Vol. 207 No. 3, November 3, 2014. Pages 335–349.
In the original version of Fig. 1 C, the No Probe Control merged color panel was a duplicate of the SAT III merged color panel. A corrected version of Fig. 1 is shown below.
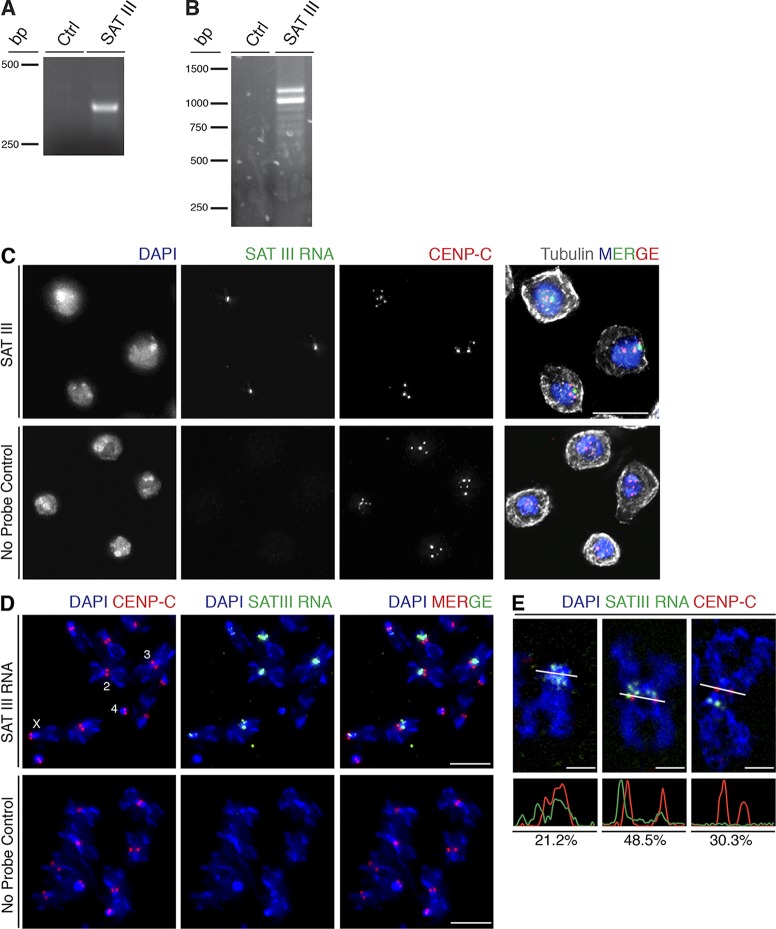
Figure 1.
SAT III is transcribed and localizes to mitotic centromeres. (A) RT-PCR using primers that amplify one repeating unit of SAT III produces a 359-bp product. Ctrl, control RT-PCR reaction with no reverse transcription. (B) SAT III 3ʹ RACE amplified transcripts consisting of multiple repeats of the 359-bp basic unit, resulting in a series of bands. Ctrl, control reaction with no reverse transcription. (C) S2 cells in interphase display one major cluster of SAT III RNA (green) within the nucleus in close proximity to centromeric clusters (labeled with CENP-C in red). Cells were counterstained with DAPI to visualize DNA (blue). Bars, 10 µm. (D) SAT III RNA localization on metaphase chromosome spreads. SAT III RNA localizes to centromeric region of chromosomes X, 2, and 3 but is not detected on the small chromosome 4. Bars, 5 µm. (E) SAT III RNA localization patterns. The bottom panels show intensity profiles of the indicated cross sections in the images (white lines). Based on the intensity profiles of SAT III RNA and CENP-C signal, the localization is characterized as overlapping (21.2%), partially overlapping (48.5%), or not overlapping (30.3%) of analyzed chromosomes (n = 33). The colocalization was measured on a single z stack.
The html and pdf versions of this article have been corrected. The error remains only in the print version.