Abstract
PURPOSE
Selection bias is a form of systematic error that can be severe in compromised study designs such as case-control studies with inappropriate selection mechanisms or follow-up studies that suffer from extensive attrition. External adjustment for selection bias is commonly undertaken when such bias is suspected, but the methods used can be overly simplistic, if not unrealistic, and fail to allow for simultaneous adjustment of associations of the exposure and covariates with the outcome, when of interest. Internal adjustment for selection bias via inverse-probability-weighting allows bias parameters to vary with levels of covariates but has only been formalized for longitudinal studies with covariate data on patients up until loss-to-follow-up.
METHODS
We demonstrate the use of inverse-probability-weighting and externally obtained bias parameters to perform internal adjustment of selection bias in studies lacking covariate data on unobserved participants.
RESULTS
The ‘true’ or selection-adjusted odds ratio for the association between exposure and outcome was successfully obtained by analyzing only data on those in the selected stratum (i.e. responders) weighted by the inverse probability of their being selected as function of their observed covariate data.
CONCLUSIONS
This internal adjustment technique using user-supplied bias parameters and inverse-probability-weighting for selection bias can be applied to any type of observational study.
Keywords: Epidemiologic methods, selection bias, casual inference
INTRODUCTION
Selection bias is a form of systematic error that can be severe in compromised study designs as in case-control studies with inappropriate selection of cases or control series (e.g., Berksonian bias or non-response bias) or in follow-up studies that suffer from extensive loss of contact with participants (e.g., loss to follow-up, follow-up bias). Adjusting for selection bias in a study requires knowledge of, or plausible assumptions about the factors that affect the selection mechanism. If the parameters of the selection mechanism are known or can be assumed reasonably, a selection factor can be used to adjust the biased measure of association, typically the sample odds ratio [1–5]. This method is formulaic, requiring external adjustment of crude and adjusted outcome models in a bias analysis [6]. In studies affected by follow-up bias (as opposed to response bias), inverse probability of censoring weighted (IPCW) fitting of the target model can be used to create a pseudo-population that mimics the underlying cohort (including those who were lost to follow up) [7–10]. This form of internal adjustment entails modeling censoring as a function of the last fully observed exposure and measured risk factor history that affect both censoring and the endpoint under study, which requires having said factors measured for both the censored and uncensored. This method generates record-level selection probabilities and their inverse can be used as a weighting factor incorporated into the analytical dataset before any outcome models are run. A distinct advantage of record-level estimation of the selection probabilities is that internal adjustment allows for the bias parameters to vary with individual covariate levels. Additionally, this approach allows end-users to conduct different analyses without and with adjustment for selection bias for different association or effect measures of interest using any statistical software and conventional regression modeling methods. In many epidemiologic studies, data on censored or non-selected participants are unknown, limiting IPCW methods to longitudinal studies that document data on everyone up until loss-to-follow-up.
In this paper, we formalize and demonstrate a method of internal adjustment for selection bias without the need for data on censored patients. This can be done using externally obtained bias parameters combined with data on respondents, or uncensored participants, to simulate or impute the corresponding selection probability for each respondent under the assumed selection and data generating mechanism, as would be depicted in a directed acyclic graph (DAG). Selection bias can then be adjusted using IPCW fitting of any planned outcome regression. Unlike IPCW, this technique is applicable to any observational study. This work is an extension of IPCW, because rather than reliance on data from a censored population, the relationships depicted in the causal diagram can be used to inform specification of selection bias parameters. Externally derived parameters (i.e., from a validation study) can also be used to generate selection probabilities. We formalize our method using probability and illustrate its use with a series of simulations.
NOTATION AND METHODS
Let X be a binary exposure, Y a binary disease outcome, Z be a set of confounding variables that are common causes of both X and Y, and S be a binary selection factor affected by both X, Y and at least one Z, such that exposure in the population can be represented by the probability of P(X=1 | Z=z), prevalence of disease among the unexposed can be represented by the probability P(Y=1 | X=0, Z=z), and those selected into the study population can be represented by the probability P(S=1 | X=x, Y=y, Z=z). Assuming no unmeasured confounding, the causal odds ratio can be represented by the conditional odds ratio, ORYX|Z.
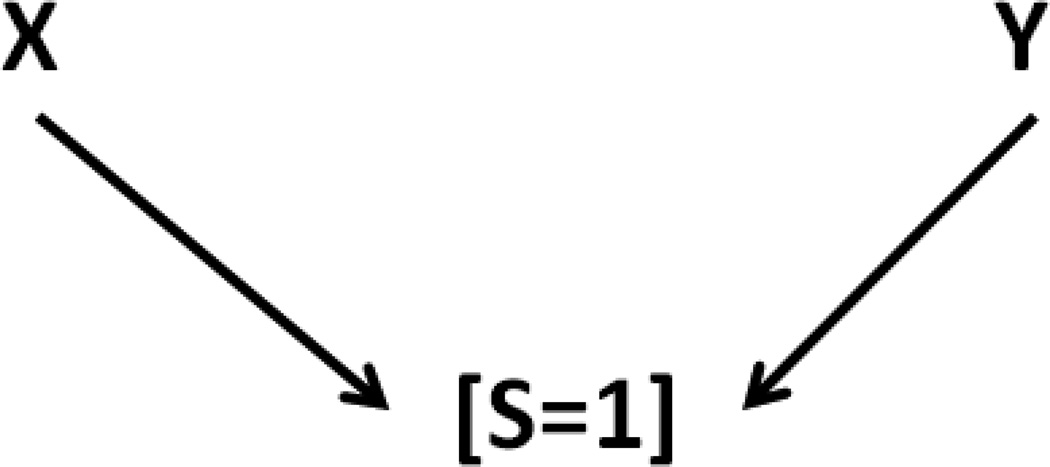
In the language of DAGs, selection bias is the result of collider bias, which occurs when the exposure (or cause of the exposure) and outcome (or cause of the outcome) both directly or indirectly affect selection into the study. The use of DAGs to express these causal relationships imparts a basic set of rules that have been extensively described elsewhere [11–16]. The minimal structure for collider bias is depicted in Figure 1.
Figure 1.
A DAG representing marginally independent but conditionally (on S=1) dependent X and Y; a simple example of collider-stratification bias.
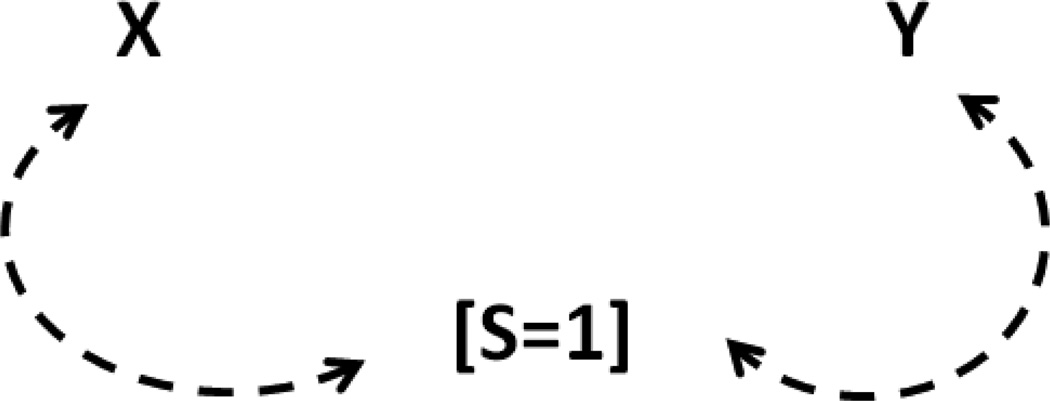
This figure shows that the marginally independent exposure X and outcome Y can become conditionally dependent given selection S=1. Figure 2 shows another example.
Figure 2.
A DAG representing marginally independent but conditionally (on S=1) dependent X and Y, another example of collider-stratification bias in the presence of uncontrolled common causes of X-S and Y-S
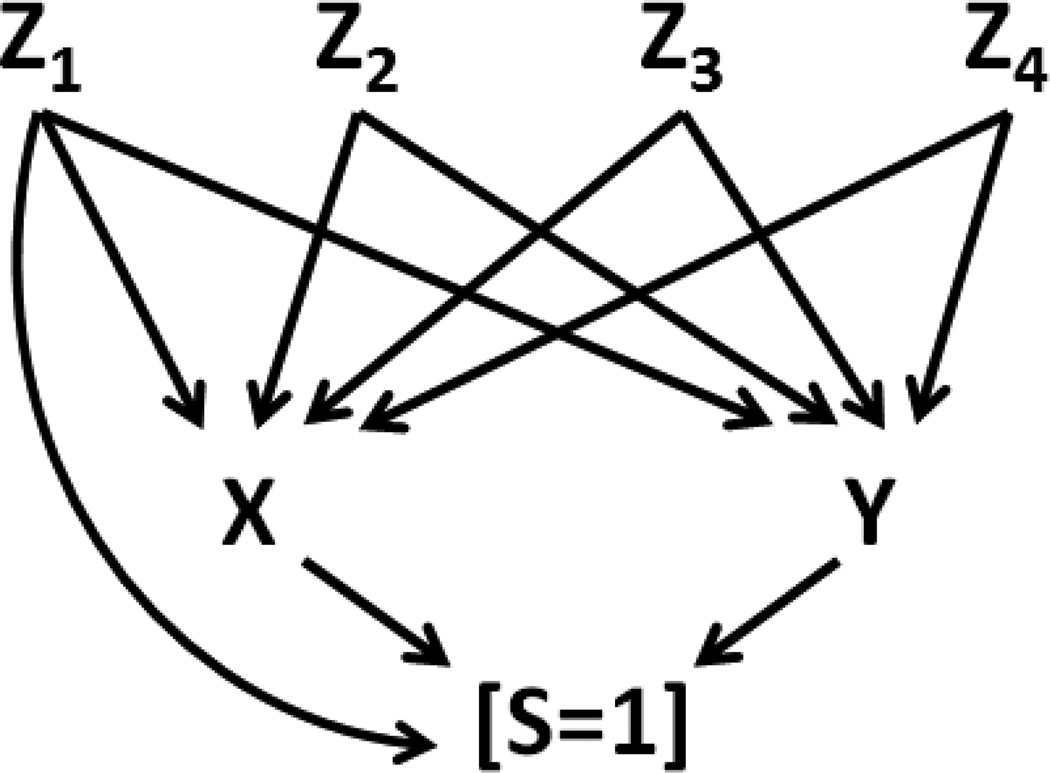
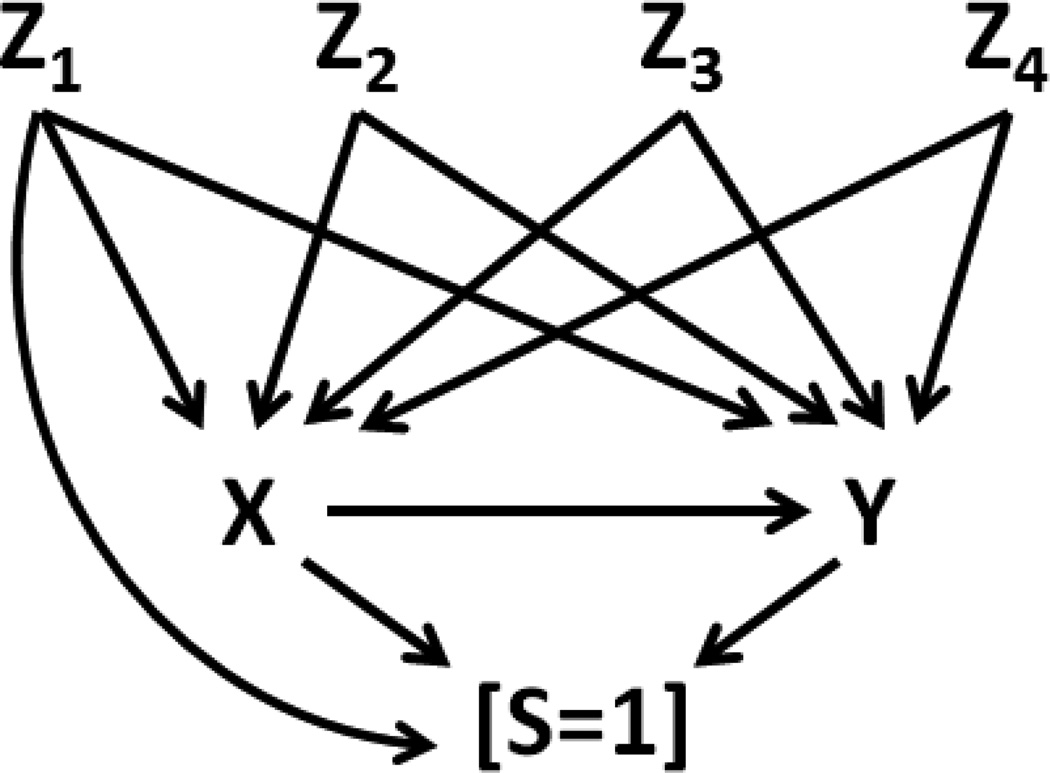
Figures 3 and 4 show scenarios 1 and 2, respectively, where selection is cause by exposure X, confounding variable set Z = [Z1, Z2, Z3, and Z4] and outcome Y. In either scenario, the joint probability of S=1, y, x, and z is given by:
| (1) |
Figure 3.
Scenario 1 – A DAG representing marginally independent but conditionally (on S=1) dependent X and Y, with four confounding variables Z1, Z2, Z3, and Z4, one of which directly affects S.
Figure 4.
Scenario 2 – A DAG representing marginally dependent X and Y with additional conditional (on S=1) dependency and four confounding variables Z1, Z2, Z3, and Z4, one of which directly affects S.
The term P(S=1|y,x,z) is the probability of selection given the observed data on Y, X and Z. To obtain the selection-bias-free joint probability P(y, x, z) or P(S=1)P(y, x, z), we re-weight the observed P(S=1, y, x, z) by the inverse of P(S=1 | y, x, z) or P(S=1 | y, x, z)/P(S=1). This entails weighting all records in the S=1 sample by either 1/P(S=1 | y, x, z) or P(S=1)/P(S=1 | y, x, z) (the latter being the stabilized version of the inverse probability weight) in a procedure known as inverse-probability-weighting. We will call this procedure inverse-probability-of-selection-weighting (IPSW), a generalization of IPCW.
The conditional probability of selection P(S=1 | y, x, z) is unknown, but it can be modeled using a logistic equation with bias parameter set β as follows:
| (2) |
Where:
βS is the log odds of selection S=1 when Y=0, X=0 and Z=0 (indicating a degree of selection that is independent of Y, X, and Z);
βSY is the log odds ratio (OR) relating selection S and Y when X=Z=0;
βSX is the log odds ratio relating S and X when Y=Z=0;
βSZ is the log odds ratio relating S and Z when Y=X=0;
βSYX is the logarithm of the ratio of (i) the odds ratio relating S and Y among X=1 and Z=0 to (ii) the odds ratio relating S and Y among X=0 and Z=0 (that is, log(ORSY|X=1,Z=0/ORSY|X=0,Z=0) = log(ORSX|Y=1,Z=0/ORSX|Y=0,Z=0), by the symmetry of the odds ratio);
βSYZ is the logarithm of the ratio of (i) the odds ratio relating S and Y when Z=1 and X=0 to (ii) the odds ratio relating S and Y when Z=0 and X=0 (that is, log(ORSY|Z=1,X=0/ORSY|Z=0,X=0) = log(ORSZ|Y=1,X=0/ORSZ|Y=0,X=0));
βSYZ is the logarithm of the ratio of (i) the odds ratio relating S and X when Z=1 and Y=0 to (ii) the odds ratio relating S and X when Z=0 and Y=0 (that is, log(ORSX|Z=1,Y=0/ORSX|Z=0,Y=0) = log(ORSZ|X=1,Y=0/ORSZ|X=0,Y=0)); and
βSYXZ is the logarithm of the ratio of two ratios, namely the ratio of (i) the ratio of the odds ratio relating S and Y when X=1 and Z=1 and the odds ratio relating S and Y when X=0 and Z=1 to (ii) the ratio of the odds ratio relating S and Y when X=1 and Z=0 and the odds ratio relating S and Y when X=0 and Z=0 (that is, log[(ORSY|X=1,Z=1/ORSY|X=0,Z=1)/(ORSY|X=1,Z=0/ORSY|X=0,Z=0)]). This βSYXZ is alternatively given by log[(ORSX|Y=1,Z=1/ORSX|Y=0,Z=1)/(ORSX|Y=1,Z=0/ORSX|Y=0,Z=0)] = log[(ORSZ|Y=1,X=1/ORSZ|Y=0,X=1)/(ORSZ|Y=1,X=0/ORSZ|Y=0,X=0)].
The expit transform, expit(logit(P(S=1|y,x, z))), yields the selection probability P(S=1|y,x,z) for each actually selected (S=1) record in the dataset conditional on their Y, X and Z values and given the externally obtained β above. An important advantage of using the logistic model to estimate the selection probability is that it will be bounded by 0 and 1, as a probability should be. In some scenarios, the product term parameters might be presumed to be null, but if a selection mechanism involves product terms this might result in insufficient bias adjustment.
Bias parameters should be defined using knowledge of the selection process, or the underlying source population. In most cases, however, these parameters will not be known, and the selection bias adjustment should use a range of plausible bias parameters to conduct robust bias analysis. We reiterate that the key difference between this technique (IPSW) and the now-conventional IPCW used in longitudinal data with censoring is that the betas, or bias parameters, are externally estimated (either using validation data, similar studies, etc.) and supplied to the dataset in our technique while they are estimated from observed data in IPCW. In most epidemiologic studies, data are rarely collected on non-respondents; hence, the specification of a bias model from a range of assumed parameters using our technique or something similar is often the only option.
ILLUSTRATION 1: PROOF OF CONCEPT SIMULATION USING “CORRECT” BIAS PARAMETERS
Illustration 1 provides a proof of principle using a valid, empirically derived set of bias parameters from a hypothetical cohort in which both strata S=1 and S=0 were simulated. Using the equation in expression (2), and IPSW techniques, we demonstrate the ability to recovery of the true ORYX in an analysis involving only the S=1 stratum. To do this, we simulated a large cohort (N=100,000) with one dichotomous exposure variable (X), two dichotomous confounders (Z1 and Z2), one continuous confounder (Z3), one trichotomous confounder (Z4), and a dichotomous outcome (Y). The data generating mechanism was based on the relationships between these variables as depicted in the causal structures in Figures 3 and 4. In scenario 1 (Figure 3), after control for the sufficient set of Z confounders, Y is marginally independent of X; in scenario 2 (Figure 4), X causes Y.
Z1 and Z2 were generated by random draws from independent Bernoulli distributions with success probability of P(Z1=1) = 0.3 and P(Z2=1) = 0.3. Z3 was generated from the normal distribution such that Z3 ~N (0, 1). Z4 was generated from two conditional Bernoulli distributions such that the resulting two indicator variables combined make an exclusive categorization with mean population distributions P(Z4=1) = 0.4, P(Z4=2) = 0.3 and P(Z4=0) = 0.3. The probability of exposure was generated as a function of variables Z1…Z4, and the exposure variable was generated from random draws from a corresponding Bernoulli distribution.
The disease variable was generated from random draws from a Bernoulli distribution as a function of the background risk of disease (P(Y=1 | X=0, Z1=0, Z2=0, Z3=0, Z4=0) = 0.3), the exposure status, and Z1…Z4.
Finally S was generated by drawing from a Bernoulli distribution as a function of X, Y, and Z1 with varying levels of P(S=1 | Y=0, X=0, Z1=0, Z2=0, Z3=0, Z4=0).
Next, we ran logistic regression of Y on X, Z1, Z2, Z3, and Z4 for the entire cohort to estimate the “true” OR relating Y and X conditional on Z1, Z2, Z3, and Z4 (ORYX|z). We then fit a binary logistic model for S=1 as a function of the other DAG variables in the full cohort, including all 2-way, 3-way, 4-way and 5-way product terms according to expression (2). We then restricted the cohort to those subjects where S=1 and ran a logistic regression of Y on X, Z1, Z2, Z3, Z4 to estimate the biased OR relating Y and X conditional on Z among the S=1 records, ORYX|z,S=1. Finally, we generated each selected records’ P(S=1 | y, x, z) using the bias parameters β estimated from the full data as described above.
We then ran logistic regression of Y on X, Z1, Z2, Z3, and Z4 using data on the S=1 records, with 1/P(S=1 | y, x, z) as the regression weight to estimate the “adjusted” ORYX|z,S-adj. We repeated this illustration for different hypothetical selection bias scenarios. Trials A1-A8 correspond to Figure 3, trials B1-B8 correspond to Figure 4 with no modification of the S-Y relationship by X, and trials C1-C4 correspond to Figure 4 with an added parameter for the modification by X on the S-Y relationship in the data generation process. We evaluated model performance by calculating bias and RMSE comparing “true” ORYX|z and “adjusted” ORYX|z,S-adj.
ILLUSTRATION 2: PERFORMANCE OF A REDUCED ALGORITHM
Illustration 2 assesses the performance of the algorithm applied in illustration 1 under less flexible equations not accounting for any 2-way, 3-way-, 4-way, or 5-way interaction coefficients other than βSYX in the bias parameter set (β). To do this, we repeated the DAG-directed simulation of our selection weights for the hypothetical population described in illustration 1, excluding all interaction terms in our modeling of P(S=1 | y, x, z) from the full cohort, using the following modified version of equation (2):
| (3) |
This resulted in a reduced bias parameter set βr which was used in the IPSW process to weight the outcome model in the S=1 stratum. As in illustration 2, we ran logistic regression of Y on X, Z1, Z2, Z3, and Z4 using data on the S=1 records, with 1/P(S=1 | y, x, z) as the regression weight to estimate the “adjusted” ORYX|z,S-adj. We repeated this illustration for the same selection bias scenarios as illustration 1, varying the effect of X and Y on selection.
Trials A1-A8 correspond to Figure 3, trials B1-B8 correspond to Figure 4 with no modification by X on the S-Y relationship, and trials C1-C4 correspond to Figure 4 with an added parameter for the modification by X on the S-Y relationship in the data generation process. We evaluated the reduced model algorithm performance by calculating bias and RMSE comparing “true” ORYX|z and “adjusted” ORYX|z,S-adj.
ILLUSTRATION 3: MISSPECIFIED PARAMETERS
Illustration 3 demonstrates the performance of the algorithm using external bias parameters that are an imperfect measure of the true bias. We repeated the DAG-directed simulation of our probability of selection weights for a hypothetical population (N=100,000) corresponding to the DAG in Figure 4, with ORYX|Z = 2. This time we applied bias parameters with slight misspecification (−20% to +20%) of the empirical bias parameters. For illustration, true prevalences in the hypothetical population were held constant as follows: P(S=1 | Y=0, X=0, Z=0) = 0.2, P(X=1 | Z=0) = 0.3 and P(Y=1 | X=0, Z=0) = 0.5. The trials were performed twice, once with a strong level of selection bias: eβSX = 5.0, eβSY = 5.0, eβSZ1 = 5.0 and eβSY X = 5.0 (trials D1-D21), and once with a weak to moderate level of selection bias: eβSX = 2.0, eβSY = 2.0, eβSZ1 = 2.0, and eβSY X = 0.8 (trials E1-E21). In both sets of trials, these parameters were “misspecified” by multiplying or dividing by 0.1 and 0.2 to represent the bias adjustment under incorrect externally applied bias parameters. As in illustrations 1 and 2, we evaluated model performance by calculating bias and RMSE comparing “true” ORYX|z and “adjusted” ORYX|z,S-adj.
RESULTS
Tables 1 and 2 include results from simulated populations based on the DAGs pictured in Figures 3 and 4 and used IPW to correct for the selection bias effect that was the result of conditioning on the collider at the S node. All bias parameters were empirically derived from the underlying hypothetical population. Generally, we observed a downward bias in any model that included a positive relationship between exposure and selection and disease and selection. If for the relationship of interest at least one of these direct effects were negative, the bias was upward. As has been demonstrated in the literature [8, 9], bias adjustment using IPSW was adequate in all models. Variation in the population characteristics P(S=1 | Y=0, X=0, Z=0), P(X=1 | Z=0), and P(Y=1 | X=0, Z=0) did not result in any discernible pattern of bias adjustment accuracy. Increasing the eβSX and eβSY resulted in slightly reduced accuracy of the bias adjustment. Addition of the interaction parameter, eβSY X, also slightly degraded bias adjustment performance.
Table 1.
Correctly specified parameters for adjustment of collider-stratification bias in a cohort (N=100,000) defined by the DAGs in Figures 31 and 41
| Trial | P(S=1|Y=0,X=0,Z=0) | eβSX | eβSY | eβSYX | True ORYX|z |
Biased ORYX|z,S=1 |
Bias adjusted ORYX|z,S-adj |
Bias | RMSE |
|---|---|---|---|---|---|---|---|---|---|
| A1 | 0.10 | 5.0 | 5.0 | 1.0 | 1.01 (0.97, 1.04) | 0.59 (0.55, 0.63) | 1.01 (0.97, 1.04) | −0.0023 | 0.0176 |
| A2 | 0.20 | 5.0 | 5.0 | 1.0 | 1.01 (0.97, 1.04) | 0.62 (0.59, 0.65) | 1.00 (0.97, 1.03) | −0.0099 | 0.0200 |
| A3 | 0.50 | 5.0 | 5.0 | 1.0 | 1.01 (0.97, 1.04) | 0.75 (0.72, 0.78) | 1.00 (0.97, 1.04) | −0.0036 | 0.0178 |
| A4 | 0.70 | 5.0 | 5.0 | 1.0 | 1.01 (0.97, 1.04) | 0.83 (0.80, 0.87) | 1.01 (0.97, 1.04) | −0.0013 | 0.0174 |
| A5 | 0.10 | 0.7 | 0.7 | 1.0 | 1.01 (0.97, 1.04) | 1.00 (0.88, 1.14) | 1.03 (0.99, 1.06) | 0.0192 | 0.0260 |
| A6 | 0.10 | 10.0 | 0.5 | 1.0 | 1.00 (0.96, 1.03) | 1.25 (1.12, 1.39) | 0.99 (0.95, 1.02) | −0.0073 | 0.0196 |
| A7 | 0.10 | 10.0 | 5.0 | 1.0 | 1.00 (0.96, 1.03) | 0.44 (0.41, 0.48) | 0.99 (0.95, 1.02) | −0.0073 | 0.0195 |
| A8 | 0.10 | 10.0 | 10.0 | 1.0 | 1.00 (0.96, 1.03) | 0.33 (0.30, 0.35) | 0.98 (0.95, 1.02) | −0.0133 | 0.0224 |
| B1 | 0.10 | 5.0 | 5.0 | 1.0 | 1.97 (1.90, 2.05) | 1.16 (1.09, 1.24) | 1.95 (1.88, 2.03) | −0.0200 | 0.0275 |
| B2 | 0.20 | 5.0 | 5.0 | 1.0 | 1.97 (1.90, 2.05) | 1.22 (1.16, 1.29) | 1.95 (1.87, 2.02) | −0.0279 | 0.0336 |
| B3 | 0.50 | 5.0 | 5.0 | 1.0 | 1.97 (1.90, 2.05) | 1.47 (1.41, 1.53) | 1.97 (1.90, 2.04) | −0.0063 | 0.0198 |
| B4 | 0.70 | 5.0 | 5.0 | 1.0 | 1.97 (1.90, 2.05) | 1.63 (1.57, 1.70) | 1.97 (1.90, 2.04) | −0.0022 | 0.0189 |
| B5 | 0.10 | 0.7 | 0.7 | 1.0 | 1.97 (1.90, 2.05) | 1.99 (1.73, 2.29) | 2.02 (1.95, 2.10) | 0.0465 | 0.0502 |
| B6 | 0.10 | 10.0 | 0.5 | 1.0 | 1.97 (1.90, 2.04) | 2.54 (2.32, 2.78) | 1.99 (1.92, 2.07) | 0.0261 | 0.0322 |
| B7 | 0.10 | 10.0 | 5.0 | 1.0 | 1.97 (1.90, 2.04) | 0.92 (0.85, 0.98) | 1.93 (1.86, 2.01) | −0.0330 | 0.0381 |
| B8 | 0.10 | 10.0 | 10.0 | 1.0 | 1.97 (1.90, 2.04) | 0.82 (0.77, 0.87) | 1.90 (1.83, 1.97) | −0.0668 | 0.0694 |
| C1 | 0.20 | 5.0 | 5.0 | 0.4 | 1.97 (1.90, 2.05) | 1.05 (1.00, 1.11) | 1.94 (1.87, 2.01) | −0.0368 | 0.0413 |
| C2 | 0.20 | 5.0 | 5.0 | 0.8 | 1.97 (1.90, 2.05) | 1.19 (1.13, 1.25) | 1.94 (1.87, 2.02) | −0.0286 | 0.0342 |
| C3 | 0.20 | 5.0 | 5.0 | 2.0 | 1.97 (1.90, 2.05) | 1.29 (1.22, 1.36) | 1.94 (1.87, 2.01) | −0.0313 | 0.0365 |
| C4 | 0.20 | 5.0 | 5.0 | 5.0 | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.95 (1.88, 2.02) | −0.0218 | 0.0288 |
Simulated prevalences in the hypothetical population were held constant as follows: P(X=1 | Z=0) = 0.3 and P(Y=1 | X=0, Z=0) = 0.5.
Table 2.
Reduced models with correctly specified parameters for adjustment of collider-stratification bias in a cohort (N=100,000) defined by the DAGs in Figures 31 and 41
| Trial | P(S=1|Y=0,X=0,Z=0) | eβSX | eβSY | eβSYX | True ORYX|z |
Biased ORYX|z,S=1 |
Bias adjusted ORYX|z,S-adj |
Bias | RMSE |
|---|---|---|---|---|---|---|---|---|---|
| A1 | 0.10 | 5.0 | 5.0 | 1.0 | 1.01 (0.97, 1.04) | 0.59 (0.55, 0.63) | 1.01 (0.98, 1.05) | 0.0055 | 0.0182 |
| A2 | 0.20 | 5.0 | 5.0 | 1.0 | 1.01 (0.97, 1.04) | 0.62 (0.59, 0.65) | 1.01 (0.98, 1.05) | 0.0019 | 0.0175 |
| A3 | 0.50 | 5.0 | 5.0 | 1.0 | 1.01 (0.97, 1.04) | 0.75 (0.72, 0.78) | 1.01 (0.97, 1.04) | −0.0001 | 0.0174 |
| A4 | 0.70 | 5.0 | 5.0 | 1.0 | 1.01 (0.97, 1.04) | 0.83 (0.80, 0.87) | 1.01 (0.97, 1.04) | −0.0008 | 0.0174 |
| A5 | 0.10 | 0.7 | 0.7 | 1.0 | 1.01 (0.97, 1.04) | 1.00 (0.88, 1.14) | 1.02 (0.98, 1.05) | 0.0067 | 0.0187 |
| A6 | 0.10 | 10.0 | 0.5 | 1.0 | 1.00 (0.96, 1.03) | 1.25 (1.12, 1.39) | 0.99 (0.96, 1.03) | −0.0027 | 0.0183 |
| A7 | 0.10 | 10.0 | 5.0 | 1.0 | 1.00 (0.96, 1.03) | 0.44 (0.41, 0.48) | 0.99 (0.96, 1.03) | −0.0012 | 0.0181 |
| A8 | 0.10 | 10.0 | 10.0 | 1.0 | 1.00 (0.96, 1.03) | 0.33 (0.30, 0.35) | 0.99 (0.96, 1.03) | −0.0018 | 0.0182 |
| B1 | 0.10 | 5.0 | 5.0 | 1.0 | 1.97 (1.90, 2.05) | 1.16 (1.09, 1.24) | 1.98 (1.91, 2.05) | 0.0077 | 0.0203 |
| B2 | 0.20 | 5.0 | 5.0 | 1.0 | 1.97 (1.90, 2.05) | 1.22 (1.16, 1.29) | 1.98 (1.90, 2.05) | 0.0030 | 0.0190 |
| B3 | 0.50 | 5.0 | 5.0 | 1.0 | 1.97 (1.90, 2.05) | 1.47 (1.41, 1.53) | 1.97 (1.90, 2.05) | 0.0000 | 0.0188 |
| B4 | 0.70 | 5.0 | 5.0 | 1.0 | 1.97 (1.90, 2.05) | 1.63 (1.57, 1.70) | 1.97 (1.90, 2.05) | −0.0014 | 0.0188 |
| B5 | 0.10 | 0.7 | 0.7 | 1.0 | 1.97 (1.90, 2.05) | 1.99 (1.73, 2.29) | 2.01 (1.94, 2.09) | 0.0365 | 0.0410 |
| B6 | 0.10 | 10.0 | 0.5 | 1.0 | 1.97 (1.90, 2.04) | 2.54 (2.32, 2.78) | 1.97 (1.90, 2.05) | 0.0062 | 0.0199 |
| B7 | 0.10 | 10.0 | 5.0 | 1.0 | 1.97 (1.90, 2.04) | 0.92 (0.85, 0.98) | 1.97 (1.90, 2.04) | 0.0002 | 0.0189 |
| B8 | 0.10 | 10.0 | 10.0 | 1.0 | 1.97 (1.90, 2.04) | 0.82 (0.77, 0.87) | 1.97 (1.90, 2.04) | 0.0027 | 0.0191 |
| C1 | 0.20 | 5.0 | 5.0 | 0.4 | 1.97 (1.90, 2.05) | 1.05 (1.00, 1.11) | 1.98 (1.90, 2.05) | 0.0023 | 0.0189 |
| C2 | 0.20 | 5.0 | 5.0 | 0.8 | 1.97 (1.90, 2.05) | 1.19 (1.13, 1.25) | 1.98 (1.90, 2.05) | 0.0033 | 0.0191 |
| C3 | 0.20 | 5.0 | 5.0 | 2.0 | 1.97 (1.90, 2.05) | 1.29 (1.22, 1.36) | 1.98 (1.90, 2.05) | 0.0029 | 0.0190 |
| C4 | 0.20 | 5.0 | 5.0 | 5.0 | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.98 (1.90, 2.05) | 0.0031 | 0.0190 |
Simulated prevalences in the hypothetical population were held constant as follows: P(X=1 | Z=0) = 0.3 and P(Y=1 | X=0, Z=0) = 0.5.
In Table 3 we carried forward the simulation from DAG 4, this time including a varying degree of misspecification of the bias parameters (−20% to +20%). We did this twice, once for a strong selection bias (trials D1-D21) and once for a moderate to weak selection bias (trials E1-E21). In both scenarios, misspecification of the βS or the eβSYX parameters did not greatly inhibit bias adjustment. Misspecification of the eβSX and eβSY resulted in inadequate bias adjustment in the presence of strong selection bias (trials D1-D21).
Table 3.
Misspecified parameters for adjustment of collider-stratification bias in a cohort (N=100,000) defined by the DAG in Figure 41
| Trial | Misspecified parameter2,3 |
Degree of misspecification |
True ORYX|z |
Biased ORYX|z,S=1 |
Bias adjusted ORYX|z,S-adj |
Bias | RMSE |
|---|---|---|---|---|---|---|---|
| D1 | None | None | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.98 (1.90, 2.05) | 0.0031 | 0.0190 |
| D2 | βS | −20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.97 (1.89, 2.05) | −0.0042 | 0.0203 |
| D3 | βS | −10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.98 (1.90, 2.05) | 0.0036 | 0.0197 |
| D4 | βS | +10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.97 (1.90, 2.04) | −0.0067 | 0.0194 |
| D5 | βS | +20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.95 (1.88, 2.02) | −0.0263 | 0.0317 |
| D6 | eβSX | −20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.75 (1.69, 1.82) | −0.2186 | 0.2194 |
| D7 | eβSX | −10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.87 (1.80, 1.94) | −0.1065 | 0.1081 |
| D8 | eβSX | +10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 2.08 (2.01, 2.16) | 0.1089 | 0.1106 |
| D9 | eβSX | +20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 2.18 (2.10, 2.27) | 0.2101 | 0.2110 |
| D10 | eβSY | −20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.75 (1.69, 1.82) | −0.2230 | 0.2238 |
| D11 | eβSY | −10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.86 (1.80, 1.93) | −0.1088 | 0.1104 |
| D12 | eβSY | +10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 2.08 (2.01, 2.16) | 0.1115 | 0.1131 |
| D13 | eβSY | +20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 2.19 (2.11, 2.27) | 0.2155 | 0.2163 |
| D14 | eβSZ1 | −20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 2.00 (1.93, 2.07) | 0.0250 | 0.0312 |
| D15 | eβSZ1 | −10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.99 (1.92, 2.06) | 0.0137 | 0.0232 |
| D16 | eβSZ1 | +10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.97 (1.89, 2.04) | −0.0067 | 0.0200 |
| D17 | eβSZ1 | +20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.96 (1.89, 2.03) | −0.0156 | 0.0245 |
| D18 | eβSYX | −20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 2.00 (1.93, 2.07) | 0.0269 | 0.0328 |
| D19 | eβSYX | −10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.99 (1.91, 2.06) | 0.0138 | 0.0233 |
| D20 | eβSYX | +10% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.97 (1.90, 2.04) | −0.0057 | 0.0197 |
| D21 | eβSYX | +20% | 1.97 (1.90, 2.05) | 1.34 (1.27, 1.41) | 1.96 (1.89, 2.03) | −0.0130 | 0.0228 |
| E1 | None | None | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.98 (1.91, 2.05) | 0.0049 | 0.0194 |
| E2 | βS | −20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.96 (1.89, 2.04) | −0.0104 | 0.0230 |
| E3 | βS | −10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.97 (1.90, 2.05) | −0.0021 | 0.0197 |
| E4 | βS | +10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.98 (1.91, 2.05) | 0.0105 | 0.0208 |
| E5 | βS | +20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.99 (1.92, 2.06) | 0.0149 | 0.0227 |
| E6 | eβSX | −20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.95 (1.88, 2.02) | −0.0269 | 0.0325 |
| E7 | eβSX | −10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.96 (1.89, 2.03) | −0.0112 | 0.0217 |
| E8 | eβSX | +10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.99 (1.92, 2.07) | 0.0212 | 0.0285 |
| E9 | eβSX | +20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 2.01 (1.94, 2.09) | 0.0378 | 0.0424 |
| E10 | eβSY | −20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.95 (1.88, 2.02) | −0.0252 | 0.0313 |
| E11 | eβSY | −10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.96 (1.89, 2.04) | −0.0103 | 0.0214 |
| E12 | eβSY | +10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.99 (1.92, 2.07) | 0.0203 | 0.0277 |
| E13 | eβSY | +20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 2.01 (1.94, 2.08) | 0.0360 | 0.0407 |
| E14 | eβSZ1 | −20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.98 (1.91, 2.06) | 0.0089 | 0.0207 |
| E15 | eβSZ1 | −10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.98 (1.91, 2.05) | 0.0068 | 0.0199 |
| E16 | eβSZ1 | +10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.98 (1.90, 2.05) | 0.0029 | 0.0191 |
| E17 | eβSZ1 | +20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.97 (1.90, 2.05) | 0.0011 | 0.0189 |
| E18 | eβSZ1 | −20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.92 (1.85, 1.99) | −0.0522 | 0.0555 |
| E19 | eβSYX | −10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 1.95 (1.88, 2.02) | −0.0241 | 0.0306 |
| E20 | eβSYX | +10% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 2.01 (1.94, 2.08) | 0.0346 | 0.0394 |
| E21 | eβSYX | +20% | 1.97 (1.90, 2.05) | 1.62 (1.52, 1.73) | 2.04 (1.96, 2.11) | 0.0652 | 0.0679 |
Simulated prevalences in the hypothetical population were held constant as follows: P(S=1|Y=0,X=0,Z=0)= 0.2, P(X=1|Z=0) = 0.3 and P(Y=1|X=0, Z=0) = 0.5. The true ORYX|Z was simulated as 2.
For trials D1-D21, each hypothetical population, the true bias parameters were specified as follows: βS = 0.2, eβSX = 5.0, eβSY = 5.0, eβSZ1 = 5.0, and eβSYX = 5.0.
For trials E1-E21 each hypothetical population, the true bias parameters were specified as follows: βS = 0.2, eβSX = 2.0, eβSY = 2.0, eβSZ1 = 2.0, and eβSYX = 0.8.
DISCUSSION
We have demonstrated a method of sensitivity analysis for selection bias adjustment using record level data augmentation, which is based on the recoverability of the joint distribution given data on the S=1 stratum and prior knowledge or beliefs about the S=0 stratum [17], and can be implemented in absence of data on the S=0 stratum. In our simulations, we used imputed probabilities with IPSW and were able to produce unbiased estimates of the causal odds ratio using only the selected stratum. This method is distinct from IPCW because it need not be based on data from censored individuals in the underlying cohort, and thus may be applicable to case-control studies. As has been done previously, we used DAGs to visualize the selection bias mechanisms and considered selection (or collider) bias to be a form of nonignorable missing data [15, 18, 19].
We found via our simulation scenarios that this method provided adequate adjustment of selection bias under empirically derived priors, but the framework of the method can be adapted to the use of external bias parameters. We found that performance was optimal using fully saturated models, but the reduced model forms performed comparably well, and with much simpler computational execution. Application of this method under misspecification demonstrated that (as would be expected intuitively) reweighting the population according to invalid bias parameters produces invalid results.
Although this method performs adequately in our simulation scenarios, it is highly dependent on plausible characterization of the magnitude and direction of the bias, most of which we derived empirically from the underlying source population. If input data are not available from empirical sources, arriving at a set of bias parameters that plausibly characterize a completely unknown population of individuals (i.e., the S=0 stratum) may be a difficult undertaking. To this end, we suggest (as others have) to always present selection bias adjustments as part of a detailed bias analysis [20]. Additionally, using this method under extreme levels of selection bias, upon even slight misspecification of these parameters the bias adjustment would degrade considerably. Although we did not present examples of it, as can be expected, gross misspecification, or misspecification of multiple parameters could result in entirely invalid adjusted estimates.
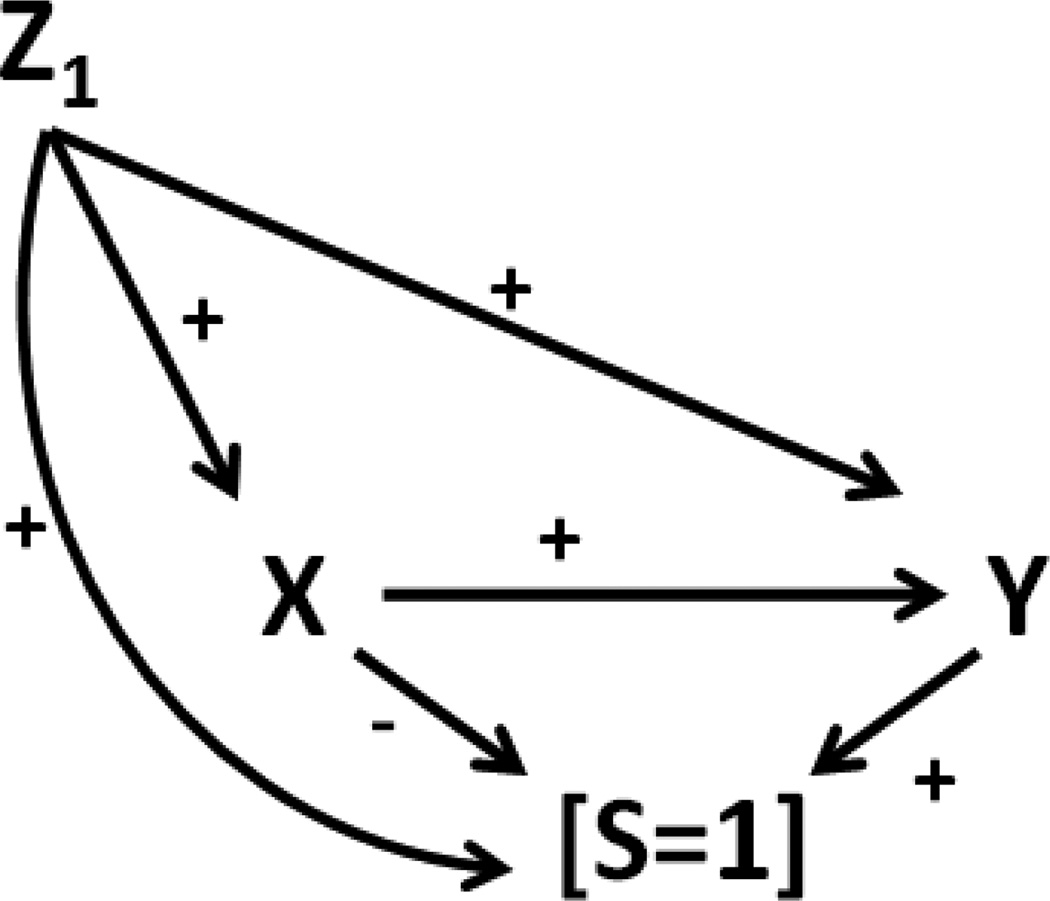
Assignment of bias parameters (i.e., in the absence of a validation sub-study) could be aided by the use of signed DAGs. In a signed DAG, edges are marked with the direction (positive or negative) of the average effect for each pair of directly connected variables, conditional on other relevant variables. The use of signed DAGs for characterizing the directionality of relationships in the diagram and in understanding confounding bias has been described in detail [21, 22]. For example, as depicted in Figure 5, if Y increased the probability of selection conditional on X and Z1, then the odds ratio eβSY would be assigned a positive value(> 1). Similarly, eβSX could be assigned a negative value (<1) on the X→S path whereby other paths connecting X and S are blocked by conditioning on Y and Z1. Assignment can proceed similarly for eβSZ1 by considering the net sign of all open paths between Z1 and S conditional on X and Y. More work is needed to formalize these insights.
Figure 5.
A hypothetical selection bias mechanism with signed edges indicating the direction of effect between each pair of connected variables.
In simulating varying scenarios of selection bias in hypothetical populations, we detected a discernible pattern of bias direction that may warrant further investigation. When both the exposure and disease were positively associated with selection, the bias direction was downward. When one was positive and the other was negative, the bias direction was upward. If the overall magnitude of bias was small, this rule of directionality was not as evident. A thorough evaluation of the expected magnitude and direction of selection bias has not yet been published in the epidemiologic literature. Suspected examples of severe Berksonian bias have been shown to cause extreme downward bias, to 10-fold decrease in effect estimate [23, 24]. Exploration of the potential impact of selection bias in the electromagnetic fields (EMF) and leukemia literature has found that this type of bias could result in a 2-fold increase in effect estimates [25]. Some theoretical work has been done to predict the magnitude of expected bias from controlling on a collider, when bias parameters are known [14]. Further research including simulation studies may be warranted in this area.
ACKNOWLEDGEMENTS
The authors wish to thank Harold Luft for his critical review of this manuscript. CAT was supported by a pre-doctoral fellowship from the National Institutes of Health, National Cancer Institute T32 CA09142. OAA was supported by a Veni career grant (# 916.96.059) from the Netherlands Organization for Scientific Research (NWO), the European Commission’s Seventh Framework Programme under grant # 241822, and grant # 1R01HD072296-01A1 from The Eunice Schriver Kennedy National Institute of Child Health and Human Development.
ABBREVIATIONS AND ACRONYMS
- DAG
directed acyclic graph
- IPCW
inverse probability of censoring weight(ed/ing)
- IPSW
inverse probability of selection weight(ed/ing)
- OR
odds ratio
- RMSE
root mean squared error
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST
None declared.
REFERENCES
- 1.Walker AM. Anamorphic analysis: sampling and estimation for covariate effects when both exposure and disease are known. Biometrics. 1982;38(4):1025–1032. [PubMed] [Google Scholar]
- 2.Kleinbaum DGKL, Morgenstern H. Epidemiologic research: principles and quantitative methods. New York: Van Nostrand Reinhold; 1982. [Google Scholar]
- 3.White JE. A two stage design for the study of the relationship between a rare exposure and a rare disease. Am J Epidemiol. 1982;115(1):119–128. doi: 10.1093/oxfordjournals.aje.a113266. [DOI] [PubMed] [Google Scholar]
- 4.Rothman KJ, Greenland S, Lash TL. Modern Epidemiology. 3rd ed. London: Lippincott Williams & Wilkins; 2008. [Google Scholar]
- 5.Lash TL, Fox MP, Flink AK. Applying Quantitative Bias Analysis to Epidemiologic Data. New York: Springer Science+Business Media; 2009. [Google Scholar]
- 6.Greenland S. Multiple-bias modelling for analysis of observational data. Journal of the Royal Statistical Society: Series A (Statistics in Society) 2005;168(2):267–306. [Google Scholar]
- 7.Robins JM, Finkelstein DM. Correcting for noncompliance and dependent censoring in an AIDS Clinical Trial with inverse probability of censoring weighted (IPCW) log-rank tests. Biometrics. 2000;56(3):779–788. doi: 10.1111/j.0006-341x.2000.00779.x. [DOI] [PubMed] [Google Scholar]
- 8.Robins JM, Rotnitzky A, Zhao LP. Analysis of Semiparametric Regression Models for Repeated Outcomes in the Presence of Missing Data. Journal of the American Statistical Association. 1995;90(429):106–121. [Google Scholar]
- 9.Scharfstein DO, Rotnitzky A, Robins JM. Adjusting for Nonignorable Drop-Out Using Semiparametric Nonresponse Models. Journal of the American Statistical Association. 1999;94(448):1096–1120. [Google Scholar]
- 10.Rotnitzky A, Robins JM, Scharfstein DO. Semiparametric Regression for Repeated Outcomes with Nonignorable Nonresponse. Journal of the American Statistical Association. 1998;93(444):1321–1339. [Google Scholar]
- 11.Pearl J. Causal diagrams for empirical research. Biometrika. 1995;82(4):669–688. [Google Scholar]
- 12.Greenland S, Pearl J, Robins JM. Causal diagrams for epidemiologic research. Epidemiology. 1999;10(1):37–48. [PubMed] [Google Scholar]
- 13.Hernan MA, Hernandez-Diaz S, Werler MM, Mitchell AA. Causal knowledge as a prerequisite for confounding evaluation: an application to birth defects epidemiology. Am J Epidemiol. 2002;155(2):176–84. doi: 10.1093/aje/155.2.176. [DOI] [PubMed] [Google Scholar]
- 14.Greenland S. Quantifying biases in causal models: classical confounding vs colliderstratification bias. Epidemiology. 2003;14(3):300–306. [PubMed] [Google Scholar]
- 15.Hernan MA, Hernandez-Diaz S, Robins JM. A structural approach to selection bias. Epidemiology. 2004;15(5):615–625. doi: 10.1097/01.ede.0000135174.63482.43. [DOI] [PubMed] [Google Scholar]
- 16.Pearl J. Causality. 2nd ed. New York: Cambridge University Press; 2009. [Google Scholar]
- 17.Bareinboim E, Pearl J, editors. Controlling for selection bias in causal inference. Fiffteenth International Conference on Artificial Intelligence and Statistics (AISTATS).2012. [Google Scholar]
- 18.Westreich D. Berkson's bias, selection bias, and missing data. Epidemiology. 2012;23(1):159–164. doi: 10.1097/EDE.0b013e31823b6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Daniel RM, Kenward MG, Cousens SN, De Stavola BL. Using causal diagrams to guide analysis in missing data problems. Statistical methods in medical research. 2012;21(3):243–256. doi: 10.1177/0962280210394469. [DOI] [PubMed] [Google Scholar]
- 20.Geneletti S, Mason A, Best N. Adjusting for selection effects in epidemiologic studies: why sensitivity analysis is the only "solution". Epidemiology. 2011;22(1):36–39. doi: 10.1097/EDE.0b013e3182003276. [DOI] [PubMed] [Google Scholar]
- 21.VanderWeele TJ, Hernan MA, Robins JM. Causal directed acyclic graphs and the direction of unmeasured confounding bias. Epidemiology. 2008;19(5):720–728. doi: 10.1097/EDE.0b013e3181810e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanderweele TJ, Tan Z. Directed acyclic graphs with edge-specific bounds. Biometrika. 2012;99(1):115–126. doi: 10.1093/biomet/asr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwitz RI, Feinstein AR. Alternative Analytic Methods for Case-Control Studies of Estrogens and Endometrial Cancer. New England Journal of Medicine. 1978;299(20):1089–1094. doi: 10.1056/NEJM197811162992001. [DOI] [PubMed] [Google Scholar]
- 24.Schwartzbaum J, Ahlbom A, Feychting M. Berkson's Bias Reviewed. European Journal of Epidemiology. 2003;18(12):1109–1112. doi: 10.1023/b:ejep.0000006552.89605.c8. [DOI] [PubMed] [Google Scholar]
- 25.Mezei G, Kheifets L. Selection bias and its implications for case–control studies: a case study of magnetic field exposure and childhood leukaemia. International Journal of Epidemiology. 2006;35(2):397–406. doi: 10.1093/ije/dyi245. [DOI] [PubMed] [Google Scholar]