Abstract
Biological membranes display distinct domains that organize membrane proteins and signaling molecules to facilitate efficient and reliable signaling. The organization of rhodopsin, a G protein-coupled receptor, in native rod outer segment disc membranes was investigated by atomic force microscopy. Atomic force microscopy revealed that rhodopsin is arranged into domains of variable size, which we refer to herein as nanodomains, in native membranes. Quantitative analysis of 150 disc membranes revealed that the physical properties of nanodomains are conserved in humans and mice and that the properties of individual disc membranes can be variable. Examining the variable properties of disc membranes revealed some of the factors contributing to the size of rod outer segment discs and the formation of nanodomains in the membrane. The diameter of rod outer segment discs was dependent on the number of rhodopsin molecules incorporated into the membrane but independent of the spatial density of rhodopsin. The number of nanodomains present in a single disc was also dependent on the number of rhodopsin molecules incorporated into the membrane. The size of the nanodomains was largely independent of the number or spatial density of rhodopsin in the membrane.
Keywords: Atomic force microscopy, G protein-coupled receptor, lipid rafts, membrane structure, photoreceptor, phototransduction
1. Introduction
Rhodopsin is a prototypical G protein-coupled receptor (GPCR) 1 with 7 transmembrane domains (Fig. 1A). This light receptor is located in the outer segment of rod photoreceptor cells and initiates vision upon photon capture. The rod outer segment (ROS) is comprised of discs that are stacked one on top of another in a highly ordered manner [1, 2] (Fig. 2A). There are 500-2000 discs in a single vertebrate ROS [3], depending on the species, encased by a plasma membrane. Rhodopsin is primarily found in the disc membranes of ROS.
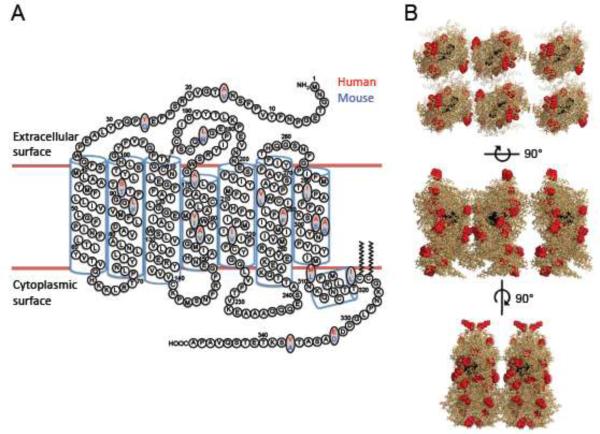
FIGURE 1. Structure of rhodopsin.
(A) The secondary structure of rhodopsin is shown. The 18 amino acid residue differences in the sequences of human (red) and mouse (blue) rhodopsin are highlighted on the secondary structure. (B) The oligomeric model of rhodopsin (PDB: 1N3M) derived from AFM studies is shown in stick representation. The location of the 18 amino acid residue differences in the sequences of human and mouse rhodopsin are highlighted as red spheres. The chromophore 11-cis retinal is shown as black spheres. The first structure is a top view of the extracellular surface. The other structures are side views with the extracellular surface on top and the cytoplasmic surface on the bottom.
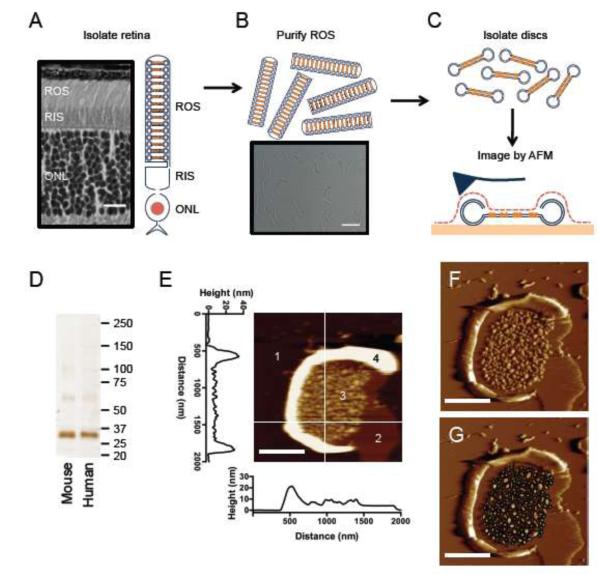
FIGURE 2. Preparation and AFM imaging of ROS disc membranes.
(A) A cross-section of a mouse retina and a cartoon of a rod photoreceptor cell are shown. The rod outer segment (ROS), rod inner segment (RIS) and outer nuclear layer (ONL) are highlighted. Scale bar, 15 μm. (B) Purified ROS are shown as a cartoon and in a light micrograph. Scale bar, 15 μm. (C) ROS discs are isolated from purified ROS by osmotic bursting and washing steps. ROS disc membranes are adsorbed onto mica for AFM imaging. In AFM, a sharp probe is raster-scanned over the sample surface to generate topographical images. (D) SDS-PAGE on ROS disc membrane preparations from mouse and human samples reveal that rhodopsin is the predominant protein species. The sizes of protein standards are indicated in kDa. (E) A height image of a ROS disc membrane. Four different surfaces are revealed: 1, mica; 2, protein-free lipid bilayer; 3, rhodopsin nanodomains; 4, rim region. The height profiles of the highlighted line scans are shown. Scale bar, 500 nm. (F) A deflection image of the same ROS disc membrane. Scale bar, 500 nm. G, The deflection image with nanodomains outlined by black ellipses. The diameters of the ellipses were measured to determine the surface area of the nanodomains.
Rhodopsin is densely packed and found in high concentrations in ROS disc membranes [1, 4]. It is estimated that rhodopsin represents about 70-90% of the protein content in ROS and greater than 90% of the protein content in the disc membranes [5-9]. The high purity and concentration of rhodopsin in the ROS has greatly facilitated the structural, biophysical, and biochemical characterization of the native receptor, making it the most thoroughly studied GPCR in native form. The high concentration of rhodopsin in retinal membranes has been advantageous for experiments, yet this high concentration of molecules creates a crowded disc membrane environment in which signaling must occur with high efficiency, sensitivity, and reliability [10-13].
A crowded membrane environment is not unique to ROS disc membranes. All biological membranes have crowded environments with most being occupied by a heterogeneous complement of membrane proteins. Crowded environments are not ideal for efficient signaling if the signaling cascade proceeds in a random manner via freely diffusing proteins, a condition originally envisioned for biological membranes [14]. More recent evidence suggests biological membranes are more organized than initially recognized forming domains such as lipid rafts [15-18], which can facilitate the efficiency, sensitivity, and reliability that is required for signaling in the ROS [19-21]. The sizes of these domains are usually less than 200 nm [22] and are, therefore, difficult to visualize and study by conventional microscopy methods.
Atomic force microscopy (AFM) is uniquely suited to visualize the nanoscale organization of proteins and domains in biological membranes [23-27]. Imaging can be performed under physiological conditions and requires minimal processing of samples compared to other structural methods. AFM has revealed that ROS disc membranes like other types of biological membranes exhibit order. Two types of packing arrangements for rhodopsin in ROS disc membranes have been observed by AFM: a densely packed paracrystalline lattice arrangement and a less densely packed nanodomain organization. In both cases, rhodopsin forms an oligomeric complex composed of rows of dimeric receptor [28](Fig. 1B).
The first AFM image of an intact single bilayer disc membrane displayed a densely packed paracrystalline lattice of rhodopsin molecules [29]. This image has received the most attention to date and has provided the highest resolution of rhodopsin in native membranes, thereby allowing the generation of an oligomeric model of rhodopsin using the geometric constraints revealed in the image [28, 30] (Fig. 1B). Subsequent AFM images have consistently revealed a nanodomain organization of rhodopsin [28, 30-32], similar to that detected in the current study. Rhodopsin appears to form similar oligomeric complexes within these nanodomains to those present in the densely packed paracrystalline lattice arrangement [28]. The nanodomain organization has received less attention than the densely packed paracrystalline lattice organization. The nanodomain organization, however, may represent the native arrangement since it is the most consistently observed arrangement in AFM studies and is also consistent with cryoelectron tomograms of disc membranes in preserved intact ROS [1].
Dimerization/oligomerization appears to be a common trait among GPCRs [33-35]. Rhodopsin dimers appear to be the basic unit of oligomeric complexes in either densely packed paracrystalline lattices or in nanodomains [28]. The role of dimeric interactions in rhodopsin is beginning to be revealed. The arrangement of rhodopsin into dimers provides a platform for binding signaling partners such as transducin and arrestin, where the stoichiometry can be 2:1 between rhodopsin and the signaling partner [36-38]. Structural and functional asymmetry exists between rhodopsin molecules in a dimeric unit when bound to either transducin or arrestin [37, 39, 40]. This asymmetry may contribute to signaling efficiency and may play a protective role in photoreceptor cells under intense lighting conditions [37, 39, 40].
In contrast to dimeric interactions in rhodopsin, less is known about the role and properties of rhodopsin nanodomains. One limitation has been that the nanodomain organization of rhodopsin has only been qualitatively characterized. Therefore, basic knowledge about the nanodomains, including their size and factors contributing to their formation, has yet to be determined but is required to better understand their role in phototransduction and photoreceptor biology. To gain some of these important insights, AFM was utilized to quantitatively assess the nanodomain organization of rhodopsin in native ROS disc membranes.
Previous AFM studies have focused on samples obtained from murine and bovine retinas. Murine and bovine rhodopsin have served as models to understand the effect of the over 100 mutations detected in the rhodopsin gene that cause retinitis pigmentosa or congenital night blindness in human patients [41]. While murine and bovine rhodopsin are predicted to be structurally indistinguishable [42], it is unclear how structurally similar they are with human rhodopsin. There are 18 amino acid residue differences between human and murine rhodopsin (Fig. 1A). A comparison of rhodopsin nanodomains in ROS disc membranes from human and murine retinas was conducted to determine whether the amino acid residue differences alters the organization of the receptor in the membrane.
2. Materials and Methods
2.1. ROS disc membrane preparation
All experimental procedures were conducted under dim red light. Murine ROS disc membranes were prepared from the retina of 10-15 C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) that were 4-6 weeks old. Eight preparations were used to collect data reported in the study. Mice were dark-adapted overnight prior to being sacrificed. Human ROS disc membranes were prepared from the retina of a single whole donor globe (Saving Sight, Columbia, MO). Five preparations were used to collect data reported in the study. The sex, age, and cause of death of donors are as follows: female, 45, septic shock; female, 63, esophageal cancer; female, 70, cerebral vascular accident; female, 75, lung cancer; male, 51, liver failure. Donors had no reported ocular history known to impact rod photoreceptor cells. Donor cadavers were cooled within 2 hours and eyes procured between 3.5-18 hours after death of the donor. Eyes were placed in Life4°C preservation media (Numedis Incorporated, Isanti, MN) and shipped on ice in a light-tight container. Eyes were received in the laboratory within 24-36 hours of donor death and processed immediately. ROS and ROS disc membranes were obtained from murine and human retina using procedures reported previously [1, 43]. ROS disc membranes were resuspended in 50 μL of Ringer's buffer (10 mM HEPES, 130 mM NaCl, 3.6 mM KCl, 2.4 mM MgCl2, 1.2 mM CaCl2, 0.02 mM EDTA, pH 7.4).
2.2. AFM imaging
All AFM procedures were conducted at ambient temperatures under dim red light. Samples were prepared for AFM by adding 40 μL of ROS disc membranes (5-10 μg/mL) onto freshly cleaved mica and incubating for 10 min. The mica was washed 5 times with 40 μL of Ringer’s buffer to remove unadsorbed material. ROS disc membranes adsorbed on mica were imaged by AFM in imaging buffer (20 mM Tris, 150 mM KCl, 25 mM MgCl2, pH 7.8). AFM was performed using a Multimode II atomic force microscope equipped with an E scanner (13 μm scan size) and silicon nitride cantilevers with a nominal spring constant of 0.06 N/m (NP-S, Bruker Corporation, Santa Barbara, CA). Samples were imaged using contact mode at a scan rate of 5.09 Hz to acquire images at a resolution of 512 lines per frame. Minimal force (< 100 pN) was applied to samples during imaging. Height and deflection images were collected and analyzed [44, 45].
2.3. Analysis of AFM images
The dimension of ROS disc membranes and nanodomains were measured using Nanoscope 5.3 software (Bruker Corporation, Santa Barbara, CA). The height of ROS disc membrane features were measured from height images that were flattened to the first order (Fig. 2E). The average height of the rim region, nanodomains, and protein-free lipid bilayer were obtained from peaks in height distribution histograms of individual images. These values were confirmed by line scan analysis. The lateral dimensions of ROS disc membranes and nanodomains were measured from deflection images. Accuracy of lateral dimensions is limited by tip convolution effects of the AFM probe [46, 47]. ROS disc membranes were presumed to be circular. The diameter of the entire disc membrane and the disc membrane excluding the rim region were measured. The latter measurement was used to compute the inner disc area. Nanodomains were presumed to be elliptical (Fig. 2G). The diameters of the ellipse were measured to compute the surface area of an individual nanodomain. To calculate the number of rhodopsin molecules in a nanodomain, rhodopsin was presumed to form an oligomer with a surface area of 84 nm2 for six rhodopsin molecules (Fig. 1B). Statistical analysis was performed using Prism 6 (GraphPad Software Incorporated, La Jolla, CA). Mean values are reported with the associated standard deviation.
2.4. SDS-PAGE
ROS disc membranes were resuspended in LDS sample buffer containing 50 mM dithiothreitol (Expedeon Incorporated, San Diego, CA). Solubilized samples (500 ng of protein) were loaded onto a 4-12% Tris-Glycine precast gel (Life Technologies, Grand Island, NY) and electrophoresis was conducted. Gels were silver-stained to detect proteins extracted from ROS disc membranes.
3. Results
3.1. AFM of native ROS disc membranes
ROS were purified from human and murine retinas and disc membranes were released from the ROS (Figs. 2A-C). The purity of ROS disc membranes was assessed by SDS-PAGE (Fig. 2D). SDS-PAGE of ROS disc membranes isolated from both human and murine retinas displayed a single major band corresponding to rhodopsin [43]. Thus, preparations of ROS disc membranes were pure and contained predominantly rhodopsin.
ROS disc membranes were adsorbed on mica and imaged by AFM (Fig. 2C). Height images contain information about the height of material adsorbed onto mica, and were used to measure vertical features in ROS disc membranes (Fig. 2E). Deflection images accentuate the edges of sample features, and were used to measure lateral features of ROS disc membranes (Figs. 2F and 2G). Measurements made from AFM images are summarized in Table 1.
Table 1.
Summary of quantitative analysis of AFM images
| Properties of a ROS disc | Parameter value |
|
|---|---|---|
| Human | Mouse | |
| Disc diameter | 1.17 ± 0.24 μm | 1.20 ± 0.25 μm |
| Inner disc area | 0.74 ± 0.37 μm2 | 0.77 ± 0.39 μm2 |
| Nanodomain height | 8.11 ± 0.21 nm | 8.10 ± 0.44 nm |
| Lipid bilayer height | 3.68 ± 0.31 nm | 3.69 ± 0.28 nm |
| Number of nanodomains | 156 ± 103 | 156 ± 99 |
| Nanodomain density | 208 ± 79 μ−2 | 207 ± 79 μ−2 |
| Mean nanodomain size | 1,109 ± 311 nm2 | 1,245 ± 379 nm2 |
| Disc area occupied by nanodomains |
22 ± 8 % | 24 ± 9 % |
| Number of rhodopsin molecules |
11,642 ± 7,108 | 13,513 ± 8,522 |
| Rhodopsin spatial density | 15,865 ± 5,894 μm−2 | 17,486 ± 6,170 μm−2 |
Parameter values were determined from individual ROS disc membranes. The mean values are reported with the associated standard deviation. The mean was determined from the analysis of 50 human ROS disc membranes or 100 murine ROS disc membranes. Significant differences between human and mouse data was assessed by a t-test. No significant differences between parameters were observed (p > 0.05) except for the mean nanodomain size in a ROS disc membrane, where there is a small but significant difference (p = 0.03).
ROS disc membranes displayed a distinct topography in AFM images (Fig. 2E). Discs contain double lamellar membranes that are circumscribed by a hairpin loop forming a rim (Fig. 2C). A majority of adsorbed discs displayed only a single lamellar membrane with a rim region. Incisures were not regularly observed in AFM images, presumably because they are disrupted during adsorption of ROS discs to the mica support. Single-bilayer disc membranes predominantly adsorb on mica exposing the extracellular surface [32, 48]. Thus, AFM images of ROS disc membranes represent the topography of the extracellular surface in most instances. Individual rhodopsin molecules cannot be resolved by AFM in these instances since covalently linked sugar groups at the amino terminal region of rhodopsin interfere with the AFM probe [28].
3.2. Features of ROS disc membranes in AFM images
The average diameter of ROS disc membranes of both human and murine retinas was 1.2 μm (Table 1). The highest features in height profiles corresponded to the rim region (Fig. 2E). The heights of the rim region were variable and corresponded to intact and partially disrupted rims. The lamellar region of disc membranes exhibited two major classes of heights (Fig. 2E). One class had a height of about 4 nm (Table 1), which corresponds to the thickness of a lipid bilayer without embedded proteins. The protein-free lipid bilayer often formed a pool, presumably at the location where the disc breaks open, which may form from lipids in the rim region. The other class of heights from the lamellar region had a height of about 8 nm (Table 1), which corresponds to the height of rhodopsin [49]. The areas in the lamellar region with this height have been shown by AFM and single-molecule force spectroscopy to consist of high concentrations of rhodopsin [28, 32, 48].
Rhodopsin predominantly resides in the lamellar region of the ROS disc and is excluded from the rim region where structural proteins such as peripherin and Rom-1 are localized [50]. In AFM images of human and murine ROS discs, the lamellar region of the disc membrane is organized into clusters of distinct domains that are surrounded by protein-free lipid bilayer (Fig. 2F). The size of the domains is heterogeneous, but almost all have dimensions of less than 100 nm; therefore, these domains are referred to as nanodomains. Both human and murine ROS discs displayed nanodomains in the lamellar region (Fig. 3). Since rhodopsin is the predominant protein species in the lamellar region of discs, rhodopsin molecules must largely form the observed nanodomains. It is possible that rhodopsin coexists with a minor population of additional proteins in these nanodomains.
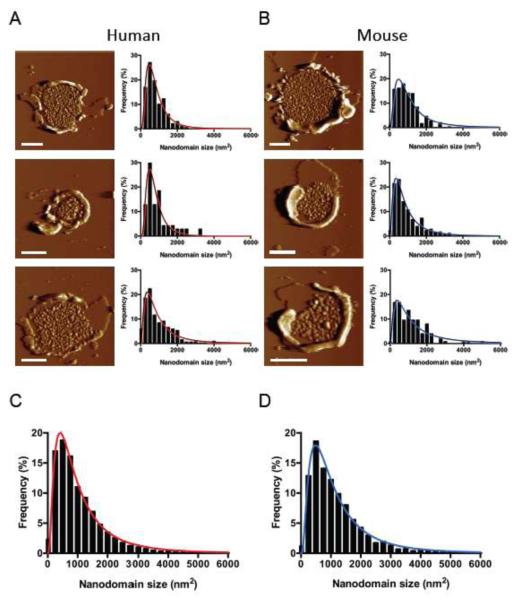
FIGURE 3. Deflection images of human and murine ROS disc membranes.
(A, B) A sampling of deflection images of human and murine ROS disc membranes is shown. A histogram of rhodopsin nanodomain surface areas measured in each ROS disc membrane is shown along with the fit of the data to a Log Gaussian function. (C) A histogram of nanodomain surface areas measured from 50 images of human ROS disc membranes is shown along with the fit of the data to a Log Gaussian function (n = 7,793). (D) A histogram of nanodomain surface areas measured from 100 images of murine ROS disc membranes is shown along with the fit of the data to a Log Gaussian function (n = 15,608).
3.3. Size of nanodomains and density of rhodopsin in disc membranes
Nanodomains have been observed in previous AFM studies on murine and bovine samples but have not been characterized quantitatively. The dimensions of the nanodomains were manually measured to determine their surface area (Fig. 2G). The surface areas of nanodomains from a single human or murine ROS disc generally displayed a skewed distribution that could be fit by a Log Gaussian function (Figs. 3A and 3B). Histograms of nanodomain surface areas obtained from all ROS disc membranes analyzed were also generated. These histograms also displayed a skewed distribution that could be fit by a Log Gaussian function (Figs. 3C and 3D).
The average size of nanodomains in human and murine ROS disc membranes was 1,109 nm2 and 1,245 nm2, respectively (Table 1). Human nanodomains occupied 22 % of the lamellar area of ROS disc membranes whereas murine nanodomains occupied 24 % of the same area. These values are consistent with previous estimates of 25 % of the ROS disc membrane area being occupied by rhodopsin [4].
The number of rhodopsin molecules present in a nanodomain was previously estimated assuming a monomeric arrangement of rhodopsin [32]. Thus, each rhodopsin molecule was estimated to occupy 15 nm2 of space within the membrane [49]. High-resolution AFM images of nanodomains, however, display oligomeric complexes of rhodopsin organized as rows of dimers [28]. The surface area of oligomeric rhodopsin was therefore used to estimate the number of rhodopsin molecules present in a nanodomain of a given size. Using the oligomeric model derived from AFM data [28, 30] (Fig. 1B), six rhodopsin molecules are estimated to occupy 84 nm2 of space on the extracellular surface. Using this value, the number and spatial density of rhodopsin in ROS disc membranes were computed (Table 1).
The average spatial density of rhodopsin in a human ROS disc membrane is 15,865/μm2 (Table 1). This value represents the density of rhodopsin if it were homogeneously distributed within the membrane. The spatial density of rhodopsin in murine ROS disc membranes is 17,486/μm2. The rhodopsin density values derived from AFM images of nanodomains are much lower than that estimated from a densely packed paracrystalline lattice arrangement, where the density was estimated to be 48,300/um2 [29]. Rhodopsin density values derived from a nanodomain arrangement are more consistent with previously reported estimates of about 25,000/um2 [1, 4], albeit a little lower. The discrepancies are small and may be due to a variety of factors. Errors associated with different methods may account for the small differences in the estimate of rhodopsin density. In the current AFM study, rhodopsin monomers may coexist with rhodopsin oligomers that form nanodomains. Diffusely distributed monomers would not be distinguishable in areas that have been classified as protein-free lipid bilayers (Fig. 2E). The presence of monomers would increase rhodopsin density estimates. In addition, computed density values may be underestimated because the estimate for the inner disc area included a proposed girdle zone separating the rim region from the area containing rhodopsin [32].
3.4. Correlation of parameters computed from AFM images
In addition to revealing the average properties of ROS discs, the analysis of 150 individual AFM images revealed the variability that exists among discs. Quantification of parameters describing features of ROS disc membranes provided insights about the properties of nanodomains and their influence on the ROS disc. The variability and correlation of parameters were similar between human and murine samples (Fig. 4).
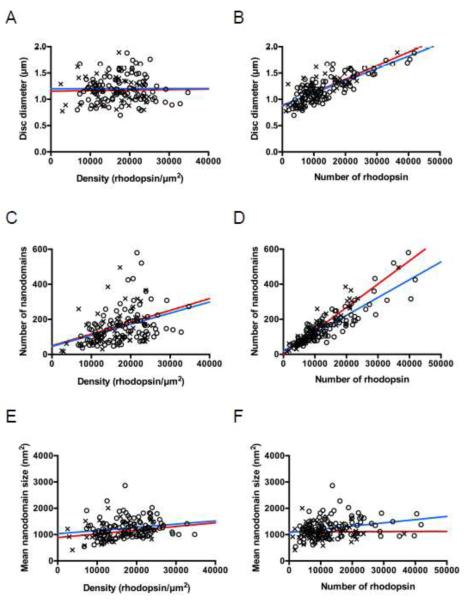
FIGURE 4. Correlation between different parameters computed from AFM images of ROS disc membranes.
Human (red, ×, n = 50) and mouse (blue, o, n = 100) data were plotted and fit by linear regression. A correlation analysis was conducted to compute the Pearson coefficient (r) and level of significance (p). (A) Disc diameter vs. density of rhodopsin. Human, r = 0.02, p = 0.87. Mouse, r = 0.002, p = 0.98. (B) Disc diameter vs. number of rhodopsin molecules. Human, r = 0.75, p < 0.0001. Mouse, r = 0.79, p < 0.0001. (C) Number of nanodomains vs. density of rhodopsin. Human, r = 0.38, p = 0.006. Mouse, r = 0.40, p < 0.0001. (D) Number of nanodomains vs. number of rhodopsin molecules. Human, r = 0.92, p < 0.0001. Mouse, r = 0.88, p < 0.0001. (E) Mean nanodomain size vs. density of rhodopsin. Human, r = 0.27, p = 0.06. Mouse, r = 0.20, p = 0.05. (F) Mean nanodomain size vs. number of rhodopsin molecules. Human, r = 0.008, p = 0.95. Mouse, r = 0.28, p = 0.006.
Although the average diameter of ROS disc membranes is consistent with previous estimates [1], variability was observed in the diameter of individual disc membranes. The diameter of ROS disc membranes ranged from 0.7 – 1.9 μm. No correlation was observed between the diameter of ROS disc membranes and the spatial density of rhodopsin in the disc membrane (Fig. 4A). In contrast, a strong positive correlation was observed between the diameter of ROS disc membranes and the number of rhodopsin molecules present in the membrane (Fig. 4B).
The number of nanodomains found in a ROS disc membrane is strongly correlated with the number of rhodopsin molecules and is more weakly correlated with the spatial density of rhodopsin (Figs. 4C and 4D). In contrast, the average size of nanodomains did not show a correlation, or only a weak correlation, with either the number or spatial density of rhodopsin in the ROS disc membrane (Figs. 4E and 4F).
4. Discussion
4.1. Nanodomain organization of rhodopsin
In the current study, investigation of ROS disc membranes from human and murine retinas by AFM revealed that rhodopsin is organized into nanodomains in both species. Nanodomains of rhodopsin have also been observed in ROS disc membranes from bovine retina [32]. The nanodomain organization of rhodopsin is the most consistently observed arrangement in ROS disc membranes by AFM and is also consistent with the heterogeneous densities observed within disc membranes in cyroelectron tomograms of preserved intact murine ROS [1, 28, 30-32]. The observed nanodomain organization differs from the densely packed paracrystalline lattice arrangement reported in the first AFM study of ROS disc membranes [29]. ROS disc membranes displaying a densely packed paracrystalline lattice were not observed in the current study or in a previous study [32]. Thus, the nanodomain organization of rhodopsin likely represents the most common or physiologically relevant arrangement in the hundreds of stacked discs of the intact ROS. The formation of rhodopsin nanodomains is conserved among humans, mice, and cows, and likely other vertebrate species as well.
4.2. Factors that impact the size of ROS discs
The analysis of individual ROS disc membranes allows for the detection of variability that exists among ROS discs. Variability is present in both the size of ROS discs and the number of rhodopsin molecules incorporated into the disc (Fig. 4). The number of rhodopsin molecules incorporated into ROS disc membranes has a strong influence on the size of ROS discs (Fig. 4B). The correlation between disc diameter and the number of rhodopsin molecules is consistent with observations where either overexpression or underexpression of rhodopsin in genetically modified mice resulted in larger or smaller diameters of ROS, respectively, which in turn is accompanied by altered electrophysiological responses [31, 51, 52].
The variability in the number of rhodopsin molecules incorporated into a ROS disc likely does not arise from differences at the level of transcription or rhodopsin synthesis, but instead, may be due, in part, to the variability in the rate of ROS disc formation induced by light [53, 54]. Constant rhodopsin synthesis and variable rates of ROS disc formation would result in different amounts of rhodopsin being incorporated into the discs. In contrast to the number of rhodopsin molecules incorporated into a ROS disc, the density of rhodopsin in the disc membrane is not a factor in determining the size of ROS discs (Fig. 4A). Thus, the size of ROS discs may adjust to maintain a constant range or average density of rhodopsin in disc membranes [55], perhaps in order to maintain signaling efficiency or sensitivity.
4.3. Factors that impact the number and size of nanodomains
Understanding the physical properties of nanodomains will be required to better understand their functional role. The incorporation of more rhodopsin molecules into disc membranes results in the formation of a greater number of nanodomains (Figs. 4C and 4D). In contrast, the size of nanodomains is largely unaffected by either the number or spatial density of rhodopsin in the membrane (Figs. 4E and 4F).
Rhodopsin likely arranges in nanodomains as an oligomeric complex consisting of rows of dimeric receptor [28]. The factors contributing to the oligomerization of rhodopsin, or other GPCRs, are poorly understood. The hydrophobic mismatch between monomeric GPCRs and the lipid bilayer appears to play a significant role in the spontaneous aggregation of GPCRs [56, 57]. Amino acid residue differences can result in different levels of hydrophobic mismatch, thereby resulting in different sizes of oligomers [56]. Since the sizes of nanodomains in human and murine ROS disc membranes are similar (Table 1), the 18 amino acid residue differences in human and murine rhodopsin do not appear to play a role in determining the size of nanodomains. This is consistent with the putative oligomeric model of rhodopsin where the amino acid residue differences occur in regions of the receptor that are not directly involved in the putative oligomeric interfaces (Fig. 1B), although some are in close proximity to these proposed interfaces.
The skewed distributions displayed in histograms of nanodomain size (Fig. 3) indicate that the variability in the size of nanodomains is random and arises from the product of multiple independent factors. The number or spatial density of rhodopsin in the ROS disc membrane is not one of the factors contributing to this variability (Figs. 4E and 4F). The variability in the average size of nanodomains observed among different ROS discs must then arise from extrinsic factors. One factor may be the lipid bilayer in which rhodopsin is embedded. Properties of the lipid bilayer can have significant influence on the structure and function of rhodopsin [58-63]. The lipid composition is heterogeneous in discs at different axial positions in ROS [64-67], and lipids can impact the oligomerization of rhodopsin [57, 62]. Thus, ROS discs with different complements of lipid in the membrane may result in variable sizes of nanodomains, which in turn may impact phototransduction.
4.4. Nanodomains and rhodopsin diffusion
Classical studies demonstrating the apparent rapid lateral and rotational diffusion of rhodopsin in the disc membranes of amphibian ROS led, in part, to the notion that rhodopsin freely diffuses within the membrane as a monomer [68-71]. These studies are inconsistent with a densely packed paracrystalline lattice arrangement of rhodopsin [72], where the receptor would be expected to be relatively immobile. Determining the size of rhodopsin nanodomains in the current AFM study allows for the assessment of whether this type of organization is consistent with reported lateral diffusion coefficients.
Since the size of a protein embedded in membranes only weakly impacts the lateral diffusion rate [73], it is difficult to differentiate between monomeric and oligomeric rhodopsin by monitoring diffusion alone. A range of lateral diffusion coefficients has been reported for rhodopsin in amphibian ROS: 0.1-0.6 μm2/s [68, 70, 71, 74-79]. Multiple factors can lead to incorrect estimates of rhodopsin lateral diffusion (e.g., [78, 79]), and improvements in experimental procedures indicate that the lower end of the reported range better describes the lateral diffusion of rhodopsin [79].
Lipid rafts in the plasma membrane of various types of cells are predicted to be heterogeneous with sizes in the range of 10-200 nm [22]. Rhodopsin nanodomains fall within the lower half of this size range. It is unclear whether rhodopsin nanodomains represent a typical lipid raft found in plasma membranes since the majority of rhodopsin molecules in ROS disc membranes are not found in the detergent-insoluble fractions typically associated with lipid rafts [80]. However, the similarity in size between rhodopsin nanodomains and lipid rafts suggest that some parallels may exist. Lipid rafts are mobile and those with sizes on the lower end of the spectrum are predicted to exhibit lateral diffusion coefficients of about 0.1 μm2/s [81]. If rhodopsin nanodomains exhibit similar properties as smaller lipid rafts, then rhodopsin nanodomains would be expected to be mobile within the ROS disc membrane and exhibit a lateral diffusion coefficient consistent with the lower end of reported values obtained from amphibian ROS.
4.5. Nanodomains and phototransduction
A functional role for nanodomains may be to provide order to the ROS disc membranes so that efficient and sensitive signaling in ROS can be achieved despite a crowded environment [13, 82]. Theoretical considerations suggest that the nanodomain organization of rhodopsin can facilitate signaling that is quantitatively consistent with classical kinetic data on the initial steps of phototransduction in ROS [83, 84]. Rhodopsin must be arranged as oligomeric complexes consisting of rows of dimeric receptor within nanodomains to achieve consistency with classical kinetic data [84, 85]. Consistency with classical kinetic data can be achieved even in the absence of rhodopsin diffusion within the membrane. Oligomeric complexes of rhodopsin in nandomains can provide a platform for transient interactions between rhodopsin and transducin in the dark state, which may contribute to the efficiency observed in the phototransduction cascade initiated upon light activation of rhodopsin [86]. Nanodomains are consistent with classical diffusion and kinetic data and therefore may represent the organizing principle utilized by ROS disc membranes to achieve signaling efficiency and sensitivity.
5. Conclusions
AFM has provided an avenue to directly visualize the organization of rhodopsin into nanodomains in native ROS disc membranes. The analysis of single ROS disc membranes has revealed the variability in the size of nanodomains and the number and spatial density of rhodopsin in the ROS disc membranes. This quantification has provided insights about some of the basic rules that define the properties of ROS discs and the organization of rhodopsin within the membrane under normal conditions. The number of rhodopsin molecules is a determinant for the size of ROS discs and the number of nanodomains formed. In contrast, the concentration of rhodopsin in the membrane is not a factor in determining the size of ROS discs. The size of nanodomains is largely independent of the number or density of rhodopsin in the disc membranes, and instead, is presumably affected by extrinsic factors. Despite 18 amino acid residue differences (Fig. 1), both human and murine rhodopsin form nanodomains with similar physical properties.
The results from this study lay the groundwork to begin dissecting out the specific factors governing nanodomain formation. The similarities in the properties of human and murine nanodomains indicate that factors contributing to the formation of nanodomains are conserved and that murine ROS disc membranes can serve as a good model for understanding the organization of human rhodopsin in ROS disc membranes. Additionally, the availability of different animal models will allow the testing of whether pathological conditions can alter the normal organization of rhodopsin in the ROS disc membrane and provide insights into the detrimental effects that may follow (e.g., [87, 88]). Although rhodopsin is unique among GPCRs in that it is expressed at high concentrations and purity in ROS disc membranes, the organization of receptors into nanodomains may be a common feature among GPCRs [89].
Highlights.
The organization of rhodopsin in native membranes was visualized by AFM
Human and murine rhodopsin form nanodomains with similar physical properties
The size of nanodomains is heterogeneous within a single rod outer segment disc
Heterogeneity was observed among individual rod outer segment disc membranes
Factors contributing to nanodomain and disc membrane properties were determined
Acknowledgements
We thank Heather Butler and Kathryn Zongolowicz for maintaining our mouse colonies. This work was funded by grants from the National Institutes of Health (R01EY021731, P30EY011373, and T32EY007157) and Research to Prevent Blindness (Unrestricted Grant and Career Development Award).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: AFM, atomic force microscopy; GPCR, G protein-coupled receptor; ROS, rod outer segment(s).
References
- [1].Nickell S, Park PS, Baumeister W, Palczewski K. Three-dimensional architecture of murine rod outer segments determined by cryoelectron tomography. J Cell Biol. 2007;177:917–925. doi: 10.1083/jcb.200612010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gilliam JC, Chang JT, Sandoval IM, Zhang Y, Li T, Pittler SJ, Chiu W, Wensel TG. Three-dimensional architecture of the rod sensory cilium and its disruption in retinal neurodegeneration. Cell. 2012;151:1029–1041. doi: 10.1016/j.cell.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Daemen FJ. Vertebrate rod outer segment membranes. Biochim Biophys Acta. 1973;300:255–288. doi: 10.1016/0304-4157(73)90006-3. [DOI] [PubMed] [Google Scholar]
- [4].Liebman PA, Parker KR, Dratz EA. The molecular mechanism of visual excitation and its relation to the structure and composition of the rod outer segment. Annu Rev Physiol. 1987;49:765–791. doi: 10.1146/annurev.ph.49.030187.004001. [DOI] [PubMed] [Google Scholar]
- [5].Papermaster DS, Dreyer WJ. Rhodopsin content in the outer segment membranes of bovine and frog retinal rods. Biochemistry. 1974;13:2438–2444. doi: 10.1021/bi00708a031. [DOI] [PubMed] [Google Scholar]
- [6].Hamm HE, Bownds MD. Protein complement of rod outer segments of frog retina. Biochemistry. 1986;25:4512–4523. doi: 10.1021/bi00364a010. [DOI] [PubMed] [Google Scholar]
- [7].Godchaux W, 3rd, Zimmerman WF. Soluble proteins of intact bovine rod cell outer segments. Exp Eye Res. 1979;28:483–500. doi: 10.1016/0014-4835(79)90123-4. [DOI] [PubMed] [Google Scholar]
- [8].Heitzmann H. Rhodopsin is the predominant protein of rod outer segment membranes. Nat New Biol. 1972;235:114. doi: 10.1038/newbio235114a0. [DOI] [PubMed] [Google Scholar]
- [9].Downer NW, Englander SW. Hydrogen exchange study of membrane-bound rhodopsin. I. Protein structure. J Biol Chem. 1977;252:8092–8100. [PubMed] [Google Scholar]
- [10].Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. J Physiol. 1979;288:613–634. [PMC free article] [PubMed] [Google Scholar]
- [11].Hecht S, Shlaer S, Pirenne MH. Energy, Quanta, and Vision. J Gen Physiol. 1942;25:819–840. doi: 10.1085/jgp.25.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dartnall HJ. The photosensitivities of visual pigments in the presence of hydroxylamine. Vision Res. 1968;8:339–358. doi: 10.1016/0042-6989(68)90104-1. [DOI] [PubMed] [Google Scholar]
- [13].Cangiano L, Dell'Orco D. Detecting single photons: a supramolecular matter? FEBS Lett. 2013;587:1–4. doi: 10.1016/j.febslet.2012.11.015. [DOI] [PubMed] [Google Scholar]
- [14].Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- [15].Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- [16].Jacobson K, Mouritsen OG, Anderson RG. Lipid rafts: at a crossroad between cell biology and physics. Nat Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- [17].Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- [18].Edidin M. The state of lipid rafts: from model membranes to cells. Annu Rev Biophys Biomol Struct. 2003;32:257–283. doi: 10.1146/annurev.biophys.32.110601.142439. [DOI] [PubMed] [Google Scholar]
- [19].Mugler A, Tostevin F, ten Wolde PR. Spatial partitioning improves the reliability of biochemical signaling. Proc Natl Acad Sci U S A. 2013;110:5927–5932. doi: 10.1073/pnas.1218301110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bethani I, Skanland SS, Dikic I, Acker-Palmer A. Spatial organization of transmembrane receptor signalling. EMBO J. 2010;29:2677–2688. doi: 10.1038/emboj.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Radhakrishnan K, Halasz A, McCabe MM, Edwards JS, Wilson BS. Mathematical simulation of membrane protein clustering for efficient signal transduction. Annals of biomedical engineering. 2012;40:2307–2318. doi: 10.1007/s10439-012-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pike LJ. Rafts defined: a report on the Keystone Symposium on Lipid Rafts and Cell Function. J Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- [23].Whited AM, Park PS. Atomic force microscopy: a multifaceted tool to study membrane proteins and their interactions with ligands. Biochim Biophys Acta. 2014;1838:56–68. doi: 10.1016/j.bbamem.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Engel A, Gaub HE. Structure and mechanics of membrane proteins. Annu Rev Biochem. 2008;77:127–148. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- [25].Muller DJ. AFM: a nanotool in membrane biology. Biochemistry. 2008;47:7986–7998. doi: 10.1021/bi800753x. [DOI] [PubMed] [Google Scholar]
- [26].Casuso I, Rico F, Scheuring S. Biological AFM: where we come from--where we are-where we may go. J Mol Recognit. 2011;24:406–413. doi: 10.1002/jmr.1081. [DOI] [PubMed] [Google Scholar]
- [27].Fotiadis D. Atomic force microscopy for the study of membrane proteins. Curr Opin Biotechnol. 2012;23:510–515. doi: 10.1016/j.copbio.2011.11.032. [DOI] [PubMed] [Google Scholar]
- [28].Liang Y, Fotiadis D, Filipek S, Saperstein DA, Palczewski K, Engel A. Organization of the G protein-coupled receptors rhodopsin and opsin in native membranes. J Biol Chem. 2003;278:21655–21662. doi: 10.1074/jbc.M302536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- [30].Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 2004;564:281–288. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Liang Y, Fotiadis D, Maeda T, Maeda A, Modzelewska A, Filipek S, Saperstein DA, Engel A, Palczewski K. Rhodopsin signaling and organization in heterozygote rhodopsin knockout mice. J Biol Chem. 2004;279:48189–48196. doi: 10.1074/jbc.M408362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Buzhynskyy N, Salesse C, Scheuring S. Rhodopsin is spatially heterogeneously distributed in rod outer segment disk membranes. J Mol Recognit. 2011;24:483–489. doi: 10.1002/jmr.1086. [DOI] [PubMed] [Google Scholar]
- [33].Milligan G, Bouvier M. Methods to monitor the quaternary structure of G protein-coupled receptors. Febs J. 2005;272:2914–2925. doi: 10.1111/j.1742-4658.2005.04731.x. [DOI] [PubMed] [Google Scholar]
- [34].Ferre S, Casado V, Devi LA, Filizola M, Jockers R, Lohse MJ, Milligan G, Pin JP, Guitart X. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol Rev. 2014;66:413–434. doi: 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Park PS-H, Filipek S, Wells JW, Palczewski K. Oligomerization of G protein-coupled receptors: past, present, and future. Biochemistry. 2004;43:15643–15656. doi: 10.1021/bi047907k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Filipek S, Krzysko KA, Fotiadis D, Liang Y, Saperstein DA, Engel A, Palczewski K. A concept for G protein activation by G protein-coupled receptor dimers: the transducin/rhodopsin interface. Photochem Photobiol Sci. 2004;3:628–638. doi: 10.1039/b315661c. [DOI] [PubMed] [Google Scholar]
- [37].Jastrzebska B, Ringler P, Palczewski K, Engel A. The rhodopsin-transducin complex houses two distinct rhodopsin molecules. J Struct Biol. 2013;182:164–172. doi: 10.1016/j.jsb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sommer ME, Hofmann KP, Heck M. Arrestin-rhodopsin binding stoichiometry in isolated rod outer segment membranes depends on the percentage of activated receptors. J Biol Chem. 2011;286:7359–7369. doi: 10.1074/jbc.M110.204941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sommer ME, Hofmann KP, Heck M. Distinct loops in arrestin differentially regulate ligand binding within the GPCR opsin. Nat Commun. 2012;3:995. doi: 10.1038/ncomms2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jastrzebska B, Orban T, Golczak M, Engel A, Palczewski K. Asymmetry of the rhodopsin dimer in complex with transducin. FASEB J. 2013;27:1572–1584. doi: 10.1096/fj.12-225383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Mechanisms of cell death in rhodopsin retinitis pigmentosa: implications for therapy. Trends Mol Med. 2005;11:177–185. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- [42].Kawamura S, Colozo AT, Muller DJ, Park PS. Conservation of molecular interactions stabilizing bovine and mouse rhodopsin. Biochemistry. 2010;49:10412–10420. doi: 10.1021/bi101345x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Park PS, Sapra KT, Jastrzebska B, Maeda T, Maeda A, Pulawski W, Kono M, Lem J, Crouch RK, Filipek S, Muller DJ, Palczewski K. Modulation of molecular interactions and function by rhodopsin palmitylation. Biochemistry. 2009;48:4294–4304. doi: 10.1021/bi900417b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Frederix PL, Bosshart PD, Engel A. Atomic force microscopy of biological membranes. Biophys J. 2009;96:329–338. doi: 10.1016/j.bpj.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Hansma HG, Hoh JH. Biomolecular imaging with the atomic force microscope. Annu Rev Biophys Biomol Struct. 1994;23:115–139. doi: 10.1146/annurev.bb.23.060194.000555. [DOI] [PubMed] [Google Scholar]
- [46].Allen MJ, Hud NV, Balooch M, Tench RJ, Siekhaus WJ, Balhorn R. Tip-radius-induced artifacts in AFM images of protamine-complexed DNA fibers. Ultramicroscopy. 1992;42-44:1095–1100. doi: 10.1016/0304-3991(92)90408-c. [DOI] [PubMed] [Google Scholar]
- [47].Chen SW, Odorico M, Meillan M, Vellutini L, Teulon JM, Parot P, Bennetau B, Pellequer JL. Nanoscale structural features determined by AFM for single virus particles. Nanoscale. 2013;5:10877–10886. doi: 10.1039/c3nr02706f. [DOI] [PubMed] [Google Scholar]
- [48].Sapra KT, Park PS, Filipek S, Engel A, Muller DJ, Palczewski K. Detecting molecular interactions that stabilize native bovine rhodopsin. J Mol Biol. 2006;358:255–269. doi: 10.1016/j.jmb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- [49].Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Molday RS, Hicks D, Molday L. A rim-specific membrane protein of rod outer segment discs. Invest Ophthalmol Vis Sci. 1987;28:50–61. Peripherin. [PubMed] [Google Scholar]
- [51].Makino CL, Wen XH, Michaud NA, Covington HI, DiBenedetto E, Hamm HE, Lem J, Caruso G. Rhodopsin expression level affects rod outer segment morphology and photoresponse kinetics. PLoS One. 2012;7:e37832. doi: 10.1371/journal.pone.0037832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wen XH, Shen L, Brush RS, Michaud N, Al-Ubaidi MR, Gurevich VV, Hamm HE, Lem J, Dibenedetto E, Anderson RE, Makino CL. Overexpression of rhodopsin alters the structure and photoresponse of rod photoreceptors. Biophys J. 2009;96:939–950. doi: 10.1016/j.bpj.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Haeri M, Calvert PD, Solessio E, Pugh EN, Jr., Knox BE. Regulation of rhodopsin-eGFP distribution in transgenic xenopus rod outer segments by light. PLoS One. 2013;8:e80059. doi: 10.1371/journal.pone.0080059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hollyfield JG, Rayborn ME, Verner GE, Maude MB, Anderson RE. Membrane addition to rod photoreceptor outer segments: light stimulates membrane assembly in the absence of increased membrane biosynthesis. Invest Ophthalmol Vis Sci. 1982;22:417–427. [PubMed] [Google Scholar]
- [55].Saxton MJ, Owicki JC. Concentration effects on reactions in membranes: rhodopsin and transducin. Biochim Biophys Acta. 1989;979:27–34. doi: 10.1016/0005-2736(89)90519-1. [DOI] [PubMed] [Google Scholar]
- [56].Mondal S, Johnston JM, Wang H, Khelashvili G, Filizola M, Weinstein H. Membrane driven spatial organization of GPCRs. Scientific reports. 2013;3:2909. doi: 10.1038/srep02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Periole X, Huber T, Marrink SJ, Sakmar TP. G protein-coupled receptors self-assemble in dynamics simulations of model bilayers. J Am Chem Soc. 2007;129:10126–10132. doi: 10.1021/ja0706246. [DOI] [PubMed] [Google Scholar]
- [58].Soubias O, Niu SL, Mitchell DC, Gawrisch K. Lipid-rhodopsin hydrophobic mismatch alters rhodopsin helical content. J Am Chem Soc. 2008;130:12465–12471. doi: 10.1021/ja803599x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Soubias O, Teague WE, Gawrisch K. Evidence for specificity in lipid-rhodopsin interactions. J Biol Chem. 2006;281:33233–33241. doi: 10.1074/jbc.M603059200. [DOI] [PubMed] [Google Scholar]
- [60].Niu SL, Mitchell DC, Litman BJ. Manipulation of cholesterol levels in rod disk membranes by methyl-beta-cyclodextrin: effects on receptor activation. J Biol Chem. 2002;277:20139–20145. doi: 10.1074/jbc.M200594200. [DOI] [PubMed] [Google Scholar]
- [61].Niu SL, Mitchell DC, Lim SY, Wen ZM, Kim HY, Salem N, Jr., Litman BJ. Reduced G protein-coupled signaling efficiency in retinal rod outer segments in response to n-3 fatty acid deficiency. J Biol Chem. 2004;279:31098–31104. doi: 10.1074/jbc.M404376200. [DOI] [PubMed] [Google Scholar]
- [62].Botelho AV, Huber T, Sakmar TP, Brown MF. Curvature and hydrophobic forces drive oligomerization and modulate activity of rhodopsin in membranes. Biophys J. 2006;91:4464–4477. doi: 10.1529/biophysj.106.082776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang Y, Botelho AV, Martinez GV, Brown MF. Electrostatic properties of membrane lipids coupled to metarhodopsin II formation in visual transduction. J Am Chem Soc. 2002;124:7690–7701. doi: 10.1021/ja0200488. [DOI] [PubMed] [Google Scholar]
- [64].Albert AD, Young JE, Paw Z. Phospholipid fatty acyl spatial distribution in bovine rod outer segment disk membranes. Biochim Biophys Acta. 1998;1368:52–60. doi: 10.1016/s0005-2736(97)00200-9. [DOI] [PubMed] [Google Scholar]
- [65].Boesze-Battaglia K, Fliesler SJ, Albert AD. Relationship of cholesterol content to spatial distribution and age of disc membranes in retinal rod outer segments. J Biol Chem. 1990;265:18867–18870. [PMC free article] [PubMed] [Google Scholar]
- [66].Caldwell RB, McLaughlin BJ. Freeze-fracture study of filipin binding in photoreceptor outer segments and pigment epithelium of dystrophic and normal retinas. J Comp Neurol. 1985;236:523–537. doi: 10.1002/cne.902360408. [DOI] [PubMed] [Google Scholar]
- [67].Andrews LD, Cohen AI. Freeze-fracture evidence for the presence of cholesterol in particle-free patches of basal disks and the plasma membrane of retinal rod outer segments of mice and frogs. J Cell Biol. 1979;81:215–228. doi: 10.1083/jcb.81.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liebman PA, Entine G. Lateral diffusion of visual pigment in photorecptor disk membranes. Science. 1974;185:457–459. doi: 10.1126/science.185.4149.457. [DOI] [PubMed] [Google Scholar]
- [69].Cone RA. Rotational diffusion of rhodopsin in the visual receptor membrane. Nat.New Biol. 1972;236:39–43. doi: 10.1038/newbio236039a0. [DOI] [PubMed] [Google Scholar]
- [70].Poo M, Cone RA. Lateral diffusion of rhodopsin in the photoreceptor membrane. Nature. 1974;247:438–441. doi: 10.1038/247438a0. [DOI] [PubMed] [Google Scholar]
- [71].Poo MM, Cone RA. Lateral diffusion of phodopsin in Necturus rods. Exp Eye Res. 1973;17:503–510. doi: 10.1016/0014-4835(73)90079-1. [DOI] [PubMed] [Google Scholar]
- [72].Chabre M, Cone R, Saibil H. Biophysics: is rhodopsin dimeric in native retinal rods? Nature. 2003;426:30–31. doi: 10.1038/426030b. [DOI] [PubMed] [Google Scholar]
- [73].Saffman PG, Delbruck M. Brownian motion in biological membranes. Proc Natl Acad Sci U S A. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wey CL, Cone RA, Edidin MA. Lateral diffusion of rhodopsin in photoreceptor cells measured by fluorescence photobleaching and recovery. Biophys.J. 1981;33:225–232. doi: 10.1016/S0006-3495(81)84883-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Wang Q, Zhang X, Zhang L, He F, Zhang G, Jamrich M, Wensel TG. Activation-dependent hindrance of photoreceptor G protein diffusion by lipid microdomains. J Biol Chem. 2008;283:30015–30024. doi: 10.1074/jbc.M803953200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gupta BD, Williams TP. Lateral diffusion of visual pigments in toad (Bufo marinus) rods and in catfish (Ictalurus punctatus) cones. J Physiol. 1990;430:483–496. doi: 10.1113/jphysiol.1990.sp018303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Drzymala RE, Weiner HL, Dearry CA, Liebman PA. A barrier to lateral diffusion of porphyropsin in Necturus rod outer segment disks. Biophys J. 1984;45:683–692. doi: 10.1016/S0006-3495(84)84210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Govardovskii VI, Korenyak DA, Shukolyukov SA, Zueva LV. Lateral diffusion of rhodopsin in photoreceptor membrane: a reappraisal. Mol Vis. 2009;15:1717–1729. [PMC free article] [PubMed] [Google Scholar]
- [79].Najafi M, Haeri M, Knox BE, Schiesser WE, Calvert PD. Impact of signaling microcompartment geometry on GPCR dynamics in live retinal photoreceptors. J Gen Physiol. 2012;140:249–266. doi: 10.1085/jgp.201210818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Elliott MH, Nash ZA, Takemori N, Fliesler SJ, McClellan ME, Naash MI. Differential distribution of proteins and lipids in detergent-resistant and detergent-soluble domains in rod outer segment plasma membranes and disks. J Neurochem. 2008;104:336–352. doi: 10.1111/j.1471-4159.2007.04971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Cicuta P, Keller SL, Veatch SL. Diffusion of liquid domains in lipid bilayer membranes. J Phys Chem B. 2007;111:3328–3331. doi: 10.1021/jp0702088. [DOI] [PubMed] [Google Scholar]
- [82].Dell'Orco D. A physiological role for the supramolecular organization of rhodopsin and transducin in rod photoreceptors. FEBS Lett. 2013;587:2060–2066. doi: 10.1016/j.febslet.2013.05.017. [DOI] [PubMed] [Google Scholar]
- [83].Pugh EN, Jr., Lamb TD. Amplification and kinetics of the activation steps in phototransduction. Biochim Biophys Acta. 1993;1141:111–149. doi: 10.1016/0005-2728(93)90038-h. [DOI] [PubMed] [Google Scholar]
- [84].Dell'Orco D, Schmidt H. Mesoscopic Monte Carlo simulations of stochastic encounters between photoactivated rhodopsin and transducin in disc membranes. J Phys Chem B. 2008;112:4419–4426. doi: 10.1021/jp709963f. [DOI] [PubMed] [Google Scholar]
- [85].Schoneberg J, Heck M, Hofmann KP, Noe F. Explicit spatiotemporal simulation of receptor-g protein coupling in rod cell disk membranes. Biophys J. 2014;107:1042–1053. doi: 10.1016/j.bpj.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Dell'Orco D, Koch KW. A dynamic scaffolding mechanism for rhodopsin and transducin interaction in vertebrate vision. Biochem J. 2011;440:263–271. doi: 10.1042/BJ20110871. [DOI] [PubMed] [Google Scholar]
- [87].Haeri M, Knox BE. Rhodopsin mutant P23H destabilizes rod photoreceptor disk membranes. PLoS One. 2012;7:e30101. doi: 10.1371/journal.pone.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Zhang N, Kolesnikov AV, Jastrzebska B, Mustafi D, Sawada O, Maeda T, Genoud C, Engel A, Kefalov VJ, Palczewski K. Autosomal recessive retinitis pigmentosa E150K opsin mice exhibit photoreceptor disorganization. J Clin Invest. 2013;123:121–137. doi: 10.1172/JCI66176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Ianoul A, Grant DD, Rouleau Y, Bani-Yaghoub M, Johnston LJ, Pezacki JP. Imaging nanometer domains of beta-adrenergic receptor complexes on the surface of cardiac myocytes. Nat Chem Biol. 2005;1:196–202. doi: 10.1038/nchembio726. [DOI] [PubMed] [Google Scholar]