Abstract
Background
The aim of this study was to investigate the feasibility of using serum miR-221 as a noninvasive prognostic biomarker for cutaneous malignant melanoma (CMM).
Material/Methods
We measured the expression levels of miR-221 in serum samples from 72 CMM patients and 54 healthy controls by real-time quantitative polymerase chain reaction (RT-PCR). The overall survival (OS) and disease-free survival (DFS) were calculated using the Kaplan-Meier method. The differences between the survival curves were tested by using the log-rank test. The COX proportional hazards regression model was used to determine the joint effects of several variables on survival.
Results
The serum miR-221 levels were significantly higher in patients with CMM than in healthy controls (p<0.0001). Patients with high serum miR-221 levels had a significantly lower 5-year OS rate (22.1% vs. 54.6%; P=0.018) and RFS rate (12.5% vs. 45.2%; P=0.008) than those with low serum miR-221 level. In a multivariate Cox model, we found that miR-221 expression was an independent predictor of poor 5-year OS (hazards ratio [HR]=3.189, 95% confidence interval [CI]=1.782–6.777, P=0.007) and 5-year DFS (HR=2.119, CI=1.962–8.552, P=0.01) in CMM patients.
Conclusions
Our data indicate that serum miR-221expression level has prognostic value in patients with CMM.
MeSH Keywords: Melanoma, MicroRNAs, Prognosis
Background
Cutaneous malignant melanoma (CMM) originates from melanocytes and is a highly aggressive disease, accounting for 80% of deaths arising from skin cancer worldwide [1]. CMM is highly characteristic of aggressive invasion, early metastasis, and resistance to chemotherapy or radiotherapy, which results in increased incidence and mortality worldwide [2,3]. Despite improvements in early CMM diagnosis, the 5-year survival rate for patients with advanced disease is still very low [4]. Therefore, better understanding of the molecular mechanisms of CMM tumorigenesis and progression will help to explore novel therapeutic agents and prognostic markers in the treatment of patients with CMM.
MiRNAs are small noncoding RNAs, usually 20–23 nucleotide long, which regulate the expression of protein-coding genes at the post-transcriptional level. Previous studies have also shown that aberrant miRNA expression is involved in the development and progression of cancer [5–10]; thus, miRNAs could be used as biomarkers for diagnosis and prognosis of cancer, and as targets for molecular cancer therapy [11–14]. miR-221 has been reported to be overexpressed in human tumor tissues such as breast cancer, colorectal cancer, and glioblastoma [15–19]. Previous in vitro and in vivo functional studies have confirmed the key role of miR-221 in regulating the progression of human melanoma [20,21]. Kanemaru et al. found that serum levels of miR-221 were significantly increased in patients with malignant melanoma and that they may be useful for the diagnosis of malignant melanoma [22]. However, they did not investigate the relationship between serum miR-221 expression level and the prognosis of malignant melanoma. The present study investigated the feasibility of using serum miR-221 as a noninvasive prognostic biomarker for CMM.
Material and Methods
Patients and serum samples
Serum samples were obtained from 72 patients with CMM from the Department of Cosmetic Surgery, the Third Xiangya Hospital, from May 2004 to June 2013. The histological diagnosis, Breslow thickness, and Clark level were re-examined in 1–5 original sections of the primary tumor by the same pathologist, who was unaware of the clinical data. Details of clinical and pathological characteristics of the patients are summarized in Table 1. Control serum samples were collected from 54 healthy volunteers. Blood samples were drawn from all patients and healthy control subjects at the beginning of treatment. A 5-mL sample of peripheral venous blood was drawn from all study participants after an overnight fast and placed at room temperature for 60 min. Then the blood samples were centrifuged at 1000 g for 10 min at 4°C to spin down the blood cells. All serum samples were stored at −80°C prior to use. Informed consent was obtained from all participants for the use of their blood samples. Informed consent was obtained from all participants for the use of their blood samples. This project was approved by the Clinical Research Ethics Committee of the Third Xiangya Hospital.
Table 1.
Patient characteristics and clinicopathologic correlation of serum miR-221 expression levels.
| Clinical variable | All cases | Serum miR-221 | P value | |
|---|---|---|---|---|
| High expression | Low expression | |||
| Age (years) | ||||
| ≥55 | 39 | 19 | 20 | 0.54 |
| <55 | 33 | 17 | 16 | |
| Sex | ||||
| Male | 35 | 17 | 18 | |
| Female | 37 | 19 | 18 | 0.71 |
| Thickness (mm) | ||||
| ≤2.0 | 27 | 3 | 24 | |
| >2.0 | 45 | 33 | 12 | 0.03 |
| Ulceration | ||||
| Absent | 32 | 13 | 19 | |
| Present | 40 | 23 | 17 | 0.08 |
| Histologic type | ||||
| SSM | 25 | 13 | 12 | |
| LMM | 29 | 14 | 15 | |
| Other | 18 | 9 | 9 | 0.42 |
| Site | ||||
| Sun exposed | 41 | 23 | 18 | 0.15 |
| Sun protected | 31 | 13 | 18 | |
| Differentiation | ||||
| Well | 34 | 18 | 26 | |
| Moderate/poor | 28 | 18 | 10 | 0.01 |
| T classification | ||||
| T1, T2 | 49 | 19 | 30 | |
| T3, T4 | 23 | 17 | 6 | 0.04 |
| N classification | ||||
| N0 | 47 | 19 | 28 | |
| N1–N3 | 25 | 17 | 8 | 0.009 |
| Metastasis | ||||
| M0 | 22 | 3 | 19 | |
| M1 | 50 | 33 | 17 | <0.001 |
| Stage | ||||
| I/II | 27 | 1 | 26 | |
| III/IV | 45 | 35 | 10 | <0.001 |
LMM – lentigo maligna melanoma; SSM – superficial spreading melanoma.
miRNA extraction and quantitative RT-PCR analysis
The isolation of miRNA from serum samples was performed using the miRNeasy™ RNA isolation kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions, with minor modifications [23]. Briefly, 200 mL of serum was supplemented with 10 mL of 0.05 μf 0.0 synthetic nonhuman miRNA (Caenorhabditis elegans miR-54, Takara Bio, Shiga, Japan) as controls, providing an internal reference for normalization of technical variations between samples. After 1 mL of QIAzol® Lysis Reagent (Qiagen) was added and well mixed (by gentle vortexing), the samples were incubated at room temperature for 5 min. Aqueous and organic phase separation was achieved by the addition of chloroform. The aqueous phase was done in an RNeasy™ spin column and RNeasy™ MinElute™ spin column (Takara Bio). The microRNA was eluted from the column with nuclease-free water.
To carry out quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) of serum miR-221 levels, cDNA was synthesized from miRNA with a MirX™ miRNA First Strand Synthesis and SYBR® green quantitative RT-PCR kit (Takara Bio) according to the manufacturer’s instructions. Quantitative RT-PCR was carried out using the Thermal Cycler Dice TP800 (Takara Bio) with primers and cDNA templates mixed with the SYBR® premix. DNA was amplified for 50 cycles of denaturation for 5 s at 95°C and annealing for 20 s at 60°C. Analysis of relative miRNA and mRNA expression was performed using the ΔΔCT method, with miR-16 as endogenous controls, following the manufacturer’s guidelines. Data analysis was performed using ABI Prism® 7300 SDS software (Applied Biosystems, Foster City, CA, USA).
Statistical analysis
Statistical analysis was performed using SPSS 17.0 (SPSS, IBM, Chicago, IL, USA). For continuous variables, the data were expressed as means ±SD. The Mann-Whitney U test was used to analyze the relationship between miR-221 expression and clinicopathological variables. The overall survival (OS) and disease-free survival (DFS) were calculated using the Kaplan-Meier method. The differences between the survival curves were tested by using the log-rank test. The COX proportional hazards regression model was used to determine the joint effects of several variables on survival. P values <0.05 were considered to be significant.
Results
Increased miR-221 expression level in CMM patients and its relationship with clinicopathological variables
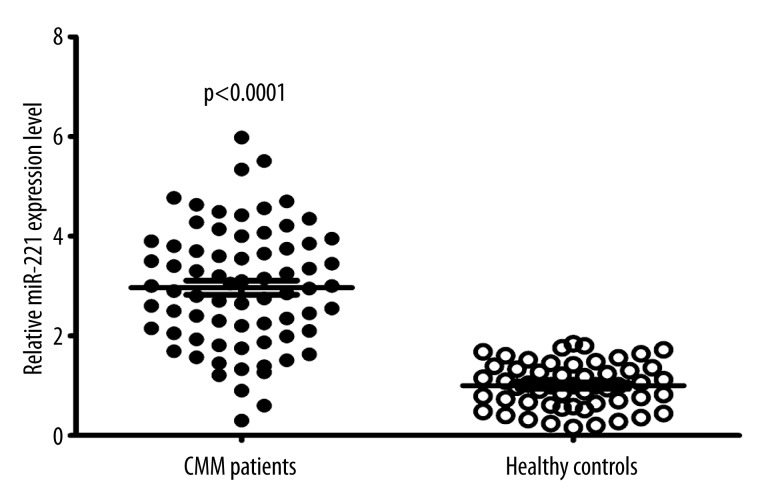
Expression levels of miR-221 were detectable in circulation in our study population. Using miR-16 to normalize samples, we observed that the serum miR-221 levels were significantly higher in patients with CMM than in healthy controls (p<0.0001, Figure 1). In this study, patients with values less than the median expression level of 2.95 were assigned to the Low Expression Group (n=36), whereas those with values ≥2.95 were assigned to the High Expression Group (n=36). As shown in Table 1, high miR-221 expression was correlated with tumor thickness (p=0.03), poor differentiation (p=0.01), higher T classification (p=0.04), higher N classification (p=0.009), metastasis (p<0.001), and advanced clinical stage (p<0.001). However, high miR-221 expression was not associated with other clinicopathological factors of patients, including age, sex, ulceration, histologic type, or tumor site (all P>0.05).
Figure 1.
Serum levels of miR-221 in healthy normal subjects (n=54) and CMM patients (n=72). Upper line, highest expression; intermediate line, mean expression; bottom line, lowest expression. Statistically significant differences were determined using the t-test.
Expression of miR-221 in serum samples in relation to prognosis of CMM patients
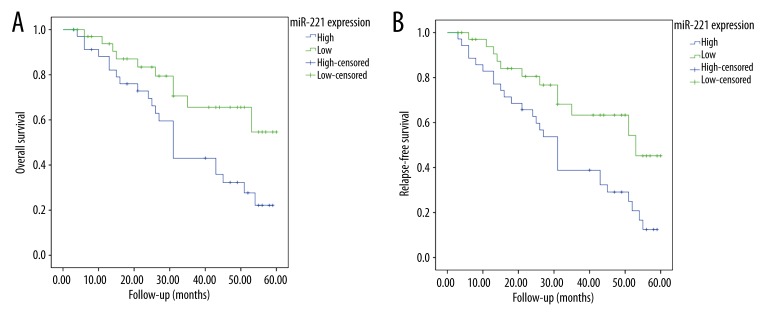
As shown in Figure 2, patients with high serum miR-221 level had a significantly lower 5-year OS rate (22.1% vs. 54.6%; P=0.018; Figure 2A) and RFS rate (12.5% vs. 45.2%; P=0.008; Figure 2B) than those with low serum miR-221 level. Table 2 shows multivariate analyses of clinical variables and serum miR-221 levels for their prognostic influence on OS and RFS. In a multivariate Cox model, we found that miR-221 expression was an independent poor prognostic factor for both 5-year OS (hazards ratio [HR]=3.189, 95% confidence interval [CI]=1.782–6.777, P=0.007) and 5-year DFS (HR=2.119, CI=1.962–8.552, P=0.01) in CMM.
Figure 2.
MiR-221 expression in serum in relation to the survival of CMM patients. High miR-221 expression correlates with poorer (A) 5-year overall survival (P=0.018) and (B) 5-year disease-free survival (P=0.008).
Table 2.
Correlation of miR-221 expression with clinical variables of patients with CMM.
| Features | Overall survival | Disease-free survival | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | 1.271 | 0.887–2.639 | 0.52 | 1.038 | 0.563–3.438 | 0.43 |
| Sex | 0.854 | 0.793–1.908 | 0.86 | 1.276 | 0.954–1.796 | 0.32 |
| Thickness | 2.189 | 0.976–6.095 | 0.06 | 3.097 | 2.178–5.887 | 0.03 |
| Ulceration | 1.965 | 0.657–4.992 | 0.12 | 2.886 | 0.875–7.354 | 0.09 |
| Histologic type | 0.782 | 0.378–7.891 | 0.44 | 0.835 | 0.278–4.009 | 0.31 |
| Site | 1.982 | 0.782–3.001 | 0.37 | 0.881 | 0.782–4.891 | 0.41 |
| Metastasis | 5.091 | 3.923–17.891 | 0.001 | 6.728 | 4.927–19.032 | <0.001 |
| Differentiation | 3.082 | 1.909–7.811 | 0.009 | 2.901 | 1.927–7.032 | 0.011 |
| Clinical stage | 5.872 | 4.072–18.112 | <0.001 | 5.891 | 3.991–19.903 | <0.001 |
| miR-221 expression | 3.189 | 1.782–6.777 | 0.007 | 2.119 | 1.962–8.552 | 0.01 |
HR – hazard ratio; 95% CI – 95% confidence interval.
Discussion
CMM is an aggressive and chemoresistant form of skin cancer characterized by malignant growth and poor patient prognosis [1]. Although many novel strategies have been used recently for the diagnosis and treatment of CMM, the prognosis is still very poor. Improvement in CMM outcomes largely depends on increasing our understanding of the molecular biology and regulatory mechanisms underlying the growth of CMM. Furthermore, there is still a need to improve early detection screening methods and to identify new prognostic biomarkers. Ideal biomarkers should be easy to measure and have a strong association with clinical outcome. miRNAs may match these proposed criteria and could be used as biomarkers for diagnosis and prognosis of cancer [24].
Among all the cancer-related miRNAs, miR-221, which is considered a micro-oncogene, was reported to be increasingly expressed in various carcinomas, including liver cancer, breast cancer, prostate cancer, colorectal carcinoma, melanoma, and acute myeloid leukemia [15–19]. Furthermore, high level of miR-221 expression is correlated with metastasis, tumor capsular infiltration, tumor stage, and poor prognosis. MiR-221 plays an important role in epithelial-to-mesenchymal transition (EMT). It has been identified as a basal-like subtype-specific miRNA that downregulates the expression of epithelial-specific genes and enhances the expression of mesenchymal-specific genes. Furthermore, miR-221 increases cell migration and invasion [25–27]. The basal-like transcription factor, FOSL1, can directly stimulate the transcription of miR-221 [26]. The abundance of miR-221 declines with the suppression of mitogen-activated or extracellular signal-regulated protein kinase (MEK) [26]. The miR-221-mediated reduction in E-cadherin is dependent on the targeting of the 3′-UTR of trichorhinophalangeal syndrome type 1 (TRPS1). TRPS1 inhibits EMT by directly repressing the expression of zinc finger E-box-binding homeobox 2 (ZEB2)[26]. Thus, miR-221 could contribute to the aggressive clinical behavior of various types of cancers. Kanemaru et al. found that serum levels of miR-221 were significantly increased in patients with malignant melanoma and that miR-221 levels may be useful for the diagnosis of malignant melanoma [22]. However, they did not investigate the relationship between serum miR-221 expression level and the prognosis of malignant melanoma. Our study aimed to investigate the feasibility of using serum miR-221 as a noninvasive prognostic biomarker for CMM.
In the present study, we showed that individuals with CMM had significantly elevated levels of tissue miR-221 expression, and that high level of miR-221 expression was significantly correlated with tumor thickness, poor differentiation, T classification, N classification, metastasis, and advanced clinical stage, suggesting that the overexpression of miR-221 might be able to play a critical role in the development of CMM. In addition, we also found that the 5-year OS and DFS rates of patients with high miR-221 expression were significantly lower than those of patients with low miR-221 expression, suggesting that miR-221 has potential for use as a molecular marker for predicting the prognosis of CMM patients. To further confirm this, multivariate regression analysis was performed by using the COX proportional hazard model. Our data indicate that miR-221 status might be an independent prognostic factor for CMM patients. To the best of our knowledge, we are the first to report that patients with higher serum miR-221 levels have a significantly lower survival rate than those with lower expression levels, revealing the usefulness of serum miR-221 expression as an independent prognosis factor in CMM.
Conclusions
In summary, our analysis shows that level of miR-221 is elevated in the circulation of CMM patients, and it is independently associated with poor survival. Based on these results, serum miR-221 may serve as a prognostic marker for CMM patients. Larger series are needed to confirm the prognostic value of miR-221 in CMM patients.
Footnotes
Conflicts of interest
None.
Source of support: Departmental sources
References
- 1.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 2.Terando A, Sabel MS, Sondak VK. Melanoma: adjuvant therapy and other treatment options. Curr Treat Options Oncol. 2003;4:187–99. doi: 10.1007/s11864-003-0020-0. [DOI] [PubMed] [Google Scholar]
- 3.Heyman BM, Chung MM, Lark AL, Shofer S. Endobronchial metastasis from primary anorectal melanoma. Am J Case Rep. 2013;14:253–57. doi: 10.12659/AJCR.889291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garbe C, Eigentler TK. Diagnosis and treatment of cutaneous melanoma: state of the art 2006. Melanoma Res. 2007;17:117–27. doi: 10.1097/CMR.0b013e328042bb36. [DOI] [PubMed] [Google Scholar]
- 5.Ma L, Weinberg RA. MicroRNAs in malignant progression. Cell Cycle. 2008;7:570–72. doi: 10.4161/cc.7.5.5547. [DOI] [PubMed] [Google Scholar]
- 6.Bullock MD, Sayan AE, Packham GK, Mirnezami AH. MicroRNAs: critical regulators of epithelial to mesenchymal (EMT) and mesenchymal to epithelial transition (MET) in cancer progression. Biol Cell. 2012;104:3–12. doi: 10.1111/boc.201100115. [DOI] [PubMed] [Google Scholar]
- 7.Fang YX, Gao WQ. Roles of microRNAs during prostatic tumorigenesis and tumor progression. Oncogene. 2014;33:135–47. doi: 10.1038/onc.2013.54. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Zhao J, Zhang PY, et al. MicroRNA-10b targets E-cadherin and modulates breast cancer metastasis. Med Sci Monit. 2012;18(8):BR299–308. doi: 10.12659/MSM.883262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang C, Lu YM, Meng LR. MicroRNA-125b down-regulation mediates endometrial cancer invasion by targeting ERBB2. Med Sci Monit. 2012;18(4):BR149–55. doi: 10.12659/MSM.882617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang Y, Fang D, Hu J. MicroRNA and its roles in esophageal cancer. Med Sci Monit. 2012;18(3):RA22–30. doi: 10.12659/MSM.882509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tousoulis D. Novel biomarkers in the prognosis, progression and treatment of cardiovascular disease: the role of microRNAs. Curr Top Med Chem. 2013;13:1491–92. doi: 10.2174/15680266113139990097. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson BA, Semus HM, Montgomery RL, et al. Plasma microRNAs serve as biomarkers of therapeutic efficacy and disease progression in hypertension-induced heart failure. Eur J Heart Fail. 2013;15:650–59. doi: 10.1093/eurjhf/hft018. [DOI] [PubMed] [Google Scholar]
- 13.Brase JC, Wuttig D, Kuner R, Sultmann H. Serum microRNAs as non-invasive biomarkers for cancer. Mol Cancer. 2010;9:306. doi: 10.1186/1476-4598-9-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Qi F, Cao Y, et al. Down-regulated microRNA-101 in bladder transitional cell carcinoma is associated with poor prognosis. Med Sci Monit. 2014;20:812–17. doi: 10.12659/MSM.890300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283:29897–903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galardi S, Mercatelli N, Giorda E, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–24. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 18.He H, Jazdzewski K, Li W, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–80. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Cui H, Wang W, et al. Construction of circular miRNA sponges targeting miR-21 or miR-221 and demonstration of their excellent anticancer effects on malignant melanoma cells. Int J Biochem Cell Biol. 2013;45:2643–50. doi: 10.1016/j.biocel.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Felicetti F, Errico MC, Bottero L, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer research. 2008;68:2745–54. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 22.Kanemaru H, Fukushima S, Yamashita J, et al. The circulating microRNA-221 level in patients with malignant melanoma as a new tumor marker. J Dermatol Sci. 2011;61:187–93. doi: 10.1016/j.jdermsci.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 23.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heneghan HM, Miller N, Lowery AJ, et al. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 25.Howe EN, Cochrane DR, Richer JK. The miR-200 and miR-221/222 microRNA families: opposing effects on epithelial identity. J Mammary Gland Biol Neoplasia. 2012;17:65–77. doi: 10.1007/s10911-012-9244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stinson S, Lackner MR, Adai AT, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4:ra41. doi: 10.1126/scisignal.2001538. [DOI] [PubMed] [Google Scholar]
- 27.Hwang MS, Yu N, Stinson SY, et al. miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One. 2013;8:e66502. doi: 10.1371/journal.pone.0066502. [DOI] [PMC free article] [PubMed] [Google Scholar]