993.1136. [DOI] [PubMed] [Google Scholar]
- Theis J.L., Bos J.M., Bartleson V.B., Will M.L., Binder J., Vatta M., Towbin J.A., Gersh B.J., Ommen S.R., Ackerman M.J. Echocardiographic-determined septal morphology in Z-disc hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2006;351:896–902. doi: 10.1016/j.bbrc.2006.10.119. [DOI] [PubMed] [Google Scholar]
- Tskhovrebova L., Trinick J. Roles of titin in the structure and elasticity of the sarcomere. J. Biomed. Biotechnol. 2010;2010:612482. doi: 10.1155/2010/612482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J., Lünsdorf H., Jockusch B.M. Flexibility and fine structure of smooth-muscle α-actinin. Eur. J. Biochem. 1997;248:193–199. doi: 10.1111/j.1432-1033.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- Ylänne J., Scheffzek K., Young P., Saraste M. Crystal structure of the α-actinin rod reveals an extensive torsional twist. Structure. 2001;9:597–604. doi: 10.1016/s0969-2126(01)00619-0. [DOI] [PubMed] [Google Scholar]
- Young P., Gautel M. The interaction of titin and α-actinin is controlled by a phospholipid-regulated intramolecular pseudoligand mechanism. EMBO J. 2000;19:6331–6340. doi: 10.1093/emboj/19.23.6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P., Ferguson C., Bañuelos S., Gautel M. Molecular structure of the sarcomeric Z-disk: two types of titin interactions lead to an asymmetrical sorting of α-actinin. EMBO J. 1998;17:1614–1624. doi: 10.1093/emboj/17.6.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P., Pinotsis N., Lange S., Song Y.H., Popov A., Mavridis I., Mayans O.M., Gautel M., Wilmanns M. Palindromic assembly of the giant muscle protein titin in the sarcomeric Z-disk. Nature. 2006;439:229–233. doi: 10.1038/nature04343. [DOI] [PubMed] [Google Scholar]
Supplemental References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Hermans J. In: Intermolecular Forces. Pullman B., editor. Reidel; Dordrecht: 1981. Interaction models for water in relation to protein hydration; pp. 331–342. [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Blanchet C.E., Zozulya A.V., Kikhney A.G., Franke D., Konarev P.V., Shang W.F., Klaering R., Robrahn B., Hermes C., Cipriani F. Instrumental setup for high-throughput small- and wide-angle solution scattering at the X33 beamline of EMBL Hamburg. J. Appl. Crystallogr. 2012;45:489–495. [Google Scholar]
- Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., Pupko T., Ben-Tal N. ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 2013;53:199–206. [Google Scholar]
- Djinovic-Carugo K., Gautel M., Ylänne J., Young P. The spectrin repeat: a structural platform for cytoskeletal protein assemblies. FEBS Lett. 2002;513:119–123. doi: 10.1016/s0014-5793(01)03304-x. [DOI] [PubMed] [Google Scholar]
- Duhr S., Braun D. Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. USA. 2006;103:19678–19682. doi: 10.1073/pnas.0603873103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans G., Polentarutti M., Djinović Carugo K., Bricogne G. SAD phasing with triiodide, softer X-rays and some help from radiation damage. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1429–1434. doi: 10.1107/s0907444903012526. [DOI] [PubMed] [Google Scholar]
- Fraley T.S., Pereira C.B., Tran T.C., Singleton C., Greenwood J.A. Phosphoinositide binding regulates α-actinin dynamics: mechanism for modulating cytoskeletal remodeling. J. Biol. Chem. 2005;280:15479–15482. doi: 10.1074/jbc.M500631200. [DOI] [PubMed] [Google Scholar]
- Fürst D.O., Osborn M., Nave R., Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J. Cell Biol. 1988;106:1563–1572. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard F.C., Ribeiro Ede.A., Jr., Albertini A.A., Gutsche I., Zaccai G., Ruigrok R.W., Jamin M. Unphosphorylated rhabdoviridae phosphoproteins form elongated dimers in solution. Biochemistry. 2007;46:10328–10338. doi: 10.1021/bi7007799. [DOI] [PubMed] [Google Scholar]
- Goldsmith S.C., Pokala N., Shen W., Fedorov A.A., Matsudaira P., Almo S.C. The structure of an actin-crosslinking domain from human fimbrin. Nat. Struct. Biol. 1997;4:708–712. doi: 10.1038/nsb0997-708. [DOI] [PubMed] [Google Scholar]
- Guinier, A. (1939). La diffraction des rayons X aux tres petits angles: application a l’etude des phenomnes ultramicroscopiques. Thesis (Paris: Université Paris).
- Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C.R., Hilger D., Jung H. DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006;30:473–498. [Google Scholar]
- Joseph C., Stier G., O’Brien R., Politou A.S., Atkinson R.A., Bianco A., Ladbury J.E., Martin S.R., Pastore A. A structural characterization of the interactions between titin Z-repeats and the alpha-actinin C-terminal domain. Biochemistry. 2001;40:4957–4965. doi: 10.1021/bi002739r. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus P.A., Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, R. (2005). Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. PhD thesis (Zürich: Institute of Molecular Biology and Biophysics, ETH Zürich).
- Konarev P.V., Volkov V.V., Sokolova A.V., Koch M.H.J., Svergun D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Krissinel E., Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lange S., Auerbach D., McLoughlin P., Perriard E., Schäfer B.W., Perriard J.C., Ehler E. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J. Cell Sci. 2002;115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meador W.E., Means A.R., Quiocho F.A. Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- Moens P.D.J., Bagatolli L.A. Profilin binding to sub-micellar concentrations of phosphatidylinositol (4,5) bisphosphate and phosphatidylinositol (3,4,5) trisphosphate. Biochim Biophys Acta. 2007;1768:439–449. doi: 10.1016/j.bbamem.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Pannier M., Veit S., Godt A., Jeschke G., Spiess H.W. Dead-time free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000;142:331–340. doi: 10.1006/jmre.1999.1944. [DOI] [PubMed] [Google Scholar]
- Petoukhov M.V., Svergun D.I. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Polyhach Y., Bordignon E., Jeschke G. Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys. 2011;13:2356–2366. doi: 10.1039/c0cp01865a. [DOI] [PubMed] [Google Scholar]
- Polyhach Y., Bordignon E., Tschaggelar R., Gandra S., Godt A., Jeschke G. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Phys. Chem. Chem. Phys. 2012;14:10762–10773. doi: 10.1039/c2cp41520h. [DOI] [PubMed] [Google Scholar]
- Pyrkov T.V., Chugunov A.O., Krylov N.A., Nolde D.E., Efremov R.G. PLATINUM: a web tool for analysis of hydrophobic/hydrophilic organization of biomolecular complexes. Bioinformatics. 2009;25:1201–1202. doi: 10.1093/bioinformatics/btp111. [DOI] [PubMed] [Google Scholar]
- Ribeiro E.A., Jr., Ramos C.H. Origin of the anomalous circular dichroism spectra of many apomyoglobin mutants. Anal. Biochem. 2004;329:300–306. doi: 10.1016/j.ab.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Schuler L.D., Daura X., van Gunsteren W.F. An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase. J. Comput. Chem. 2001;22:1205–1218. [Google Scholar]
- Seidel S.A., Dijkman P.M., Lea W.A., van den Bogaart G., Jerabek-Willemsen M., Lazic A., Joseph J.S., Srinivasan P., Baaske P., Simeonov A. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J.A., Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- Svergun D., Barberato C., Koch M.H.J. CRYSOL - a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- Uversky V.N. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry. 1993;32:13288–13298. doi: 10.1021/bi00211a042. [DOI] [PubMed] [Google Scholar]
- Walsh J.P., Suen R., Glomset J.A. Arachidonoyl-diacylglycerol kinase. Specific in vitro inhibition by polyphosphoinositides suggests a mechanism for regulation of phosphatidylinositol biosynthesis. J. Biol. Chem. 1995;270:28647–28653. doi: 10.1074/jbc.270.48.28647. [DOI] [PubMed] [Google Scholar]
- Walter T.S., Meier C., Assenberg R., Au K.F., Ren J., Verma A., Nettleship J.E., Owens R.J., Stuart D.I., Grimes J.M. Lysine methylation as a routine rescue strategy for protein crystallization. Structure. 2006;14:1617–1622. doi: 10.1016/j.str.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shaner N., Mittal B., Zhou Q., Chen J., Sanger J.M., Sanger J.W. Dynamics of Z-band based proteins in developing skeletal muscle cells. Cell Motil. Cytoskeleton. 2005;61:34–48. doi: 10.1002/cm.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M.S., Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J. Appl. Crystallogr. 1997;30:203–205. [Google Scholar]
- Xia Y., Legge G., Jun K.Y., Qi Y., Lee H., Gao X. IP-COSY, a totally in-phase and sensitive COSY experiment. Magn. Reson. Chem. 2005;43:372–379. doi: 10.1002/mrc.1558. [DOI] [PubMed] [Google Scholar]
- Xie X., Harrison D.H., Schlichting I., Sweet R.M., Kalabokis V.N., Szent-Györgyi A.G., Cohen C. Structure of the regulatory domain of scallop myosin at 2.8 A resolution. Nature. 1994;368:306–312. doi: 10.1038/368306a0. [DOI] [PubMed] [Google Scholar]
- Zhang M., Tanaka T., Ikura M. Calcium-induced conformational transition revealed by the solution structure of apo calmodulin. Nat. Struct. Biol. 1995;2:758–767. doi: 10.1038/nsb0995-758. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.