Abstract
Background
The association between the HLA-B-associated transcript 3 polymorphisms and lung cancer risk is a subject of debate. We conducted a meta-analysis to evaluate the association between these polymorphisms and lung cancer susceptibility.
Material/Methods
A systematic search of electronic databases (PubMed, EMBASE, Wanfang, and China National Knowledge Infrastructure) was performed. Data were extracted and pooled odds ratios (OR) with 95% confidence intervals (CI) were calculated.
Results
Ten case-control studies with 37 945 and 56 807 controls were included in this meta-analysis. Overall, a significant association between rs1052486 polymorphism and lung cancer susceptibility was observed (OR=1.07, 95% CI 1.01–1.12, P=0.01). In addition, a significant association was found for rs3117582 polymorphism (OR=1.29, 95% CI 1.22–1.37, P<0.01).
Conclusions
This meta-analysis suggested that HLA-B-associated transcript 3 polymorphisms are risk factors for lung cancer.
MeSH Keywords: Lung Neoplasms; Meta-Analysis; Polymorphism, Genetic
Background
Lung cancer is the most common cancer in the world and represents a major public health problem, accounting for ~1.2 million cancer-related deaths worldwide each year. Although >80% of the population-attributable risk of lung cancer can be ascribed to tobacco smoking, several lines of evidence indicate that inherited genetic factors influence the development and progression of lung cancer.
HLA-B-associated transcript 3 is a member of the Bcl-2-associated anthanogene (BAG) family of proteins. HLA-B-associated transcript 3 was first discovered as a member of a group of genes located within the Class III region of the human major histocompatibility complex on chromosome 6, and has been extensively studied for its role in regulating apoptosis under various stress conditions such as DNA damage and endoplasmic reticulum-related stress [1,2]. Several studies have investigated the association between HLA-B-associated transcript 3 polymorphisms and lung cancer risk. However, the results were inconclusive [3–12]. Meta-analysis is a useful method for investigating associations between genetic factors and diseases, because a quantitative approach is used to combine the results from different studies on the same topic, thereby providing more reliable conclusions. Thus, we performed a meta-analysis to assess the association of HLA-B-associated transcript 3 polymorphisms with lung cancer. To our knowledge, this is the first meta-analysis of the association between HLA-B-associated transcript 3 polymorphisms and the risk of lung cancer.
Material and Methods
Publication search
In this meta-analysis, we searched the articles using the search terms „HLA-B-associated transcript 3”, “lung cancer” and “polymorphism” in the PubMed, EMBASE, and Chinese National Knowledge Infrastructure (CNKI) databases, and the last search was updated March 2014. Additional studies were identified by a hand search of references of original studies or review articles on the association between HLA-B-associated transcript 3 polymorphisms and lung cancer. No publication date or language restrictions were imposed.
Inclusion and exclusion criteria
The following inclusion criteria were used: (1) evaluation of the HLA-B-associated transcript 3 polymorphisms and lung cancer risk, (2) using a case-control design, and (3) genotype distributions in both cases and controls should be available for estimating an odds ratio (OR) with 95% confidence interval (CI).
Studies were excluded if any of the following existed: (1) not relevant to lung cancer or HLA-B-associated transcript 3 polymorphisms, (2) not designed as case-control studies, (3) genotype frequencies or number not included, (4) animal studies, and (5) editorials, reviews, and abstracts. If more than 1 study used the same cases, the study with the most comprehensive population was included.
Data extraction
The following data were collected from each study: first author’s surname, year of publication, ethnicity, histology of cancer, smoking status, and sample size.
Statistical analysis
The strength of the associations between the HLA-B-associated transcript 3 polymorphisms and lung cancer risk was measured by ORs and 95% CIs. The random-effects model was used. The statistical significance of summary OR was determined with the Z test. The Q statistic and the I2 statistic were used to assess the degree of heterogeneity among the studies included in the meta-analysis. Subgroup analyses were carried out by ethnicity, histology, and smoking. Sensitivity analysis was performed through sequentially excluding individual studies to assess the stability of the results. The potential publication bias was examined visually in a funnel plot of log [OR] against its standard error (SE), and the degree of asymmetry was tested using Egger’s test [13]. All statistical tests were performed using STATA 11.0 software (Stata Corporation, College Station, TX, USA). A P value <0.05 was considered statistically significant.
Results
Study characteristics
A total of 10 case-control studies with 37 945 and 56 807 controls on the association between HLA-B-associated transcript 3 polymorphisms and lung cancer risk were included in this meta-analysis [3–12]. There were 6 studies of rs1052486 and 5 studies of rs3117582. The characteristics of each case-control study are listed in Table 1.
Table 1.
Studies and data included in this meta-analysis.
| First author | Year | Ethnity | Histology | Smoking | Case | Control | Polymorphism |
|---|---|---|---|---|---|---|---|
| Rudd | 2006 | Caucasian | Mixed | Mixed | 1529 | 2707 | rs1052486 |
| Ter-Minassian | 2008 | Caucasian | NSCLC* | Mixed* | 2135 | 1492 | rs1052486 |
| Broderick | 2009 | Caucasian | Mixed* | Mixed* | 7560 | 8205 | rs3117582 |
| Truong | 2010 | Mixed* | Mixed | Mixed | 5214 | 6620 | rs1052486 |
| Wang | 2010 | Caucasian | Mixed* | Nonsmoker | 239 | 553 | rs3117582 |
| Young | 2011 | Caucasian | Mixed | Mixed | 454 | 488 | rs1052486 |
| Timofeeva | 2012 | Caucasian | Mixed | Mixed | 14845 | 29389 | rs3117582 |
| Wang | 2012 | Asian | Mixed | Mixed | 784 | 782 | rs1052486 |
| Brenner | 2013 | Caucasian | Mixed | Mixed | 4441 | 5094 | rs1052486, rs3117582 |
| Doherty | 2013 | Caucasian | Mixed | Smoker | 744 | 1477 | rs3117582 |
Different data can be extracted.
NSCLC – non-small cell lung cancer.
Overall and subgroup meta-analysis results
rs1052486
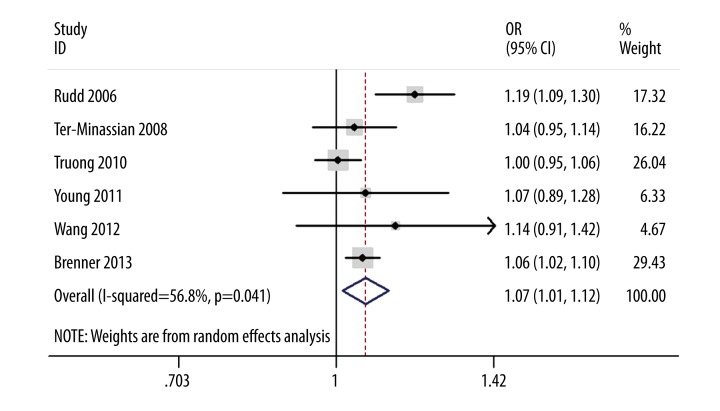
The association between HLA-B associated transcript 3 rs1052486 polymorphism and lung cancer risk was investigated in 6 case-control studies with a total of 14 557 cases and 17 183 controls. The result suggested that this polymorphism was associated with lung cancer risk (OR=1.07, 95% CI 1.01–1.12, P=0.01). In the subgroup analysis by ethnicity, a significant association was found among whites (OR=1.07, 95% CI 1.01–1.12, P=0.02), but not among Asians (OR=1.15, 95% CI 0.97–1.38, P=0.11). The sensitivity analysis did not influence the result excessively by omitting any single study (data not shown). Funnel plot and Egger’s test were both performed to access the publication bias of this meta-analysis. The shape of the funnel plot appeared symmetrical (data not shown). Egger’s test showed no evidence of publication bias (P=0.48) (Figure 1).
Figure 1.
Meta-analysis of the association between HLA-B-associated transcript 3 rs1052486 polymorphism and lung cancer risk.
rs3117582
The association between HLA-B associated transcript 3 rs3117582 polymorphism and lung cancer risk was investigated in 5 case-control studies with a total of 27 829 cases and 44 718 controls. The result suggested that this polymorphism was associated with lung cancer risk (OR=1.29, 95% CI 1.22–1.37, P<0.01). In the subgroup analysis by ethnicity, a significant association was found among whites (OR=1.29, 95% CI 1.22–1.37, P<0.01). Subgroup analysis was also performed by the type of lung cancer. The significant association was observed among squamous carcinoma patients (OR=1.30, 95% CI 1.11–1.52, P<0.01). In the subgroup analysis according to smoking status, increased lung cancer risk was found among smokers (OR=1.24, 95% CI 1.06–1.47, P<0.01). A summary of results is listed in Table 2. Statistically similar results were obtained after sequentially excluding each study (data not shown). The shape of the funnel plot was symmetrical (data not shown). Egger’s test did not find evidence of publication bias (P=0.34) (Figure 2).
Table 2.
Detailed results of meta-analysis.
| Association | Heterogeneity | |||
|---|---|---|---|---|
| OR (95% CI) | P Value | P Value | I2 (%) | |
| rs1052486 | ||||
| Overall | 1.07 (1.02–1.12) | 0.01 | 0.04 | 57.0 |
| Caucasian | 1.07 (1.01–1.12) | 0.02 | 0.04 | 61.0 |
| Asian | 1.15 (0.97–1.38) | 0.11 | 0.85 | 0.0 |
| rs3117582 | ||||
| Overall | 1.29 (1.22–1.37) | <0.01 | 0.16 | 39.0 |
| Caucasian | 1.29 (1.22–1.37) | <0.01 | 0.16 | 39.0 |
| Adenocarcinoma | 1.01 (0.82–1.24) | 0.93 | 1.00 | 0.0 |
| Squamous carcinoma | 1.30 (1.11–1.52) | <0.01 | 0.91 | 0.0 |
| Small cell lung cancer | 1.05 (0.92–1.20) | 0.49 | 0.65 | 0.0 |
| Smoker | 1.24 (1.06–1.47) | <0.01 | 0.17 | 47.0 |
Figure 2.
Meta-analysis of the association between HLA-B-associated transcript 3 rs3117582 polymorphism and lung cancer risk.
Discussion
The main finding of this meta-analysis was that rs1052486 and rs3117582 polymorphisms were potential risk factors for developing lung cancer. In the subgroup analysis of rs1052486 by ethnicity, a significant association was found in whites. However, no study with Asians was included in this meta-analysis. Thus, more studies with Asians should be conducted to determine the association between rs1052486 polymorphism and lung cancer. In the subgroup analysis of rs3117582 by ethnicity, no significant association was found in Asians, but lung cancer risk was increased in whites. It is possible that different lifestyles, diets, and environments may account for this apparent discrepancy. These issues should be investigated in future studies. In the subgroup analysis by histology, we observed that there was a significant association between this polymorphism and squamous carcinoma risk, suggesting that rs3117582 polymorphism might influence the etiology of squamous carcinoma. In the subgroup analysis stratified by smoking, rs3117582 polymorphism was associated with increased lung cancer risk in smokers. Wang et al. [7] suggested that this polymorphism plays no role in the development of lung cancer. This result indicated that even the same variant in the same gene may have a different effect on the pathogenesis of lung cancer in different individuals.
HLA-B-associated transcript 3 is necessary for p300-mediated p53 acetylation, and functions as a novel positive regulator of p53-mediated apoptosis induced by genotoxic stress in vivo and in vitro [14]. Since dysregulation of the p300–p53 pathway has been observed in multiple cancers [15] and a biallelic inactivating mutation of HLA-B-associated transcript 3 has been found in a colon cancer cell line [16], it will be important to investigate whether mutations of HLA-B-associated transcript 3 are associated with lung cancer. Tsukahara et al. suggested that HLA-B-associated transcript 3 regulated apoptotic cell death induced by papillomavirus-binding factor in human osteosarcoma [17]. However, the role of polymorphisms in the HLA-B-associated transcript 3 gene in the development of lung cancer is still uncertain and the exact mechanism should be elucidated.
Some limitations should be acknowledged. First, only published studies that were included in the selected electronic databases were identified. It is possible that some relevant published or unpublished studies may have been missed. Second, the effects of gene-gene and gene-environment interactions were not addressed in this meta-analysis, because of limited available data. Third, our meta-analysis was based on unadjusted OR estimates because not all published studies presented adjusted ORs.
Conclusions
This meta-analysis found significant associations between HLA-B-associated transcript 3 polymorphisms and lung cancer risk. Further studies in more ethnic groups are warranted to validate these results.
Footnotes
Conflicts of interest
None.
Source of support: Self financing
References
- 1.De Flora S, Izzotti A, Walsh D, et al. Molecular epidemiology of atherosclerosis. FASEB J. 1997;11:1021–31. [PubMed] [Google Scholar]
- 2.Martinet W, Knaapen MW, De Meyer GR, et al. Elevated levels of oxidative DNA damage and DNA repair enzymes in human atherosclerotic plaques. Circulation. 2002;106:927–32. doi: 10.1161/01.cir.0000026393.47805.21. [DOI] [PubMed] [Google Scholar]
- 3.Rudd MF, Webb EL, Matakidou A, et al. Variants in the GH-IGF axis confer susceptibility to lung cancer. Genome Res. 2006;16:693–701. doi: 10.1101/gr.5120106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ter-Minassian M, Zhai R, Asomaning K, et al. Apoptosis gene polymorphisms, age, smoking and the risk of non-small cell lung cancer. Carcinogenesis. 2008;29:2147–52. doi: 10.1093/carcin/bgn205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broderick P, Wang Y, Vijayakrishnan J, et al. Deciphering the impact of common genetic variation on lung cancer risk: a genome-wide association study. Cancer Res. 2009;69:6633–41. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Truong T, Sauter W, McKay JD, et al. International Lung Cancer Consortium: coordinated association study of 10 potential lung cancer susceptibility variants. Carcinogenesis. 2010;31:625–33. doi: 10.1093/carcin/bgq001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Broderick P, Matakidou A, et al. Role of 5p15.33 (TERT-CLPTM1L), 6p21.33 and 15q25.1 (CHRNA5-CHRNA3) variation and lung cancer risk in never-smokers. Carcinogenesis. 2010;31:234–38. doi: 10.1093/carcin/bgp287. [DOI] [PubMed] [Google Scholar]
- 8.Young RP, Hopkins RJ, Whittington CF, et al. Individual and cumulative effects of GWAS susceptibility loci in lung cancer: associations after sub-phenotyping for COPD. PLoS One. 2011;6:e16476. doi: 10.1371/journal.pone.0016476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timofeeva MN, Hung RJ, Rafnar T, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21:4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, Li G, Zhang X, et al. BAT3 gene polymorphisms and susceptibility to lung cancer in Han Chinese. Communication on Contemporary Anthropology. 2012;6:165–73. [Google Scholar]
- 11.Brenner DR, Brennan P, Boffetta P, et al. Hierarchical modeling identifies novel lung cancer susceptibility variants in inflammation pathways among 10,140 cases and 11,012 controls. Hum Genet. 2013;132:579–89. doi: 10.1007/s00439-013-1270-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty JA, Sakoda LC, Loomis MM, et al. DNA repair genotype and lung cancer risk in the beta-carotene and retinol efficacy trial. Int J Mol Epidemiol Genet. 2013;4:11–34. [PMC free article] [PubMed] [Google Scholar]
- 13.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sasaki T, Gan EC, Wakeham A, et al. HLA-B-associated transcript 3 (Bat3)/Scythe is essential for p300-mediated acetylation of p53. Genes Dev. 2007;21:848–61. doi: 10.1101/gad.1534107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–77. [PubMed] [Google Scholar]
- 16.Ivanov I, Lo KC, Hawthorn L, et al. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene. 2007;26:2873–84. doi: 10.1038/sj.onc.1210098. [DOI] [PubMed] [Google Scholar]
- 17.Tsukahara T, Kimura S, Ichimiya S, et al. Scythe/BAT3 regulates apoptotic cell death induced by papillomavirus binding factor in human osteosarcoma. Cancer Sci. 2009;100:47–53. doi: 10.1111/j.1349-7006.2008.00991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]