CDK5 activates the tumor suppressor DLC1 by phosphorylating and diminishing the binding of an autoinhibitory region of DLC1 to its Rho-GAP domain and allows it to localize to focal adhesions.
Abstract
DLC1 is a tumor suppressor protein whose full activity depends on its presence at focal adhesions, its Rho–GTPase activating protein (Rho-GAP) function, and its ability to bind several ligands, including tensin and talin. However, the mechanisms that regulate and coordinate these activities remain poorly understood. Here we identify CDK5, a predominantly cytoplasmic serine/threonine kinase, as an important regulator of DLC1 functions. The CDK5 kinase phosphorylates four serines in DLC1 located N-terminal to the Rho-GAP domain. When not phosphorylated, this N-terminal region functions as an autoinhibitory domain that places DLC1 in a closed, inactive conformation by efficiently binding to the Rho-GAP domain. CDK5 phosphorylation reduces this binding and orchestrates the coordinate activation DLC1, including its localization to focal adhesions, its Rho-GAP activity, and its ability to bind tensin and talin. In cancer, these anti-oncogenic effects of CDK5 can provide selective pressure for the down-regulation of DLC1, which occurs frequently in tumors, and can contribute to the pro-oncogenic activity of CDK5 in lung adenocarcinoma.
Introduction
Tumor development is a multistep process that involves the activation of genes that promote neoplastic growth, such as oncogenes and anti-apoptotic genes, together with the down-regulation of anti-oncogenic factors, such as tumor suppressor genes and pro-apoptotic genes (Vogelstein and Kinzler, 2004). The tumor suppressor Deleted in Liver Cancer 1 (DLC1), which encodes a Rho–GTPase activating protein (Rho-GAP), is down-regulated, via genetic or epigenetic mechanisms, in a variety of malignancies, including cancers of the lung, breast, prostate, and liver (Durkin et al., 2007; Kim et al., 2009; Vigil et al., 2010; Lukasik et al., 2011, Kim et al., 2013; Ko and Ping Yam, 2014). Rho-GTPases, which include Rho, Cdc42, and Rac, regulate many physiological functions, such as the actin cytoskeleton, focal adhesions, and cell migration, and their up-regulation occurs frequently in cancer (Ellenbroek and Collard, 2007; Vega and Ridley, 2008; Vigil et al., 2010; Rathinam et al., 2011). Dlc1 is required for mouse embryogenesis (Durkin et al., 2005; Sabbir et al., 2010), and high RhoGTP results from its conditional inactivation in mouse embryonic fibroblasts (MEFs; Qian et al., 2012). DLC1 protein influences focal adhesion turnover, and its Rho-GAP activity strongly inactivates RhoA, -B, and -C, and weakly inactivates Cdc42 (Wong et al., 2003; Healy et al., 2008; Qian et al., 2012).
The full tumor suppressor activity of DLC1 depends on its presence at focal adhesions, its Rho-GAP function, and its ability to bind several ligands, including tensin, talin, and FAK (Yam et al., 2006; Liao et al., 2007; Qian et al., 2007, 2009; Li et al., 2011). However, the mechanisms that regulate and coordinate these activities remain poorly understood. Human DLC1 encodes a 1,091–amino acid protein whose Rho-GAP domain has been genetically localized to amino acids 609–878 (Kim et al., 2008). The DLC1 protein contains two well-recognized domains in addition to its Rho-GAP domain: an N-terminal SAM domain (amino acids 1–78; Qiao and Bowie, 2005) and a C-terminal START domain (Ponting and Aravind, 1999). Deletion mapping of DLC1 has suggested that amino acids N-terminal to the Rho-GAP domain can negatively regulate its Rho-GAP activity (Healy et al., 2008), but the mechanisms remain unclear. Although tensin, talin, and FAK bind to sequences N-terminal to the Rho-GAP domain, the Rho-GAP activity of DLC1 mutants deficient for binding these proteins appears to be similar to that of wild-type (WT) DLC1 (Qian et al., 2007; Li et al., 2011), which suggests that other putative N-terminal functions may account for its Rho-GAP regulation. In this regard, our preliminary in silico analysis identified several consensus motifs for cyclin-dependent kinase 5 (CDK5) in the N terminus of DLC1, which raised the possibility, investigated in this report, that CDK5 might be a previously unidentified regulator of DLC1.
CDK5, a predominantly cytoplasmic proline-directed serine/threonine kinase activated by p35 or p39, can regulate cytoskeletal organization and cell adhesion, contraction, and migration (Kawauchi et al., 2006; Tripathi and Zelenka, 2009; Su and Tsai, 2011; Arif, 2012). Although its pro-differentiation (Cicero and Herrup, 2005; Miyamoto et al., 2007) physiologic activities may be anti-oncogenic, CDK5 may be pro-oncogenic in some cancers (Lin et al., 2007; Feldmann et al., 2010). Here, we report that CDK5 coordinately activates multiple DLC1 functions, elucidate the mechanism underlying this activation, and identify a role for DLC1 inactivation in the pro-oncogenic activity CDK5.
Results
Enzymatically active CDK5 forms a protein complex with DLC1
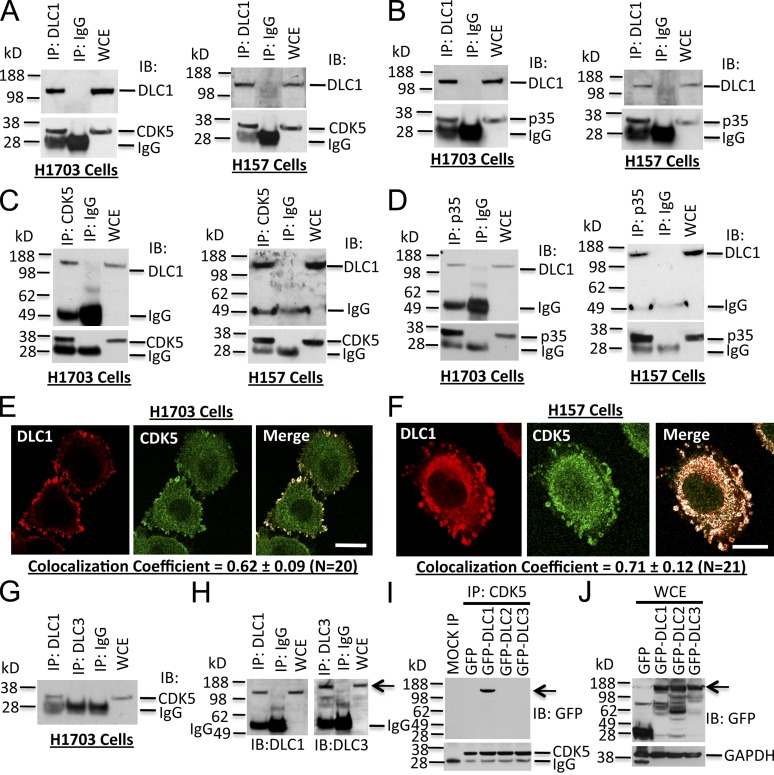
To establish whether an endogenous protein complex containing DLC1 and CDK5 exists in vivo, we performed co-immunoprecipitation (co-IP) experiments from two non-small cell lung cancer (NSCLC) lines, H1703 and H157, which expressed both proteins. DLC1 and CDK5 formed a protein complex in both lines (Fig. 1 A) when cell lysates were immunoprecipitated with DLC1 antibody and immunoblotted for CDK5. The CDK5 activator p35 appears to be part of this complex, as positive results were obtained when cell lysates were immunoprecipitated with DLC1 antibody followed by immunoblotting (IB) for p35 (Fig. 1 B). Reciprocal co-IP with CDK5 or p35 antibodies and immunoblotting with DLC1 antibodies was also positive (Fig. 1, C and D). The presence of p35 in the complex implied that the CDK5 associated with DLC1 is enzymatically active. Confocal microscopy and quantitative colocalization in both lines confirmed the presence of both CDK5 and DLC1 in focal adhesions, with overlapping colocalization coefficients >0.60 between CDK5 and DLC1 (Fig. 1, E and F), and ∼0.65 between DLC1 and Vinculin, a focal adhesion marker (Fig. S1, A–C).
Figure 1.
DLC1, CDK5, and its activator p35 form a protein complex in human cell lines. (A) Protein complex between DLC1 and CDK5. Cell lysates were immunoprecipitated (IP) with DLC1 antibody followed by IB with DLC1 (top) or CDK5 (bottom) antibodies. WCE, whole cell extract. H1703 and H157 are NSCLC lines. (B) Protein complex between DLC1 and CDK5 activator p35. Cell lysates were immunoprecipitated with DLC1 antibody followed by IB with DLC1 (top) or p35 (bottom) antibodies. (C) The protein complex between DLC1 and CDK5 was confirmed by reciprocal co-IP. Cell lysates were immunoprecipitated with CDK5 antibody followed by IB with DLC1 (top) or CDK5 (bottom) antibodies. (D) The protein complex between DLC1 and p35 was confirmed by reciprocal co-IP. Cell lysates were immunoprecipitated with p35 antibody followed by IB with DLC1 (top) or p35 (bottom) antibodies. (E) Colocalization of endogenous DLC1 with CDK5 in H1703 cells. Cells were stained with DLC1 (red) and CDK5 (green) antibodies. The colocalization of DLC1 and CDK5 is highlighted in white in the merge image. The images are representative of the majority of cells. An averaged overlapping colocalization coefficient ± SD was calculated from 20 cells randomly selected from several fields, and is shown at the bottom of each panel. (F) Colocalization of endogenous DLC1 with CDK5 in H157 cells. (G–I) CDK5 interacts with DLC1, but not with DLC2 or DLC3. (G) Cell extracts were immunoprecipitated with DLC1 or DLC3 antibodies followed by IB with CDK5 antibody. (H) IP reaction was confirmed by IB with DLC1 or DLC3 antibodies (arrow). (I) Lysates from HEK 293T cells transfected with GFP-tagged DLC1, DLC2, or DLC3 were immunoprecipitated with CDK5 antibody followed by IB with GFP antibody (arrow). (J) Expression of various DLC constructs (arrow) for I. See also Fig. S1. Bars, 20 µm.
We also confirmed complex formation and colocalization between DLC1 and CDK5 in non-transformed human bronchial epithelial cells (HBECs; Fig. S1, D–F), human foreskin fibroblasts (Fig. S1, G–I), and human skin cells (Fig. S1, J–L), which indicates that this interaction also occurs in non-transformed cells, implying that it is physiologically relevant. DLC1 is the prototypic member of a three gene family that also includes DLC2 and DLC3 (Durkin et al., 2007; Lukasik et al., 2011). Although they are closely related to DLC1, no interaction was detected between CDK5 and DLC3 (Fig. 1, G–J) or DLC2 (Fig. 1, I and J; and Fig. S2 A) in transfected cells or NSCLC lines in vivo, which indicates that complex formation with CDK5 is exclusive to DLC1.
To examine the influence of CDK5 kinase activity on its interaction with DLC1, the CDK5 inhibitors Olomoucine and Roscovitine (Tripathi et al., 2008; Huber and O’Day, 2012) were used. Both drugs induced a reduction in complex formation between CDK5 and DLC1 (Fig. S2, B and C) and in punctate structures (Fig. S2 D), which implies that CDK5 kinase activity may be required for efficient complex formation.
CDK5 interacts with an 80–200-amino-acid region of DLC1
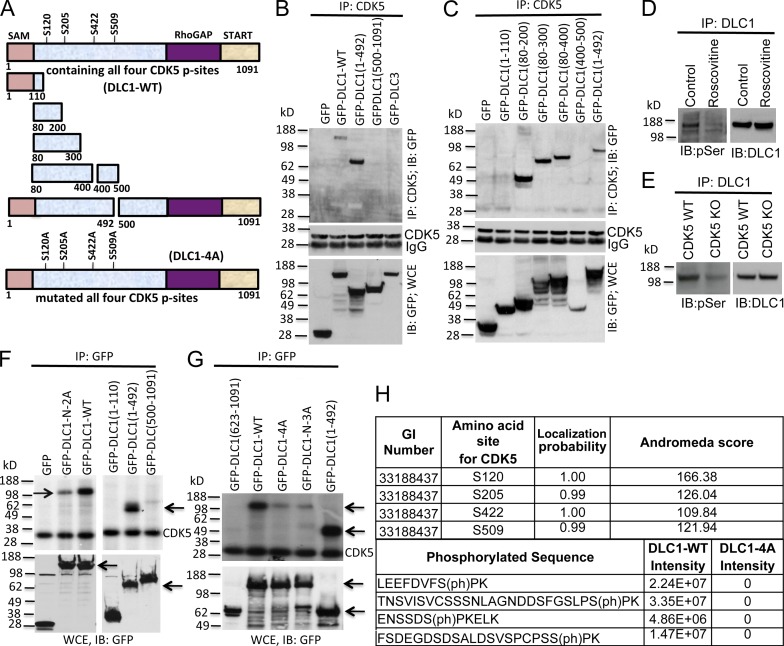
To map the DLC1 amino acids required for CDK5 binding, lysates from HEK 293T cells expressing various GFP-tagged DLC1 fragments (Fig. 2 A) were coimmunoprecipitated with CDK5 antibody followed by immunoblotting with GFP antibody. CDK5 bound the 80–200-amino-acid DLC1 fragment and other fragments that included these amino acids (Fig. 2, B and C). Binding of CDK5 to DLC1(1–110), 400–500, and 500–1,091 was negative (Fig. 2, B and C), which demonstrates that DLC1(80–200) is necessary and sufficient for CDK5 binding.
Figure 2.
Mapping the DLC1 region required for CDK5 binding; four serines in DLC1 are CDK5 substrates. (A) Schematic representation of full-length WT DLC1 with the location of four serines phosphorylated by CDK5 (DLC1-WT), various DLC1 fragments, and DLC1-4A mutant with the four CDK5 serines mutated to alanine. All constructs were GFP tagged. (B) DLC1 amino acids 1–492 are sufficient for complex formation between DLC1 and CDK5. (B, top) Lysates of 293T cells transfected with the indicated DLC1 construct were immunoprecipitated with CDK5 antibody followed by IB with GFP antibody. (B, middle) IP reaction to show the equal IP for CDK5 protein. (B, bottom) Expression of transfected DLC1 constructs. (C) DLC1 amino acids 80–200 are necessary and sufficient for complex formation between DLC1 and CDK5. Experimental conditions and data display are as in B, except that we used smaller DLC1 fragments to map the minimal DLC1 fragment required for CDK5 binding. (D) Roscovitine treatment of H1703 cells reduces serine phosphorylation (pSer) of DLC1 (left) without reducing the level of DLC1 (right). (E) DLC1 from CDK5−/− MEFs has less phosphoserine (pSer) than DLC1 from WT MEFs (left), while the DLC1 level is unchanged (right). (F) In vitro CDK5 kinase assay. (top) Immunoprecipitated DLC1-WT was strongly phosphorylated by recombinant CDK5 (left, top arrow), as detected with 32P autoradiography. DLC1-N-2A mutant (with an S-to-A mutation of S120 and S205) is less strongly phosphorylated (left, top arrow). DLC1(1–492) is strongly phosphorylated (right, bottom arrow). (bottom) Expression of GFP, GFP-DLC1-WT, and GFP-DLC1 fragments. (G, top) In vitro phosphorylation signal by recombinant CDK5 depends on the number of Serines present in WT and mutant DLC1 fragments (top arrow). DLC1-N-3A has S-to-A mutations of S102, S205, and S422. DLC1(1–492) is a positive control (bottom arrow). (G, bottom) Expression of GFP and GFP-tagged DLC1 constructs (top arrow) and DLC1(1–492) (bottom arrow). (H) Phosphorylation of DLC1 by CDK5. DLC1 phosphopeptides were detected by mass spectrometry. All CDK5 phosphorylation sites were fully localized to the indicated serine residues, and were absent from the DLC1-4A mutant protein. See also Figs. S2 and S3.
Four serines in DLC1 are CDK5 substrates
Using in silico analysis of the CDK5 consensus sequence, (S/T)PX(K/H/R), we identified four serines in DLC1 as candidates for phosphorylation by CDK5: S120, S205, S422, and S509 (Fig. 2 A). Experimentally, phosphorylation of some serines in DLC1 was found to depend on CDK5, as determined by the reduction of DLC1 phosphoserine induced by Roscovitine in cells containing WT CDK5 (Fig. 2 D) or by comparing the level of DLC1 phosphoserine in WT MEFs and CDK5-null MEFs (Fig. 2 E). Furthermore, DLC1 is a direct substrate for CDK5, as partially purified full-length transfected DLC1 (Fig. 2 F, left, lane 3) and endogenous DLC1 (Fig. S2 E) were phosphorylated in vitro by recombinant CDK5/p35, while neither DLC2 nor DLC3, which had not formed a complex with CDK5, were phosphorylated under the same conditions (Fig. S2 F).
When DLC1 fragments were analyzed in cells, the N-terminal 492 amino acids (GFP-DLC1(1–492)) were highly phosphorylated (Fig. 2 F, right, lane 2), which is consistent with this fragment containing the amino acids (80–200) found to be sufficient for CDK5 binding and the three most N-terminal candidate serines (S120, S205, and S422). In contrast, a fragment that contained most of the remainder of the DLC1 and had only one candidate serine (S509; GFP-DLC1(500–1,091)) was weakly phosphorylated (Fig. 2 F, right, lane 3), whereas DLC1 fragments lacking the candidate serines, GFP-DLC1(1–110) and GFP-DLC1(623–1,091), were not phosphorylated (Fig. 2 F, right, lane 1; and Fig. 2 G, lane 1).
To determine if the four serines are the major CDK5 sites in DLC1, they were mutated to alanine (GFP-DLC1-4A), resulting in a drastically reduced in vitro phosphorylation signal (Fig. 2 G, lane 3). An analogous attenuation of the signal was seen when the three most N-terminal amino acids were mutated (Fig. 2 G, lane 4; GFP-DLC1-N-3A). In contrast, mutation of the two N-terminal serines to alanine (GFP-DLC1-N-2A) produced a stronger phosphorylation signal (Fig. 2 F, left, lane 2) than GFP-DLC1-N-3A (in which the three N-terminal serines were mutated to alanine), as did the single mutation of each serine (not depicted). Table 1 provides a list of the primers used in this study.
Table 1.
Primers used for engineering DLC1 plasmids
| Primer | Sequence (5′–3′) |
| DLC1-S120A-F | GAGTTTGATGTCTTTGCTCCAAAACAAGAC |
| DLC1-S120A-R | GTCTTGTTTTGGAGCAAAGACATCAAACTC |
| DLC1-S120D-F | GAGTTTGATGTCTTTGATCCAAAACAAGAC |
| DLC1-S120D-R | GTCTTGTTTTGGATCAAAGACATCAAACTC |
| DLC1-S205A-F | GGCAGCCTGCCCGCTCCCAAGGAACTG |
| DLC1-S205A-R | CAGTTCCTTGGGAGCGGGCAGGCTGCC |
| DLC1-S205D-F | GGCAGCCTGCCCGATCCCAAGGAACTG |
| DLC1-S205D-R | CAGTTCCTTGGGATCGGGCAGGCTGCC |
| DLC1-S422A-F | GAAAACAGTAGCGACGCCCCCAAGGAACTG |
| DLC1-S422A-R | CAGTTCCTTGGGGGCGTCGCTACTGTTTTC |
| DLC1-S422D-F | GAAAACAGTAGCGACGATCCCAAGGAACTG |
| DLC1-S422D-R | CAGTTCCTTGGGATCGTCGCTACTGTTTTC |
| DLC1-S509A-F | CCCTGCCCGTCCGCTCCAAAACAGATACAC |
| DLC1-S509A-R | GTGTATCTGTTTTGGAGCGGACGGGCAGGG |
| DLC1-S509D-F | CCCTGCCCGTCCGATCCAAAACAGATACAC |
| DLC1-S509D-R | GTGTATCTGTTTTGGATCGGACGGGCAGGG |
At least some of these serines were also phosphorylated in vivo, as transfected GFP-DLC1-WT had a much stronger anti-phospho-serine signal compared to transfected GFP-DLC1-4A mutant (Fig. S2 G). To confirm that all four serines in DLC1 are phosphorylated in vivo, partially purified GFP-DLC1-WT and GFP-DLC1-4A mutants were analyzed by liquid chromatography–mass spectrometry (LC-MS). In DLC1-WT, phosphorylation of the relevant peptides was specifically and solely detected on S120, S205, S422, and S509, while the DLC1-4A mutant protein was not phosphorylated on these residues (Figs. 2 H and S3).
CDK5 negatively regulates RhoGTP in a DLC1-dependent manner
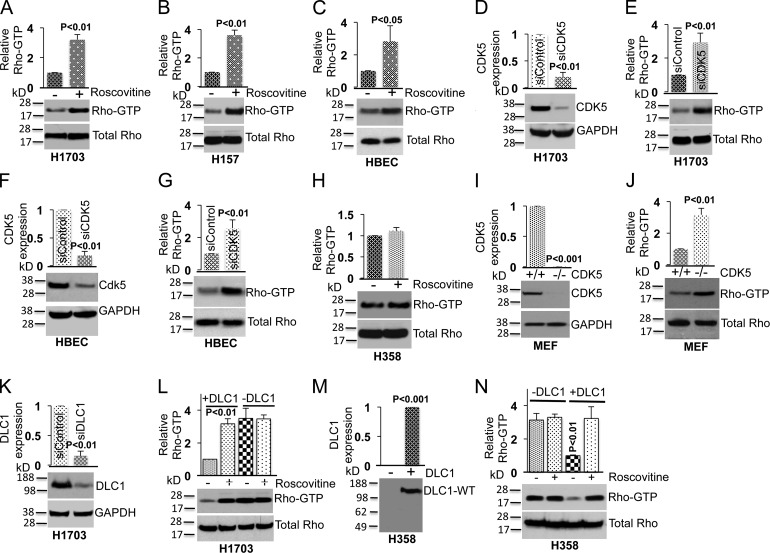
To determine if the CDK5 regulation of RhoGTP was mediated by its effects on DLC1, we studied the effects of endogenous CDK5 in lines that did, or did not, express endogenous DLC1. CDK5 negatively regulated RhoGTP in DLC1-positive NSCLC lines (H1703 and H157) or in DLC1-positive non-transformed lung epithelial HBEC cells, as Roscovitine treatment increased RhoGTP in these lines (Fig. 3, A–C), but not in a DLC1-negative NSCLC H358 line (Fig. 3 H). As an alternate approach, transfection with a CDK5 siRNA was used to reduce the expression of endogenous CDK5 in H1703 and HBEC lines. Knockdown of CDK5 by the siRNA (Fig. 3, D and F) resulted in higher RhoGTP in both lines unlike control cells transfected with a scrambled siRNA (Fig. 3, E and G). Similarly, a higher RhoGTP level was seen in CDK5-null MEFs compared to WT MEFs (Fig. 3, I and J). Thus, CDK5 reduces RhoGTP in cells expressing DLC1.
Figure 3.
CDK5 regulates RhoGTP levels through DLC1. (A) Treatment of a DLC1-positive H1703 line with Roscovitine (10 µM) increases RhoGTP (top) but not total Rho (bottom). The graph represents the relative RhoGTP ± SD from three independent experiments. (B and C) Experimental conditions were similar for H157, which is a second DLC1-positive NSCLC line (B), and for the HBEC cell line, which is an untransformed human lung epithelial cell line (C). (D) Knockdown of CDK5 by siRNA (top); GAPDH was used as a loading control (bottom). The graph represents the relative CDK5 expression ± SD from three independent experiments. (E) Knockdown of CDK5 increases RhoGTP (top) but not total Rho (bottom). The graph is as in D. (F and G) Knockdown of CDK5 expression in HBEC cells leads to an increase in RhoGTP. (H) Roscovitine does not alter RhoGTP in the DLC1-negative H358 NSCLC line. (I) Expression of CDK5 in MEFs cells isolated from WT and CDK5−/− embryos (top). GAPDH was used as loading control (bottom). (J) RhoGTP is higher in CDK5−/− MEFs than in WT MEFs (top). Total RhoGTP is similar in both MEFs (bottom). (K and L) Knockdown of DLC1 in H1703 renders the RhoGTP in the cells unresponsive to Roscovitine. (K) Knockdown of DLC1 expression (top). GAPDH was used as a loading control (bottom). (L) RhoGTP (top) and total Rho (bottom) in DLC1 siRNA– or control siRNA–transfected H1703 cells in the absence or presence of Roscovitine. Knockdown of DLC1 abrogates the ability of Roscovitine to affect the level of RhoGTP. (M and N) Transfection of DLC1 into DLC1-negative H358 cells enables RhoGTP to be regulated by CDK5. (M) Expression of DLC1-WT in stably transfected H358. (N) RhoGTP (top) and total Rho (bottom) in H358 cells, with or without transfected DLC1, incubated in the absence or presence of Roscovitine. Data in each panel are representative of three independent experiments. Error bars indicate ± SD.
To confirm that the observed effects of CDK5 activity on RhoGTP are mediated by DLC1, H1703 cells with endogenous DLC1 were treated with Roscovitine in the absence or presence of DLC1. Although reduced DLC1 expression by siRNA (Fig. 3 K) was associated with an increase in RhoGTP comparable to that of Roscovitine treatment (Fig. 3 L), Roscovitine treatment produced no additional effects on RhoGTP in the cells with reduced DLC1 expression, in contrast to its effects on RhoGTP in the parental cells expressing DLC1 (Fig. 3 L). Furthermore, when DLC1 was transfected into the DLC1-negative H358 cells (Fig. 3 M), it reduced their RhoGTP, and this reduction in RhoGTP was reversed upon treatment with Roscovitine, unlike the parental H358 cells, whose RhoGTP was not altered by Roscovitine (Fig. 3 N). Thus, CDK5 regulates RhoGTP through DLC1.
CDK5-dependent phosphorylation of DLC1 activates its Rho-GAP and other tumor suppressor activities
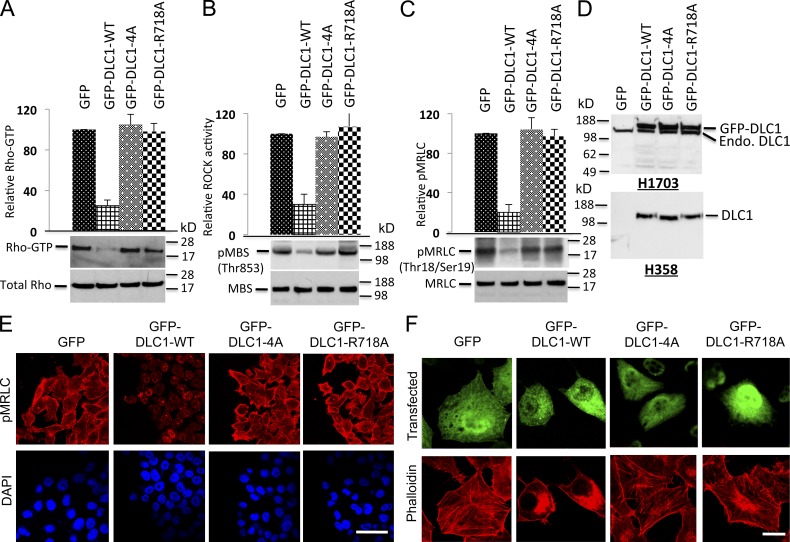
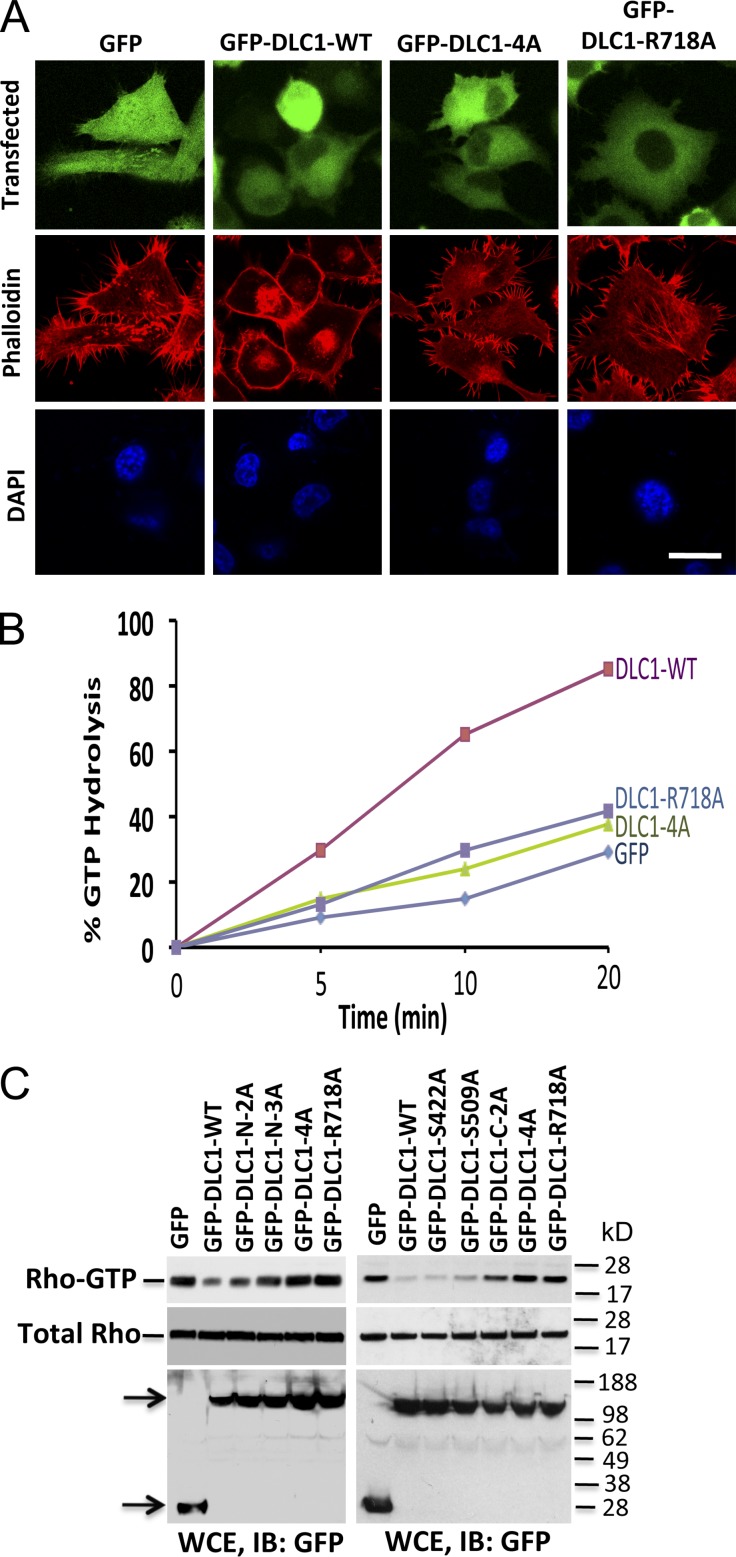
Given the ability of CDK5 to regulate RhoGTP in a DLC1-dependent manner, we examined RhoGTP in stable H1703 and H358 lines expressing similar levels of the various DLC1 S-to-A mutants (Fig. 4, A and D). The RhoGTP levels in DLC1-4A transfectants in H1703 cells were indistinguishable from those of the “GAP-dead” mutant (DLC1-R718A), unlike the reduced RhoGTP level induced by DLC1-WT. Analogous results were seen when the DLC1 mutants were analyzed for in vivo Rho-kinase (ROCK) activity, a major downstream effector of RhoGTP, and phosphorylation of its substrate, myosin regulatory light chain (pMRLC; Fig. 4, B and C). By immunofluorescence, cells transfected with GFP, GFP-DLC1-4A, or the “GAP dead” mutant showed similar strong staining of pMRLC (Fig. 4 E), with well-formed stress fibers (Fig. 4 F for H1703 and Fig. 5 A for H358). In contrast, cells transfected with GFP-DLC1-WT showed less pMRLC staining (Fig. 4 E), fewer stress fibers, and few if any concave boundaries, which is consistent with reduced Rho-ROCK signaling and reduced contraction (Figs. 4 F and 5 A). Similar results were seen when the DLC1 mutants were analyzed for GTP hydrolysis for in vitro Rho-GAP activity (Fig. 5 B). In particular, the Rho-GAP activity of the DLC1-4A mutant was low, and indistinguishable from the “GAP-dead” mutant DLC1-R718A, in contrast to the high Rho-GAP activity of the DLC1-WT.
Figure 4.
DLC1-4A mutant behaves similarly to the “GAP-dead” mutant. (A) The GFP-DLC1-WT–positive control reduced RhoGTP, but the GFP-DLC1-4A mutant was as defective as a “GAP-dead” GFP-DLC1-R718A mutant for negative regulation of RhoGTP. (top) Densitometry quantification ± SD (error bars) of three experiments (P < 0.001) for GFP-DLC1-WT compared with indicated stable transfectants in H1703 cells. The graph represents the RhoGTP/total Rho ratio (n = 3). (B) Experimental conditions and data displays are similar to A but for ROCK activity. (C) Experimental conditions and data displays similar to A but for pMRLC. (D) Expression of stable DLC1 transfectants in H1703 (top) and in H358 cells (bottom). (E) GFP-DLC1-WT transfectants have less pMRLC immunofluorescence and fewer concave boundaries, which is consistent with reduced contraction, unlike the other transfectants. DAPI (blue) represent nuclei. The confocal images are representative of the majority of cells. Bar, 100 µm. (F) DLC1-WT has fewer stress fibers (as measured by red phalloidin), especially in the center region of the cells compared with the other transfectants. GFP (green) fluorescence identifies transfected cells. The confocal images are representative of the majority of cells. Bar, 20 µm.
Figure 5.
Rho-GAP activity of DLC1 is inversely related to the number of S-to-A mutations in DLC1. (A) H358 cells transfected with GFP, GFP-WT-DLC1, GFP-DLC1-4A, or GFP-DLC1-R718A mutants were immunostained with phalloidin (red). Cells transfected with GFP-DLC1-WT show fewer stress fibers. In contrast, cells transfected with GFP-DLC1-4A mutant show almost identical morphology as “GAP-dead” GFP-DLC1-R718A or GFP-negative control for stress fibers formation. DAPI (blue) represent nuclei. The confocal images are representative of the majority of cells. Bar, 20 µm. (B) In vitro GTP hydrolysis of RhoGTP was significantly increased by GFP-DLC1-WT compared to GFP alone, GFP-DLC1-4A, or “GAP-dead” GFP-DLC1-R718A mutant. Unlike the GFP-DLC1-WT–positive control, GFP-DLC1-4A mutant is as defective as “GAP-dead” mutant for Rho-GAP activity. (C) RhoGTP (top) and total Rho (middle) in individual or combined mutants of DLC1 in DLC1-negative A549 cells. The RhoGTP levels are inversely related to the number of S-to-A mutations, and the degree of Rho-GAP reduction is additive for each mutation. Expression of indicated DLC1 constructs was detected by GFP antibody in WCE (bottom). Arrows indicate the expressed proteins.
When the RhoGTP levels for less drastic individual or combined mutants were analyzed in DLC1-negative A549 cells (Fig. 5 C), the RhoGTP levels were inversely related to the number of S-to-A mutations in DLC1. The RhoGTP levels in cells expressing the three most N-terminal S-to-A mutants (GFP-DLC1-N-3A) were almost as high as those with GFP-DLC1-4A, and those of the single mutants were similar to those of GFP-DLC1-WT, while those of the double mutants were intermediate, whether it concerned the two most N-terminal serines (GFP-DLC1-N-2A) or the two most C-terminal serines (GFP-DLC1-C-2A). The simplest interpretation of these results is that the effects of the S-to-A mutations are additive, analogous to what was seen with the phosphorylation results (Fig. 2, F and G).
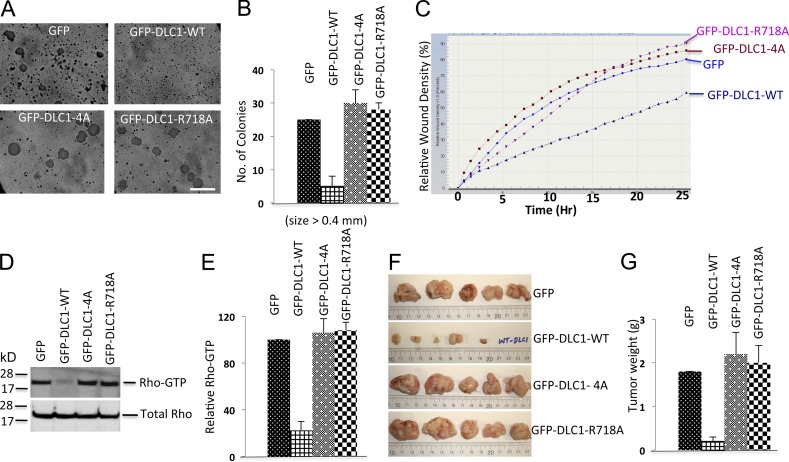
Next, we evaluated the DLC1-4A mutant in several cancer-related bioassays in order to assess the biological significance of DLC1 phosphorylation by CDK5. As with the functional assays, the DLC1-4A mutant was found to be as deficient as the “GAP-dead” DLC1-R718A mutant in these bioassays. Stable transfection of DLC1-WT into the DLC1-negative H358 NSCLC line reduced anchorage-independent growth, as determined by colony formation in soft agar (Fig. 6, A and B); reduced the cell migration rate, as measured by an IncuCyte wound healing assay (Fig. 6 C); reduced the level of RhoGTP (Fig. 6, D and E); and reduced the size of tumors in a mouse xenograft assay (Fig. 6, F and G). In contrast, the DLC1-4A mutant was markedly deficient for these activities in each of these bioassays, similar to that of the “GAP-dead” DLC1-R718A mutant.
Figure 6.
DLC1-4A mutant lacks the Rho-GAP and tumor suppressor activities of DLC1-WT. (A) Anchorage-independent growth of H358 cells stably transfected with GFP, GFP-DLC1-WT, GFP-DLC1-4A, or GFP-DLC1-R718A mutant. Bar, 2 mm. (B) Quantification of agar colonies (>0.4 mm) in the indicated groups from three independent experiments. (C) Relative cell migration rate and wound density (%) were calculated by IncuCyte assay. Multiple scratch wounds were made in confluent cells stably transfected with DLC1 constructs. Scratch wounds were allowed to heal for 24 h, and the cell migration rate was plotted in the graph. (D) RhoGTP (top) and total Rho (bottom) in stably transfected H358 cells expressing the indicated DLC1 constructs. (E) Immunoblots for RhoGTP and total Rho as in D were quantified by densitometry, and the ratio of RhoGTP to total Rho was normalized. Statistical analysis demonstrated a significant decrease (P < 0.001) in RhoGTP/total Rho in DLC1-WT–transfected cells compared to GFP control, DLC1-4A–transfected, or DLC1-R718A–transfected cells. The graph represent means ± SD of three independent experiments. (F) Xenographic tumors from nude mice after injecting H358 cells, which stably expressed GFP, DLC1-WT, DLC1-4A, or DLC1-R718A mutant. Tumors were excised after 6 wk, and representative tumors are shown. (G) Average tumor weight ± SD (g) for each group is shown in graph. Unlike the stable DLC1-WT–positive control in H358 cells, the stable DLC1-4A mutant is as defective as a “GAP-dead” DLC1-R718A mutant for regulating anchorage-independent growth, cell migration rate, RhoGTP, and xenographic tumor growth in nude mice. Error bars indicate ± SD.
CDK5 activates DLC1 Rho-GAP by reducing an autoinhibitory interaction
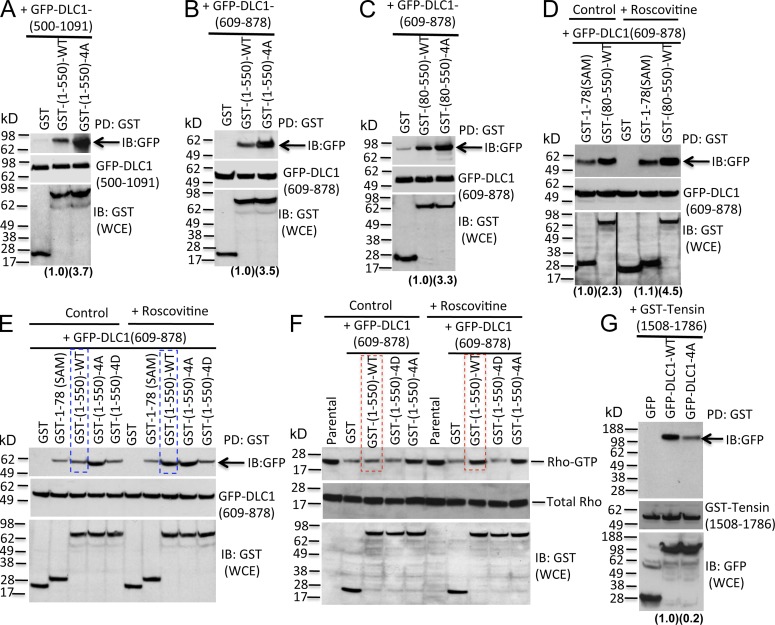
Given that some N-terminally truncated DLC1 mutants have greater Rho-GAP activity than full-length DLC1 (Healy et al., 2008), we speculated that the CDK5 phosphorylation of the four serines, which are N-terminal to the Rho-GAP domain, might regulate a putative interaction between the Rho-GAP domain and N-terminal DLC1 sequences. In an initial test of this hypothesis, we cotransfected a C-terminal DLC1 fragment (GFP-DLC1(500–1,091)), which includes the Rho-GAP domain, together with a WT N-terminal DLC1 fragment (GST-(1–550)-WT) or the 4A mutant version of this fragment (GST-(1–550)-4A). GST pull-down followed by IB with GFP antibody indicated that GST-(1–550)-WT formed a complex with GFP-DLC1(500–1,091), but the cotransfected GST-(1–550)-4A mutant fragment bound more efficiently (Fig. 7 A).
Figure 7.
CDK5 activates the Rho-GAP activity of DLC1 by phosphorylating the four DLC1 Serines, which reduces an autoinhibitory interaction between the Rho-GAP domain and the region of DLC1 containing the four Serines. (A–E) Top panels: Pull-down of GST-tagged DLC1 fragments by GST antibody, followed by IB with GFP antibody (arrows), to measure binding between an N-terminal GST-tagged DLC1 fragment and a C-terminal GFP-tagged DLC1 fragment cotransfected into DLC1-negative A549 cells. Compared with GST-(1–550)-WT, GST-(1–550)-4A mutant binds more efficiently with GFP-DLC1-(500–1,091) (A) and the Rho-GAP domain GFP-DLC1-(609–878) (B, top). Relative binding was calculated by densitometry, and is shown at the bottom of each panel in parentheses. Middle panels show expression of GFP-DLC1 constructs in WCE. Bottom panels show GST-DLC1 polypeptide in WCE. (C) Similar results are seen when binding with isolated Rho-GAP domain GFP-DCL1-(609–878) is compared for the GST-DLC1(80–550)-WT (which lacks the DLC1 SAM domain) and the GST-DLC1(80–550)-4A mutant. (D) Roscovitine treatment increases binding between GST-DLC1(80–550)-WT and the Rho-GAP domain [GFP-DLC1(609–878)], but not binding between DLC1 SAM domain (GST-1–78 (SAM)) and the Rho-GAP domain. The black line indicates the removal of an intervening lane for presentation purposes. (E) Without Roscovitine treatment, GST-(1–550)-WT and GST-(1–550)-4D mutant bind inefficiently to the Rho-GAP domain, while GST-(1–550)-4A binds more efficiently (top, left side). Roscovitine treatment increases binding for GST-(1–550)-WT to the Rho-GAP domain (compare lanes outlined in blue), but not the other constructs (right). (F) Binding efficiency between the N-terminal 550 DLC1 amino acids and the Rho-GAP domain is associated with a concomitant increase in RhoGTP. (top) RhoGTP; (middle) total Rho in WCE; (bottom) expression of indicated DLC1 constructs in WCE. Without Roscovitine, GST-(1–550)-WT and GST-(1–550)-4D do not inhibit the Rho-GAP activity of the Rho-GAP domain, whereas GST-(1–550)-4A does inhibit the activity (left). With Roscovitine, the GST-(1–550)-WT does inhibit the Rho-GAP activity of the Rho-GAP domain (compare lanes outlined in red), but Roscovitine does not influence the other constructs. (G) GST-tensin(1,508–1,786) binds more efficiently to GFP-DLC1-WT than to GFP-DLC1-4A mutant. IB with GFP (middle) or GST (bottom) antibodies represent the expression of the DLC1 constructs in WCE. Data are representative of three independent experiments. See also Fig. S4.
The interaction with the C-terminal fragment is attributable to the Rho-GAP domain, as a fragment composed of this domain (amino acids 609–878) bound efficiently (Fig. 7 B), while fragments composed of residues 899–996 and 996–1,091 did not bind (Fig. S4, A and B). The interaction with the N-terminal fragment does not require the SAM domain (residues 1–78), as DLC1(80–550)-4A mutant bound the Rho-GAP domain more efficiently than did the SAM-deleted WT fragment DLC1(80–550)-WT (Fig. 7 C), and binding to the WT fragment was increased by the CDK5 inhibitor Roscovitine (Fig. 7 D). The effect of Roscovitine on the binding of the Rho-GAP domain depended on the presence of the four serines in the N-terminal fragment, as Roscovitine treatment did not influence the binding between the Rho-GAP domain and GST-(1–550)-4A or GST-(1–550)-4D (a phosphomimetic mutant that bound somewhat less efficiently than GST-(1–550)-WT; Fig. S4 C), but it did increase the binding of the GST-(1–550)-WT (Fig. 7 E).
The above observations suggested that, compared with the WT DLC1 N terminus, the 4A mutant would interfere more efficiently with the reduced RhoGTP induced by the Rho-GAP domain. Consistent with this prediction, cotransfection of the Rho-GAP domain with the GST-(1–550)-4A mutant induced an increase in RhoGTP level, compared with the GST control, whereas GST-(1–550)-WT and GST-(1–550)-4D, which had not bound efficiently to the Rho-GAP domain, had little inhibitory effect on RhoGTP compared with the GST control (Fig. 7 F, left control panel). To examine whether CDK5 activity was responsible for the limited effect of the WT N terminus on RhoGTP, the cells were treated with Roscovitine, leading to an increase in RhoGTP level (Fig. 7 F, right; Roscovitine treated). As had been true of the binding to the Rho-GAP domain (Fig. 7 E), Roscovitine had no effect on RhoGTP in cells expressing the 4A or 4D mutants, which are insensitive to CKD5 (Fig. 7 F, right; Roscovitine treated).
The inactive closed conformation of DLC1 interferes with tensin, talin binding, and localization to focal adhesions
The closed conformation induced by strong binding of the non-phosphorylated N terminus of DLC1 to the Rho-GAP domain suggested that this conformation might also interfere with the binding of talin and tensin to DLC1 (Yam et al., 2006; Liao et al., 2007; Qian et al., 2007; Li et al., 2011), as tensin binds tyrosine-442, whereas talin (and FAK) binds an LD-like motif that spans residues 469–476. Consistent with this possibility, the relevant regions of tensin and talin interacted with DLC1 less efficiently when cotransfected with the full-length DLC1-4A mutant compared to DLC1-WT (Figs. 7 G and S4 D). To verify that the effects on binding tensin and talin depended on the presence of sequences C-terminal to DLC1 amino acid 550, we confirmed that DLC1 amino acids 80–550, which are in an “open” conformation because they are not bound to the Rho-GAP domain, bound tensin and talin with similar efficiency whether the four serines were WT or the 4A mutant (Fig. S4, E and F).
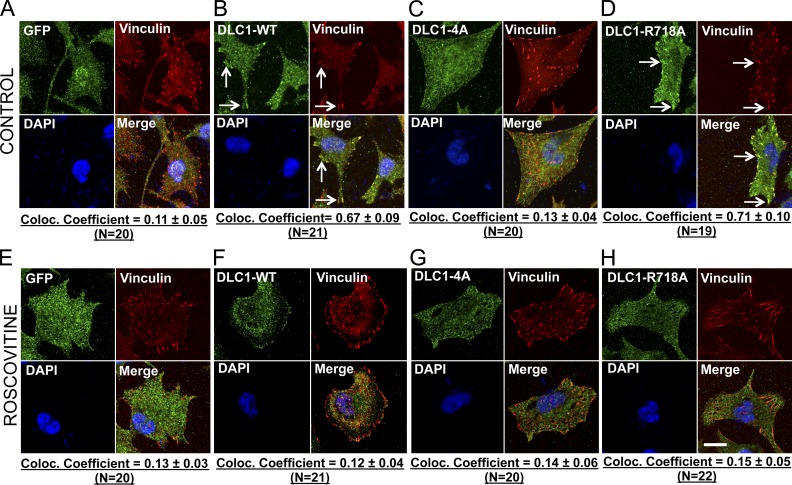
These results suggested that the closed conformation of DLC1 might also affect its subcellular localization, as some investigators have designated sequences located between the SAM and Rho-GAP domains as the focal adhesion targeting (FAT) domain (Liao et al., 2007). Indeed, unlike DLC1-WT or DLC1-R718A mutant (Fig. 8, B and D), the DLC1-4A mutant did not colocalize with the focal adhesions (Fig. 8 C). Remarkably, a similar DLC1-4A phenotype was observed when cells with DLC1-WT or DLC1-R718A mutant were treated with the Roscovitine (Fig. 8, F and H). However, as expected, Roscovitine did not influence the localization of the GFP control or the DLC1-4A mutant (Fig. 8, E and G).
Figure 8.
The closed conformation of DLC1-4A affects its subcellular localization. (A–D) Colocalization of GFP-tagged DLC1 constructs (green) with the focal adhesion protein Vinculin (red). Unlike the stable GFP control in H1703 cells (A), the stable DLC1-WT (B) and “GAP-dead” DLC1-R718A (D) efficiently colocalize to focal adhesions. In contrast, DLC1-4A mutant poorly colocalizes to focal adhesions (C). (E–H) Experimental conditions were similar to as above, except that cells were treated with Roscovitine. Treatment with Roscovitine changes the subcellular localization of DLC1-WT and “GAP-dead” DLC1-R718A proteins away from focal adhesions so that they no longer colocalize with focal adhesions, which appears similar to DLC1-4A mutant protein. The images are representative of a majority of cells. An averaged overlapping colocalization coefficient ± SD was calculated from 18–22 cells per condition randomly selected from several fields, and is shown at the bottom of each panel. Bar, 20 µm.
CDK5 is more pro-oncogenic when DLC1 expression is low
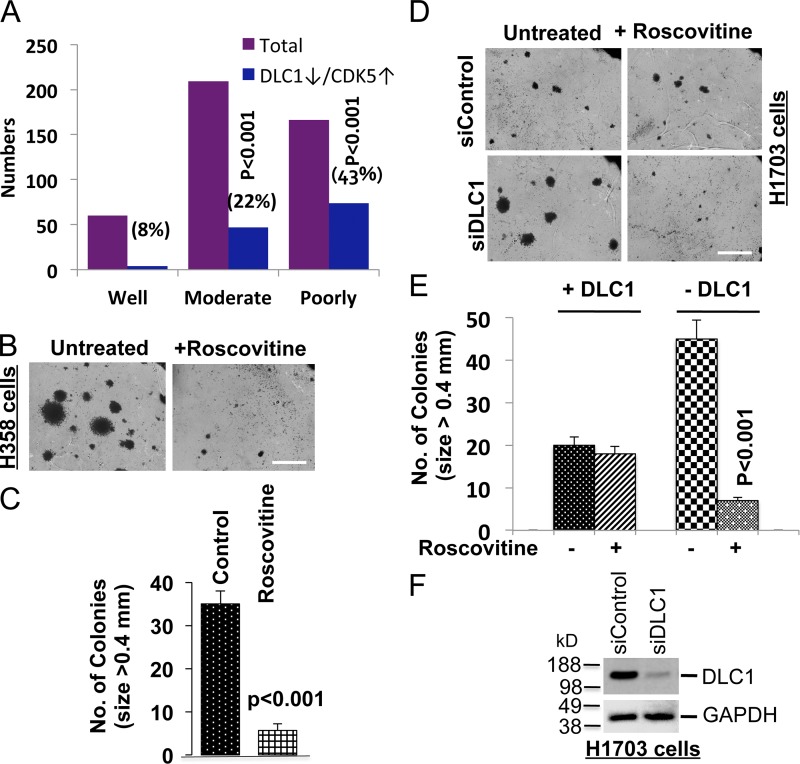
One way to reconcile the high CDK5 levels found in some human tumors, including NSCLC, with the findings reported here would be to assume that cancer progression was associated with the combination of low DLC1 and high CDK5 expression. Indeed, examination of an NSCLC dataset that contains mRNA expression profiles of CDK5 and DLC1 (Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma et al., 2008) indicated an inverse relationship between the degree of tumor differentiation and the combination of low DLC1 and high CDK5 expression. Although only 8% of well-differentiated tumors had this combination, it was found in 22% of moderately differentiated and 43% of poorly differentiated tumors (P < 0.001; Fig. 9 A).
Figure 9.
CDK5 stimulates tumor suppressor activities of DLC1, and is pro-oncogenic when the DLC1 level is reduced. (A) Analysis of a Jacob dataset of 442 human NSCLC cases that were well, moderately, or poorly differentiated. The graph represents the number and percentage of cases in each differentiation category that had both high CDK5 and low DLC1 mRNA expression. (B) Anchorage-independent growth of DLC1-negative H358 cells in the absence and presence of Roscovitine. Roscovitine treatment inhibits colony formation and growth in soft agar. Bar, 2 mm. (C) Quantification of agar colonies (>0.4 mm) in the Roscovitine-treated and untreated group from three independent experiments as shown in B. (D) Anchorage-independent growth of H1703 cells in the absence and presence of DLC1 and with or without Roscovitine treatment. Roscovitine does not inhibit anchorage-independent growth of parental H1703 cells, which express endogenous DLC1, but does inhibit growth when DLC1 expression is knocked down. Bar, 2 mm. (E) Quantification of agar colonies in the presence and absence of DLC1, and with and without Roscovitine treatment. Roscovitine treatment of DLC1-positive H1703 parental cells did not alter the number of soft agar colonies >0.4 mm, but it reduced the number of colonies after siRNA knockdown of endogenous DLC1. n = 3. (F) Knockdown of DLC1 expression by DLC1 siRNA (top). GAPDH was used as a loading control (bottom). Error bars indicate ± SD.
These findings suggested that CDK5 might promote neoplastic growth when DLC1 expression was low. Remarkably, Roscovitine was found to reduce anchorage-independent growth in DLC1-negative H358 cells (Fig. 9, B and C), but not in DLC1-positive H1703 cells (Fig. 9 D, compare upper panels; and Fig. 9 E). Furthermore, when DLC1 was knocked down by siRNA in H1703 (Fig. 9 F), it led, as expected, to an increase in the size (Fig. 9 D, lower left panel) and number of colonies (Fig 9 E); more informatively, Roscovitine now reduced the size (Fig. 9 D, lower right panel) and number of colonies (Fig. 9 E), in contrast to when the cells expressed DLC1. Thus, knockdown of DLC1 increased the pro-oncogenic effects of CDK5.
Discussion
In this study, we have identified CDK5 as a major regulator of DLC1 activities. In the absence of active CDK5, DLC1 has low Rho-GAP activity, low binding of tensin and talin, and does not colocalize to focal adhesions. These conditions are associated with high-RhoGTP, high-Rho/ROCK–dependent activities, such as contraction and well-formed stress fibers, and an attenuated tumor suppressor activity of DLC1, including efficient cell migration, anchorage-independent growth, and tumor formation. In contrast, high CDK5 activity resulted in the opposite phenotypes. Although the negative regulation of RhoGTP by CDK5 was readily seen in both transformed and non-transformed cells that express DLC1, this function was absent in DLC1-negative cells.
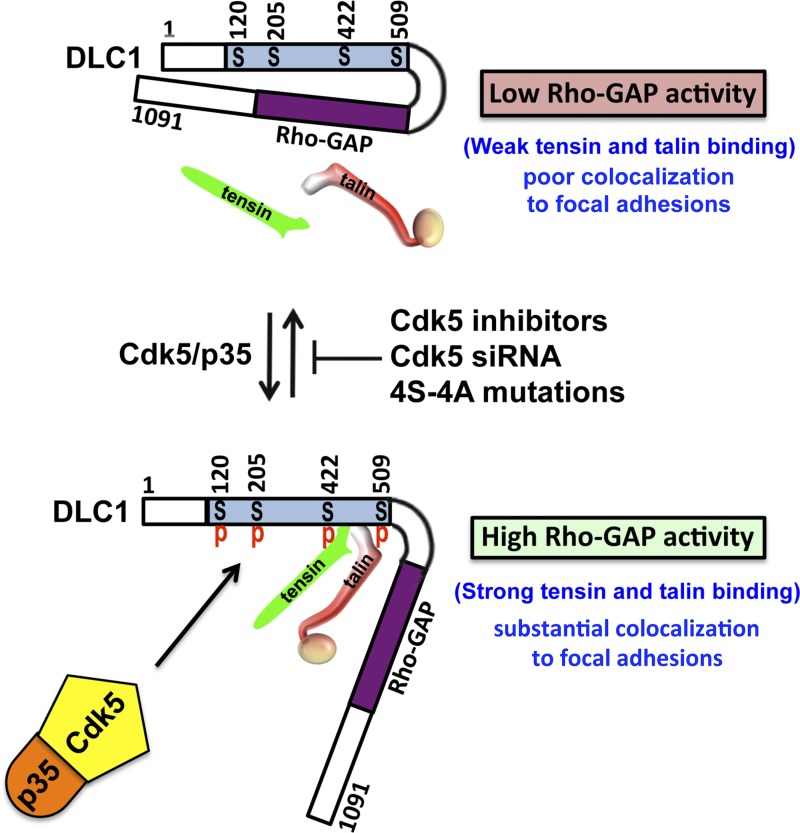
The observed attenuation of several DLC1 activities when CDK5 is enzymatically inactive suggested that this phenotype might result from a closed, autoinhibitory conformation, whereas high CDK5 activity would induce an open conformation with high Rho-GAP activity, high tensin and talin binding, and colocalization to focal adhesions (Fig. 10). Our findings support this model of coordinate regulation of DLC1 by CDK5. Complex formation between CDK5 and DLC1 depended on the CDK5 activator p35 being part of the complex and on CDK5 being enzymatically active. We identified four serines (S120, S205, S422, and S509) in DLC1 that are direct CDK5 substrates in vitro and that are phosphorylated in vivo in a CDK5-dependent manner. The N-terminal DLC1 region with the four serines binds to the Rho-GAP domain. Furthermore, the binding efficiency was much higher in the absence of CDK5 kinase activity or when all four of the serines phosphorylated by CDK5 were mutated to alanine, compared with its binding when the serines were phosphorylated or mutated to the phosphomimetic aspartate. Remarkably, the Rho-GAP and biological activities of full-length DLC1 harboring the four serine-to-alanine mutations were similar to that of a “GAP-dead” DLC1 mutant, as was also the case when CDK5 activity was inhibited pharmacologically or genetically. Single alanine mutants of each serine resulted in activities close to those of the WT, while each additional serine to alanine mutation appeared to result in a mutant with progressively increased binding efficiency and reduced Rho-GAP activity. These results suggest that the electrostatic charge resulting from phosphorylation of the serines may be the principal mechanism by which this posttranslational modification leads to a reduced interaction with the Rho-GAP domain.
Figure 10.
Model of the regulation of DLC1 by CDK5. The upper part of model shows that DLC1 has a closed conformation with low Rho-GAP activity, weak tensin, and talin binding, as well as poor colocalization to focal adhesions, in the absence of Serine phosphorylation (S120, S205, S442, and S509) by CDK5. The lower part of the model shows that DLC1 has an open conformation with high Rho-GAP activity, strong tensin, and talin binding, and substantial colocalization to focal adhesions, in the presence of Serine phosphorylation by CDK5. The middle part of model indicates that activation of DLC1 can be inhibited by CDK5 inhibitors, CDK5 siRNA, or 4S-4A mutations of DLC1.
Our previous analysis of DLC1 mutants deficient for binding tensin, talin, or both ligands have indicated that the non-overlapping binding sites for both proteins in DLC1 make independent contributions to the localization of DLC1 to focal adhesions (Li et al., 2011). The CDK5-dependent conformational change in DLC1 described here provides a physiologic mechanism for regulating the efficiency with which tensin and talin bind DLC1, thus contributing to whether DLC1 does, or does not, colocalize with focal adhesions. This localization is independent of the Rho-GAP activity, as the “GAP-dead” mutant is found at the focal adhesions.
The CDK5-dependent activation of DLC1 described here differs from a mechanism proposed from the analysis of the non-transformed MCF10A cells (Katz et al., 2007; Cao et al., 2012), a phenomenon that occurs in response to relatively long-term EGF stimulation, and depends on which member of the tensin gene family is bound to DLC1. If tensin-3 is bound, DLC1 Rho-GAP activity is high, whereas if Cten, which lacks the N-terminal actin-binding sequences present in tensin-3, is bound, DLC1 Rho-GAP activity is low. We speculate that this result depends on the binding of the unique tensin-3 sequences to the SAM domain of DLC1, which in turn may decrease a putative binding of the SAM domain to the Rho-GAP domain of DLC1. In contrast, the CDK5 mechanism reported here is independent of the SAM domain, does not require EGF stimulation, and does not seem to depend on which tensin molecule binds DLC1. When we analyzed MCF10A cells, we found that the basal activity of CDK5 was very low, as determined by the low expression of its p35 activator (Fig. S5 A) and low phosphorylation of tyrosine-15 in CDK5 (Fig. S5, A–C), a finding that was correlated with low expression of c-Abl (Fig. S5 C), which stimulates the CDK5 kinase activity by phosphorylating CDK5 tyrosine-15 (Zukerberg et al., 2000). siRNA-induced depletion of tensin-3 in multiple lines did not result in a substantial change in RhoGTP (Fig. S5 D). Thus far, MCF10A is the only cell line we have examined in which CDK5 is not active and, therefore, in which the CDK5 mechanism we describe here is not seen. In contrast, we have observed the CDK5 mechanism in several lung and breast cancer cell lines, as well as in non-transformed MEFs and a human HBEC line (Fig. 3).
Although our studies indicate that CDK5 promotes the Rho-GAP and tumor suppressor activities of DLC1, which is consistent with the ability of CDK5 to stimulate differentiation of normal cells (Zhang et al., 2002; Yang et al., 2013), CDK5 expression is reported to be increased in some cancers, including NSCLC and breast cancer, where it may affect prognosis (Liu et al., 2011; Levacque et al., 2012) and promote neoplastic cell growth (Feldmann et al., 2010). These observations suggest that CDK5 has context-dependent anti-oncogenic and pro-oncogenic activities, analogous to other genes, such as TGF-β (Massagué, 2012).
CDK5 targets many activities (Contreras-Vallejos et al., 2012), which implies that its pro-oncogenic phenotype in cancer would arise from an increase in the pro-oncogenic pathways it targets and/or a decrease in the anti-oncogenic pathways it targets. The pro-oncogenic targets of CDK5 include Ral (Feldmann et al., 2010) and FAK (Park et al., 2009), while its anti-oncogenic targets include the tumor suppressors p53 (Ajay et al., 2010) and Dab1 (Sato et al., 2007). In addition, CDK5 expression in cancer may promote cell cycle progression; pRB inactivation is one mechanism (Pozo et al., 2013). Here we demonstrate directly that down-regulation of DLC1, which occurs frequently in NSCLC and other human tumors presumably because of selection against its tumor suppressor function, can contribute to the pro-oncogenic activity of CDK5. This observation appears to be biologically relevant, as a substantially higher percentage of poorly differentiated NSCLC have low DLC1 expression together with high CDK5 expression compared with well-differentiated NSCLC (43% vs. 8%, respectively; P < 0.001).
One intriguing difference between DLC1 and the other two DLC family members is that DLC1 knockout mice are embryonic lethal (Durkin et al., 2005; Sabbir et al., 2010), whereas DLC2 and DLC3 knockouts are viable (Yau et al., 2009; Lin et al., 2010; Sanger Institute Mouse Resources Portal colony prefix MEGB). However, most activities thus far identified in one DLC family member seem to be shared qualitatively by the other members, although eukaryotic elongation factor-1A-1 (EF1A1) is reported to bind the SAM domain of DLC1, but not DLC2, and to increase the ability of DLC1 to suppress cell migration (Zhong et al., 2009). In contrast, we have found that although CDK5 strongly activates DLC1, the four DLC1 serines phosphorylated by CDK5 are not conserved in DLC2 and DLC3, and CDK5 does not regulate DLC2 or DLC3. We speculate that the effects of CDK5 on DLC1 may contribute to the stronger biological phenotype of DLC1 compared with that of DLC2 and DLC3.
In summary, we have determined that CDK5 is a major regulator of DLC1 activity. It negatively regulates Rho activity in both untransformed and transformed cells, and this regulation is attributable to CDK5 stimulating the Rho-GAP activity of DLC1, which occurs after the CDK5-dependent phosphorylation of several serines in DLC1 located between the SAM and Rho-GAP domains. Phosphorylation of DLC1 by CDK5 changes DLC1 from a closed conformation to an open one, which facilitates tensin and talin binding and its localization to focal adhesions, in addition to activating its Rho-GAP. Inactivation of DLC1 in tumors can contribute to the pro-oncogenic function of CDK5, and these observations are relevant to NSCLC.
Materials and methods
Plasmid constructs
The GFP-tagged WT DLC1 (GFP-DLC1-WT), the GAP-dead mutant of DLC1 (GFP-DLC1-R718A), and fragments encoding DLC1 residues 1–492 (GFP-DLC1(1–492)), 500–1,091 (GFP-DLC1(500–1,091)), and GFP-DLC3 were constructed by PCR and subcloned into a modified pEGFP-C1 vector (Takara Bio Inc.) through Kpn1–NotI sites, as described previously (Qian et al., 2007). The GFP-DLC2 construct was a gift from M. Mowat (Manitoba Institute of Cell Biology, Winnipeg, Manitoba, Canada). GFP-tagged full-length Dlc2 cDNA was cloned in PEGFP-C1 (Promega) vector using Xho1 and Sma1 restriction enzymes. PEGFP-C1 vector uses a CMV promoter. DH5-α stain was used for cloning this plasmid. A series of individual and compound serine-to-alanine (S-A) and serine-to-aspartate (S-D) mutations were introduced into full-length DLC1 and in the fragments encoding GST-DLC1(1–550) and GST-DLC1(80–550) using a site-directed mutagenesis kit (Agilent Technologies) or by overlapping PCR for a small DNA fragment followed by subcloning into PEGFP-DLC1 vector. The DLC1(1–550) and SAM domain–deleted DLC1(80–550) fragments with or without 4A mutations were engineered into the PEBG vector by BamHI and NotI, resulting in GST-tagged DLC1 constructs. The other GFP- or GST-tagged DLC1 fragments were made by PCR cloning into the PEGFP-C1 vector or PEBG vector, respectively. All PCR regions were confirmed by sequencing to verify their accuracy.
Antibodies and fluorescent probes
CDK5 mouse monoclonal (J-3; sc-6247), CDK5 rabbit polyclonal (C-8; sc-173), p35 rabbit polyclonal (C-19; sc-820), phospho-specific CDK5-pY15 rabbit polyclonal (sc-12918-R), DLC2/STARD13 goat polyclonal (sc-67843), and c-ABL rabbit polyclonal (sc-131) antibodies were purchased from Santa Cruz Biotechnology, Inc. DLC3/STARD8 rabbit polyclonal antibody (13899-1-AP) was obtained from Proteintech. Two antibodies for DLC1 from different sources were used in this study to confirm the findings. One was generated in our laboratory (DLC1 antibody; clone 428), as described previously (Li et al., 2011), and other commercial mouse monoclonal DLC1 (612021) was purchased from BD. To make the anti–DLC1-specific antibody (clone 428), the cDNA encoding a human DLC1 polypeptide (amino acids 82–251) was subcloned into the bacterial expression vector PGEX-4T-1 (Pharmacia) using EcoRI and XhoI, and its encoded GST fusion protein was induced by IPTG in bacteria, purified by a Glutathione Sepharose 4B column, and used to immunize rabbits. Antibodies were purified by peptide affinity chromatography. MRLC mouse monoclonal (ab11082-100), GFP mouse monoclonal (ab1218), and GFP rabbit polyclonal (ab290) were purchased from Abcam. pMRLC goat polyclonal (sc-12896) and phosphorylated Thr18/Ser19 pMRLC rabbit polyclonal (3674S) were purchased from Santa Cruz Biotechnology and, Inc. Cell Signaling Technology, respectively. The specificity of the pMRLC antibody was confirmed by using blocking peptide. Tensin3 rabbit polyclonal (SAB4200204) was from Sigma-Aldrich, GAPDH (14C10) rabbit monoclonal (2118) and p39 rabbit polyclonal (3275S) were from Cell Signaling Technology, and phospho-serine mouse monoclonal antibody (612547) was from BD. Anti–rabbit and anti–mouse IgG horseradish peroxidase–linked secondary antibodies were from GE Healthcare. Alexa Fluor 568–goat anti–rabbit IgG, Alexa Fluor 488–donkey anti–mouse IgG, Alexa Fluor 488–phalloidin, Alexa Fluor 568–phalloidin, and DAPI were from Invitrogen.
Cell lines, culture conditions, and transfection
HEK 293T (293T), MCF10A, and human skin fibroblastic cells were cultured in DMEM supplemented with 10% FBS. NSCLC cell lines with endogenous DLC1 expression (H1703 and H157) or undetectable DLC1 expression (A549 and H358; a gift from C. Harris, National Cancer Institute [NCI], National Institutes of Health) were cultured in RPMI-1640 supplemented with 10% FBS. HBEC cells were purchased from Cell Applications, Inc., and cultured in bronchial/tracheal epithelial cell growth medium provided by Cell Applications, Inc. All cells were cultured at 37°C in a humidified atmosphere of 95% air/5% CO2. Where indicated, transient transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions and cultured for 48 h before use. Stable clones expressing GFP, GFP-DLC1-WT, GFP-DLC1-4A, and GFP-DLC1-R718A were made by transfection of H1703 or H358 cells with Lipofectamine 2000 followed by selection with G418 (0.9 µg/ml).
siRNA transfection and pharmacologic inhibitors
To suppress the expression of DLC1, CDK5, or the other indicated genes, cells were transfected with 160 nM of the appropriate antisense oligonucleotides or with scrambled control siRNA, and were harvested after 48 h of transfection. Suppression of protein expression was confirmed by immunoblotting with the appropriate antibodies, with at least two different siRNAs in each case. Validated siRNAs for human DLC1 (siRNA-5 and siRNA-11) and human tensin 3 (FlexiTube GeneSolution GS64759 and SI00134372) were from QIAGEN. Validated siRNAs for human CDK5 were from QIAGEN (FlexiTube GeneSolution GS1020) and from Santa Cruz Biotechnology, Inc. (sc-29263). Negative control siRNAs (Control siRNA 1, catalogue no. 1027280; and Control siRNA 2, catalogue no. 1027310) were procured from QIAGEN. For inhibition of CDK5, kinase activity cells were treated with CDK5 inhibitor Roscovitine (10 µM) or Olomoucine (15 µM), which were purchased from EMD Millipore, for 24 h.
In vitro CDK5 kinase assay
Lysates from transfected cells were immunoprecipitated with GFP antibody and immunopellets were sequentially washed once with high-salt HNTG buffer (20 mM Hepes, 500 mM NaCl, 0.1% Triton X-100, and 10% glycerol), twice with low-salt HNTG buffer (20 mM Hepes, 150 mM NaCl, 0.1% Triton X-100, and 10% glycerol), and once with kinase reaction buffer (35 mM Hepes, pH 7.4, 10 mM MgCl2, 1 mM EGTA, 1% Tween 20, 0.1 mM sodium vanadate, and 1 mM DTT). The kinase reaction was carried out in 30 µl of reaction buffer containing 15 µM cold ATP, 2.5 µCi [32P]γ-ATP, and 100 ng of recombinant active CDK5–p35 complex (EMD Millipore) at 30°C for 45 min. The reaction was stopped by adding 10 µl of 4× Laemmli sample buffer and heating at 95°C for 5 min. Proteins were separated by gel electrophoresis and autoradiographed to detect 32P incorporation.
RhoGTP (Rhotekin-RBD pull-down) assay
Measurement of GTP-bound Rho was assessed using the Rho activation assay kit (EMD Millipore). In brief, cells were lysed in ice-cold lysis buffer (50 mM Tris, pH 7.2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 500 mM NaCl, and 10 mM MgCl2) containing a protease inhibitor mixture (Roche). Equal amounts (500 µg) of each cell lysate were incubated with 30 µg of GST-Rhotekin Rho-binding domain coupled to glutathione-agarose beads on a rocker platform at 4°C for 45 min. After incubation, beads were washed three times with the washing buffer (50 mM Tris-HCl, pH 7.2, 150 mM NaCl, 10 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride, and 1% Triton X-100). Bound RhoGTP was eluted with Laemmli’s SDS-sample buffer (BP-110R; Boston BioProducts) containing DTT and boiled for 5 min. The samples were subjected to 4–12% SDS-PAGE, transferred onto nitrocellulose membranes (Invitrogen), and detected by IB, using Rho antibody (clone 55; catalogue no. 05-778; EMD Millipore).
ROCK assay
Cells were fixed and harvested in 10% trichloroacetic acid containing 10 mM DDT. Pellets were dissolved in 10 µl of 1 M Tris base and mixed with 100 µl of extraction buffer (8 M urea, 2% SDS, 5% sucrose, and 5% 2-mercaptoethanol). Equal amounts of protein from each cell extract were subjected to 10% SDS-PAGE, transferred onto nitrocellulose membranes, and incubated with antibody specific for phospho-myosin binding subunit (phospho-Thr853-MBS) or myosin binding subunit (MBS), and bands were visualized by enhanced chemiluminescence. ROCK activity was expressed as the ratio of phospho-MBS to total MBS.
Rho-GAP activity assay
GFP-tagged DLC1 constructs were purified by IP using GFP rabbit polyclonal (ab290) antibody from transfected cells using a high-stringency buffer (20 mM Tris-HCl, pH 8.0, 100 mM NaCl, 5 mM MgCl2, NP-40 [0.5%], 1 mM DTT, and protease and phosphatase inhibitor). Highly purified Rho was bound to γ-labeled [P32]GTP. The GTPase accelerating activity (Rho-GAP activity) of various DLC1 mutants at each time point was determined by incubating with GTP-bound Rho at 18°C with shaking while removing samples at the indicated time points. The guanidine nucleotides were separated by chromatography on cellulose filter paper dissolved in buffer. We then measured the γ-P32 signal, and the net GTP hydrolysis was calculated.
Mouse embryo fibroblasts
CDK5−/− mice are nonviable, and die either during embryogenesis or perinatally (Ohshima et al., 1996). Genomic clones of the mouse Cdk5 gene were isolated from the 129/SvJ mouse genomic library, and the gene structure was characterized. These genomic fragments were used to construct the targeting vector, which carried a 0.8-kb deletion including a part of exon III, and exons IV and V. This deletion region was replaced by a neomycin resistance gene as a positive selection marker. Embryonic stem cells were electroporated with the targeting vector DNA, and, subsequently, 238 resistant clones were isolated and screened. Selected clones were injected into blastocysts to generate −/− chimeric mice (Ohshima et al., 1996). To obtain CDK5−/− MEFs, CDK5+/− mice were interbred and the uterine horns dissected from pregnant mice on embryonic day 14.5 of gestation (a gift from A. Kulkarni, National Institutes of Health, Bethesda, MD). MEFs were derived from CDK5−/− embryos, and the lack of CDK5 protein expression in these cells was verified.
In vivo tumorigenesis study
The mouse studies were approved by the NCI Animal Care and Use Committee and conducted in compliance with the approved protocols. The GFP-tagged WT DLC1 (GFP-DLC1-WT), GFP-DLC-4A mutant, and the GAP-dead mutant of DLC1 (GFP-DLC1-R718A) were constructed by PCR and site-directed mutagenesis, and subcloned into a modified pEGFP-C1 vector (Takara Bio Inc.) through Kpn1–NotI sites, as described previously (Qian et al., 2007). For the tumor xenograft, H358 stable clones expressing GFP, GFP-DLC1-WT, GFP-DLC1-4A, and GFP-DLC1-R718A were trypsinized, washed with cold PBS, diluted to 108/ml with serum-free medium/Matrigel basement membrane matrix (BD) at a ratio of 3:1, and injected subcutaneously into NOD/SCID mice (107 cells/injection). The animals were monitored for tumor growth, and tumor mass was weighed (in grams) 6 wk after injection.
Publicly available microarray data analysis
The gene expression microarray analyses reported in this study used data from caArray (https://array.nci.nih.gov/caarray/home.action) of the NCI. A cohort from caArray contains lung adenocarcinomas (jacob-00182) for Affymetrix GeneChip U133A Arrays. The CEL files containing the raw data from the experiment were directly downloaded from the NCI website. Data were then normalized with CEL file quality control evaluation using 3′ Expression MAS5 from the Affymetrix software Expression Console (http://www.affymetrix.com). Each individual gene (DLC1 and CDK5) expression value higher or lower than its corresponding median was considered to be high or low expression, respectively, in the analysis. The χ2 test (chisq.test) was carried out using the open source statistical tool R (version 2.14.1).
Mass spectrometry analysis
HEK 293T cells transiently expressing GFP, GFP-DLC1-WT, or GFP-DLC1-4A were lysed with golden lysis buffer (20 mM Tris, pH 7.9, 137 mM NaCl, 10% glycerol, 1% Triton, 5 mM EDTA, 1 mM EGTA, 1 mM Na3VO4, 10 mM NaF, 1 mM sodium pyrophosphate, 0.5 mM β-glycerophosphate, protease inhibitor mixture tablet, and phosphatase inhibitor) and subjected to IP with GFP antibody. The immunopellets were sequentially washed once with golden lysis buffer, once with high-salt HNTG, and twice with low-salt HNTG. Immunoprecipitated proteins were eluted by boiling for 5 min in 30 µl Laemmli sample buffer containing 10% (vol/vol) 2-mercaptoethanol and resolved on a NuPage 4–12% BisTris gel. The Coomassie-stained DLC1 gel band was digested with Trypsin or LysC as described previously (Shevchenko et al., 2006), and digested peptides were further desalted using StageTip C18 columns (Rappsilber et al., 2007). Desalted peptides were analyzed on a Q-Exactive instrument (Thermo Fisher Scientific) equipped with a Proxeon EASY-nLC 1,000 UHPLC (Thermo Fisher Scientific). Liquid chromatography–tandem mass spectrometry (LC-MS/MS) data was searched against a human Refseq database using MaxQuant v1.3.0.5 (Cox and Mann, 2008; Cox et al., 2011) with carbamidomethylation as a fixed modification and the following variable modifications: Oxidation of methionine, acetylation of protein N termini, deamidation of asparagine and phosphorylation of serine, threonine, and tyrosine residues. For peptide identification, we have applied a 1% FDR using a target-decoy search strategy (Elias and Gygi, 2010).
In vivo pull-down, co-IP, and IB
Cells were transiently cotransfected with plasmids expressing GST or the indicated GST fusion constructs together with GFP or the indicated GFP-DLC1 constructs. 48 h after transfection, cells were lysed with golden lysis buffer. The cleared supernatants were collected, and a small portion of supernatants was taken to determine the protein concentration using the DC protein assay (Bio-Rad Laboratories). For the pull-down assay, 1.0 mg of total protein from each cell extracts was used, to which 30 µl of glutathione Sepharose-4B slurry (GE Healthcare) was added, with continuous rotation for 3 h at 4°C. The pellets were sequentially washed once with golden lysis buffer, once with high-salt HNTG buffer, and twice with low-salt HNTG buffer. The beads were incubated with 30 µl Laemmli sample buffer, subjected to 10% SDS-PAGE, transferred onto nitrocellulose membranes (Invitrogen), and detected by IB using specific antibodies. A portion of the cell extracts was used as a loading control to verify expression of the GFP fusion proteins and the GPF control. For co-IP experiments, equal amounts of protein from the each cell lysates were pre-cleared with protein G slurry (Thermo Fisher Scientific) and then incubated with the indicated antibodies or control IgG for 1 h at room temperature. After incubation, 30 µl of protein G slurry was added to each immune reaction and rotated at 4°C overnight. The immunopellets were washed four times as above. Coimmunoprecipitated proteins were eluted by boiling for 5 min in 30 µl Laemmli sample buffer containing 5% (vol/vol) 2-mercaptoethanol. Eluted proteins were resolved on a NuPage 4-12% BisTris gel and detected by IB using specific antibodies. Immunoreactive bands were detected by enhanced chemiluminescence (ECL-Plus; GE Healthcare) using horseradish peroxidase–linked anti–rabbit or anti–mouse secondary antibodies (1:5,000 dilutions).
Immunofluorescent staining
Transiently or stably transfected cells were seeded in the glass chambers, incubated for 24 h, and fixed with 4% paraformaldehyde at room temperature for 20 min. Fixed cells were permeabilized with 0.25% Triton X-100 in PBS and then blocked with 3% BSA in PBS for 2 h. The cells were incubated with a 1:200 dilution of the indicated primary antibodies in PBS at 4°C overnight. After being thoroughly washed in PBS, the cells were incubated with the appropriate 1:250 Alexa Fluor–conjugated secondary antibodies for 1 h. To visualize actin or nuclei, cells were incubated with phalloidin (1:50) or DAPI (1:2,500) for 1 h. After being stained, cells were thoroughly washed with PBS and mounted with gel mounting solution (Biomeda Corporation).
Fluorescent confocal microscopy
Confocal microscopy of fluorescent-labeled cells was carried out using a microscope (LSM 780; Carl Zeiss) with an excitation wavelength of 488 nm to detect transfected GFP fusion proteins. Fluorescent Alexa Fluor probes were viewed with excitation wavelengths of 488 nm (Alexa Fluor 488) and 568 nm (Alexa Fluor 568). Images were made at room temperature using photomultiplier tubes (PMTs) with a Plan-Apochromat 63×/1.4 NA oil differential interference contrast objective lens with a 2× magnifier to produce a 125× magnification. The colocalization of two proteins was analyzed by confocal software (ZEN 2012; Carl Zeiss). For quantification of representative morphology in each group, ∼20 cells per condition randomly selected from several fields were analyzed. The average Mander’s overlapping colocalization coefficient ± SD was calculated, and is shown at the bottom of each panel. The overlapping colocalization coefficients can range from 0 to 1, where 0 means no colocalization and 1 means full colocalization of two proteins.
Wound healing, soft agar, and anchorage-independent growth
For wound healing assays, 105 cells/well were evenly plated (in triplicate) in the 12-well plates, grown for 24 h to become confluent, and subsequently scratched in the center of the well with standard pipette tips. After the suspended cells were washed away, the cultures were re-fed with fresh medium. The migrated cells within the scratched area were observed and photographed at fixed times using IncuCyte. For soft agar assays, a 0.6% agar (BD) base in RPMI-1640 medium was placed in 60-mm dishes for 1 h at room temperature. 105 cells were mixed with complete medium containing 0.4% agar and placed over 0.6% basal agar in 60-mm dishes. Cells were grown for 3 wk, and colonies were photographed microscopically and quantified with a colony counter. For clonogenic assays, 0.6 × 105 cells were seeded in 6-well plates and cultured in 0.9 µg/ml G418 RPMI-1640 medium with 10% FBS for 3 wk. Colonies were fixed, stained with 4% crystal violet, and counted.
Data analysis
At least two independent experiments were performed for all in vitro experiments. Immunoblots were quantified by densitometric scanning using ImageQuant software. Results are expressed as mean densities ± SD of three independent experiments. All experiments were designed with matched control conditions within each experiment. For statistical analysis, a Student’s t test was performed using SigmaPlot software (Systat), and P < 0.05 was considered statistically significant.
Online supplemental material
Fig. S1 shows that an efficient protein complex is formed between the tumor suppressor DLC1 and the serine/threonine kinase CDK5, and that DLC1 colocalizes with CDK5 at focal adhesions (with Vinculin) in various human cell lines. Fig. S2 shows that CDK5 forms a protein complex with DLC1 but not with other members of the DLC family, DLC2 or DLC3, and the interaction of CDK5 and DLC1 requires CDK5 kinase activity. CDK5 kinase phosphorylates DLC1 but not DLC2 or DLC3 protein. Fig. S3 shows phosphorylation of four serines in DLC1 (S120, S205, S422, and S509). They are consensus phosphorylation sites for CDK5 kinase, and their phosphorylation is confirmed by mass spectrometry. Fig. S4 shows that the Rho-GAP domain of DLC1 binds more strongly to the DLC1(1–550)-4A mutant than to DLC1(1-550)-WT, and that talin binds more strongly to GFP-DLC1-WT compared to the GFP-DLC1-4A mutant. Fig. S5 shows that the basal CDK5 kinase activity is low in the MCF10A cell line, and that depletion of tensin-3 does not result in a substantial change in RhoGTP level. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201405105/DC1.
Supplementary Material
Acknowledgments
We are grateful to Jana Qiao, William Vass, and Richard Braverman for technical assistance, and to Drs. Marian Durkin, Nicholas Popescu, and Peggy Zelenka for critically reading the manuscript. We thank NCI Center for Cancer Research Imaging Core Facility, for confocal microscopy, and Drs. Ashok Kulkarni (National Institute of Dental and Craniofacial Research) for CDK5−/− embryos, Michael Mowat for the GFP-DLC2 construct, Xioali Du (NCI) for DLC1 constructs, and Curt Harris (NCI) for H1703 and H358 cell lines.
This research is supported by the Intramural Research Program, National Institutes of Health, National Cancer Institute, and Center for Cancer Research.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- HBEC
- human bronchial epithelial cell
- IB
- immunoblotting
- IP
- immunoprecipitation
- MEF
- mouse embryonic fibroblast
- NCI
- National Cancer Institute
- NSCLC
- non-small cell lung cancer
- pMRLC
- phosphorylated myosin regulatory light chain
- Rho-GAP
- Rho–GTPase activating protein
- ROCK
- Rho-kinase
- WCE
- whole cell extract
- WT
- wild type
References
- Ajay A.K., Upadhyay A.K., Singh S., Vijayakumar M.V., Kumari R., Pandey V., Boppana R., and Bhat M.K.. 2010. Cdk5 phosphorylates non-genotoxically overexpressed p53 following inhibition of PP2A to induce cell cycle arrest/apoptosis and inhibits tumor progression. Mol. Cancer. 9:204–218 10.1186/1476-4598-9-204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif A.2012. Extraneuronal activities and regulatory mechanisms of the atypical cyclin-dependent kinase Cdk5. Biochem. Pharmacol. 84:985–993 10.1016/j.bcp.2012.06.027 [DOI] [PubMed] [Google Scholar]
- Cao X., Voss C., Zhao B., Kaneko T., and Li S.S.. 2012. Differential regulation of the activity of deleted in liver cancer 1 (DLC1) by tensins controls cell migration and transformation. Proc. Natl. Acad. Sci. USA. 109:1455–1460 10.1073/pnas.1114368109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cicero S., and Herrup K.. 2005. Cyclin-dependent kinase 5 is essential for neuronal cell cycle arrest and differentiation. J. Neurosci. 25:9658–9668 10.1523/JNEUROSCI.1773-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Vallejos E., Utreras E., and Gonzalez-Billault C.. 2012. Going out of the brain: non-nervous system physiological and pathological functions of Cdk5. Cell. Signal. 24:44–52 10.1016/j.cellsig.2011.08.022 [DOI] [PubMed] [Google Scholar]
- Cox J., and Mann M.. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26:1367–1372 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Director’s Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma, Shedden K., Taylor J.M., Enkemann S.A., Tsao M.S., Yeatman T.J., Gerald W.L., Eschrich S., Jurisica I., Giordano T.J., Misek D.E., et al. 2008. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat. Med. 14:822–827 10.1038/nm.1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin M.E., Avner M.R., Huh C.G., Yuan B.Z., Thorgeirsson S.S., and Popescu N.C.. 2005. DLC-1, a Rho GTPase-activating protein with tumor suppressor function, is essential for embryonic development. FEBS Lett. 579:1191–1196 10.1016/j.febslet.2004.12.090 [DOI] [PubMed] [Google Scholar]
- Durkin M.E., Ullmannova V., Guan M., and Popescu N.C.. 2007. Deleted in liver cancer 3 (DLC-3), a novel Rho GTPase-activating protein, is downregulated in cancer and inhibits tumor cell growth. Oncogene. 26:4580–4589 10.1038/sj.onc.1210244 [DOI] [PubMed] [Google Scholar]
- Elias J.E., and Gygi S.P.. 2010. Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol. Biol. 604:55–71 10.1007/978-1-60761-444-9_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenbroek S.I., and Collard J.G.. 2007. Rho GTPases: functions and association with cancer. Clin. Exp. Metastasis. 24:657–672 10.1007/s10585-007-9119-1 [DOI] [PubMed] [Google Scholar]
- Feldmann G., Mishra A., Hong S.M., Bisht S., Strock C.J., Ball D.W., Goggins M., Maitra A., and Nelkin B.D.. 2010. Inhibiting the cyclin-dependent kinase CDK5 blocks pancreatic cancer formation and progression through the suppression of Ras-Ral signaling. Cancer Res. 70:4460–4469 10.1158/0008-5472.CAN-09-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healy K.D., Hodgson L., Kim T.Y., Shutes A., Maddileti S., Juliano R.L., Hahn K.M., Harden T.K., Bang Y.J., and Der C.J.. 2008. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol. Carcinog. 47:326–337 10.1002/mc.20389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R.J., and O’Day D.H.. 2012. The cyclin-dependent kinase inhibitor roscovitine inhibits kinase activity, cell proliferation, multicellular development, and Cdk5 nuclear translocation in Dictyostelium discoideum. J. Cell. Biochem. 113:868–876 10.1002/jcb.23417 [DOI] [PubMed] [Google Scholar]
- Katz M., Amit I., Citri A., Shay T., Carvalho S., Lavi S., Milanezi F., Lyass L., Amariglio N., Jacob-Hirsch J., et al. 2007. A reciprocal tensin-3-cten switch mediates EGF-driven mammary cell migration. Nat. Cell Biol. 9:961–969 10.1038/ncb1622 [DOI] [PubMed] [Google Scholar]
- Kawauchi T., Chihama K., Nabeshima Y., and Hoshino M.. 2006. Cdk5 phosphorylates and stabilizes p27kip1 contributing to actin organization and cortical neuronal migration. Nat. Cell Biol. 8:17–26 10.1038/ncb1338 [DOI] [PubMed] [Google Scholar]
- Kim T.Y., Healy K.D., Der C.J., Sciaky N., Bang Y.J., and Juliano R.L.. 2008. Effects of structure of Rho GTPase-activating protein DLC-1 on cell morphology and migration. J. Biol. Chem. 283:32762–32770 10.1074/jbc.M800617200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.Y., Vigil D., Der C.J., and Juliano R.L.. 2009. Role of DLC-1, a tumor suppressor protein with RhoGAP activity, in regulation of the cytoskeleton and cell motility. Cancer Metastasis Rev. 28:77–83 10.1007/s10555-008-9167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.Y., Jackson S., Xiong Y., Whitsett T.G., Lobello J.R., Weiss G.J., Tran N.L., Bang Y.J., and Der C.J.. 2013. CRL4A-FBXW5-mediated degradation of DLC1 Rho GTPase-activating protein tumor suppressor promotes non-small cell lung cancer cell growth. Proc. Natl. Acad. Sci. USA. 110:16868–16873 10.1073/pnas.1306358110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko F.C., and Ping Yam J.W.. 2014. Regulation of deleted in liver cancer 1 tumor suppressor by protein-protein interactions and phosphorylation. Int. J. Cancer. 135:264–269 10.1002/ijc.28505 [DOI] [PubMed] [Google Scholar]
- Levacque Z., Rosales J.L., and Lee K.Y.. 2012. Level of cdk5 expression predicts the survival of relapsed multiple myeloma patients. Cell Cycle. 11:4093–4095 10.4161/cc.21886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., Du X., Vass W.C., Papageorge A.G., Lowy D.R., and Qian X.. 2011. Full activity of the deleted in liver cancer 1 (DLC1) tumor suppressor depends on an LD-like motif that binds talin and focal adhesion kinase (FAK). Proc. Natl. Acad. Sci. USA. 108:17129–17134 10.1073/pnas.1112122108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y.C., Si L., deVere White R.W., and Lo S.H.. 2007. The phosphotyrosine-independent interaction of DLC-1 and the SH2 domain of cten regulates focal adhesion localization and growth suppression activity of DLC-1. J. Cell Biol. 176:43–49 10.1083/jcb.200608015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Chen M.C., Chiu C.Y., Song Y.M., and Lin S.Y.. 2007. Cdk5 regulates STAT3 activation and cell proliferation in medullary thyroid carcinoma cells. J. Biol. Chem. 282:2776–2784 10.1074/jbc.M607234200 [DOI] [PubMed] [Google Scholar]
- Lin Y., Chen N.T., Shih Y.P., Liao Y.C., Xue L., and Lo S.H.. 2010. DLC2 modulates angiogenic responses in vascular endothelial cells by regulating cell attachment and migration. Oncogene. 29:3010–3016 10.1038/onc.2010.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.L., Wang X.Y., Huang B.X., Zhu F., Zhang R.G., and Wu G.. 2011. Expression of CDK5/p35 in resected patients with non-small cell lung cancer: relation to prognosis. Med. Oncol. 28:673–678 10.1007/s12032-010-9510-7 [DOI] [PubMed] [Google Scholar]
- Lukasik D., Wilczek E., Wasiutynski A., and Gornicka B.. 2011. Deleted in liver cancer protein family in human malignancies (Review). Oncol Lett. 2:763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J.2012. TGFβ signalling in context. Nat. Rev. Mol. Cell Biol. 13:616–630 10.1038/nrm3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y., Yamauchi J., Chan J.R., Okada A., Tomooka Y., Hisanaga S., and Tanoue A.. 2007. Cdk5 regulates differentiation of oligodendrocyte precursor cells through the direct phosphorylation of paxillin. J. Cell Sci. 120:4355–4366 10.1242/jcs.018218 [DOI] [PubMed] [Google Scholar]
- Ohshima T., Ward J.M., Huh C.G., Longenecker G., Veeranna H.C., Pant H.C., Brady R.O., Martin L.J., and Kulkarni A.B.. 1996. Targeted disruption of the cyclin-dependent kinase 5 gene results in abnormal corticogenesis, neuronal pathology and perinatal death. Proc. Natl. Acad. Sci. USA. 93:11173–11178 10.1073/pnas.93.20.11173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A.Y., Shen T.L., Chien S., and Guan J.L.. 2009. Role of focal adhesion kinase Ser-732 phosphorylation in centrosome function during mitosis. J. Biol. Chem. 284:9418–9425 10.1074/jbc.M809040200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., and Aravind L.. 1999. START: a lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24:130–132 10.1016/S0968-0004(99)01362-6 [DOI] [PubMed] [Google Scholar]
- Pozo K., Castro-Rivera E., Tan C., Plattner F., Schwach G., Siegl V., Meyer D., Guo A., Gundara J., Mettlach G., et al. 2013. The role of Cdk5 in neuroendocrine thyroid cancer. Cancer Cell. 24:499–511 10.1016/j.ccr.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Li G., Asmussen H.K., Asnaghi L., Vass W.C., Braverman R., Yamada K.M., Popescu N.C., Papageorge A.G., and Lowy D.R.. 2007. Oncogenic inhibition by a deleted in liver cancer gene requires cooperation between tensin binding and Rho-specific GTPase-activating protein activities. Proc. Natl. Acad. Sci. USA. 104:9012–9017 10.1073/pnas.0703033104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Li G., Vass W.C., Papageorge A., Walker R.C., Asnaghi L., Steinbach P.J., Tosato G., Hunter K., and Lowy D.R.. 2009. The Tensin-3 protein, including its SH2 domain, is phosphorylated by Src and contributes to tumorigenesis and metastasis. Cancer Cell. 16:246–258 10.1016/j.ccr.2009.07.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Durkin M.E., Wang D., Tripathi B.K., Olson L., Yang X.Y., Vass W.C., Popescu N.C., and Lowy D.R.. 2012. Inactivation of the Dlc1 gene cooperates with downregulation of p15INK4b and p16Ink4a, leading to neoplastic transformation and poor prognosis in human cancer. Cancer Res. 72:5900–5911 10.1158/0008-5472.CAN-12-2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao F., and Bowie J.U.. 2005. The many faces of SAM. Sci. STKE. 2005:re7. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Mann M., and Ishihama Y.. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- Rathinam R., Berrier A., and Alahari S.K.. 2011. Role of Rho GTPases and their regulators in cancer progression. Front Biosci (Landmark Ed). 16:2561–2571 10.2741/3872 [DOI] [PubMed] [Google Scholar]
- Sabbir M.G., Wigle N., Loewen S., Gu Y., Buse C., Hicks G.G., and Mowat M.R.. 2010. Identification and characterization of Dlc1 isoforms in the mouse and study of the biological function of a single gene trapped isoform. BMC Biol. 8:17–37 10.1186/1741-7007-8-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Taoka M., Sugiyama N., Kubo K., Fuchigami T., Asada A., Saito T., Nakajima K., Isobe T., and Hisanaga S.. 2007. Regulation of the interaction of Disabled-1 with CIN85 by phosphorylation with Cyclin-dependent kinase 5. Genes Cells. 12:1315–1327 10.1111/j.1365-2443.2007.01139.x [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Tomas H., Havlis J., Olsen J.V., and Mann M.. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860 10.1038/nprot.2006.468 [DOI] [PubMed] [Google Scholar]
- Su S.C., and Tsai L.H.. 2011. Cyclin-dependent kinases in brain development and disease. Annu. Rev. Cell Dev. Biol. 27:465–491 10.1146/annurev-cellbio-092910-154023 [DOI] [PubMed] [Google Scholar]
- Tripathi B.K., and Zelenka P.S.. 2009. Cdk5-dependent regulation of Rho activity, cytoskeletal contraction, and epithelial cell migration via suppression of Src and p190RhoGAP. Mol. Cell. Biol. 29:6488–6499 10.1128/MCB.01098-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi B.K., Stepp M.A., Gao C.Y., and Zelenka P.S.. 2008. The Cdk5 inhibitor olomoucine promotes corneal debridement wound closure in vivo. Mol. Vis. 14:542–549. [PMC free article] [PubMed] [Google Scholar]
- Vega F.M., and Ridley A.J.. 2008. Rho GTPases in cancer cell biology. FEBS Lett. 582:2093–2101 10.1016/j.febslet.2008.04.039 [DOI] [PubMed] [Google Scholar]
- Vigil D., Cherfils J., Rossman K.L., and Der C.J.. 2010. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat. Rev. Cancer. 10:842–857 10.1038/nrc2960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., and Kinzler K.W.. 2004. Cancer genes and the pathways they control. Nat. Med. 10:789–799 10.1038/nm1087 [DOI] [PubMed] [Google Scholar]
- Wong C.M., Lee J.M., Ching Y.P., Jin D.Y., and Ng I.O.. 2003. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 63:7646–7651. [PubMed] [Google Scholar]
- Yam J.W., Ko F.C., Chan C.Y., Jin D.Y., and Ng I.O.. 2006. Interaction of deleted in liver cancer 1 with tensin2 in caveolae and implications in tumor suppression. Cancer Res. 66:8367–8372 10.1158/0008-5472.CAN-05-2850 [DOI] [PubMed] [Google Scholar]
- Yang Y., Wang H., Zhang J., Luo F., Herrup K., Bibb J.A., Lu R., and Miller R.H.. 2013. Cyclin dependent kinase 5 is required for the normal development of oligodendrocytes and myelin formation. Dev. Biol. 378:94–106 10.1016/j.ydbio.2013.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau T.O., Leung T.H., Lam S., Cheung O.F., Tung E.K., Khong P.L., Lam A., Chung S., and Ng I.O.. 2009. Deleted in liver cancer 2 (DLC2) was dispensable for development and its deficiency did not aggravate hepatocarcinogenesis. PLoS ONE. 4:e6566 10.1371/journal.pone.0006566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Krishnamurthy P.K., and Johnson G.V.. 2002. Cdk5 phosphorylates p53 and regulates its activity. J. Neurochem. 81:307–313 10.1046/j.1471-4159.2002.00824.x [DOI] [PubMed] [Google Scholar]
- Zhong D., Zhang J., Yang S., Soh U.J., Buschdorf J.P., Zhou Y.T., Yang D., and Low B.C.. 2009. The SAM domain of the RhoGAP DLC1 binds EF1A1 to regulate cell migration. J. Cell Sci. 122:414–424 10.1242/jcs.027482 [DOI] [PubMed] [Google Scholar]
- Zukerberg L.R., Patrick G.N., Nikolic M., Humbert S., Wu C.L., Lanier L.M., Gertler F.B., Vidal M., Van Etten R.A., and Tsai L.H.. 2000. Cables links Cdk5 and c-Abl and facilitates Cdk5 tyrosine phosphorylation, kinase upregulation, and neurite outgrowth. Neuron. 26:633–646 10.1016/S0896-6273(00)81200-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.