Abstract
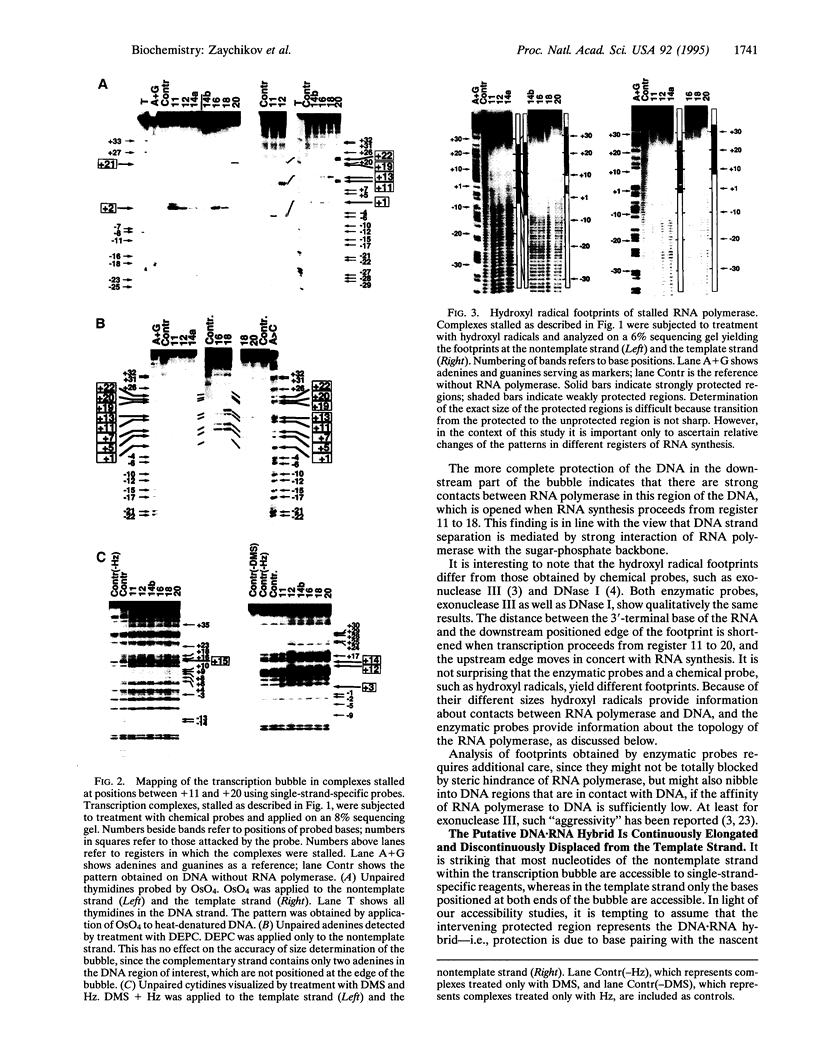
Translocation of DNA-dependent RNA polymerase along the DNA template during RNA synthesis encompasses continuous as well as discontinuous steps. This is demonstrated by chemical probing of transcription complexes stalled in consecutive registers of RNA synthesis at base positions +11, +12, +14, +16, +18, and +20. The "transcription bubble" translocates by continuous opening of the downstream edge in tandem with the growing RNA chain and discontinuous closing at the upstream edge after at least nine steps of RNA synthesis. The position of the enzyme remains unchanged during extension of the transcription bubble and "jumps" 10 bp downstream simultaneously with collapse of the transcription bubble.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borukhov S., Polyakov A., Nikiforov V., Goldfarb A. GreA protein: a transcription elongation factor from Escherichia coli. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8899–8902. doi: 10.1073/pnas.89.19.8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borukhov S., Sagitov V., Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993 Feb 12;72(3):459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- Buckle M., Buc H. Fine mapping of DNA single-stranded regions using base-specific chemical probes: study of an open complex formed between RNA polymerase and the lac UV5 promoter. Biochemistry. 1989 May 16;28(10):4388–4396. doi: 10.1021/bi00436a040. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Gralla J. D. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980 Jul 8;19(14):3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Gralla J. D. Interaction of RNA polymerase with lacUV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation, and rifampicin-sensitivity changes accompanying transcription initiation. J Mol Biol. 1985 May 25;183(2):165–177. doi: 10.1016/0022-2836(85)90210-4. [DOI] [PubMed] [Google Scholar]
- Chamberlin M. J. New models for the mechanism of transcription elongation and its regulation. Harvey Lect. 1992 1993;88:1–21. [PubMed] [Google Scholar]
- Grachev M. A., Zaychikov E. F. Initiation by Escherichia coli RNA-polymerase: transformation of abortive to productive complex. FEBS Lett. 1980 Jun 16;115(1):23–26. doi: 10.1016/0014-5793(80)80718-6. [DOI] [PubMed] [Google Scholar]
- Heumann H., Ricchetti M., Werel W. DNA-dependent RNA polymerase of Escherichia coli induces bending or an increased flexibility of DNA by specific complex formation. EMBO J. 1988 Dec 20;7(13):4379–4381. doi: 10.1002/j.1460-2075.1988.tb03336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. RNA polymerase marching backward. Science. 1993 Feb 12;259(5097):944–945. doi: 10.1126/science.7679800. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Zentner P. G., Geiduschek E. P. Transcription at bacteriophage T4 variant late promoters. An application of a newly devised promoter-mapping method involving RNA chain retraction. J Biol Chem. 1986 Oct 25;261(30):14256–14265. [PubMed] [Google Scholar]
- Kirkegaard K., Buc H., Spassky A., Wang J. C. Mapping of single-stranded regions in duplex DNA at the sequence level: single-strand-specific cytosine methylation in RNA polymerase-promoter complexes. Proc Natl Acad Sci U S A. 1983 May;80(9):2544–2548. doi: 10.1073/pnas.80.9.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry. 1989 Sep 19;28(19):7829–7842. doi: 10.1021/bi00445a045. [DOI] [PubMed] [Google Scholar]
- Krummel B., Chamberlin M. J. Structural analysis of ternary complexes of Escherichia coli RNA polymerase. Deoxyribonuclease I footprinting of defined complexes. J Mol Biol. 1992 May 20;225(2):239–250. doi: 10.1016/0022-2836(92)90918-a. [DOI] [PubMed] [Google Scholar]
- Lee D. N., Landick R. Structure of RNA and DNA chains in paused transcription complexes containing Escherichia coli RNA polymerase. J Mol Biol. 1992 Dec 5;228(3):759–777. doi: 10.1016/0022-2836(92)90862-e. [DOI] [PubMed] [Google Scholar]
- Levin J. R., Krummel B., Chamberlin M. J. Isolation and properties of transcribing ternary complexes of Escherichia coli RNA polymerase positioned at a single template base. J Mol Biol. 1987 Jul 5;196(1):85–100. doi: 10.1016/0022-2836(87)90512-2. [DOI] [PubMed] [Google Scholar]
- Metzger W., Schickor P., Heumann H. A cinematographic view of Escherichia coli RNA polymerase translocation. EMBO J. 1989 Sep;8(9):2745–2754. doi: 10.1002/j.1460-2075.1989.tb08416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger W., Schickor P., Meier T., Werel W., Heumann H. Nucleation of RNA chain formation by Escherichia coli DNA-dependent RNA polymerase. J Mol Biol. 1993 Jul 5;232(1):35–49. doi: 10.1006/jmbi.1993.1368. [DOI] [PubMed] [Google Scholar]
- Mustaev A., Kashlev M., Zaychikov E., Grachev M., Goldfarb A. Active center rearrangement in RNA polymerase initiation complex. J Biol Chem. 1993 Sep 15;268(26):19185–19187. [PubMed] [Google Scholar]
- Nudler E., Goldfarb A., Kashlev M. Discontinuous mechanism of transcription elongation. Science. 1994 Aug 5;265(5173):793–796. doi: 10.1126/science.8047884. [DOI] [PubMed] [Google Scholar]
- Palecek E. Probing DNA structure with osmium tetroxide complexes in vitro. Methods Enzymol. 1992;212:139–155. doi: 10.1016/0076-6879(92)12010-n. [DOI] [PubMed] [Google Scholar]
- Rees W. A., Keller R. W., Vesenka J. P., Yang G., Bustamante C. Evidence of DNA bending in transcription complexes imaged by scanning force microscopy. Science. 1993 Jun 11;260(5114):1646–1649. doi: 10.1126/science.8503010. [DOI] [PubMed] [Google Scholar]
- Schickor P., Metzger W., Werel W., Lederer H., Heumann H. Topography of intermediates in transcription initiation of E.coli. EMBO J. 1990 Jul;9(7):2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky A. Passage of RNA polymerase from open complex to elongation mode at the Escherichia coli lacUV5 promoter: nucleolytic hypersensitivity as a probe for complex conformational changes. Biochemistry. 1992 Nov 3;31(43):10502–10509. doi: 10.1021/bi00158a013. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. A stressed intermediate in the formation of stably initiated RNA chains at the Escherichia coli lac UV5 promoter. J Mol Biol. 1987 Jan 20;193(2):267–278. doi: 10.1016/0022-2836(87)90218-x. [DOI] [PubMed] [Google Scholar]
- Straney D. C., Crothers D. M. Intermediates in transcription initiation from the E. coli lac UV5 promoter. Cell. 1985 Dec;43(2 Pt 1):449–459. doi: 10.1016/0092-8674(85)90175-8. [DOI] [PubMed] [Google Scholar]
- Suh W. C., Ross W., Record M. T., Jr Two open complexes and a requirement for Mg2+ to open the lambda PR transcription start site. Science. 1993 Jan 15;259(5093):358–361. doi: 10.1126/science.8420002. [DOI] [PubMed] [Google Scholar]
- Surratt C. K., Milan S. C., Chamberlin M. J. Spontaneous cleavage of RNA in ternary complexes of Escherichia coli RNA polymerase and its significance for the mechanism of transcription. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):7983–7987. doi: 10.1073/pnas.88.18.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Hydroxyl radical "footprinting": high-resolution information about DNA-protein contacts and application to lambda repressor and Cro protein. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5469–5473. doi: 10.1073/pnas.83.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]







