PIP2 binds vinculin and directs its oligomerization, which promotes proper focal adhesion structure and function.
Abstract
Adherens junctions (AJs) and focal adhesion (FA) complexes are necessary for cell migration and morphogenesis, and for the development, growth, and survival of all metazoans. Vinculin is an essential regulator of both AJs and FAs, where it provides links to the actin cytoskeleton. Phosphatidylinositol 4,5-bisphosphate (PIP2) affects the functions of many targets, including vinculin. Here we report the crystal structure of vinculin in complex with PIP2, which revealed that PIP2 binding alters vinculin structure to direct higher-order oligomerization and suggests that PIP2 and F-actin binding to vinculin are mutually permissive. Forced expression of PIP2-binding–deficient mutants of vinculin in vinculin-null mouse embryonic fibroblasts revealed that PIP2 binding is necessary for maintaining optimal FAs, for organization of actin stress fibers, and for cell migration and spreading. Finally, photobleaching experiments indicated that PIP2 binding is required for the control of vinculin dynamics and turnover in FAs. Thus, through oligomerization, PIP2 directs a transient vinculin sequestration at FAs that is necessary for proper FA function.
Introduction
Inositol phospholipids are important regulators of cell physiology, and their head group interactions play fundamental roles in controlling membrane–cytosol interfaces. In addition to affecting signal transduction at the cell surface, these lipids regulate membrane traffic, nuclear events, the permeability and transport functions of membranes, and the cytoskeleton (Di Paolo and De Camilli, 2006). Phosphatidylinositol 4,5-bisphosphate (PIP2) plays roles in the recruitment and activation of a wide variety of actin regulatory proteins at the plasma membrane in order to regulate the release of cytokines, and to affect cell shape and motility, as well as several other processes (Ling et al., 2002, 2006; McLaughlin et al., 2002; Yin and Janmey, 2003). Binding of PIP2 is also critical for the function of actin-binding proteins that serve to link cell–cell or cell–matrix adhesion complexes at the plasma membrane with the actin cytoskeleton. There currently is a major conceptual gap in our understanding of how PIP2 binds to and alters the function of these adaptor/scaffold proteins, such as vinculin, at the molecular level.
Vinculin functions as a central regulator of adherens junctions (AJs) and focal adhesions (FAs) and is comprised of five loosely packed, antiparallel α-helix bundle domains. Four of these helix bundles are organized in a head (VH) domain that is connected to a five-helix bundle vinculin tail (Vt) domain by a disordered proline-rich region (Borgon et al., 2004), which functions as a hinge for the molecule. The inactive closed state of vinculin is characterized by extensive intramolecular hydrophobic head–tail interactions of its N-terminal helix bundle domain (Vh1) with Vt, and physiological triggers such as talin that activate vinculin disrupt these interactions, and allow the tail domain of the open conformer to bind to other ligands, including PIP2 (but not other lipids; Weekes et al., 1996; Palmer et al., 2009) and F-actin (Johnson and Craig, 1995; Peng et al., 2011). PIP2 levels are locally elevated at cell adhesion sites and PIP2 has been proposed to prevent F-actin binding (Steimle et al., 1999), to regulate FA turnover (Chandrasekar et al., 2005; Saunders et al., 2006), and to activate vinculin (Bakolitsa et al., 1999). This latter model was challenged by mutational analyses, which revealed that vinculin mutants that cannot bind to PIP2 still localize to FAs (Chandrasekar et al., 2005) and by the structure of full-length vinculin, which established that the basic patch thought to bind PIP2 is occluded by an extended coil of Vt that lies over the face of the C terminus and α-helix H1 (Bakolitsa et al., 2004; Borgon et al., 2004). More convincingly, mechanical tension emanating from integrin receptors has been shown to activate vinculin at FAs via its effects on the structure of integrin-bound talin, which releases amphipathic vinculin binding sites (VBSs) from their buried locales in helix bundle domains of the central rod domain of talin to then bind to Vh1 and displace Vt from a distance (Galbraith et al., 2002; Ziegler et al., 2006; Grashoff et al., 2010).
Activated vinculin is found as higher-order oligomers in cells (Milam, 1985; Molony and Burridge, 1985; Hüttelmaier et al., 1997; Johnson and Craig, 2000; Witt et al., 2004), and this is mediated by Vt–Vt interactions (Bakolitsa et al., 1999; Palmer et al., 2008; Abé et al., 2011). Indeed, Vt can dimerize at high concentrations (Bakolitsa et al., 1999; Palmer et al., 2008), even in the presence of F-actin (Hüttelmaier et al., 1997; Bakolitsa et al., 1999; Janssen et al., 2012; Tolbert et al., 2013). However, oligomerization is markedly augmented by PIP2, which drives the formation of dimers, trimers, and higher-order oligomers (Hüttelmaier et al., 1998; Bakolitsa et al., 1999; Witt et al., 2004; Palmer et al., 2009). Although cross-linking and limited proteolysis analyses have suggested that self-association, PIP2-induced dimerization, and F-actin–induced dimerization of vinculin are distinct (Bakolitsa et al., 1999; Johnson and Craig, 2000), extensive mutagenesis studies (Weekes et al., 1996; Johnson et al., 1998; Bakolitsa et al., 1999; Johnson and Craig, 2000; Saunders et al., 2006; Diez et al., 2008, 2009; Palmer et al., 2009; Wirth et al., 2010) have failed to reveal the PIP2 binding sites.
Here we report the oligomeric structure of human vinculin in complex with PIP2 at a 3.2-Å resolution. This structure and analyses of bona fide PIP2-binding defective mutants establish essential functions for this signaling lipid in driving protein oligomerization, and in modulating interactions with the actin cytoskeleton and RNA-binding protein raver1. Importantly, this structure lays the foundation for understanding the roles of signaling lipids in other cytoskeletal proteins.
Results
Architecture of PIP2-bound vinculin oligomers
Full-length vinculin is monomeric in solution, and, once activated, vinculin oligomerizes at sites of FAs (Hüttelmaier et al., 1998). Binding of the inositol head group and the hydrophobic acyl chain poses difficulties in generating PIP2-bound protein crystals. Indeed, the Vt–PIP2 complex crystal structure reported here represents only the third nonmembrane protein–PIP2 structure (out of five in total; Saad et al., 2006; Hansen et al., 2011; Kono et al., 2013; Poon et al., 2014). Further, the propensity of Vt to self-associate posed difficulties. We initially obtained ∼5 Å diffraction, which could only be improved to 3.2 Å (space group P3212; a = b = 102.58 Å, c = 190.77 Å) by using an R1060A mutation in the presence of a shorter chain (C8) derivative of PIP2 (Tables 1 and 2). The Arg-1060 side chain is solvent exposed and resides >13 Å from the PIP2 binding site (Fig. 1 A), and this alanine substitution has no effect on the conformation of vinculin nor upon the data and conclusions presented herein. Consistent with proteolysis studies (Bakolitsa et al., 1999), in the Vt–PIP2 structure, one PIP2 ligand is sandwiched between two Vt molecules by interacting with the loop connecting the first α-helices, H1 and H2 (Fig. 1 B). Dimer formation is mediated by the interactions of the 4′ and 5′ phosphates with Vt (molecule “I”) Lys-915 and Lys-1061, and with Ser-913 through a water-mediated interaction. Lys-915 is also within hydrogen bonding distance to the carbonyl of the inositol. PIP2 bridges one Vt (I) molecule to another Vt (II), where the carbonyl of the glycerol backbone and of the inositol reside within hydrogen bonding distance of Lys-915 and where there is a water-mediated interaction of the carbonyl of the glycerol backbone with Ser-913 (Fig. 1, C–E). Further, PIP2 directs trimer formation, where the sandwiched PIP2 interacts with α-helix H3 of a third Vt (III) molecule (Fig. 1 F) from an adjacent dimer via distinct interactions of Lys-944 and Arg-945 with the hydroxyl groups of the inositol (Fig. 1, C–E). Significantly, Lys-944 is strictly conserved across all 25 known species. The lipid acyl chains are mainly solvent exposed with limited hydrophobic interactions to the Lys-915 side chain. This architecture allows for vinculin to anchor in the lipid bilayer.
Table 1.
Vt–PIP2 x-ray data reduction statistics
| Statistics | Overall | Last shell |
| Resolution | 45.17–3.20 Å | 3.37–3.20 Å |
| Total measurements | 142,775 | 20,959 |
| No. of unique reflections | 19,218 | 2,744 |
| Rp.i.ma | 0.02 | 0.33 |
| CC1/2b | 0.999 | 0.725 |
| CC*c | 1.0 | 0.914 |
| Average I/σ(I) | 22.1 | 2.4 |
| Completeness | 0.997 | 1.0 |
| Redundancy | 7.4 | 7.6 |
CC, correlation coefficient.
Precision-indicating merging R factor (Weiss and Hilgenfeld, 1997; Weiss, 2001).
Pearson CC calculated between the average intensities of each random half of measurements of unique reflections (Karplus and Diederichs, 2012).
CC* estimates the value of the true level of signal CCtrue (the correlation of the averaged and thus less noisy data set with the noise-free true signal), based on a finite-size sample (Karplus and Diederichs, 2012).
Table 2.
Vt–PIP2 crystallographic refinement statistics
| Statistics | Overall | Last shell |
| Resolution | 45.17–3.20 Å | 3.37–3.2 Å |
| No. reflections (working set) | 18,212 | 2,594 |
| No. of reflections (test set) | 967 | 144 |
| Rwork/Rfree | 0.206/0.253 | 0.235/0.277 |
| No. atoms | ||
| Protein | 8,090 | |
| PIP2 | 141 | |
| Solvent | 75 | |
| B-factors | ||
| Protein | 147 Å2 | |
| PIP2 | 235 Å2 | |
| Solvent | 92 Å2 | |
| Root mean square deviations | ||
| Bond lengths | 0.010 Å | |
| Bond angles | 1.11° | |
| Ramachandran plot | ||
| Favored | 96.95% | |
| Allowed | 3.05% | |
| Generously allowed | 0% | |
| Outliers | 0% |
Figure 1.
PIP2 directs vinculin oligomerization by unfurling the N terminus of Vt. (A) The R1060A mutation used for crystallization does not affect PIP2 (oxygen, phosphor, and carbon atoms are shown as red, orange, and white sticks, respectively) binding. Vt α-helices are colored sequentially across the color spectrum from red (H1) to blue (H5). PIP2 and residues 1,060 and 1,061 are depicted, and residues 1,060 and 1,061 and Vt α-helices H1–H5 are labeled. (B) PIP2 directed dimerization of the vinculin five-helix bundle tail domain (α-helices are colored sequentially across the color spectrum from red (H1) to blue (H5), monomers are labeled as I and II) by binding to the α1-α2 loops and the C terminus of Vt. (C–E) Close-up view of the PIP2-directed oligomerization of Vt (α-helices and bonds are colored sequentially across the color spectrum from red to blue). Residues Lys-944 and Arg-945 are contributed from a third (III) monomer within the trimer. (D and E) View of the second (D; shown in blue) and third (E; shown in green) PIP2 in the asymmetric unit in the same orientation as in C, which indicates the specificity. (F) The PIP2-directed Vt trimer. Vt α-helices are colored sequentially across the color spectrum from red to blue, PIP2 is shown as spheres, and the three Vt molecules are labeled I–III. (G) With the exception of the loop connecting the last α-helix H5 to its C-terminal extended coil, unbound Vt (shown in cyan) superposes onto PIP2-bound Vt with a root-mean-square deviation (RMSD) of <0.5 Å for ∼970 atoms (depending on the monomer in the asymmetric unit). However, the PIP2 binding site coincides with the N-terminal extended coil (in particular residues 881–886, indicated by a red “N”), which is unfurled in the PIP2-bound structure.
PIP2 binding alters vinculin structure
In the unbound structure, the N-terminal extended coil of Vt occupies the PIP2 binding site, and PIP2 binding unfurls this region (Fig. 1 G). In addition to the side chain movements accompanied by PIP2 binding, lipid-directed dimerization replaces the His-906 stacking with Phe-885 with the more stable cation–Π interaction of the bulkier Trp-1064 indole ring stacking with His-906, which orders this otherwise disordered region in all known vinculin structures (Bakolitsa et al., 1999, 2004; Borgon et al., 2004; Izard et al., 2004; Lee et al., 2009; Rangarajan et al., 2011). In the apo structure, the N-terminal extended coil, in particular Pro-886, is located where Trp-1064 lies in the PIP2-bound structure. In contrast, in the closed conformation of full-length vinculin, Phe-885 stacks against His-906, and Asp-882 and Glu-880 engage in electrostatic interactions with Lys-924 or with Lys-924 and Lys-1061, respectively.
PIP2 binding to vinculin differs from other canonical PIP2 binding motifs such as the KK(X)nK/RK motif in pleckstrin homology (PH) domain containing proteins, epsin N-terminal homology (ENTH) domains, band 4.1 protein, ezrin, radixin and moesin (FERM) domains, Bin-Amphiphysin-Rvs167 (BAR) domain containing proteins, or myristoylated alanine-rich C-kinase substrate (MARCKS), where a cluster of positively charged residues are arranged next to each other to electrostatically sequester the local PIP2 concentrations (McLaughlin et al., 2002). However, in the case of our vinculin–PIP2 interface, there is no canonical sequence motif.
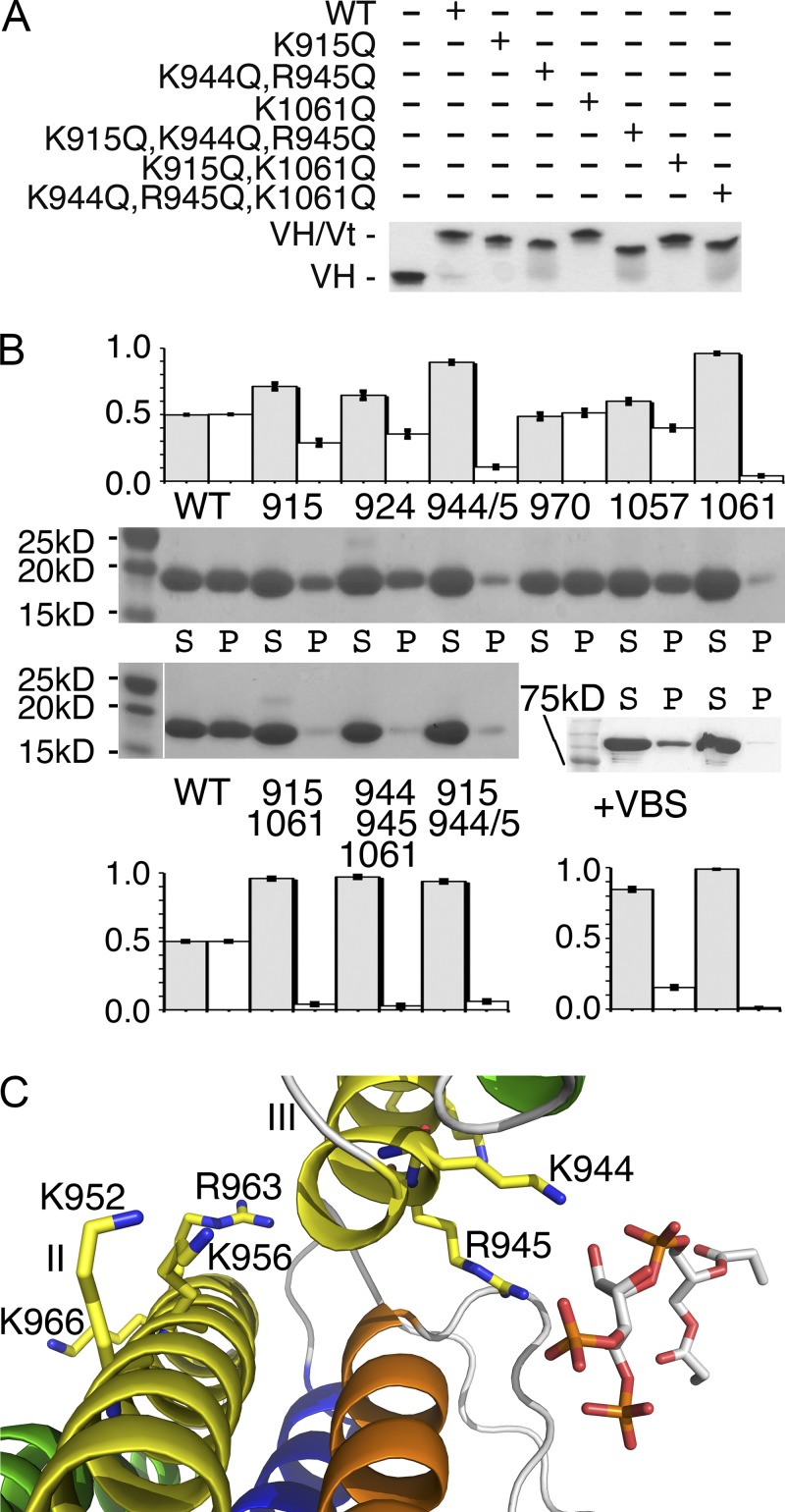
To validate the PIP2-bound structure, we performed vesicle pull-down assays of wild-type and mutant forms of vinculin-bearing mutations in key lysine and arginine residues that, according to their structure, mediate interactions with PIP2. While Vt truncations destabilize the Vt five-helix bundle (Palmer et al., 2009), our Vt point mutations do not interfere with binding of Vt to the VH domain (Fig. 2 A and Fig. S1 A) or to F-actin (Fig. 4 A and Fig. S2 A). These studies revealed an almost complete loss of lipid binding for the K1061Q mutant (Fig. 2 B and Fig. S1, B and C). Thus, Lys-1061 plays key roles in vinculin binding to the lipid bilayer.
Figure 2.
Pull-down assay to analyze the lipid-binding capacity of vinculin. (A) The interaction of wild-type and mutant Vt with VH was analyzed by electrophoretic mobility shift assays on a native gel, incubating the respective Vt proteins with VH. All proteins formed a Vt–VH complex. Data shown are representative of three independent experiments. (B) Unilamellar vesicles of the phospholipids PC and 15% PIP2 were incubated with wild-type and mutant Vt (top: K915Q, K924Q, K944Q/R945Q, K970Q, R1057Q, and K1061Q; bottom: K915Q/K1061Q, K944Q/R945Q/K1061Q, and K915Q/K944Q/R945Q) and sedimented. Supernatant (S) and pellet (P) fractions were detected on Coomassie-stained SDS-PAGE gel. Inactive and talin-VBS3 (residues 1,945–1,970) activated full-length (FL) vinculin were run as the nonbinding and binding controls, respectively. Densitometric values averaged over three experiments together with their standard deviations are shown above or below the respective experiment (gray bars, supernatant; white bars, pellet) and are as follows. (top) Wild type, 49.9% unbound in the supernatant with a standard deviation of 0.19; K915Q, 71.2%, 2.5; K924Q, 64.5%, 3.03; K944Q/R945Q, 89.3%, 1.35; K970Q, 48.7%, 2.5; R1057Q, 60%, 2.1; R1061Q, 96%, 0.72; (bottom left) wild type, 50%, 0.4; K915Q/K1061Q, 95.9%, 0.9; K944Q/R945Q/K1061Q, 97%, 0.9; K915Q/K944Q/R945Q, 93.7%, 1.1; (bottom right) full-length vinculin with VBS, 84.6%, 1.3; full-length vinculin alone, 99%, 0.1. The white line in one panel indicates the removal of unrelated lanes for presentation purposes. The corresponding full-length gels are shown in Fig. S1 B. Error bars indicate SD. (C) Previously identified residues on α-helix H3 reside near the PIP2-induced Vt–Vt interface. Shown is a close-up view of the PIP2-directed Vt (α-helices are colored sequentially across the color spectrum from red [H1] to blue [H5]) interface between subunits II and III. Residues Lys-952, Lys-956, and Arg-963, which were previously thought to bind to PIP2, reside near or at the Vt–Vt interface, which might explain the decreased binding of the potentially oligomer-defective mutant (K952Q, K956Q, and R963Q) Vt compared with wild-type Vt.
Figure 4.
The lipid-binding–deficient vinculin mutants bind and bundle F-actin. (A) Wild-type and mutant Vt (top: K915Q, K944Q/R945Q “944,” K1061Q, K915Q/K944Q/R945Q “915 944,” K915Q/K1061Q; K944Q/R945Q/K1061Q “944 1061”; bottom: K924Q, K970Q, and R1057Q) were cosedimented with F-actin and analyzed on Coomassie-stained SDS-PAGE. Full-length (FL) vinculin was run as the nonbinding control. S, supernatant; P, pellet. The white lines indicate the removal of unrelated lanes for presentation purposes. Data shown are representative of three independent experiments. The corresponding full-length gels are shown in Fig. S2 (A–C). (B) Confocal images of TRITC-phalloidin–labeled F-actin in the absence or presence of wild-type or mutant (K1061Q or K915Q/K944Q/R945Q/K1061Q) Vt domain. Data shown are representative of three independent experiments. Larger micrographs corresponding to these panels are shown in Fig. S3. Bars, 20 µm. (C) Model based on the superposition (not depicted) of the cryo-EM Vt–F-actin structure (Thompson et al., 2014; F-actin subunits are shown in gray or black) onto the PIP2-bound Vt dimer (α-helices H1–H5 are colored sequentially across the color spectrum from red [H1] to blue [H5]; PIP2 are represented as spheres), which suggests that vinculin has distinct F-actin and lipid binding sites. Residues Ile-997 and Val-1001 residing on α-helices H3 (green) that exhibit a significant decreased affinity for F-actin (Thompson et al., 2014) are also shown as spheres.
In the context of the full-length vinculin structure, the PIP2 binding site is on the surface of the protein and is distinct from the head–tail interface. However, PIP2 binding to full-length vinculin is impaired (Fig. 2 B and Fig. S1 D) because the N-terminal extended coil occludes the binding site (Fig. 3 A). Indeed, truncation of residues 879–897 increases Vt binding to PIP2 approximately twofold (Palmer et al., 2009). In contrast, talin VBS-activated full-length vinculin is fully permissive for lipid binding (Fig. 2 B and Fig. S1 D). Our structural and biochemical data fully support a PIP2-directed full-length vinculin dimer as a building block for higher-order vinculin oligomers (Fig. 3 B).
Figure 3.
PIP2 binding in the context of full-length vinculin. (A) The PIP2 binding site is distinct from the head–tail interface. Superposition of Vt–PIP2 onto full-length vinculin highlights the steric hindrance of the Vt N-terminal extended coil in the full-length vinculin–PIP2 model (Vt α-helices are colored sequentially across the color spectrum from red [H1] to blue [H5]; Vh1, residues 1–258 are shown in dark gray; the remainder of VH, residues 259–843, is shown in light gray; PIP2 is shown as spheres). The disordered regions (residues 844–876 and 1,063–1,066) are indicated. PIP2 binding orders the C terminus, and the Vt–PIP2 structure is the only vinculin structure that has interpretable electron density for the last residues. (B) PIP2-directed oligomerization of full-length vinculin. Once vinculin is activated and its head and tail domain severed, PIP2 directs vinculin oligomerization by binding to three Vt molecules. For clarity, only one PIP2 molecule, interacting with Lys-944 and Arg-945 of vinculin (III), is shown, whereas the PIP2 molecules bound to Lys-944 and Arg-945 of vinculin (I) and (II), which form a higher-order oligomer, are not shown. Vt α-helices are colored sequentially across the colorspectrum from red (H1) to blue (H5), VH are shown in gray, and PIP2 is represented with spheres.
The PIP2 and F-actin binding sites of vinculin are distinct
Activated vinculin binds directly to F-actin (Jockusch and Isenberg, 1981; Wilkins and Lin, 1982) and caps actin filaments to promote their polymerization (Ramarao et al., 2007; Le Clainche et al., 2010). This is thought to play critical roles at the leading edge of migrating cells (Dominguez and Holmes, 2011). Both PIP2 and F-actin bind to the Vt domain, and PIP2 binding has been reported to inhibit F-actin binding (Steimle et al., 1999). We therefore tested the structure-based PIP2-binding–deficient mutants of vinculin for their ability to interact with F-actin. Notably, all PIP2-binding deficient mutants cosedimented with F-actin (Fig. 4 A and Fig. S2), and also bundled F-actin, as seen with wild-type vinculin (Fig. 4 B and Fig. S3). Although the Vt F-actin binding sites have been hotly debated (Shen et al., 2011; Janssen et al., 2012; Thompson et al., 2013, 2014), the recently characterized I997A and V1001A mutants do show decreased binding to F-actin (Thievessen et al., 2013). These residues reside >12 Å from the PIP2-binding site (Fig. 4 C). Thus, the PIP2 and F-actin binding sites of vinculin are distinct.
The raver1–vinculin complex does not bind to PIP2
The RNA-binding protein raver1 simultaneously binds to vinculin mRNA and the Vt domain of activated vinculin, and this interaction has thus been postulated to control local production of vinculin to facilitate FA assembly (Lee et al., 2009, 2012; Madl and Sattler, 2009; Rangarajan et al., 2011). Interestingly, Arg-945 that binds PIP2 is also involved in electrostatic interactions with the raver1 residue Glu-120 (Fig. 5, A and B). To test if the lipid-binding–deficient K944Q/R945Q mutant binds raver1 and if the raver1–Vt complex binds PIP2, we ran the wild-type and mutant Vt–raver1 complexes on a size exclusion chromatography column (Fig. 5, C and D) and performed lipid cosedimentation assays (Fig. 5 E). Interestingly, although the K944Q/R945Q vinculin mutant still bound to raver1, the complex eluted earlier than the wild-type Vt–raver1 complex. Further, the crystal structures suggested that the raver1 and PIP2-binding sites are overlapping (Fig. 5, A and B), and indeed the Vt–raver1 complex did not bind to PIP2 (Fig. 5 E). Thus, although Vt–raver1 can form a ternary complex with F-actin (Lee et al., 2009), the Vt–raver1 complex does not bind to the membrane.
Figure 5.
Raver1 and PIP2 binding to vinculin is mutually exclusive. (A) Superposition of the Vt–raver1 structure onto the Vt–PIP2 structure. Vt α-helices are colored sequentially across the color spectrum from red (H1) to blue (H5), monomers are labeled I–III, and raver1 is shown in gray. Superposition of lipid-bound Vt (I) onto the raver1-bound Vt shows overlapping binding of Vt (III) and raver1. (B) Close-up view of some interface residues. In its raver1-bound state, vinculin residue Arg-945 engages in interactions with raver1 residues Glu-120 and Arg-117, whereas in its lipid-bound state, Arg-945 points toward PIP2. (C) Size exclusion chromatogram (SEC) of Vt–raver1 (blue line; peak #4 eluting at 14.56 ml; apparent mol wt = 54.6 kD; polypeptide chain mol wt = 51.2 kD), Vt-K944Q/R945Q/raver1 (black line; peak #5, 15 ml; apparent mol wt = 45 kD; polypeptide chain mol wt = 51.2 kD), raver1 (dotted line; peak #3, 15.77 ml; apparent mol wt = 32.19 kD; polypeptide chain mol wt = 31.5 kD), and Vt (gray line; peak #1, 17.2 ml; apparent mol wt =17.22 kD; polypeptide chain mol wt = 19.7 kD). (D) SDS-PAGE of peak fractions from the SEC shown in the previous panel. The lane numbers correspond to the peak numbering in the previous panel. (E) Lipid cosedimentation assay of the Vt–raver1 complex. The Vt–raver1 complex does not cosediment with PIP2 and remains in the soluble (S) fraction. P, pellet.
The vinculin–PIP2 interaction affects the actin cytoskeleton, cell migration, and FA turnover
PIP2 binding to vinculin has been proposed to play roles in the turnover of FAs (Chandrasekar et al., 2005; Saunders et al., 2006). Vinculin-null mouse embryonic fibroblasts (MEFs) have few and very unstable FAs, and those present fail to form links to the actin cytoskeleton, which is disorganized in these cells (Ezzell et al., 1997). Confocal immunofluorescence studies demonstrated that exogenously expressed wild-type GFP-tagged vinculin colocalized with paxillin in FAs in vinculin-null MEFs (Fig. S4) and led to reestablishment of a well-organized actin network (Fig. 6). In contrast, the expression of similar levels of GFP-vinculin–bearing mutations that block PIP2 binding resulted in colocalization with paxillin but did not rescue the chaotic actin network that is manifest in vinculin-null MEFs (Fig. 6).
Figure 6.
PIP2-directed oligomerization of vinculin contributes to the stabilization of FAs. Vinculin−/− MEFs engineered to express wild-type GFP-vinculin or mutant GFP-vinculin fusions (K944Q/R945Q and R1061Q) were analyzed by confocal laser-scanning microscopy. MEFs were grown on gelatin-coated coverslips, fixed after 24 h, and stained with TRITC-phalloidin to visualize the actin cytoskeleton. Representative images, which define the localization of GFP-vinculin (green) at FAs decorating F-actin (red), along with the merged channels, are shown. Also shown are nuclei stained with DAPI. Data shown are representative of three independent experiments. Bars, 100 µm.
Although vinculin−/− MEFs can assemble a modicum of FAs, they have defects in cell spreading, and their migration is chaotic and rapid. In wound healing assays, these cells close a wound faster than paired vinculin+/+ cells (Xu et al., 1998; Saunders et al., 2006). To assess the effects of the vinculin–lipid interaction on FAs in these cells, we performed scratch-wound healing assays (Fig. 7). Expression of wild-type vinculin suppressed the enhanced but chaotic wound closure typical of vinculin-null cells and led to a tighter closure of the wound. In contrast, forced expression of lipid-binding–deficient vinculin mutants led to marked suppression of wound closure, which indicates that the lipid-binding–deficient mutants of vinculin have defects in cell migration. Collectively, these findings were consistent with the notion that the PIP2–vinculin interaction might play roles in FA turnover.
Figure 7.
MEFs expressing the PIP2-binding–deficient mutant of vinculin are impaired in wound closure. Vinculin-null cells (left) that were engineered to express comparable levels of wild-type vinculin (center left) or K944Q/R945Q or K1061Q mutant vinculin (center right and right) were plated onto fibronectin-coated 12-well plates. Monolayers were then wounded, and the extent of cell migration into the wound was recorded at 0, 4, 8, and 12 h. Vinculin-null cells migrate in a rapid yet chaotic fashion, whereas wound closure in these cells expressing wild-type vinculin is an ordered process. Vinculin-null cells expressing K944Q/R945Q or K1061Q mutant vinculin had reduced migration and wound closure versus cells expressing wild-type vinculin. Data shown are representative of three independent experiments. Bars, 20 µm.
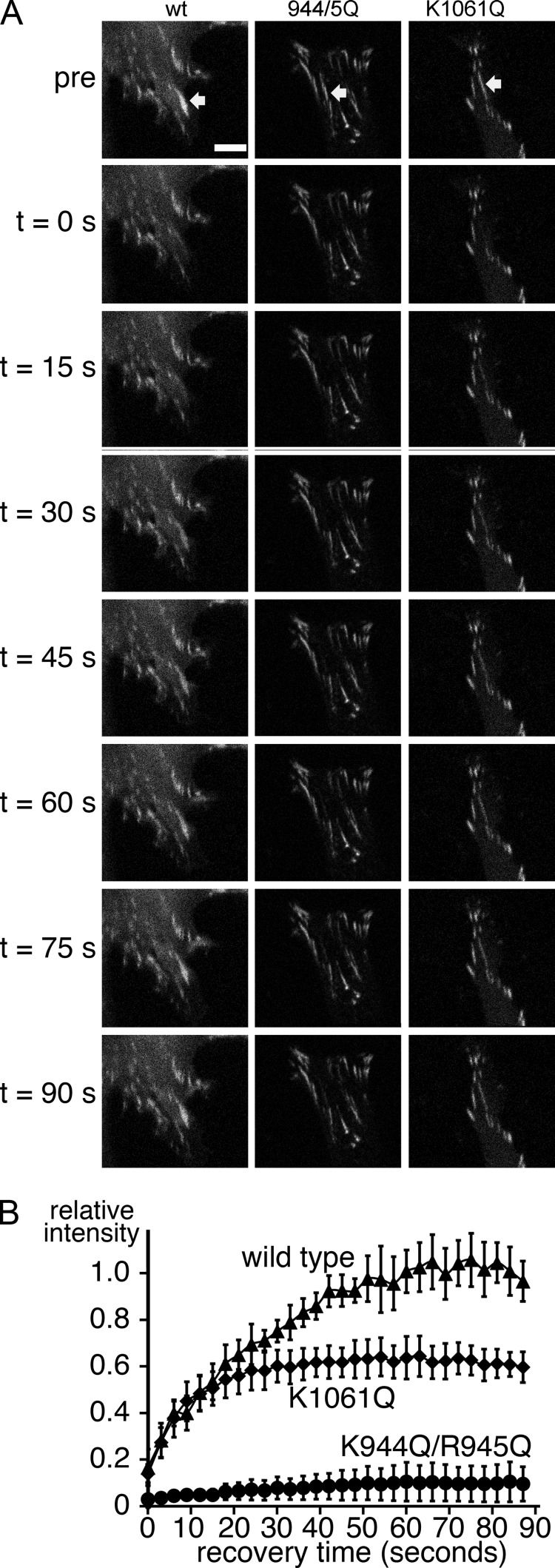
Direct measurement of wild-type GFP-vinculin turnover in FAs by FRAP revealed that the entire population within the FA is turning over with a half-time of ∼20 s (Fig. 8), a value in the range of what was previously reported (Chandrasekar et al., 2005). However, FRAP analysis of the lipid-binding–deficient vinculin mutants revealed that a large fraction (∼45% for the K1061Q mutant and >90% for the K944Q/R945Q mutant) of the GFP-vinculin within these FAs is immobile or much more tightly bound in that it did not recover within the time frame that results in complete recovery for the wild-type protein (Fig. 8). Notably, the mobile fraction of the K1061Q mutant vinculin in FAs displayed recovery kinetics that were significantly faster than wild-type vinculin, which is indicative of a much lower binding affinity. The immobile or tightly bound fraction may represent static nonfunctional interactions within the FA, which is consistent with the inability of these cells to organize the actin network and suggest that the dynamic turnover of vinculin is compromised in the PIP2-binding–deficient mutants. We conclude that the PIP2–vinculin interaction plays important roles in the stabilization and dynamic turnover of FAs.
Figure 8.
FRAP analyses of wild-type and mutant GFP-vinculin in FAs. (A) Representative images of FRAP recovery. The arrows indicate the bleached FA. (B) FRAP recovery curves for the mean of at least eight independent measurements for each cell line are displayed. The t50 values for wild-type (19 s) and the K1061Q mutant (7 s) were determined by fitting the curves to a single exponential. A t50 value for the K944Q/R945Q mutant could not be determined due to the absence of significant recovery. The immobile fraction is represented by the difference between the normalized plateau value and the prebleach fluorescence, and was estimated to be <5% for the wild-type cells, ∼45% for the K1061Q mutant, and >90% for the K944Q/R945Q mutant. Error bars indicate SD.
Discussion
The binding of integrins to extracellular matrix components at FAs triggers mechanical force on associated talin molecules, which opens up helix bundles and exposes VBSs that then bind to and activate vinculin to stabilize nascent cell–matrix adhesions via its associations with F-actin (Galbraith et al., 2002; Giannone et al., 2003; Zaidel-Bar et al., 2003). The role of PIP2 binding in the regulation and function of vinculin has been hotly debated (Gilmore and Burridge, 1996; Bakolitsa et al., 2004; Izard et al., 2004; Bois et al., 2006; Chen et al., 2006). Our studies establish that the PIP2 binding site is occluded in full-length vinculin and they demonstrate that PIP2 binding is necessary to mediate interactions of vinculin with F-actin at FAs. Our studies also suggest that F-actin and PIP2 binding to vinculin is mutually permissive, as they clearly have distinct binding sites on vinculin. These findings are consistent with a model whereby PIP2 binding triggers the formation of higher-order oligomers of activated vinculin that are competent to make multiple contacts with F-actin that would stabilize FAs, particularly because the cryo-EM structure suggests that one molecule of vinculin can be sandwiched between two molecules of F-actin (Janssen et al., 2006).
The crystal structure reveals that PIP2 principally affects vinculin function by directing vinculin oligomerization. Specifically, PIP2 directs the formation of vinculin dimers by releasing a flexible N-terminal extended coil present in the Vt five-helix bundle, and by bridging the flexible C terminus of one Vt molecule with α-helix H1 of a second Vt molecule, thereby restraining the conformation of the C terminus and stabilizing the dimer. Further, this signaling lipid directs vinculin trimerization via interactions with two lysine residues on α-helix H3 that are at the head–tail interface in inactive vinculin. Other basic residues on H3 have been proposed to direct the vinculin–lipid interaction (Chandrasekar et al., 2005), and examination of our crystal structure reveals that these residues are rather near the lipid-induced Vt–Vt interface (Fig. 2 C). Thus, mutating these residues might have prevented this Vt–Vt association by hindering vinculin oligomerization, and this might explain the decreased lipid binding. Finally, the crystal structures explain how higher-order vinculin oligomers can occur after PIP2 binding, a response that allows amplification of the interactions of vinculin with talin, and with F-actin, at FAs.
The RNA-binding protein raver1 has been proposed to regulate the local assembly of nascent FAs by binding to mRNAs encoding components of FAs in order to allow their localized translation at these sites. Specifically, raver1 binds to both Vt and vinculin mRNA, and the ternary Vt–raver1–vinculin mRNA complex is permissive for binding to F-actin. In contrast, the raver1 and PIP2 binding sites on vinculin overlap, and our studies have shown that the vinculin–raver1 complex does not bind to PIP2. This is consistent with a model where raver1 binding to vinculin is spatiotemporally regulated and where the F-actin–vinculin–raver1 complex bound to vinculin mRNA is not attached to the plasma membrane.
While the F-actin binding sites on vinculin are also controversial (Shen et al., 2011; Janssen et al., 2012; Thompson et al., 2013; Thompson et al., 2014), the recently identified I997A and V1001A mutants showed decreased F-actin binding. Notably, the crystal structure of the vinculin–PIP2 complex establishes that Ile-997 and Val-1001 are not involved in the PIP2 binding sites of vinculin. Further, PIP2-binding–deficient mutants of vinculin still bind F-actin in vitro; thus, binding of PIP2 and F-actin to vinculin seems mutually permissive. Finally, these findings suggest similar mechanisms of action for this signaling lipid in controlling the structure and functions of other lipid-and-actin-binding cytoskeletal proteins, where we propose that PIP2 may play roles in oligomerization of these targets that are also permissive for binding to F-actin.
We present three observations suggesting that binding of PIP2 by vinculin is an integral component of proper FA function. The exogenous expression of wild-type GFP vinculin, but not lipid-binding–deficient mutants, in vinculin-null MEFs resulted in the reestablishment of a well-organized actin network. Vinculin-null MEFs expressing lipid-binding–deficient GFP-vinculin displayed a severe impairment of cell migration in wound closure assays. Finally, direct measurement of vinculin recruitment to FAs by photobleaching revealed that wild-type vinculin displays dynamic turnover, with the entire population being replaced within the time frame of minutes. However, we find that a large portion of the PIP2-binding–deficient mutant vinculin is immobile or tightly bound, possibly due to static nonfunctional interactions within the FAs. Thus, although lipid-binding–deficient vinculin does localize to FAs, the observations noted here would suggest that the interactions within these FAs are distinct from those of wild-type vinculin and that these differences compromise FA function, which is consistent with the inability of these cells to organize the actin network or to close a wound. Previously, a modified FRAP procedure was used to show that a PIP2-binding–defective vinculin mutant was recruited to FAs, but disassembly was depressed (Chandrasekar et al., 2005). In a subsequent study, interference reflection microscopy was used to demonstrate that FAs in cells expressing a lipid-binding–deficient mutant displayed reduced turnover and translocation (Saunders et al., 2006). The results from these studies are in agreement with our conclusion that the PIP2–vinculin interaction is critical for the dynamic turnover of FAs and that this interaction is compromised in the PIP2-binding–deficient mutants. Our vinculin–PIP2 structure and functional data will aid future studies to understand how lipid-binding–deficient vinculin mutants affect the actomyosin contractile forces as these mutants significantly disturb the F-actin network.
Materials and methods
Protein preparation
Full-length human vinculin cDNA (residues 1–1,066) was cloned into a pET3 expression vector, and a noncleavable His8 tag was added downstream to the vinculin coding sequence. Modified pET-3 vector was used for cloning the head domain of vinculin (VH; residues 1–843) with a PreScission protease cleavable C-terminal octa-histidine tag. This expression vector contains a T7 promoter and contains an ampicillin resistance gene. For vinculin and VH purification, cell pellets were resuspended in 20 mM Tris-HCl, pH 8, and 150 mM NaCl, lysed by sonication, and clarified by ultracentrifugation (100,000 g for 45 min). The cell lysate was loaded onto a nickel affinity column (GE Healthcare), which was equilibrated with 20 mM Tris-HCl, pH 8, and 150 mM NaCl, and eluted the proteins using 0.5 M imidazole gradient. Further, proteins were purified by gel filtration using a Superdex 200 column (GE Healthcare), then equilibrated in 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl. Purified vinculin protein was further dialyzed against 10 mM Tris-HCl, pH 8, 0.1 mM EDTA, and 3 mM DTT buffer. Vinculin and VH were concentrated to 29 mg/ml and 10 mg/ml, respectively.
Site-directed mutants of vinculin (K915Q, K924Q, K944Q/R945Q, K970Q, R1057Q, R1060A, K1061Q, K915Q/K944Q/R945Q, K915Q/K1061Q, and K944Q/R945Q/K1061Q) were generated from the tail domain (Vt) of wild-type vinculin using Agilent Technology’s QuikChange site-directed mutagenesis kit. These mutants were cloned into the pGEX-6P1 expression vector (EMD Millipore) to obtain N-terminal GST-tagged proteins. All Vt constructs were cloned into a pGEX-6P1 expression vector (GE Healthcare). The basic features of these plasmids are a tac promoter and lac operator upstream of the coding sequence of GST followed by a PreScission cleavage site (LEVLFQGP). Vt and its mutant proteins were cloned between BamHI and XhoI restriction enzyme sites, which are downstream to the PreScission cleavage site and GST tag. The coding sequence is terminated by a stop codon. This expression vector contains an ampicillin resistance gene. All mutations were confirmed by sequencing. For GST-Vt and the Vt mutant protein purification, the cell pellets were resuspended in 20 mM Tris-HCl, pH 8, and 400 mM NaCl, lysed by sonication, and clarified by ultracentrifugation (100,000 g for 45 min). Subsequently, the supernatant was applied onto a GSTrap FF column, and, after extensive washing with the buffer (20 mM Tris-HCl, pH 8, and 400 mM NaCl), the bound GST-Vt was eluted with 20 mM of reduced glutathione, pH 8. GST affinity tags were removed by overnight cleavage with PreScission protease at 4°C and dialyzed into 20 mM Tris, pH 8, 400 mM NaCl, 1 mM DTT, and 0.1 mM EDTA. All proteins were further purified by gel filtration using a 26/60 Superdex 75 column (GE Healthcare) that was preequilibrated with 20 mM Tris-HCl, pH 8, 400 mM NaCl, 1 mM DTT, and 0.1 mM EDTA. Peak fractions were collected and concentrated to 10 mg/ml.
VH and full-length vinculin were purified as described previously (Rangarajan et al., 2010) and dialyzed against 20 mM Tris, pH 8, 10 mM DTT, 1 mM EDTA, and 1 mM EGTA, and concentrated to ∼29 mg/ml. Talin-VBS3 (residues 1,945–1,970) was synthesized.
The RRM1-3 (residues 39–321) domains of human raver1 were cloned into the pET-28a vector (EMD Millipore) to produce N-terminal hexahistidine-tagged proteins. This expression vector has a T7 promoter and six-histidine residue coding sequence upstream of the RRM1-3 (residues 39–321) domain coding sequence. This bacterial expression vector contains a kanamycin resistance gene. RRM1-3 was expressed in Escherichia coli strain BL21-DE3 (Invitrogen). Bacteria were grown at 37°C in Luria-Bertani medium containing 20 µg/ml kanamycin. Bacterial cultures were induced at OD600 = 0.8 by adding isopropyl-d-thiogalactopyranoside to 0.5 mM and incubating at 20°C, and cells were harvested after 24 h. The cell pellet was resuspended in 20 mM Tris-HCl, pH 8, and 150 mM NaCl, lysed by sonication, and clarified by ultracentrifugation (100,000 g for 45 min). The cell lysate was loaded onto a HisTrap chelating nickel affinity column (GE Healthcare), which was equilibrated with 20 mM Tris-HCl, pH 8, and 150 mM NaCl, and we eluted the proteins using an imidazole gradient. Further, it was purified by gel filtration using a Superdex 75 column (GE Healthcare) equilibrated in 20 mM Tris-HCl, pH 8.0, and 150 mM NaCl. Protein fractions were collected and concentrated to 15 mg/ml.
Vt–PIP2 crystallization
Human Vt (residues 891–1,066) was incubated with 20 µM PIP2diC8 (Avanti Polar Lipids) on ice for 30 min. Excess precipitate was removed by centrifugation (15,800 g for 25 min at 4°C). We screened ∼1,000 commercially available crystallization conditions and obtained a crystallization hit from the PEG/Ion kit of Hampton Research. Crystals were grown within 2 d by hanging drop vapor diffusion from 50 mM Hepes, pH 7, 12% (wt/vol) PEG 3350, and 2% (wt/vol) Tryptone, at 20°C. They diffracted x-rays at various beam lines (11-1 at Stanford Synchrotron Radiation Laboratory [SSRL], or 22ID and 22BM at the Advanced Photon Source of the Argonne National Laboratory [APS/ANL]) to ∼5-Å Bragg spacings. All crystallization strategies and truncations failed to improve the diffraction, but ultimately crystals of the PIP2-bound R1060A complex diffracted to 3-Å Bragg spacings. The mother liquor was supplemented with 5% increments of glycerol to 25% and transferred and stored in liquid nitrogen at 100 K.
X-ray data collection and processing
X-ray diffraction data were collected at the Advanced Photon Source 22-ID beam line (wavelength of 1 Å) and integrated and scaled using XDS and SCALA as implemented in autoPROC (Vonrhein et al., 2011). Data reduction statistics are provided in Table 1.
Structure determination and crystallographic refinement
Phases were obtained by molecular replacement using the program MOLREP (Vagin and Teplyakov, 1997), where the search model consisted of Vt from the Vh1–Vt complex (Protein Data Bank [PDB] accession no. 1RKE). A clear solution with six molecules in the asymmetric unit was obtained in space group P3212, while for the enantiomer in P3112, the R factor remained >0.5 during refinement. An acceptable packing density, VM (Matthews, 1968), was calculated at 2.45 Å3/Da, corresponding to a solvent content of ∼0.495.
Initial crystallographic refinement used a 3-Å cutoff, but best results were obtained using data up to 3.2-Å Bragg spacings. Maximum likelihood crystallographic refinement was performed using autoBUSTER (Bricogne et al., 2011) by imposing target restraints using the high-resolution structures. The model was improved by local noncrystallographic symmetry restraints through local structure similarity restraints (LSSR; Smart, O.S., C. Brandl, P. Flensburg, W. Keller, C. Paciorek, C. Vonrhein, T. Womack, and G. Bricogne. 2008. Proceedings of the annual meeting of the American Crystallographic Association. Abstr. TP139). Iterative cycles of model building were performed using Coot (Emsley and Cowtan, 2004), and model bias was minimized by building into composite omit maps. Optimized ligand coordinates and ligand restraints were obtained from the Grade web server (grade.globalphasing.org). The electron density map was sharpened using Coot (Emsley and Cowtan, 2004) to ensure the directionality and identity of the α-helices. The quality of the final model was assessed using MolProbity (Davis et al., 2004), which revealed no outliers with >97% of the amino acid residues in the favored region of the Ramachandran plot. Crystallographic refinement statistics are provided in Table 2. The final model contains six Vt polypeptide chains and three PIP2 molecules. Residues 1,051–1,053 and 1,066 are missing due to weak electron density in this region.
Lipid cosedimentation assay
PIP2 binding to Vt mutants was assayed as described previously (Palmer et al., 2009). In brief, lipid vesicles of phosphatidylcholine (PC) and PIP2 were prepared by mixing together in chloroform to have a final composition of 85% PC and 15% PIP2. The lipid mixture was vacuum dried and resuspended in 20 mM Tris, pH 8, 400 mM NaCl, and 3 mM DTT. Unilamellar vesicles were then produced by sonication after incubating the vesicles at 37°C for 30 min. Samples containing 50 µg of total lipid in 10 µl suspension and 40 µM Vt (wild type or mutants) were incubated at 4°C for 1 h followed by centrifugation at 75,815 g for 30 min. The supernatant and pellet were separated, and the pellet was washed two times with buffer A and resuspended in 12.5 µl. The supernatant and pellet samples were analyzed by SDS-PAGE.
Native PAGE assay of Vt proteins binding to the VH domain
About 5 µM VH (residues 1–840) was mixed with 2–3-fold excess of wild-type or mutant (K915Q, K944Q/R945Q, K1061Q, K915Q/K944Q/R945Q, K915Q/K1061Q, and K944Q/R945Q/K1061Q) Vt (residues 891–1,066) in 20 mM Tris, pH 8, 400 mM NaCl, and 1 mM DTT, then incubated at room temperature for 15 min. The samples were then analyzed on 10–15% gradient PHAST gel using native buffer strips, and the bands were visualized with Coomassie blue.
F-actin cosedimentation assay
F-actin cosedimentation assays were performed as described previously (Rangarajan and Izard, 2012). In brief, 15 µM of F-actin was incubated with ∼8 µM Vt (or full-length vinculin) for 20 min at room temperature and the reaction mixture was pelleted at 125,000 g for 15 min at 20°C. The supernatant and pellet were further analyzed on SDS-PAGE and stained for protein bands with Coomassie blue.
F-actin bundling assay
Fluorescence microscopic analyses of F-actin bundling by Vt domains were performed as described previously (Rangarajan et al., 2010), with slight modifications. In brief, G-actin (1 mg/ml) was prepolymerized in either Tris buffer (20 mM Tris-HCl, pH 8, 0.1 M KCl, 2 mM MgCl2, 1 mM ATP, and 0.4 mM CaCl2) or imidazole buffer (25 mM imidazole, pH 7.4, 0.1 M KCl, 2 mM MgCl2, and 1 mM ATP) for 1 h at 37°C. Reactions containing 2.7 µM prepolymerized F-actin with 2.3 µM of Vt or its PIP2-binding–deficient mutants were incubated for 1 h at room temperature. F-actin was subsequently labeled with an equimolar concentration (2.7 µM) of TRITC-phalloidin (Life Technologies) and incubated another half hour before mounting on poly-l-lysine–coated coverslips (Thermo Fisher Scientific). The confocal images of TRITC-phalloidin–labeled F-actin were captured at ambient temperature using a confocal laser scanning microscope (LSM 780; Karl Zeiss), equipped with a gallium arsenide phosphide (GaAsP) detector and Zen software, using a 63×/1.3 NA Apochromat multi-immersion objective lens using water as the imaging medium, and further adjusted for brightness/contrast with Fiji (ImageJ).
Size exclusion chromatography
Purified raver1 (100 µM in 20 mM Tris, pH 8, and 150 mM NaCl) was incubated for 1 h at 4°C with wild-type or K944Q/R945Q mutant vinculin (20 mM Tris, pH 8, 400 mM NaCl, and 1 mM DTT) in a 1:2 molar ratio and run on an analytical S200 size exclusion chromatography column (equilibrated in 20 mM Tris, pH 8, 400 mM NaCl, and 3 mM DTT) with a flow rate of 0.5 ml/min. Vt and raver1 proteins, both at 100 µM, were also run separately. Fractions were analyzed on a 10–15% gradient PHAST gel with SDS-PAGE buffer strips.
Cell culture and immunofluorescence
Vinculin-deficient (Vin−/−) MEFs were provided by K. Burridge (University of North Carolina, Chapel Hill, NC). They were originally prepared by E. Adamson (Sanford-Burnham Medical Research Institute, La Jolla, CA): vinculin-null cells were isolated by targeting both alleles of the vinculin gene by homologous recombination in which exon 3 has been replaced by a neomycin resistance gene (Xu et al., 1998). Vinculin-null cells were cultured in DMEM containing 4.5 g/liter glucose and 2 mM l-glutamine (Invitrogen) supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37°C and 5% CO2 in a humidified incubator, and passaged every 3–4 d. Vin−/− MEFs were transfected with the pcDNA4.1 plasmid DNA encoding full-length human wild-type or mutant vinculin as an N-terminal His10-EGFP fusion, or with the vector control, using Lipofectamine LTX plus reagent (Invitrogen) according to the manufacturer’s protocol. All human vinculin constructs and PIP2-binding–deficient mutants used for cell biology studies were cloned into pcDNA4.1/TO/Myc-HisB vector (Life Technologies). The basic feature of these plasmids is a CMV promoter and two tandem tetracycline (2× TetO2) operator sequences upstream to the coding sequence of EGFP. EGFP coding sequence and a PreScission cleavage site (LEVLFQGP) downstream to it was introduced between the TET operator and the coding sequence of HV or the mutants. Further, a three-alanine linker was introduced in-between the PreScission cleavage site and the coding sequence of vinculin. The vinculin coding sequence is terminated by a stop codon (TGA) before the myc and hexahistidine tag. This mammalian expression vector contains Zeocin and ampicillin resistance genes. Stables were prepared using Zeocin. Cells were grown on six-well tissue culture treated plates for 48 h and were initially selected with medium containing 800 µg/ml Zeocin for 1–2 wk, then passaged every 3–4 d with fresh media containing 200 µg/ml Zeocin.
Preparation of cells for immunofluorescence detection for localization of vinculin in FAs was performed by plating the cells stably expressing vinculin (or mutants of vinculin) on a gelatin-coated coverslip. 24 h after plating, the cells were fixed for 10 min with 4% paraformaldehyde in PBS, pH 7.4, and permeabilized with 0.2% Triton X-100 in PBS. Blocking was performed with 1% BSA in PBS for 20 min and cells were stained with TRITC-phalloidin (5 µg/ml) for actin and with DAPI (0.1 µg/ml) for detecting nuclei. Additionally, cells (7 × 104) were also seeded on 12-mm coverslips coated with 20 µg/ml fibronectin (Sigma-Aldrich) and incubated for 6 h. To visualize colocalization of vinculin and paxillin (another FA marker), cells were fixed and stained sequentially with mouse anti-paxillin antibody (BD) and Alexa Fluor 568 conjugated anti–mouse IgG (Life Technologies) followed by staining with DAPI for visualizing the nucleus. For imaging, the coverslips were mounted on a glass slide using Vectashield mounting medium (Vector Laboratories).
Fluorescence images were collected on confocal laser scanning microscope (LSM 780; Zeiss), controlled with Zen software, using a GaAsP detector. Cells were randomly selected and imaged at room temperature (23°C) using either a 63×/1.3 NA Apochromat multi-immersion objective lens using water as the imaging medium (for FA analysis) or a 20×/0.8 NA dry objective lens (for cell spreading). Excitations of the individual fluorophores in the stained cells were achieved using an argon laser (488 nm, for EGFP), a HeNe laser (633 nm for Alexa Fluor 633), and solid-state lasers (405 nm and 561 nm for DAPI and Alexa Fluor 568, respectively). Images were further processed using Fiji (ImageJ) for brightness/contrast adjustments for confocal images. Additionally, for quantification, the images were low-band-pass filtered, an auto-threshold function was applied, and manual masks were defined before using the “analyze particle” function for cell or area counting and FA analysis.
Immunoblotting
Cells showing stable and comparable levels of expression of vinculin or vinculin mutants were cultured to 70–80% confluency on gelatin-coated 10-cm Petri dishes in media supplemented with 200 µg/ml Zeocin, and were collected through trypsinization. The cells were washed twice with ice-cold PBS and resuspended in RIPA buffer (10 mM Tris-HCl, pH 8, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, and protease inhibitor cocktail [Roche]) and briefly sonicated. The lysed samples were then separated by SDS-polyacrylamide gel, electroblotted on to Immobilon membrane (EMD Millipore) using a semidry transfer apparatus (Bio-Rad Laboratories), and incubated sequentially with anti-vinculin primary antibody and HRP-conjugated secondary antibody. The blots were then developed after treatment with chemiluminescence substrate (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Wound assays
Vinculin-null MEFs and the cells stably expressing full-length wild-type or mutant vinculin were seeded at a density of 2 × 105 cells/well on a 12-well plate, precoated with 20 mg/ml fibronectin. The cells were then incubated overnight at 37°C and 5% CO2 to achieve confluency the following day. The wound healing assay was initiated by scraping a line on the culture dish using a 200-µl pipette tip on the confluent cell monolayer. The culture dish was then gently washed with PBS to remove any cell debris and replenished with 1 ml of fresh media (DMEM with 10% bovine calf serum, 100 U/ml penicillin, and 100 µg/ml streptomycin). Cells were imaged on an EVOS FL cell imaging system (Life Technologies) equipped with a color charge-coupled device image acquisition system (ICX285AQ; Sony) through software built in to the onboard microprocessor. A 10×/0.25 dry objective lens was used for bright field imaging immediately afterwards in order to represent the zero time point, and the wound healing was monitored every 4 h, returning plates for incubation after every imaging session. All assays were repeated in triplicate. Each time, images were taken at six different points and the positions that were similar in wound distance were used for direct comparison.
FRAP assays
MEF cells were plated and observed in fibronectin-coated 35-mm2 glass-bottom culture dishes (MatTek Corporation). FRAP was performed on a laser scanning confocal microscope (TCS SP8; Leica) using the 488-nm line of an argon laser essentially as described previously (George et al., 2010). All experiments were performed at 37°C, and imaging was performed with a Plan-Apochromat 63×/1.3 NA objective lens using the FRAP module of the LAS AF software (Leica). Bleaching of a single FA per cell was achieved using a single bleach scan of 273-ms duration at maximal laser power. Recovery was monitored at 500–575 nm and recorded for 90 s. Although movement of cells and/or FAs precluded longer measurements, a recovery plateau was reached within this time period for most of the imaged cells, except where noted. Identical imaging settings (5% laser intensity) were used for all pre- and post-bleach images. Scanning was bidirectional at the highest possible rate using a 4× zoom with a pinhole of 1 Airy unit. Image analysis was performed within the LAS AF software. Datasets were imported into Origin 6.0 and normalized recovery rates were fitted to a single exponential recovery model in OriginLab software using the equation F(t) = 1 − e(−kx), where F(t) is the fluorescence at time t, k is the rate constant, and the t1/2 of recovery was calculated from the rate constant k = ln 2/t1/2. All datasets consisted of at least eight cells per experiment.
Online supplemental material
Fig. S1 shows the full gels shown in Fig. 2. Fig. S2 shows the full gels shown in Fig. 4. Fig. S3 shows larger panels compared with those shown in Fig. 4 B as well as additional confocal images of TRITC-phalloidin–labeled F-actin in the presence of Tris-HCl, pH 8. Fig. S4 shows confocal immunofluorescence studies of exogenously expressed wild-type and lipid-binding–deficient mutant GFP-tagged vinculin colocalized with paxillin in FAs in vinculin-null MEFs, as well as immunoblots obtained with vinculin antibody after SDS-PAGE to analyze the expression of wild-type and mutant vinculin proteins in vinculin-null cells using the primary anti-vinculin antibody and a secondary HRP–conjugated anti–mouse IgG. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201404128/DC1.
Supplementary Material
Acknowledgments
We are indebted to John Cleveland (Moffitt Cancer Center) for discussions and critical review of the manuscript. We are grateful to the staff at the SER-CAT (ID22) for synchrotron support. We are indebted to Dr. Keith Burridge (University of North Carolina) for generously providing vinculin-null cells. We thank Drs James R. Sellers and Sarah M. Heissler (National Heart, Lung, and Blood Institute) for TIRF assays. Confocal images used in this article were generated at the light microscopy facility at the Max Planck Florida Institute.
T. Izard is supported by grants from the National Institute of General Medical Sciences, the National Institutes of Health, the US Department of Defense, and by start-up funds provided to Scripps Florida from the state of Florida. This is publication no. 26096 from The Scripps Research Institute. The coordinates and structure factors for the Vt–PIP2 complex have been deposited with the Protein Data Bank (accession no. 4PR9). FRAP assays were performed at the University of Mississippi Medical Center Confocal Microscopy Core Facility, which is supported by National Institutes of Health grants S10RR031728 and HL51971.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in the paper:
- CC
- correlation coefficient
- FA
- focal adhesion
- MEF
- mouse embryonic fibroblast
- PC
- phosphatidylcholine
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- VBS
- vinculin binding site
- VH
- vinculin head
- Vt
- vinculin tail
References
- Abé C., Dietrich F., Gajula P., Benz M., Vogel K.P., van Gastel M., Illenberger S., Ziegler W.H., and Steinhoff H.J.. 2011. Monomeric and dimeric conformation of the vinculin tail five-helix bundle in solution studied by EPR spectroscopy. Biophys. J. 101:1772–1780 10.1016/j.bpj.2011.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakolitsa C., de Pereda J.M., Bagshaw C.R., Critchley D.R., and Liddington R.C.. 1999. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 99:603–613 10.1016/S0092-8674(00)81549-4 [DOI] [PubMed] [Google Scholar]
- Bakolitsa C., Cohen D.M., Bankston L.A., Bobkov A.A., Cadwell G.W., Jennings L., Critchley D.R., Craig S.W., and Liddington R.C.. 2004. Structural basis for vinculin activation at sites of cell adhesion. Nature. 430:583–586 10.1038/nature02610 [DOI] [PubMed] [Google Scholar]
- Bois P.R., O’Hara B.P., Nietlispach D., Kirkpatrick J., and Izard T.. 2006. The vinculin binding sites of talin and α-actinin are sufficient to activate vinculin. J. Biol. Chem. 281:7228–7236 10.1074/jbc.M510397200 [DOI] [PubMed] [Google Scholar]
- Borgon R.A., Vonrhein C., Bricogne G., Bois P.R., and Izard T.. 2004. Crystal structure of human vinculin. Structure. 12:1189–1197 10.1016/j.str.2004.05.009 [DOI] [PubMed] [Google Scholar]
- Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek P., Roversi P., Sharff A., Smart O.S., Vonrhein C., and Womack T.O.. 2011. BUSTER version 2.9. Global Phasing Ltd., Cambridge, UK: https://www.globalphasing.com/buster/ (accessed 18 November 2014) [Google Scholar]
- Chandrasekar I., Stradal T.E., Holt M.R., Entschladen F., Jockusch B.M., and Ziegler W.H.. 2005. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 118:1461–1472 10.1242/jcs.01734 [DOI] [PubMed] [Google Scholar]
- Chen H., Choudhury D.M., and Craig S.W.. 2006. Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 281:40389–40398 10.1074/jbc.M607324200 [DOI] [PubMed] [Google Scholar]
- Davis I.W., Murray L.W., Richardson J.S., and Richardson D.C.. 2004. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32:W615–W619 10.1093/nar/gkh398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G., and De Camilli P.. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature. 443:651–657 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- Diez G., List F., Smith J., Ziegler W.H., and Goldmann W.H.. 2008. Direct evidence of vinculin tail-lipid membrane interaction in β-sheet conformation. Biochem. Biophys. Res. Commun. 373:69–73 10.1016/j.bbrc.2008.05.182 [DOI] [PubMed] [Google Scholar]
- Diez G., Kollmannsberger P., Mierke C.T., Koch T.M., Vali H., Fabry B., and Goldmann W.H.. 2009. Anchorage of vinculin to lipid membranes influences cell mechanical properties. Biophys. J. 97:3105–3112 10.1016/j.bpj.2009.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R., and Holmes K.C.. 2011. Actin structure and function. Annu. Rev. Biophys. 40:169–186 10.1146/annurev-biophys-042910-155359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Ezzell R.M., Goldmann W.H., Wang N., Parashurama N., and Ingber D.E.. 1997. Vinculin promotes cell spreading by mechanically coupling integrins to the cytoskeleton. Exp. Cell Res. 231:14–26 (published erratum appears in Exp. Cell Res. 2008. 314:2163) 10.1006/excr.1996.3451 [DOI] [PubMed] [Google Scholar]
- Galbraith C.G., Yamada K.M., and Sheetz M.P.. 2002. The relationship between force and focal complex development. J. Cell Biol. 159:695–705 10.1083/jcb.200204153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George E.M., Izard T., Anderson S.D., and Brown D.T.. 2010. Nucleosome interaction surface of linker histone H1c is distinct from that of H1(0). J. Biol. Chem. 285:20891–20896 10.1074/jbc.M110.108639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G., Jiang G., Sutton D.H., Critchley D.R., and Sheetz M.P.. 2003. Talin1 is critical for force-dependent reinforcement of initial integrin-cytoskeleton bonds but not tyrosine kinase activation. J. Cell Biol. 163:409–419 10.1083/jcb.200302001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore A.P., and Burridge K.. 1996. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature. 381:531–535 10.1038/381531a0 [DOI] [PubMed] [Google Scholar]
- Grashoff C., Hoffman B.D., Brenner M.D., Zhou R., Parsons M., Yang M.T., McLean M.A., Sligar S.G., Chen C.S., Ha T., and Schwartz M.A.. 2010. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 466:263–266 10.1038/nature09198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.B., Tao X., and MacKinnon R.. 2011. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 477:495–498 10.1038/nature10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S., Bubeck P., Rüdiger M., and Jockusch B.M.. 1997. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur. J. Biochem. 247:1136–1142 10.1111/j.1432-1033.1997.01136.x [DOI] [PubMed] [Google Scholar]
- Hüttelmaier S., Mayboroda O., Harbeck B., Jarchau T., Jockusch B.M., and Rüdiger M.. 1998. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr. Biol. 8:479–488 10.1016/S0960-9822(98)70199-X [DOI] [PubMed] [Google Scholar]
- Izard T., Evans G., Borgon R.A., Rush C.L., Bricogne G., and Bois P.R.. 2004. Vinculin activation by talin through helical bundle conversion. Nature. 427:171–175 10.1038/nature02281 [DOI] [PubMed] [Google Scholar]
- Janssen M.E., Kim E., Liu H., Fujimoto L.M., Bobkov A., Volkmann N., and Hanein D.. 2006. Three-dimensional structure of vinculin bound to actin filaments. Mol. Cell. 21:271–281 10.1016/j.molcel.2005.11.020 [DOI] [PubMed] [Google Scholar]
- Janssen M.E., Liu H., Volkmann N., and Hanein D.. 2012. The C-terminal tail domain of metavinculin, vinculin’s splice variant, severs actin filaments. J. Cell Biol. 197:585–593 10.1083/jcb.201111046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch B.M., and Isenberg G.. 1981. Interaction of α-actinin and vinculin with actin: opposite effects on filament network formation. Proc. Natl. Acad. Sci. USA. 78:3005–3009 10.1073/pnas.78.5.3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.P., and Craig S.W.. 1995. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature. 373:261–264 10.1038/373261a0 [DOI] [PubMed] [Google Scholar]
- Johnson R.P., and Craig S.W.. 2000. Actin activates a cryptic dimerization potential of the vinculin tail domain. J. Biol. Chem. 275:95–105 10.1074/jbc.275.1.95 [DOI] [PubMed] [Google Scholar]
- Johnson R.P., Niggli V., Durrer P., and Craig S.W.. 1998. A conserved motif in the tail domain of vinculin mediates association with and insertion into acidic phospholipid bilayers. Biochemistry. 37:10211–10222 10.1021/bi9727242 [DOI] [PubMed] [Google Scholar]
- Karplus P.A., and Diederichs K.. 2012. Linking crystallographic model and data quality. Science. 336:1030–1033 10.1126/science.1218231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono N., Ohto U., Hiramatsu T., Urabe M., Uchida Y., Satow Y., and Arai H.. 2013. Impaired α-TTP-PIPs interaction underlies familial vitamin E deficiency. Science. 340:1106–1110 10.1126/science.1233508 [DOI] [PubMed] [Google Scholar]
- Le Clainche C., Dwivedi S.P., Didry D., and Carlier M.F.. 2010. Vinculin is a dually regulated actin filament barbed end-capping and side-binding protein. J. Biol. Chem. 285:23420–23432 10.1074/jbc.M110.102830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Rangarajan E.S., Yogesha S.D., and Izard T.. 2009. Raver1 interactions with vinculin and RNA suggest a feed-forward pathway in directing mRNA to focal adhesions. Structure. 17:833–842 10.1016/j.str.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Rangarajan E.S., Vonrhein C., Bricogne G., and Izard T.. 2012. The metavinculin tail domain directs constitutive interactions with raver1 and vinculin RNA. J. Mol. Biol. 422:697–704 10.1016/j.jmb.2012.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K., Doughman R.L., Firestone A.J., Bunce M.W., and Anderson R.A.. 2002. Type I γ phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 420:89–93 10.1038/nature01082 [DOI] [PubMed] [Google Scholar]
- Ling K., Schill N.J., Wagoner M.P., Sun Y., and Anderson R.A.. 2006. Movin’ on up: the role of PtdIns(4,5)P2 in cell migration. Trends Cell Biol. 16:276–284 10.1016/j.tcb.2006.03.007 [DOI] [PubMed] [Google Scholar]
- Madl T., and Sattler M.. 2009. Adhesion dance with raver. Structure. 17:781–783 10.1016/j.str.2009.05.004 [DOI] [PubMed] [Google Scholar]
- Matthews B.W.1968. Solvent content of protein crystals. J. Mol. Biol. 33:491–497 10.1016/0022-2836(68)90205-2 [DOI] [PubMed] [Google Scholar]
- McLaughlin S., Wang J., Gambhir A., and Murray D.. 2002. PIP2 and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31:151–175 10.1146/annurev.biophys.31.082901.134259 [DOI] [PubMed] [Google Scholar]
- Milam L.M.1985. Electron microscopy of rotary shadowed vinculin and vinculin complexes. J. Mol. Biol. 184:543–545 10.1016/0022-2836(85)90301-8 [DOI] [PubMed] [Google Scholar]
- Molony L., and Burridge K.. 1985. Molecular shape and self-association of vinculin and metavinculin. J. Cell. Biochem. 29:31–36 10.1002/jcb.240290104 [DOI] [PubMed] [Google Scholar]
- Palmer S.M., Schaller M.D., and Campbell S.L.. 2008. Vinculin tail conformation and self-association is independent of pH and H906 protonation. Biochemistry. 47:12467–12475 10.1021/bi801764a [DOI] [PubMed] [Google Scholar]
- Palmer S.M., Playford M.P., Craig S.W., Schaller M.D., and Campbell S.L.. 2009. Lipid binding to the tail domain of vinculin: specificity and the role of the N and C termini. J. Biol. Chem. 284:7223–7231 10.1074/jbc.M807842200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., Nelson E.S., Maiers J.L., and DeMali K.A.. 2011. New insights into vinculin function and regulation. Int. Rev. Cell Mol. Biol. 287:191–231 10.1016/B978-0-12-386043-9.00005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon I.Kh., Baxter A.A., Lay F.T., Mills G.D., Adda C.G., Payne J.A., Phan T.K., Ryan G.F., White J.A., Veneer P.K., et al. 2014. Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. eLife. 3:e01808 10.7554/eLife.01808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramarao N., Le Clainche C., Izard T., Bourdet-Sicard R., Ageron E., Sansonetti P.J., Carlier M.F., and Tran Van Nhieu G.. 2007. Capping of actin filaments by vinculin activated by the Shigella IpaA carboxyl-terminal domain. FEBS Lett. 581:853–857 10.1016/j.febslet.2007.01.057 [DOI] [PubMed] [Google Scholar]
- Rangarajan E.S., and Izard T.. 2012. The cytoskeletal protein α-catenin unfurls upon binding to vinculin. J. Biol. Chem. 287:18492–18499 10.1074/jbc.M112.351023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan E.S., Lee J.H., Yogesha S.D., and Izard T.. 2010. A helix replacement mechanism directs metavinculin functions. PLoS ONE. 5:e10679 10.1371/journal.pone.0010679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan E.S., Lee J.H., and Izard T.. 2011. Apo raver1 structure reveals distinct RRM domain orientations. Protein Sci. 20:1464–1470 10.1002/pro.664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad J.S., Miller J., Tai J., Kim A., Ghanam R.H., and Summers M.F.. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA. 103:11364–11369 10.1073/pnas.0602818103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders R.M., Holt M.R., Jennings L., Sutton D.H., Barsukov I.L., Bobkov A., Liddington R.C., Adamson E.A., Dunn G.A., and Critchley D.R.. 2006. Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 85:487–500 10.1016/j.ejcb.2006.01.014 [DOI] [PubMed] [Google Scholar]
- Shen K., Tolbert C.E., Guilluy C., Swaminathan V.S., Berginski M.E., Burridge K., Superfine R., and Campbell S.L.. 2011. The vinculin C-terminal hairpin mediates F-actin bundle formation, focal adhesion, and cell mechanical properties. J. Biol. Chem. 286:45103–45115 10.1074/jbc.M111.244293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle P.A., Hoffert J.D., Adey N.B., and Craig S.W.. 1999. Polyphosphoinositides inhibit the interaction of vinculin with actin filaments. J. Biol. Chem. 274:18414–18420 10.1074/jbc.274.26.18414 [DOI] [PubMed] [Google Scholar]
- Thievessen I., Thompson P.M., Berlemont S., Plevock K.M., Plotnikov S.V., Zemljic-Harpf A., Ross R.S., Davidson M.W., Danuser G., Campbell S.L., and Waterman C.M.. 2013. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202:163–177 10.1083/jcb.201303129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.M., Tolbert C.E., and Campbell S.L.. 2013. Vinculin and metavinculin: oligomerization and interactions with F-actin. FEBS Lett. 587:1220–1229 10.1016/j.febslet.2013.02.042 [DOI] [PubMed] [Google Scholar]
- Thompson P.M., Tolbert C.E., Shen K., Kota P., Palmer S.M., Plevock K.M., Orlova A., Galkin V.E., Burridge K., Egelman E.H., et al. 2014. Identification of an actin binding surface on vinculin that mediates mechanical cell and focal adhesion properties. Structure. 22:697–706 10.1016/j.str.2014.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert C.E., Burridge K., and Campbell S.L.. 2013. Vinculin regulation of F-actin bundle formation: what does it mean for the cell? Cell Adhes. Migr. 7:219–225 10.4161/cam.23184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A., and Teplyakov A.. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30:1022–1025. [Google Scholar]
- Vonrhein C., Flensburg C., Keller P., Sharff A., Smart O., Paciorek W., Womack T., and Bricogne G.. 2011. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67:293–302 10.1107/S0907444911007773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weekes J., Barry S.T., and Critchley D.R.. 1996. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem. J. 314:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M.2001. Global indicators of X-ray quality. J. Appl. Crystallogr. 34:130–135. [Google Scholar]
- Weiss M.S., and Hilgenfeld R.. 1997. On the use of the merging R factror as a quality indicator for X-ray data. J. Appl. Crystallogr. 30:203–205 10.1107/S0021889897003907 [DOI] [Google Scholar]
- Wilkins J.A., and Lin S.. 1982. High-affinity interaction of vinculin with actin filaments in vitro. Cell. 28:83–90 10.1016/0092-8674(82)90377-4 [DOI] [PubMed] [Google Scholar]
- Wirth V.F., List F., Diez G., and Goldmann W.H.. 2010. Vinculin’s C-terminal region facilitates phospholipid membrane insertion. Biochem. Biophys. Res. Commun. 398:433–437 10.1016/j.bbrc.2010.06.094 [DOI] [PubMed] [Google Scholar]
- Witt S., Zieseniss A., Fock U., Jockusch B.M., and Illenberger S.. 2004. Comparative biochemical analysis suggests that vinculin and metavinculin cooperate in muscular adhesion sites. J. Biol. Chem. 279:31533–31543 10.1074/jbc.M314245200 [DOI] [PubMed] [Google Scholar]
- Xu W., Baribault H., and Adamson E.D.. 1998. Vinculin knockout results in heart and brain defects during embryonic development. Development. 125:327–337. [DOI] [PubMed] [Google Scholar]
- Yin H.L., and Janmey P.A.. 2003. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65:761–789 10.1146/annurev.physiol.65.092101.142517 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Ballestrem C., Kam Z., and Geiger B.. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613 10.1242/jcs.00792 [DOI] [PubMed] [Google Scholar]
- Ziegler W.H., Liddington R.C., and Critchley D.R.. 2006. The structure and regulation of vinculin. Trends Cell Biol. 16:453–460 10.1016/j.tcb.2006.07.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.