Abstract
The gut microbiota plays an essential role in regulating intestinal homeostasis through its capacity to modulate various biological activities ranging from barrier, immunity and metabolic function. Not surprisingly, microbial dysbiosis is associated with numerous intestinal disorders including inflammatory bowel diseases (IBD) and colorectal cancer (CRC). In this piece, we will review recent evidence that gut microbial dysbiosis can influence intestinal disease, including colitis and CRC. We will discuss the biological events implicated in the development of microbial dysbiosis and the emergence of CRC-associated microorganisms, focusing on E.coli and F. nucleatum. Finally, the mechanisms by which E.coli and F. nucleatum exert potentially carcinogenic effects on the host will be reviewed.
Introduction
Through evolution, humans have acquired and maintained a complex relationship with their rich microbial surroundings. Microbes colonize virtually all body surfaces, but the most complex and abundantly populated microbial communities can be found in the gastro-intestinal tract, which comprise ∼99% of the microbial biomass. From the mouth to the anus, the diverse microbial communities present in the GI tract should be thought of as intrinsic parts of the human body. In general, the gut microbiota can be found living as planktonic organisms (chiefly within the gut lumen) or may also be found closely associated with the mucosal epithelium where biofilm formation can provide growth advantages. Taxonomic (16S rRNA gene based) and metagenomic (pan-genomic based) microbial sequence analyses have provided a clear link between bacteria, inflammation and CRC [1,2]. In particular, the phyla Proteobacteria and Fusobacteria are often overrepresented in patients with intestinal inflammation and cancer [3,4]. Although various microorganisms such as Helicobacter spp, enterotoxigenic Bacteroides fragilis and enterococcus feacalis have been shown to trigger CRC in pre-clinical models, their involvement in human CRC remains unclear [4,5]. As opposed, clinical isolates of Escherichia coli and Fusobacterium nucleatum have been obtained from human CRC and functional impact on CRC development has been documented using experimental models [6-10]. Consequently, in this review we will focus on these two bacterial strains as possible driver of human CRC. Evidence from functional studies indicates that E.coli and F.nucleatum utilize a complex arsenal of virulence factors to colonize and persist in the intestine. Some of these virulence factors such as the genotoxin, colibactin (E.coli) and the adhesin, FadA (F. nucleatum) have been found to promote colorectal cancer in experimental models [11-13]. It is clear that there is a complex interplay between the host and their resident microbes; certain bacterial genes have been implicated in the development of disease and thus understanding the basic elements of these interactions could lead to important advances in disease detection and management.
Microbial-host interaction
The GI tract is lined by a single-cell thickness epithelium, which has complex functions that include absorption/secretion, immune regulation and the provision of a physical barrier against the abundant microbial community that the gut houses. For example, specialized intestinal epithelial cells, such as Paneth and goblet cells generated from multipotent intestinal epithelial stem cells, closely monitor bacterial location/numbers within the gut. Indeed, these cells are active participants in regulating host-microbe interaction through their production of anti-microbial peptides (e.g. defensins) and mucins (e.g mucin-2). In addition, with their ability to form tight junctions with other epithelial cells, both Paneth and goblet cells contribute to the formation of efficient intestinal barrier function. The importance of these cell types is illustrated by the finding that defective endoplasmic reticulum stress and autophagy responses from Paneth cells are sufficient to trigger ileitis in a mouse model [14].
The intestine is also home to a rich and complex immune system that actively participates in bacterial host interaction. For example, secretory immunoglobulin A (sIgA) produced by B-lymphocytes is transported through the epithelium to the lumen by the polymeric immunoglobulin receptor (pIgR), where it forms a protective barrier against bacteria. Regulation of production is strongly influenced by luminal bacteria since expression of this important immunoglobulin is strongly reduced in germ-free mice [15]. Conventionally-raised SPF mice defective for IEC-derived MyD88 signalling showed reduced PIgR expression and down-regulation of antimicrobial activities resulting in differences in gut microbial community structure compared to WT mice [16]. The united action of the epithelial barrier and immune system prevent the excessive growth of microorganisms at the mucosal surface and create a safe-guarded zone where microorganisms are prevented from gaining direct access to the epithelium, except in dedicated and specialized structures such as Payer's patch and M cells which participate in immune homeostasis [17].
Direct interaction and adherence of microorganisms to the epithelium is often associated with deleterious host responses, and the presence of certain bacteria has been associated with inflammatory bowel diseases (IBD) and CRC (discussed below). The intricate relationship between intestinal barrier function and the mucosal immune system in cancer development is elegantly illustrated by studies using cdx2-driven cre recombinase deletion of the adenomatous polyposis coli (APC) gene in mice. These mice display defective intestinal barrier function at tumor-initiated sites, which facilitate the translocation of bacteria and bacterial products across the epithelial barrier, a process that leads to the activation of the myeloid cell-derived IL-23/IL-17 cytokine network and promotion of tumor growth [18]. In addition, defective cecal barrier function, leading to increased invasion of bacteria belonging to the Clostridiales/Lachnospiraceae family and subsequent activation of immune cells is associated with site-specific development of serrated polyps in mice [19].
The crosstalk between bacteria and the immune system is critical in the maintenance of intestinal homeostasis [12,16,20-24]. Moreover, microbial-derived production of metabolites such as short-chain fatty acids (SCFA) is essential for the maintenance of mucosal immune homeostasis. Defective production of these metabolites and/or improper host-mediated signalling through specific receptors (e.g Gpr43, Gpr109a) lead to an imbalance in the ratio of T regulatory and T effector cells, the combined function of which control gut inflammatory tones [25-29]. The importance of the interplay between the host immune system, intestinal barrier and microbiota is highlighted in studies showing that many of the IBD genetic risk alleles are implicated in regulation of the epithelial barrier, and the innate and adaptive immune responses [30].
As mentioned previously, the microbiota in the GI tract is representative of the most abundantly and diversely populated microbial community of the human body. This is likely due to the unique environmental conditions offered by the gut where various factors such as oxygen concentration, pH range, dietary nutrients and complex secondary metabolites all contribute to shape microbial composition. Interestingly, out of the 50 known bacterial phyla, 95% of the entire GI ecosystem is largely composed of only 2 phyla (Firmicutes and Bacteroides) [31,32]. Other contributors to the microbiota are Proteobacteria, Actinobacteria, Fusobacteria, Verrucomicrobia and Cyanobacteria. Diversity increases dramatically as the taxonomy is refined through class, family and genus, however, and in particular there are pronounced interindividual differences [33]. Assembly of this microbial community and niche succession is not completely understood but numerous studies have shown that the first 3 years of life represents a dynamic phase of microbial community assembly [34]. Although mode of delivery (vaginal vs. cesarian section), as well as early diet (breast milk vs. formula) influences the seeding of the microbial community, the early-life gut microbiome communities share a predominantly facultative anaerobic lifestyle which contributes to the reduction of the oxygen tension in the gut and thus eventually favors establishment of strictly anaerobic bacteria (usually by the 90th day of life) [35]. Interestingly, the facultative anaerobe, E.coli, carrying the genotoxin pks (discussed below) is able to persistently colonize the gut of infants, suggesting that specific microbial genes confer a colonization advantage in the developing intestine [36]. Although microbial communities differ taxonomically between individuals, there is a core metagenome underlying the functional output which is conserved across individuals [37].
Development of dysbiotic states during inflammation/carcinogenesis
Although longitudinal studies have shown that the human microbiota is relatively stable over time if the host environment is not perturbed [38], dietary changes, inflammation, infection and probiotic intake represent forces that can shape the microbial community [33]. Next generation sequencing methods have provided an unprecedented view of the microbiome, which in turn has indicated changes in the ecosystem in intestinal disorders. The plasticity or robustness of the healthy microbiome (the amount of perturbation that the ecosystem can absorb) before the host experiences a disease state, is currently unknown. However, studies performed in healthy and case controls have reported divergence in microbial composition between the two conditions, suggesting disease association with microbial dysbiosis.
Duration and severity of intestinal inflammation significantly enhances the risk of developing colorectal cancer, which is ∼60% higher in inflammatory bowel disease (IBD) patients than in healthy controls [39]. Chronic inflammation, as experienced by IBD patients, represents a powerful environmental factor affecting microbial composition. Indeed, the diversity of the microbiota is decreased in the intestines of IBD patients and 16S rRNA analysis using deep sequencing has shown a reduced abundance of Bacteroidetes and Firmicutes, particularly clostridial clusters IV (Clostridium leptum subgroup) and XlVa (Clostridium coccoides subgroup) [40]. A higher representation of Actinobacteria and gammaproteobacteria was also observed in patients compared to controls. At the family level, expansion of Enterobacteriaceae/E.coli is also found in IBD patients [41,42] and in different experimental models of intestinal inflammation [12,43-45]. In addition, culture-based studies show increased prevalence of adherent-invasive E. coli [6,8,46-50].
Regarding F.nucleatum, several factors, including the extremely low %GC content of the species [51,52], and its preference for colonizing the mucosa [53] mean that sequence-based surveys of fecal samples have likely under-estimated the true numbers of this species in the human gut. F.nucleatum can be readily cultured from gut biopsy specimens collected using methods to preserve anaerobic conditions for the tissue samples [53]. Interestingly, F.nucleatum was recovered more often from biopsy specimens taken from Crohn's Disease patients compared to healthy control (colon cancer screen) patients [25]. Perhaps more remarkably, isolates recovered from IBD patients were demonstrated to be significantly more invasive and proinflammatory in cultured epithelial cell assays than those strains that were isolated from healthy individuals [25,54].
Similar to IBD studies, numerous laboratories have compared the microbiome of patients at various states of CRC with that of healthy controls and found evidence of microbial dysbiosis. A higher prevalence of Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus, and Peptostreptococcus was observed in the luminal compartment of CRC patients compared to controls in two separate Chinese cohorts [1,55]. The same studies also showed decreased abundance of butyrate-producing bacteria (e.g Lachnospiraceae/Roseburia), compared to controls [1]. In a French cohort, Sobhani et al. reported that the genus group Bacteroides-Prevotella are overrepresented in the luminal microbiota of CRC patients compared with normal controls [3]. Another study using an American cohort showed that the stools of patients with adenocarcinoma revealed a lower abundance of clostridia, while Fusobacterium, Prophyromonas and Atopobium were increased compared to controls [56]. Although the above investigations have all identified decreased microbial diversity between cases and controls, the divergent microbial profile between these studies suggests that more functional analysis (e.g. metagenomics/metatranscriptomics) and the inclusion of larger cohorts with well defined histories may be necessary to gain a better understanding of the role of microorganisms in CRC.
As mentioned previously, adherent and planktonic microbial communities are distinct from each other, and because of the likely importance of microbial proximity to host cells in disease, investigators have focused their attention on characterizing mucosal adherent bacteria in CRC patients. Since CRC develops over a long period of time with specific neoplastic phases, studies have investigated microbial communities longitudinally across various cancer phases. In adenoma biopsies from an American patient cohort, an increased abundance of Firmicutes, Bacteroidetes, and Proteobacteria was observed compared with non-adenoma subjects when the V1-V2 r16S region was analyzed [11]. Perhaps the strongest link between bacteria and CRC development came from the study on Fusobacteria. Kostic et al, using a patient cohort from Spain showed that Fusobacterium prevalence was higher in adenocarcinoma tissues compared to non-affected tissues when the V3-V5 region was examined [57]. Using RNA-sequencing approaches, Castellarin et al also observed increased expansion of Fusobacterium in colonic tissues of CRC patients from a Canadian cohort compared with normal controls [58]. Finally, an examination of the V1-V3 r16S gene in the communities derived from rectal swabs of a Chinese cohort showed an expansion of Fusobacterium from CRC patients compared with healthy controls [59]. Since these studies utilized different technologies and geographically disparate subjects, the finding that abundance of tissue-associated Fusobacterium increased in CRC patients is the most robust microbiome observation so far made.
The events leading to increased abundance of Fusobacterium or Enterobacteriaceae are unclear, and longitudinal analysis and functional studies using experimental models will be necessary to address this question effectively. Interestingly, a western-type diet (high fat/high sugar) promoted development of inflammation, microbial dysbiosis and increased colonization of adherent-invasive E.coli (AIEC) in mice, suggesting that diet may be an important environmental driver of microbial activity in this context [60]. Nevertheless, recent developments using animal models have shed new light on the functional impact as well as the mechanisms of action of these microorganisms in CRC development.
Host response to E.coli/F. nucleatum and CRC
Although considered commensal bacteria, genomic and culture-based analyses suggest that E.coli and F. nucleatum are associated with development of IBD and CRC. The relationship between presence of mucosal-adherent E.coli/F. nucleatum strains and development of human pathology is still unclear, but experimental models have provided important insight into the functional impact of these bacteria on disease states. For example, an AIEC strain isolated from a patient with CRC was able to promote tumor development in Apcmin/+ mice [9]. The capacity of E.coli to induce experimental pathology is not dependent on the host origin since the AIEC strain NC101 isolated from a healthy WT mouse was able to trigger colitis in colitogenic susceptible IlI0-/ mice [61,62]. Moreover, the same strain (NC101) induced colitis- associated colorectal cancer in Il10-/- mice [12].
In the case of F.nucleatum, Kostic et al. also used the ApcMin/+ mouse model to show that a highly invasive, Crohn's disease-associated F.nucleatum isolate, EAVG_002 [25] was able to promote tumor progression through the recruitment of tumor-infiltrating myeloid cells [10]. However, in contrast to E.coli NC101, this F.nucleatum strain was not found to promote inflammation-associated intestinal carcinogenesis per se, because infection did not enhance colitis in mice [10]. In fact, on is own, EAVG_002 could not effectively colonize the ApcMin/+ or Il10-/- mice and a microbial bolus (108 cfu) was administered daily for a period of 8 weeks to assess promotion of tumorigenesis [10]. It remains to be shown whether F.nucleatum isolates with less invasive phenotypes are able to promote tumor progression; i.e. whether it is the invasive capacity of the organism that drives tumorigenesis, and/or whether F.nucleatum can behave synergistically with other organisms to colonize tissues and enhance carcinogenesis. Gnotobiotic experiments will help address the synergistic potential of various microbial species on F.nucleatum induced CRC.
Mechanisms by which E.coli and F. nucleatum promote Cancer
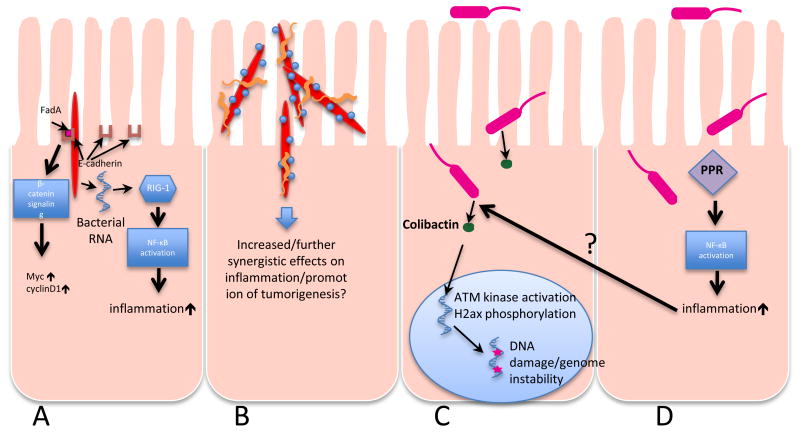
If E.coli and F.nucleatum can indeed behave as keystone microorganisms in CRC, the question of mechanism of promotion of carcinogenesis arises. Both species are known to possess various attributes that could play potential roles in the promotion of dysplasia which, given a susceptible host genotype, may result in CRC. Mechanisms of promotion of pathogenesis can be divided into 2 main themes: inflammation- and virulence determinant- associated. These mechanisms are not mutually exclusive and could work in synergy to promote host pathological response (Fig.1).
Figure 1. Schematic overview of the mechanisms by which E.coli and F. nucleatum can promote carcinogenesis.
A: F.nucleatum (red) is an invasive organism that utilizes a surface adhesin, FadA, to gain entry to host cells. FadA binds to E-cadherin on the epithelial cell surface and activates β-catenin signaling pathways that can in turn up-regulate oncogene expression. As an invasive organism that can survive inside host cells, F.nucleatum is also capable of releasing RNA into the host cell cytoplasm that is detected by cytosolic RIG-1, triggering NF-kB activation and upregulation of inflammation.
B: F.nucleatum is notable for its aggregation with other, unrelated microbes, in particular Streptococcus (blue) and Campylobacter (orange) spp., either of which may have pro-inflammatory activity themselves. F.nucleatum can also shuttle otherwise non-invasive bacteria into the host cell cytoplasm. There is thus great potential for synergism in mixed species associations with F.nucleatum as a foundation. C: AIEC E.coli producing genotoxins such as colibactin cause damage to double-stranded DNA which can lead to neoplastic transformation. D: AIEC induce an inflammatory response through various pattern recognition receptors (PRRs), a process that enhances neoplastic progression, possibly by regulating host and microbial gene expression.
i) Inflammation
Healthy epithelial cells exist in an equilibrium whereby their innate antioxidant enzymes are able to cope with a reasonable turnover of reactive oxygen and nitrogen species from infiltrating immune cells such as lymphocytes and macrophages. During inflammation, when the actions of chemo-attractants such as cytokines and growth factors recruit larger numbers of immune cells to a particular site, this equilibrium is upset and damage to epithelial cells can accrue with associated defective barrier function [63]. Should inflammation become chronic (perhaps as a result of gut microbial dysbiosis allowing persistent or increased colonization of proinflammatory microbial species), the inflammatory/oxidative microenvironment can result in pathologic damage that may also be directed at host DNA, resulting in carcinogenesis [63]. Consequently, it is of interest to determine how CRC-associated microbes exert their pro-inflammatory effects. For F.nucleatum, the abundance of the organism and increased expression of cytokines correlate well; F.nucleatum is a well- known oral commensal and its relative numbers have been shown to increase during progression from oral health to gingivitis [64]. In colorectal adenomas, the abundance of F.nucleatum as assessed by FISH was found to positively associate with cytokine gene expression, in particular for IL-10 and TNF-α [65] Similarly, experimental models have indicated an increased abundance of Enterobacteriaceae in the inflamed intestine compared to un-inflamed controls [12,43,66]. More importantly, the abundance of AIEC was shown to increase in the inflamed intestine of IBD and CRC patients compared to healthy controls [6,8,46-50]. The events leading to expansion of Enterobacteriaceae spp., including E.coli, are unclear but may relate to the ability for these microbes to utilize host-derived inflammatory by-products (e.g. nitrate) as energy sources, a capacity not shared by competing bacteria [67]. In addition, intracellular survival of the IBD strain AIEC LF82 in macrophages is dependent on TNF production, a predominant cytokine in IBD pathogenesis [68]. Interestingly, AIEC appear to attenuate autophagy response in intestinal epithelial cells by increasing expression microRNA 30C and 130A, two critical microRNAs regulating expression of proteins implicated in autophagy [69]. The capacity of Enterobacteriaceae/E.coli to induce inflammation in the host gut epithelium [48,70], in conjunction with the ability of these microbes to harness this inflammatory environment, highlights a formidable evolutionary and adaptive feature. As inflammation has been a well documented risk factor for various form of cancer [71], it is not surprising that Enterobacteriaceae/E.coli are associated with development of CRC. However, the inflammatory potential of AIEC in relation with development of CRC was recently uncoupled [12], suggesting the existence of additional mechanisms (discuss below).
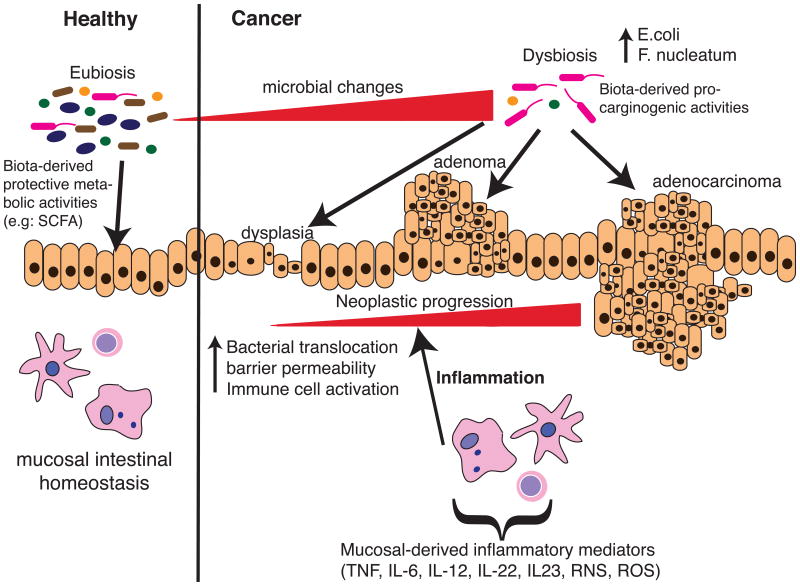
The pro-colitogenic ability of F.nucleatum and its relation to CRC is still unclear. Similar to E.coli, F.nucleatum is a genotypically and phenotypically highly variable species with several recognised subspecies and remarkably complex serotype heterogeneity [53,72,73]. We have shown that isolates even within the same subspecies vary in their ability to promote a pro-inflammatory immune response in gut epithelial cells, and pro-inflammatory activity, in turn, is tightly correlated with the ability for a given strain to invade and persist within host cells [25,54]. This suggests that invasive strains may be activating pro-inflammatory pathways independently of TLR-mediated events. Whilst it has been shown that intracellular F.nucleatum does not activate pro-inflammatory pathways via a classical NOD-1 or NOD-2 driven pathway [74], recently Lee and Tan have demonstrated that the cytosolic pattern recognization receptor (PRR), RIG-1, normally associated with the sensing of RNA viruses, can also respond to intracellular F.nucleatum in a human periodontal ligament fibroblast cell model [75]. Further investigation revealed that F.nucleatum RNA was required for optimal RIG-1 sensing and downstream activation of NF-κB, indicating that invasive, persistent (and thus transcriptionally active) F.nucleatum cells could pose a significant pro-inflammatory threat to the host. However, Kostic et al. showed that inflammation is not enhanced in F.nucleatum- colonized Il10-/- mice compared to control, neither did CRC develop in these mice [10]. These findings suggest that mechanisms other than inflammation are responsible for F.nucleatum-induced tumor development in mice. Nevertheless, with the deleterious effect of chronic inflammation on intestinal barrier (increase permeability and bacterial translocation) and impact on microbial composition [12,13,76], inflammation plays a critical role in bacteria mediated tumorigenesis (Fig.2).
Figure 2. Schematic overview of the interplay between the microbiota and the host on tumorigenesis.
At healthy state, the intestinal microbiota stands at an eubiosis stage, a phase which contributes to the maintenance of intestinal homeostasis through production of various metabolites and bacterial products (e.g SCFA), which promote immune balance. Various environmental factors such as diet, inflammation, stress or host genetics influence microbial composition and cause microbial dysbiosis (e.g increase abundance of AIEC and fusobacteria). This cancer-promoting biota may favour neoplastic progression through various carcinogenic activities (toxins, metabolites), which ultimately affect epithelial cell DNA integrity and cellular transformation. In conjunction with these changes, epithelial barrier integrity is compromised, further enhancing bacterial uptake and activation of mucosal immune cells (releases of inflammatory mediators), thereby contributing to neoplastic progression.
ii) Virulence determinant-associated mechanisms
As a fastidious anaerobic organism that is difficult to culture, the nature of F.nucleatum's repertoire of virulence determinants has only recently begun to be fully explored, aided by the ongoing availability of sequenced genomes. Approximately 50% of F.nucleatum predicted ORFs have no known function and thus there is much to be learned [51,52]. However, recently there has been some progress in unravelling the functions of a subset of F.nucleatum virulence determinants. The best characterized of these is the surface adhesin, FadA. This adhesin has a dual role in adhesion and invasion of host cells, and is conserved in F.nucleatum and its close relative, F.periodonticum [77]. FadA undergoes an intricate mechanism of secretion and assembly, with an unprocessed pre-FadA joining with a secreted, truncated mature FadA to form an active complex that has been shown to promote F.nucleatum invasion into host cells [78]. Rubinstein et al. used fadA knock out mutants to demonstrate that this adhesin/invasin complex mediates the binding to and invasion of epithelial cells through an interaction with E-cadherin, and by doing so initiated signalling cascades via the β-catenin pathway that led to, among other effects, upregulation of oncogenes Myc and cyclin D1 [13]. This work further demonstrated the relevance of FadA in the promotion of CRC by analyzing the expression levels of fadA in gut epithelial tissues obtained from either healthy individuals or from CRC cases, noting a >2log increase in fadA expression levels in the CRC tissue compared to controls.
Several outer membrane proteins of F.nucleatum have demonstrated activity against cultured human cells. The adhesins Fap2 and RadD have been shown to induce apoptosis in T-lymphocytes [79]. These proteins share homology with type Va (autotransporter) secretion systems, however, cell-free F.nucleatum membranes alone were sufficient to induce cell death suggesting that the effect seen was not mediated by a secreted effector [79]. Whether these proteins have any effect on gut epithelial cells remains to be determined.
Comparative genomic analysis of various IBD and CRC E.coli isolates has shown the presence of numerous virulence factors implicated in bacterial adhesion, invasion and survival in the host, but the functional role of these factors is still unclear [80,81]. Although some E.coli B2 strains possesses cytolethal distending toxin (CDT), a bacterial product capable of inducing direct DNA damage responses and genomic instability [82], a natural product called Colibactin was recently also identified in E.coli isolated from IBD and CRC patients [12,81,83]. This hybrid peptide-polyketide genotoxin is the product of a multi-enzymatic factory encoded by the 54kb polyketide synthase (pks) genotoxicity island [84,85]. The functional relevance of this island on tumorigenesis was shown by a genetic approach where an E. coli NC101 pks deleted strain failed to promote CRC in mono-associated Il10 mice [12,33]. Interestingly, the ability of NC101 to induce chronic intestinal inflammation was not related to the presence of pks, suggesting that bacteria-induced inflammation is not the sole driver of tumorigenesis [12]. As discussed above, inflammation does not seem to promote F. nucleatum-induced tumor in Il10-/- mice, highlighting the complex interplay between microorganisms, environment and CRC development. Key information regarding production, regulation and function of colibactin is missing as the natural product has not yet been purified or characterized. However, recent evidence suggests that colibactin is released by the action of the peptidase activity of the editing enzyme, clbP, present on the pks island, suggesting that the natural compound is generated as a prodrug [86,87]. In addition, the pks- associated gene clbA promotes synthesis of both colibactin and yersiniabactin [88]. The fact that siderophores such as yersiniabactin are essential for iron acquisition and maintenance of E.coli survival/growth suggest that a high level of interaction exists between these virulence factors. Indeed, increased numbers of E.coli observed in the mucosa of ulcerative colitis patients were associated with a higher prevalence of genes implicated in iron acquisition (e.g chuA, iutA) [46]. Identification of the mechanisms leading to colibactin synthesis, transport, activation and crosstalk with other virulence factors would undeniably open new research avenues and potentially lead to novel approaches to modulate E.coli activity.
Conclusions/Perspective
Although the field of CRC microbiome research is relatively young, the past 3 years have seen important discoveries regarding the implication of microbes in disease pathology (Fig.2). These findings range from identification of cluster of microorganisms associated with various phases of CRC, to the validation of microbial candidates such as AIEC and F.nucleatum in experimental models. Despite this progress, a number of questions still remain with respect to the events leading to microbial dysbiosis, factors controlling virulence gene expression and interplay between microbes/virulence genes. For example, F.nucleatum rarely acts alone within its host; its highly aggregative nature allows it to behave as a bridge organism in oral plaque, where it promotes hierarchical and structured biofilm formation [89]. In the gut there are tantalizing clues that the same may be true; Warren et al. have used co-occurrence network analysis to study metagenomic signatures of microbes associated with CRC tissues and compared these to matched healthy tissues, revealing a subset of microbes which significantly associate with F.nucleatum in CRC [90]. Intriguingly, these include an uncommon Campylobacter species, C.showae, a CRC- associated isolate of which was demonstrated to aggregate with F.nucleatum in vitro, and to have a genome sequence containing genes homologous to known virulence determinants, including components of the vir operon of Helicobacter pylori [90]. Since it is known that F.nucleatum can specifically associate with, and promote invasion of, other microbes, as has clearly been demonstrated for Streptococcus cristatus [91], and that such associations can modify the host response to infection [92,93], it is imperative to broaden our view of infection in CRC to consider more than one microbe at a time. The influence of the surrounding gut microbiota on E.coli pathogenesis, as for F.nulceatum, is likely important in CRC pathogenesis. For example, diet-driven changes in the gut microbiota may influence AIEC gene expression to favour colonization of the microorganisms [60], and/or the presence of flagellated microbes may work synergistically with AIEC to activate innate immune pathways [94]. The adhesive alliances that F.nucleatum naturally makes with a wide range of Streptococcus spp. [89] for example, should be considered in the context of the finding that Streptococcus bovis has already been shown to be associated with colorectal tumors [95].
Are these cancer-associated microorganisms working in parallel or sequentially to promote tumor progression? In essence, could specific microbial subcommunities be predominantly implicated in cancer initiation whilst other groupings are involved in the promotion of cancer progression (adenomas to adenocarcinoma) (Fig.2)? The fact that Enterococcus faecalis infection promotes CRC in non-initiated Il10-/- mice [96], while F. nucleatum induces colon cancer in the initiated Apcmm/+ mice but not IlI0-/- mice [10] highlights the complexity of the microbe-host relationship. Dissecting the interplay/synergy between CRC-associated microbes and the host will be critical to our understanding of the role of bacteria in tumorigenesis. Many of the studies that have been carried out to date to elucidate the mechanisms that AIEC or F.nucleatum utilizes to promote CRC have only focused on either one or a small subset of virulence determinants. Furthermore, usually a single strain or isolate is generally studied. This is a particular problem for AIEC and F.nucleatum, which are known to be highly heterogenic, and thus conclusions should not be generalized until the same effects are seen in a range of different isolates. For example, the probiotic E.coli strain Nissle 1917 possesses the pks island, yet is not known to induce CRC in mice [97]. Moreover, the presence of pks appears essential for the anti-inflammatory property of Nissle 1917 in mice [97]. Clearly, factors controlling expression/activity of potential genotoxic factors such as pks need to be defined. Additionally, murine models may not the fully capture the extent of biological activities elicited bv human clinical isolates. For example, microbiota transplantation in mice showed that the elicited immune response was strongly dependent on the origin of the microbiota (humans, rats or mice) [24]. A combination of experimental models would likely be necessary to document the modulatory impact of human clinical isolates. Nevertheless, development of CRC following colonization of mice with F. nucleatum or E.coli clinical isolates [9,10] predict that studies in this animal species could provide useful information regarding carcinogenic mechanisms, at least with these microbial strains. The potential to harness the new knowledge from microbiome-host relationship in CRC is immense. The intestinal microbiota has been implicated in cancer drug toxicity, therapeutic efficacy and tumor development, and thus it is important to see the ‘equation’ of CRC as two-sided [98]. As scientists continue to solve this complex equation, new research paradigms will likely emerge from microbiome studies, allowing the design of innovative strategies to detect, treat, and manage cancer.
Highlights.
-
-
Microbial dysbiosis is associated with colorectal cancer
-
-
Specific microbes such as E. coli and F. nucleatum promotecolorectal cancer.
-
-
Microbial activities are important for colorectal cancer development.
-
-
Complex interplay between microbes and host immune system dictate colorectal cancer susceptibility
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. Isme J. 2011;6:320–9. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jobin C. Colorectal Cancer: Looking for Answers in the Microbiota. Cancer Discov. 2013;3:384–7. doi: 10.1158/2159-8290.CD-13-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwabe RF, jobin C. The microbiome and cancer. Nature Reviews. 2013;13:800–12. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sears CL, Garrett WS. Microbes, Microbiota, and Colon Cancer. Cell Host and Microbe. 2014;15:317–28. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 7.Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–47. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swidsinski A, Khilkin M, Kerjaschki D, Schreiber S, Ortner M, Weber J, et al. Association between intraepithelial Escherichia coli and colorectal cancer. Gastroenterology. 1998;115:281–6. doi: 10.1016/s0016-5085(98)70194-5. [DOI] [PubMed] [Google Scholar]
- 9.Bonnet M, Buc E, Sauvanet P, Darcha C, Dubois D, Pereira B, et al. Colonization of the Human Gut by E. coli and Colorectal Cancer Risk. Clinical Cancer Research. 2013;20:859–67. doi: 10.1158/1078-0432.CCR-13-1343. [DOI] [PubMed] [Google Scholar]
- 10.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Short Article. Cell Host and Microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F, et al. Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. Isme J. 2012;6:1858–68. doi: 10.1038/ismej.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arthur JC, Perez-Chanona E, Muhlbauer M, Tomkovich S, Uronis JM, Fan TJ, et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science. 2012;338:120–3. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/b- Catenin Signaling via its FadA Adhesin. Cell Host and Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, et al. Paneth cells as a site of origin for intestinal inflammation. Nature. 2013;503:272–6. doi: 10.1038/nature12599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hapfelmeier S, Lawson MAE, Slack E, Kirundi JK, Stoel M, Heikenwalder M, et al. Reversible Microbial Colonization of Germ-Free Mice Reveals the Dynamics of IgA Immune Responses. Science. 2010;328:1705–9. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frantz AL, Rogier EW, Weber CR, Shen L, Cohen DA, Fenton LA, et al. Targeted deletion of MyD88 in intestinal epithelial cells results in compromised antibacterial immunity associated with downregulation of polymeric immunoglobulin receptor, mucin-2, and antibacterial peptides. Mucosal Immunol. 2012;5:501–12. doi: 10.1038/mi.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4607–14. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17- mediated tumour growth. Nature. 2012:1–7. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bongers G, Pacer ME, Geraldino TH, Chen L, He Z, Hashimoto D, et al. Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. The Journal of Experimental Medicine. 2014;211:457–72. doi: 10.1084/jem.20131587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slack E, Hapfelmeier S, Stecher B, Velykoredko Y, Stoel M, Lawson MAE, et al. Innate and Adaptive Immunity Cooperate Flexibly to Maintain Host- Microbiota Mutualism. Science. 2009;325:617–20. doi: 10.1126/science.1172747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoeberg J, Amir E, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nature Immunology. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20858–63. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL, et al. Gammadelta intraepithelial lymphocytes are essential mediators of host- microbial homeostasis at the intestinal mucosal surface. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:8743–8. doi: 10.1073/pnas.1019574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, et al. Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell. 2012;149:1578–93. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–8. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 26.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–39. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature Medicine. 2014;20:159–66. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 28.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2014;504:446–50. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 29.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–17. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dethlefsen L, Eckburg PB, Bik EM, Relman DA. Assembly of the human intestinal microbiota. Trends Ecol Evol (Amst) 2006;21:517–23. doi: 10.1016/j.tree.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Backhed F. Host-Bacterial Mutualism in the Human Intestine. Science. 2005;307:1915–20. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 33.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominguez-Bello MG, Blaser MJ, Ley RE, Knight R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–9. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mitsou EK, Kirtzalidou E, Oikonomou I, Liosis G, Kyriacou A. Fecal microflora of Greek healthy neonates. Anaerobe. 2008;14:94–101. doi: 10.1016/j.anaerobe.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 36.Nowrouzian FL, Oswald E. Escherichia coli strains with the capacity for long-term persistence in the bowel microbiota carry the potentially genotoxic pks island. Microbial Pathogenesis. 2012;53:180–2. doi: 10.1016/j.micpath.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 37.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Caporaso JG, Lauber CL, Costello EK, Berg-Lyons D, Gonzalez A, Stombaugh J, et al. Moving pictures of the human microbiome. Genome Biology. 2011;12:R50. doi: 10.1186/gb-2011-12-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Herrinton LJ, Liu L, Levin TR, Allison JE, Lewis JD, Velayos F. Incidence and mortality of colorectal adenocarcinoma in persons with inflammatory bowel disease from 1998 to 2010. Gastroenterology. 2012;143:382–9. doi: 10.1053/j.gastro.2012.04.054. [DOI] [PubMed] [Google Scholar]
- 40.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–99. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. Isme J. 2007;1:403–18. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 42.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biology. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host & Microbe. 2007;2:119–29. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 44.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garrett WS, Gallini CA, Yatsunenko T, Michaud M, DuBois A, Delaney ML, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host & Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilarczyk-Zurek M, Chmielarczyk A, Gosiewski T, Tomusiak A, Adamski P, Zwolinska-Wcislo M, et al. Possible role of Escherichia coli in propagation and perpetuation of chronic inflammation in ulcerative colitis. BMC Gastroenterol. 2013;13:61. doi: 10.1186/1471-230X-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134:577–94. doi: 10.1053/j.gastro.2007.11.059. [DOI] [PubMed] [Google Scholar]
- 48.Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflammatory Bowel Diseases. 2007;13:1277–83. doi: 10.1002/ibd.20176. [DOI] [PubMed] [Google Scholar]
- 49.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–21. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 50.Darfeuille-Michaud A, Neut C, Bamich N, Lederman E, Di Martino P, Desreumaux P, et al. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology. 1998;115:1405–13. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 51.Karpathy SE, Qin X, Gioia J, Jiang H, Liu Y, Petrosino JF, et al. Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable fusobacterium. PLoS One. 2007;2:e659. doi: 10.1371/journal.pone.0000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kapatral V, Ivanova N, Anderson I, Reznik G, Bhattacharyya A, Gardner WL, et al. Genome analysis of F. nucleatum sub spp vincentii and its comparison with the genome of F. nucleatum ATCC 25586. Genome Research. 2003;13:1180–9. doi: 10.1101/gr.566003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strauss J, White A, Ambrose C, McDonald J, Allen-Vercoe E. Phenotypic and genotypic analyses of clinical Fusobacterium nucleatum and Fusobacterium periodonticum isolates from the human gut. Anaerobe. 2008;14:301–9. doi: 10.1016/j.anaerobe.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Dharmani P, Strauss J, Ambrose C, Allen-Vercoe E, Chadee K. Fusobacterium nucleatum infection of colonic cells stimulates MUC2 mucin and tumor necrosis factor alpha. Infection and Immunity. 2011;79:2597–607. doi: 10.1128/IAI.05118-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66:462–70. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 56.Ahn J, Sinha R, Pei Z, Dominianni C, Wu J, Shi J, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105:1907–11. doi: 10.1093/jnci/djt300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Research. 2012;22:292–8. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients with Colorectal Cancer. PLoS One. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martinez-Medina M, Denizot J, Dreux N, Robin F, Billard E, Bonnet R, et al. Western diet induces dysbiosis with increased E coli in CEABAC10 mice, alters host barrier function favouring AIEC colonisation. Gut. 2014;63:116–24. doi: 10.1136/gutjnl-2012-304119. [DOI] [PubMed] [Google Scholar]
- 61.Different host genetic backgrounds determine diesease phenotype induced by selective bacterial colonization. 2005;128:A512. [Google Scholar]
- 62.Karrasch T, Kim JS, Muhlbauer M, Magness ST, Jobin C. Gnotobiotic IL-10-/-;NF-kappa B(EGFP) mice reveal the critical role of TLR/NF-kappa B signaling in commensal bacteria-induced colitis. The Journal of Immunology. 2007;178:6522–32. doi: 10.4049/jimmunol.178.10.6522. [DOI] [PubMed] [Google Scholar]
- 63.Wang LS, Kuo CT, Huang YW, Stoner GD, Lechner JF. Gene-Diet Interactions on Colorectal Cancer Risk. Curr Nutr Rep. 2012;1:132–41. doi: 10.1007/s13668-012-0023-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kistler JO, Booth V, Bradshaw DJ, Wade WG. Bacterial community development in experimental gingivitis. PLoS One. 2013;8:e71227. doi: 10.1371/journal.pone.0071227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCoy AN, Araujo-Perez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium Is Associated with Colorectal Adenomas. PLoS One. 2013;8:e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carvalho FA, Koren O, Goodrich JK, Johansson MEV, Nalbantoglu I, Aitken JD, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host & Microbe. 2012;12:139–52. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. Host-Derived Nitrate Boosts Growth of E coli in the Inflamed Gut. Science. 2013;339:708–11. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bringer MA, Billard E, Glasser AL, Colombel JF, Darfeuille-Michaud A. Replication of Crohn's disease-associated AIEC within macrophages is dependent on TNF-a secretion. Lab Invest. 2012;92:411–9. doi: 10.1038/labinvest.2011.156. [DOI] [PubMed] [Google Scholar]
- 69.Nguyen HTT, Dalmasso G, Muller S, Carriére J, Seibold F, Darfeuille- Michaud A. Crohn's disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce. Gastroenterology. 2014;146:508–19. doi: 10.1053/j.gastro.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 70.Loh G, Blaut M. Role of commensal gut bacteria in inflammatory bowel diseases. Gut Microbes. 2012;3:544–55. doi: 10.4161/gmic.22156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation- induced cancer:crosstalk between tumours,immune cells and microorganisms. Nature Reviews. 2013;13:759–71. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 72.Thurnheer T, Guggenheim B, Gruica B, Gmür R. Infinite serovar and ribotype heterogeneity among oral Fusobacterium nucleatum strains. Anaerobe. 1999;5:79–92. [Google Scholar]
- 73.Citron DM. Update on the taxonomy and clinical aspects of the genus fusobacterium. Clin Infect Dis. 2002;35:S22–7. doi: 10.1086/341916. [DOI] [PubMed] [Google Scholar]
- 74.Quah SY, Bergenholtz G, Tan KS. Fusobacterium nucleatum induces cytokine production through Toll-like-receptor-independent mechanism. Int Endod J. 2014;47:550–9. doi: 10.1111/iej.12185. [DOI] [PubMed] [Google Scholar]
- 75.Lee P, Tan KS. Fusobacterium nucleatum Activates the Immune Response through Retinoic Acid-Inducible Gene I. J Dent Res. 2014;93:162–8. doi: 10.1177/0022034513516346. [DOI] [PubMed] [Google Scholar]
- 76.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. 2014:1–15. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, Li M, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187:5330–40. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. The Journal of Biological Chemistry. 2007;282:25000–9. doi: 10.1074/jbc.M611567200. [DOI] [PubMed] [Google Scholar]
- 79.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder Haake S, et al. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infection and Immunity. 2010;78:4773–8. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vejborg RM, Hancock V, Petersen AM, Krogfelt KA, Klemm P. Comparative genomics of Escherichia coli isolated from patients with inflammatory bowel disease. BMC Genomics. 2011;12:316. doi: 10.1186/1471-2164-12-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prorok-Hamon M, Friswell MK, Alswied A, Roberts CL, Song F, Flanagan PK, et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut. 2014;63:761–70. doi: 10.1136/gutjnl-2013-304739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nesić D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature. 2004;429:429–33. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- 83.Buc E, Dubois D, Sauvanet P, Raisch J, Delmas J, Darfeuille-Michaud A, et al. High Prevalence of Mucosa-Associated E coli Producing Cyclomodulin and Genotoxin in Colon Cancer. PLoS One. 2013;8:e56964. doi: 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–51. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 85.Guerra L, Guidi R, Frisan T. Do bacterial genotoxins contribute to chronic inflammation, genomic instability and tumor progression? FEBS Journal. 2011;278:4577–88. doi: 10.1111/j.1742-4658.2011.08125.x. [DOI] [PubMed] [Google Scholar]
- 86.Brotherton CA, Balskus EP. A Prodrug Resistance Mechanism Is Involved in Colibactin Biosynthesis and Cytotoxicity. J Am Chem Soc. 2013;135:3359–62. doi: 10.1021/ja312154m. [DOI] [PubMed] [Google Scholar]
- 87.Bian X, Fu J, Plaza A, Herrmann J, Pistorius D, Stewart AF, et al. In Vivo Evidence for a Prodrug Activation Mechanism during Colibactin Maturation. ChemBioChem. 2013;14:1194–7. doi: 10.1002/cbic.201300208. [DOI] [PubMed] [Google Scholar]
- 88.Martin P, Marcq I, Magistro G, Penary M, Garcie C, Payros D, et al. Interplay between Siderophores and Colibactin Genotoxin Biosynthetic Pathways in Escherichia coli. PLoS Pathogens. 2013;9:e1003437. doi: 10.1371/journal.ppat.1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kolenbrander PE, Palmer RJ, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 90.Warren RL, Freeman DJ, Pleasance S, Watson P, Moore RA, Cochrane K, et al. Co-occurrence of anaerobic bacteria in colorectal carcinomas. Microbiome. 2013;1:16. doi: 10.1186/2049-2618-1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Edwards AM, Grossman TJ, Rudney JD. Fusobacterium nucleatum transports noninvasive Streptococcus cristatus into human epithelial cells. Infection and Immunity. 2006;74:654–62. doi: 10.1128/IAI.74.1.654-662.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang G, Chen R, Rudney JD. Streptococcus cristatus attenuates Fusobacterium nucleatum-induced interleukin-8 expression in oral epithelial cells. J Periodont Res. 2008;43:408–16. doi: 10.1111/j.1600-0765.2007.01057.x. [DOI] [PubMed] [Google Scholar]
- 93.Zhang G, Chen R, Rudney JD. Streptococcus cristatus modulates the Fusobacterium nucleatum-induced epithelial interleukin-8 response through the nuclear factor-kappa B pathway. J Periodont Res. 2011;46:558–67. doi: 10.1111/j.1600-0765.2011.01373.x. [DOI] [PubMed] [Google Scholar]
- 94.Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2013 doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Abdulamir AS, Hafidh RR, Abu Bakar F. The association of Streptococcus bovis/gallolyticus with colorectal tumors: the nature and the underlying mechanisms of its etiological role. J Exp Clin Cancer Res. 2011;30:11. doi: 10.1186/1756-9966-30-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang X, Yang Y, Moore DR, Nimmo SL, Lightfoot SA, Huycke MM. 4- hydroxy-2-nonenal mediates genotoxicity and bystander effects caused by Enterococcus faecalis-infected macrophages. Gastroenterology. 2012;142:543–7. doi: 10.1053/j.gastro.2011.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Olier M, Marcq I, Salvador-Cartier C, Secher T, Dobrindt U, Boury M, et al. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes. 2012;3:501–9. doi: 10.4161/gmic.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perez-Chanona E, jobin C. From promotion to management: The wide impact of bacteria on cancer and its treatment. Bioessays. 2014 doi: 10.1002/bies.201400015. [DOI] [PMC free article] [PubMed] [Google Scholar]