Abstract
Immunodeficiencies can lead to alterations of the gut microbiome that render it pathogenic and capable of transmitting disease to naïve hosts. Here we review the role of Toll-like receptor (TLR) 5, the innate receptor for bacterial flagellin, in immune responses to the normal gut microbiota with a focus its role on adaptive immunity. Loss of TLR5 has profound effects on the microbiota that include greater temporal instability of major lineages and upregulation of flagellar motility genes that may be linked to the reduced levels of anti-flagellin antibodies in the TLR5−/− host. A variety of human TLR5 gene alleles exist that also associated with inflammatory conditions and may do so via effects on the gut microbiome and altered host-microbial crosstalk.
Introduction
The gastrointestinal tract serves as one of the most dynamic interfaces between the host immune system and commensal, as well as pathogenic, microorganisms. Due to the vast diversity of the microbiota in the gut and of the potential presence of pathogens, the intestinal immune response must be capable of (i) maintaining a basal, appropriate level of antimicrobial activity without pronounced inflammation and, (ii) mounting a pro-inflammatory response to invasive organisms when physical barriers are breached [1,2]. From recent studies we now know that genetic deletion of microbial sensors can lead to changes in cross-talk between the host and the microbiota [3–5]. Experiments in which gut microbiota of immunodeficient animals are transferred into germfree hosts have shown that alterations of gut microbial communities can be sufficient to cause disease in a new host [6–8]. Since one of the defining characteristics of inflammatory diseases including Crohn’s disease and Ulcerative colitis is alteration of the microbiome [9–15], targeting gut microbiotas is an attractive strategy to ameliorate such microbially-mediated disease phenotypes [16]. Understanding how immunity shapes the composition and behavior of gut microbes remains an important task if treatments targeting the microbiota are to be developed.
Innate immune detection of microbes occurs rapidly through specific receptors that recognize conserved molecular structures found on or within the microbe [17]. These pattern recognition receptors include two key families the gut: transmembrane Toll-like receptors (TLRs), and cytosolic nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs). TLR expression has been primarily associated with innate immune cells; however, non-immune cells such as intestinal epithelial cells express TLRs and are equipped to respond to TLR engagement through the production of antimicrobial peptides and cytokines [18,19]. Disruptions in NLR and TLR expression have been associated with alterations in the microbiota composition and intestinal dysbiosis [5,6,20,21]. These studies suggest a critical and highly responsive interplay between the host and the microbiota.
This mini-review focuses on TLR5, the innate immune receptor for flagellin, the principal protein component of bacterial flagella. TLR5 response to flagellin appears to promote both innate and adaptive immune functions and to interact with bacteria in ways that may be fundamental to gut homeostasis and health. TLR5-deficient mice can exhibit either chronic intestinal inflammation, or an obese, metabolic syndrome profile, although there is some variability in penetrance of these phenotypes between colonies [6,20,22,23]. The phenotypic variability may relate to differences in mouse microbiomes between facilities, and may result from dysregulated host-microbial interactions. When a phenotype is apparent, mice deficient in TLR5 are prone to developing two forms of inflammation, namely colitis and metabolic syndrome [6,20]. Inflammation in the TLR5-deficeint host is associated with alterations in gut microbial community composition in conventionally-raised animals, yet is completely eliminated in the germfree state [3]. Transplantation of the gut microbiome of a TLR5-deficient host to wildtype (WT) germfree host is sufficient to transfer metabolic syndrome to the new host [6].
These observations have raised several questions: How is immune homeostasis impacted by loss of TLR5 signaling? What aspects of the TLR5-deficient microbiome are altered and which are required to transfer the phenotype? How does allelic variation in TLR5 genes in humans relate to microbially mediated host phenotypes? Here, we review recent research on the interactions between TLR5, adaptive immunity and the microbiota, and we discuss how variation in the TLR5 gene may alter these interactions to impact host inflammatory phenotypes in humans.
Location and function of TLR5
TLR5 is a receptor for bacterial flagellin [24], and is structurally composed of leucine rich repeats in the ectodomain, a transmembrane domain, and a Toll/IL-1 Receptor-like domain in the cytoplasmic tail that transmits a signal to the cell [25,26]. Part of the TLR5 ectodomain (amino acids 386–407) directly associates with recombinant flagellin [27]. The region of the flagellin molecule that is recognized by TLR5 is a 13 amino acid residue that likely participates in flagellin multimerization, and thus is not accessible for TLR5 recognition in the polymerized form [28]. However, flagellated bacteria, such as Salmonella typhimurium, shed monomeric flagellin upon infection of epithelial cells providing a potential source of monomeric TLR5 ligand in the intestine [29]. Similar to other TLRs [30–33], restricted localization of TLR5 may be a mechanism to regulate cellular response through TLR5 and thus prevent continuous inflammatory response to bacterial flagellin in the gut [19]. Parenteral injection of flagellin in vivo induces a variety of immune outcomes including proinflammatory, Th2, IL-17/IL-22, and Treg cell activation [34–36]. In addition to this complex innate immune-activating capacity, flagellin is also a protein that elicits a flagellin-specific adaptive immune response.
The role of TLR5 in anti-flagellin antibody production
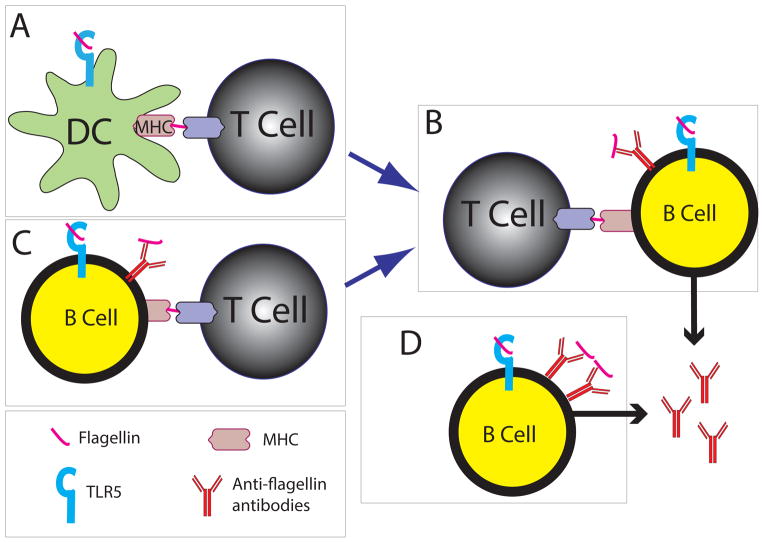
Since flagellin has both antigen and adjuvant activity, it has the capacity to drive anti-flagellin responses (Figure 1). Dendritic cells receive an innate immune signal from flagellin through TLR5, and present flagellin peptides in MHC II to activate naïve flagellin-specific T cells [37] (Figure 1A). The activated T cells will proliferate and assist B cells in producing antibodies (Figure 1B). These B cells are likely stimulated in Peyer’s patches, in a T-dependent manner, prior to trafficking to the lamina propria where they are fully differentiated, class-switched, plasma cells [38]. In this dendritic cell- and T cell-dependent scenario, B cells must also endocytose and present flagellin to receive T cell help, but there is no requirement for the B cell to express and respond via TLR5 since the dendritic cell received it and activated T cells. However, B cells could also endocytose and present flagellin in MHC II directly to T cells, and receive innate signaling from flagellin through TLR5 licensing them to activate naïve T cells (Figure 1C). The T cells will then in turn provide help to the B cells to make antibodies. However, other TLRs could provide the innate signal to B cells, and again, there is no specific requirement for the B cell to express TLR5 [39,40]. Both of these T cell-dependent mechanisms likely account for the majority of flagellin-specific IgA found in the gut.
Figure 1. Possible mechanisms for the production of anti-flagellin antibodies.
A: Dendritic cells (DCs) present flagellin to T cells via MHCII. DCs in the lamina propria express TLR5, but whether this is necessary for antigen presentation is uncertain. (B) Flagellin-specific B cells endocytose flagellin and present it to flagellin-specific T cells that have received activation signals from a DC (A). The T cell provides the necessary cytokine and costimulation for B cells to produce flagellin-specific antibody. B cells express TLR5 in the lamina propria, but whether stimulation of TLR5 by flagellin is necessary for a normal anti-flagellin antibody response is uncertain. (C) B cells can directly present flagellin in MHCII to flagellin-specific T cells after endocytosing flagellin and receiving stimulation through TLR5. In this scenario, there is no requirement for DCs since the T cells can directly provide costimulation and cytokines to the B cells. In (D), flagellin-specific B cells can engage repeating units of flagellin through the B cell receptor and TLR5, which together provide all the stimulation that is needed for the B cell to produce anti-flagellin antibodies. This would occur independently of T cells. Whether this T-independent mechanism can result in production of anti-flagellin antibody is not known.
A third mechanism to induce flagellin specific antibodies does not require dendritic cells or T cells. Shed, monomeric, flagellin could engage both the B cell receptor and TLR5 on the same B cell and elicit antigen-specific antibody production without the need for T cell help. This is known to occur for anti-RNA and anti-DNA antibodies through engagement of TLR7 and TLR9 [41] (Figure 1D). In this scenario, TLR5 must be expressed and respond to flagellin on the B cell. However, expression of TLR5 on B cells is controversial [42,43], and a role for dendritic cells in flagellin-specific antibody production has been proposed [44]. Therefore, it remains unknown whether flagellin-specific antibodies could be generated in the gut without the need for T cells. Regardless of how flagellin-specific antibody producing B cells are generated, these cells will home to the lamina propria and produce secretory IgA, which will be translocated to the lumen where it can bind to bacteria [37,45], and in the case of flagellin specific antibody production can lead to bacterial immobilization [46].
Loss of TLR5 is associated with impaired anti-flagellin antibody production
TLR5 deficient mice, and humans with a 75% reduction in TLR5 expression, exhibit reduced levels of naturally acquired systemic anti-flagellin antibodies [47]. These data suggest that there is a requirement for TLR5 expression on an antigen presenting cell, likely B cells, or dendritic cells. In support of this, B cell deficient (μMT) mice reconstituted with MyD88 deficient B cells, and thus lacking TLR signaling, were unable to generate flagellin specific IgM and IgG1, and had much reduced levels of IgG3 [48], suggesting that there is a requirement TLR response, possibly TLR5, in the B cell. Systemically, the requirement for TLR5 in anti-flagellin antibody production can be overcome upon injection of a bolus of purified flagellin because the flagellin-responsive inflammasome can compensate for absence of TLR5 [49]. It remains possible that a similar alternative flagellin response pathway occurs in B cells.
TLR5-deficient mice also had reduced levels of cecal and fecal anti-flagellin IgA and IgG despite higher overall levels of antibodies [46]. In support of the notion that loss of TLR5 signaling is related to the altered antibody repertoire rather than inflammation, reduced levels of flagellin-specific antibodies were also observed in MyD88-deficient mice but not in mice with chemically-induced inflammation [46]. Since the intestines of TLR5 and MyD88-deficient mice contain many bacteria and bacterial components such as LPS, it is unlikely that additional flagellin, or flagellin mixed with a TLR-stimulating adjuvant would increase the levels of anti-flagellin antibodies in these mice. Thus, these observations suggest that the adjuvant activity of flagellin is dispensable for systemic immune responses, but that TLR5 is critical for an appropriate antibody response against flagellin in the gut.
TLR5-deficiency can alter the nature of the microbiome
In an initial study of mouse microbiome diversity in TLR5 deficient mice, we observed a shift in the overall bacterial diversity present in gut [6]. In contrast, using the same method of analysis (UniFrac analysis of 16S rRNA gene sequence diversity), Ubeda and colleagues did not observe difference in composition between TLR-deficient and WT littermates and suggested that previously observed differences in microbiota composition were due to separate housing histories of the two genotypes [23]. While maternal and housing history can drive differences in certain aspects of microbial diversity, these factors are unlikely to underlie other aberrant microbiome phenotypes that have emerged with subsequent analyses of the TLR5-deficient mouse microbiome and that are potentially relevant inflammation.
Diversity described using 16S rRNA gene sequences derived at a single time point provides one snapshot of the microbiome that can be highly informative but is far from a complete view. To expand our understanding of the TLR5-deficient associated microbiome, we performed (i) time-series analysis of diversity profiles, (ii) metagenomics to characterize the functional gene repertoire of the microbiome, and (iii) metatranscriptomics, which provided a view of the gene expression of the microbiota. These analyses have revealed that loss of TLR5 leads to greater beta-diversity (i.e., greater between-mouse differences), reduced alpha-diversity (i.e., species richness), greater volatility over time, and dramatically altered patterns of gene expression by functionally equivalent microbiomes (patterns observed in data papers are summarized in cartoon form in Figure 2). Based on these observations, it is increasingly clear that lineage content, commonly referred to as “microbiome diversity”, sampled at one point in time is not the most dramatic manifestation of the TLR5-deficient effect on the microbiome, but that temporal dynamics of microbiome population and gene expression are greatly disrupted. The aspects of the microbiome that are most impacted by loss of TLR5 signaling, and that could contribute to development of disease, include greater bacterial motility, greater production of inflammatory molecules such as flagellin and LPS, and possibly also temporal instability. These are discussed in more detail below.
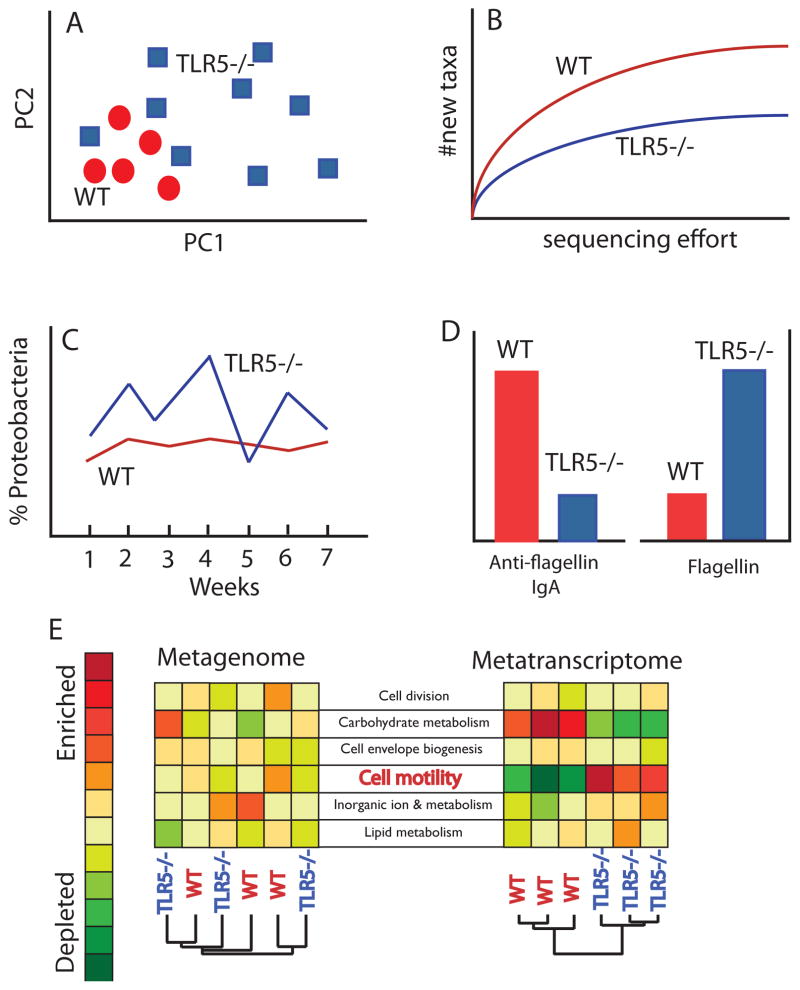
Figure 2. TLR5 deficiency alters the gut microbiome in multiple ways.
Cartoon summary of alterations to the microbiome observed in TLR5-deficient mice (TLR5−/−) compared to wild type (WT), based on real data observations. Panels A–C refer to hypothetical 16S rRNA gene sequence survey data. (A) Beta-diversity (between-sample) patterns for WT and TLR5−/− gut microbiomes in a hypothetical principal coordinates analysis of unweighted UniFrac distances calculated for a set of TLR5−/− and WT mouse gut microbiomes. Each symbols refers to a hypothetical microbiome characterized by sequencing thousands of 16S rRNA genes. When symbols cluster closely, the microbiomes for those samples are more similar. The pattern shown here is one of greater beta-diversity for TLR5−/− mice compared to WT. Note that for certain individual TLR5−/− mice, their microbiome composition may be quite similar to WT, but overall the TLR5−/− are more different from one another than are the WT to one another. (B) Hypothetical alpha-diversity patterns (within-sample diversity) showing lower overall taxonomic richness for TLR5−/− versus WT microbiomes. As sequencing effort increases, the number of new taxa discovered is lowest for the TLR5−/− microbiome, indicating reduced species richness compared to the WT microbiome. (C) Hypothetical data showing greater volatility of proteobacterial populations over time in the TLR5−/− host. Plotted are relative abundances of sequences classified as Proteobacteria over 7 weeks. Levels in the TLR5−/− host are more variable over time. (D) Hypothetical data showing inverse patterns of anti-flagellin antibody levels in the gut versus protein flagellin for TLR5−/− and WT hosts. (E) Hypothetical metagenomes and metatranscriptomes for three TLR5−/− and three WT microbiomes. Metagenomes consist of randomly sampled genes from the composite genomes of the microbes in the microbiome; metatranscriptomes consist of randomly sampled gene transcripts (expressed genes) in the microbiome. Six gene categories are shown for both the metagenomes and the metatranscriptomes, and the relative levels of functional genes or their expression across the six microbiomes are shown using a color scale, where red indicates greater capacity (metagenomes) or greater upregulation (metatranscritpomes). The expression of flagellar motility genes is far greater in the TLR5−/− compared to the WT mouse, despite equivalent coding capacity (similar levels of these genes in the metagenome). The gene expression levels of the functional genes can be used to differentiate the host genotype of the six mice (as indicated by clustering in the right dendrogram) but the metagenome, which represents the functional gene capacity of the microbiomes, cannot distinguish the genotypes (left dendrogram).
TLR5-deficiency destabilizes the microbiome
An important aspect of microbial ecology that is disrupted in the TLR5-deficient host is the temporal stability of relative abundances of taxa making up the microbiome. In both TLR5-deficient phenotypes (severe inflammation (colitic) and low-grade inflammation associated with metabolic syndrome), when the microbiota are sampled over time (days/weeks), a distinctive pattern of instability emerges that is not apparent from single snapshots (Figure 2C) [3,50]. The fluctuations of the relative abundances of the major phyla in the TLR5-deficient microbiome contrast to relative stable levels in WT hosts. Stability is an important aspect of a healthy microbiome as it confers resilience to stress [51,52]. The high volatility of the TLR5-deficient microbiota may, therefore, reduce its resilience and renders the host more susceptible to disturbances that can trigger inflammation.
Another potentially critical aspect the TLR5-deficient gut microbiome is that it exhibits sporadic bursts of abnormally high levels of Proteobacteria [3]. High levels of Proteobacteria are very often associated with inflammation [53–56]. TLR5-deficient mice are unable to effectively clear adherent-invasive E. coli species associated with ileal Crohn’s disease in human [57,58], indicating that TLR5 is essential for the control of this potentially harmful bacterium. When naturally IgA coated bacterial populations from TLR5-deficient mice were enriched by anti-IgA and FACS sorting, and examined by RNAseq, far fewer Proteobacteria were in the IgA-positive fraction compared to the IgA-negative fraction [46]. This was the opposite from WT mice where there were more Proteobacteria in the IgA negative fraction. The reduced representation of the Proteobacteria in the IgA-coated fraction of the TLR5-deficient microbiota indicates that these bacteria generally evade antibody coating in the TLR5-deficeint, but not the WT, gut. Evasion of IgA-coating by Protebacteria may relate to the reduced levels of anti-flagellin antibodies observed in the gut of the TLR5-deficient host, and to the transient bursts in the relative levels of these bacteria.
TLR5-deficiency is associated with upregulation of flagellin genes and higher flagellin protein in the gut
Shotgun metagenomic analysis, in which the genes encoded by the aggregate of microbial genomes in the microbiome are randomly sampled, revealed that there is little overall difference in the functional gene content of the microbiome between WT and TLR5-deficient mice [46](Figure 2). In contrast the gene expression profiles of these functionally similar microbiomes were dramatically different (Figure 2E). Most intriguingly, flagellated bacteria upregulated flagellin expression more in TLR5-deficient, compared to WT, mice and as a result, levels of flagellin capable of eliciting a TLR5 response were also greater in the TLR5-deficient gut [46]. Higher levels of flagellin were present in mice raised in four different facilities, suggesting that this aspect of the microbiome is robust to differences in microbiome composition that can exist between facilities [46]. Mapping flagellin gene transcripts back to reference genomes showed that multiple phyla exhibited this behavior, primarily Firmicutes and Proteobacteria (note that gut-dwelling Bacteroidetes do not encode flagella; [46]). Taken together, these results suggested that loss of TLR5 function renders the normal gut microbiome (i) prone to transient overgrowth of Proteobacteria, (ii) far more motile than normal, and (iii) a source of a high load of the pro-inflammatory protein flagellin in the gut.
Gut bacteria downregulate flagellin gene expression in response to anti-flagellin IgA
Studies in pure culture (using E. coli and Salmonella) have previously shown that anti-flagella antibodies can inhibit motility [59,60]. Similarly, we showed that E. coli downregulates flagellin gene expression in the presence of anti-flagellin antibodies but not anti-LPS antibodies [46]. Furthermore, anti-flagellin antibodies severely inhibited the motility of the gut commensal Firmicute Roseburia hominisI [61], and the motility of bacteria directly obtained from the small intestines of the TLR5-deficient mice [46]. Because the flagellin peptide sequence that stimulates TLR5 signaling is widely conserved across bacterial taxa, antibodies raised against this target of the peptide may also in principle be cross-reactive across bacterial species. As such, anti-flagellin antibodies may act as a signal to downregulate the flagellin gene expression of a wide range of bacterial species, or at a minimum to inhibit their motility.
Bacteria that have a capacity to produce flagella do so under specific environmental conditions related to substrate availability [62], competition [61], and the immune milieu [46,63]. It appears to be a general phenomenon that bacteria sense specific antibodies and respond by altering gene expression of the epitope [64,65]. This may explain why despite a broad capacity to produce flagella, levels of flagellin, when measured by shotgun proteomics, are low in the feces of healthy humans [66]. Increased motility associated with enhanced flagellin expression may contribute to the disease phenotypes, colitis and metabolic syndrome, observed in the absence of TLR5.
Bacterial flagellins differ in their ability to stimulate TLR5
In keeping with innate immune receptors recognizing conserved features among pathogens, TLR5 recognizes a highly conserved region of flagellin. However, all flagellins are not equally able to induce inflammatory responses via TLR5. For example, the stomach-dwelling Helicobacter pylori produces a flagellin that is 1000 fold less potent than Salmonella typhimurium flagellin in its ability to stimulate TLR5 [67]. Several groups have identified amino acid residues that are required for TLR5 stimulation and multiple sequence alignments of flagellin genes obtained from different species of gut bacteria can be used to identify those predicted to stimulate TLR5 [27,28,68]. Structural studies have definitively mapped the interaction domains on TLR5 and flagellin [26]. Interestingly, a single species of gut commensal bacterium can encode in its genome different genes that result in flagellins with differing abilities to activate TLR5 (e.g., Roseburia hominis) [61,62]. How and when such species would transcribe these different flagellin genes remains enigmatic, but their co-existence in the genome suggests that they have specific biological functions and that deliberate detection or evasion by TLR5 is an important component of those functions. Since flagellin is also detected by the cytosolic receptor NLRC4/IPAF, there is also the potential for modulation of flagellin expression in different locations to avoid detection by different receptors [49].
Allelelic Variation in TLR5 genes
TLR5 shows evidence of positive selection in primates, indicating it may experience some of the same evolutionary pressures previously described for adaptive immunity genes [69]. Several studies have identified single nucleotide polymorphisms in the gene for TLR5 that may be associated with disease and/or an inability to recognize its ligand (Table 1). Of particular interest is a mutation in TLR5 with a relatively high minor allele frequency, which results in a stop codon at amino acid 392 and produces a truncated form of the protein (TLR5392STOP) [70]. PMBCs harvested from individuals heterozygous for TLR5392STOP exhibited a significant reduction in IL-1, IL-6, and TNF- production in response to flagellin [70]. This suggests that expression of the truncated form of TLR5 acts in a dominant fashion to suppress signaling response through the functional allele, which is consistent with TLR5 signaling as a homodimer. Additionally, we have shown a reduction in anti-flagellin serum IgA and IgG in individuals heterozygous for the same mutation [47], which is also consistent with recent reports characterizing serum immunoglobulin profiles in TLR5-deficient mice [40]. Recently, Al-Dagrhi et al. reported that the TLR5WT/TLR5392STOP genotype was protective for obesity but predisposed women to Type 2 Diabetes in an Arabic population [71], a finding that is remarkably consistent with the metabolic syndrome phenotype observed in TLR5-deficient mice. Whether humans with this allele harbor more flagellated microbiota remains to be determined.
Table 1.
Known SNPs in the hTLR5 gene with allele characteristics
| dbSNP_ID | Mutation Type | Protein Domain | AA Variation | mRNA NT | Phenotypes in Disease and Human Health | Minor allele freqencies (%) [NHLBI]
|
|
|---|---|---|---|---|---|---|---|
| MAF_EA | MAF_AA | ||||||
| rs5744168 | Nonsense | LRR | R392* | C1174T | Dominant-negative effect in heterozygotes characterized by a reduction in IL-6 production by PBMCs (1) Increase in susceptibility to Legionnaire’s disease (1) Protection from the development of systemic lupus erythematosus (2) Dominant-negative effect in heterzygotes further characterized by a reduction in TNF-α and IL-1β production by PBMCs (2) Reduction in serum anti-flagellin IgA and IgG; no change in total IgA and IgG (3) Inability to recognize flagellin in a transfection assay (CHO-K1) (4) Increase in susceptibility to recurrent UTIs; increase in recurrent UTI disease intensity (5) |
5.4651 | 2.4058 |
| rs2072493 | Missense | LRR | N592S | A1775G | Increase in susceptibility to Legionnaire’s disease (1) Inability to recognize flagellin in a transfection assay (CHO-K1) (4) |
15.9651 | 3.2683 |
| rs5744176 | Missense | TIR | D694G | T1846C | Inability to recognize flagellin in a transfection assay (CHO-K1) (4) | 41.9767 | 16.8407 |
| rs7512943 | Missense | TIR | L822F | C3105T | Inability to recognize flagellin in a transfection assay (CHO-K1) (4) | n.d. | n.d. |
References
- Hawn T, et al. 2003. A common dominant TLR5 stop codon polymorphsim abolishes flagellin signaling and is associated with susceptibility to Legionnaire’s disease. J Exp Med 198(10):1563–1572
- Hawn T, et al. 2005. A stop codon polymorphism of Toll-like receptor 5 is associated with resistance to systemic lupus erythematous. PNAS 102(30):10593–10597
- Gewirtz A, et al. 2006. Dominant negative TLR5 polymorphism reduces adaptive immune response to flagellin and negatively associates with Crohn’s disease. Am J Physiol Gastrointest Liver Physiol 290:G1157–G1163
- Merx S, Zimmer W, Neumaier M, Ahmad-Nejad P. 2006. Characterization and functional investigation of single nucleotide polymorphisms (SNPs) in the human TLR5 gene. Hum Mut 880: 1–7. Online.
- Hawn T, et al. Toll-like receptor polymorphisms and susceptibility to urinary tract infections in adult women. PLoS One 4(6): 1–10
In addition to truncation mutants, functional TLR5 variants are associated with disease outcomes. For example, in colorectal cancer, the TLR5 variant TLR5F616L was associated with improved survival, but poor prognosis was associated with TLR5N592S [72]. The early truncation mutant and these two SNP variants lie in section of the gene encoding the ectodomain region near leucine rich repeat 22 and the C-terminal cap, but not in the region for flagellin binding or either of the receptor dimerization interfaces [26]. The truncation mutant likely functions by retaining ligand binding ability and dimerization ability, thereby interfering with productive TLR5 signaling dimers. Results from truncation and allelic variants are partially consistent with the TLR5-deficient mouse phenotypes, but future work will be required to determine the impact of mutants such as TLR5392STOP on the composition and gene expression profiles of the gut microbes, and how these alterations in gut microbes contribute to disease.
Conclusion
TLR5 is integral to mucosal barrier protection and its role extends, via its effect on adaptive immunity, to regulation of the behavior and composition of the gut microbes. Loss of TLR5 leads to inflammation that if contained can manifest as metabolic syndrome, but can progress to colitis and disease. On the host side, TLR5 genes are under positive selection, which indicates that they are a rapidly evolving component of host-microbial interactions; on the microbial side, bacteria have evolved mechanisms to modulate stimulation of TLR5 though variation in flagellin peptide sequence and levels of gene expression. Variation in human alleles for TLR5 is associated with disease susceptibility, suggesting that this variation could impact host-microbial interactions that are important to health. A number of questions remain to be answered before we have a full understanding of how TLR5, the microbiome, and disease state interact. How does loss of TLR5 signaling lead to reduced anti-flagellin antibody levels? Why is the phenotype of TLR5 deficient animals variable, and can it be predicted from environmental and microbiome factors? What benefit does the TLR5WT/TLR5392STOP genotype confer? Dissecting how variation in flagellins interacts with TLR5 variants and how these interactions modulate host phenotype will yield greater insights into the human-microbiome co-evolution and disease risk in persons carrying the different TLR5 alleles.
Highlights.
The innate receptor TLR5 recognizes flagellin
TLR5 has a role in antibody production against flagellin
Loss of TLR5 results in greater flagellin gene expression by the microbiome
Allelic variation in humans is consistent with a microbial effect on phenotype
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Cerutti A, Chen K, Chorny A. Immunoglobulin responses at the mucosal interface. Annual Review of Immunology. 2011;29:273–93. doi: 10.1146/annurev-immunol-031210-101317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Waaij LA, Limburg PC, Mesander G, van der Waaij D. In vivo IgA coating of anaerobic bacteria in human faeces. Gut. 1996;38:348–54. doi: 10.1136/gut.38.3.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho FA, Koren O, Goodrich JK, Johansson MEV, Nalbantoglu I, Aitken JD, et al. Transient Inability to Manage Proteobacteria Promotes Chronic Gut Inflammation in TLR5-Deficient Mice. Cell Host & Microbe. 2012;12:139–52. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, et al. Nod2 is required for the regulation of commensal microbiota in the intestine. Proceedings of the National Academy of Sciences. 2009;106:15813–8. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 Inflammasome Regulates Colonic Microbial Ecology and Risk for Colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–31. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–131. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 8.Garrett W, Lord G, Punit S, Lugo-Villarino G, Mazmanian S, Ito S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2002;52:237–42. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prindiville T, Cantrell M, Wilson KH. Ribosomal DNA Sequence Analysis of Mucosa-Associated Bacteria in Crohn’s Disease. Inflamm Bowel Dis. 2004;10:824–33. doi: 10.1097/00054725-200411000-00017. [DOI] [PubMed] [Google Scholar]
- 11.Lepage P, Seksik P, Sutren M. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflammatory Bowel …. 2004 doi: 10.1097/01.mib.0000159662.62651.06. [DOI] [PubMed] [Google Scholar]
- 12.Manichanh C. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–11. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gophna U, Sommerfeld K, Gophna S. Differences between Crohn’s disease and ulcerative colitis patients in tissue-associated intestinal microflora. Journal of Clinical …. 2005 doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scanlan PD, Shanahan F, O’Mahony C, Marchesi JR. Culture-Independent Analyses of Temporal Variation of the Dominant Fecal Microbiota and Targeted Bacterial Subgroups in Crohn’s Disease. Journal of Clinical Microbiology. 2006;44:3980–8. doi: 10.1128/JCM.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mangin IN, Bonnet RG, Seksik P, Rigottier-Gois L, Sutren MN, Bouhnik Y, et al. Molecular inventory of faecal microflora in patients with Crohn’s disease. FEMS Microbiology Ecology. n.d;50:25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 16.van Nood, Dijkgraaf, Keller Duodenal Infusion of Feces for Recurrent Clostridium difficile. N Engl J Med. 2013;368:2143–5. doi: 10.1056/NEJMc1303919. [DOI] [PubMed] [Google Scholar]
- 17.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Marques R, Boneca IG. Expression and functional importance of innate immune receptors by intestinal epithelial cells. Cell Mol Life Sci. 2011;68:3661–73. doi: 10.1007/s00018-011-0829-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2000;167:1882–5. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 20.Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, et al. Deletion of TLR5 results in spontaneous colitis in mice. J Clin Invest. 2007;117:3909–21. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carvalho FA, Aitken JD, Vijay-Kumar M, Gewirtz AT. Toll-Like Receptor-Gut Microbiota Interactions: Perturb at Your Own Risk! Annu Rev Physiol. 2012;74:177–98. doi: 10.1146/annurev-physiol-020911-153330. [DOI] [PubMed] [Google Scholar]
- 22.Letran SE, Lee S-J, Atif SM, Flores-Langarica A, Uematsu S, Akira S, et al. TLR5-deficient mice lack basal inflammatory and metabolic defects but exhibit impaired CD4 T cell responses to a flagellated pathogen. J Immunol. 2011;186:5406–12. doi: 10.4049/jimmunol.1003576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. Journal of Experimental Medicine. 2012;209:1445–56. doi: 10.1084/jem.20120504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 25.Bell JK, Mullen GED, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–33. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- 26.Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, et al. Structural Basis of TLR5-Flagellin Recognition and Signaling. Science. 2012;335:859–64. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizel SB, West AP, Hantgan RR. Identification of a Sequence in Human Toll-like Receptor 5 Required for the Binding of Gram-negative Flagellin. Journal of Biological Chemistry. 2003;278:23624–9. doi: 10.1074/jbc.M303481200. [DOI] [PubMed] [Google Scholar]
- 28.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SLR, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–53. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 29.Gewirtz AT, Simon PO, Schmitt CK, Taylor LJ, Hagedorn CH, O’Brien AD, et al. Salmonella typhimurium translocates flagellin across intestinal epithelia, inducing a proinflammatory response. J Clin Invest. 2001;107:99–109. doi: 10.1172/JCI10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.CAL, JCB . Regulation of Nucleic Acid Sensing Toll-Like Receptors in Systemic Lupus Erythematosus. 2011. [Google Scholar]
- 31.Leifer CA, Kennedy MN, Mazzoni A, Lee C, Kruhlak MJ, Segal DM. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J Immunol. 2004;173:1179–83. doi: 10.4049/jimmunol.173.2.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leifer CA, Brooks JC, Hoelzer K, Lopez J, Kennedy MN, Mazzoni A, et al. Cytoplasmic targeting motifs control localization of toll-like receptor 9. J Biol Chem. 2006;281:35585–92. doi: 10.1074/jbc.M607511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chockalingam A, Brooks JC, Cameron JL, Blum LK, Leifer CA. TLR9 traffics through the Golgi complex to localize to endolysosomes and respond to CpG DNA. Immunol Cell Biol. 2009;87:209–17. doi: 10.1038/icb.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bobat S, Flores-Langarica A, Hitchcock J, Marshall JL, Kingsley RA, Goodall M, et al. Soluble flagellin, FliC, induces an Ag-specific Th2 response, yet promotes T-bet-regulated Th1 clearance of Salmonella typhimurium infection. Eur J Immunol. 2011;41:1606–18. doi: 10.1002/eji.201041089. [DOI] [PubMed] [Google Scholar]
- 35.Van Maele L, Carnoy C, Cayet D, Songhet P, Dumoutier L, Ferrero I, et al. TLR5 Signaling Stimulates the Innate Production of IL-17 and IL-22 by CD3(neg)CD127(+) Immune Cells in Spleen and Mucosa. J Immunol. 2010;185:1177–85. doi: 10.4049/jimmunol.1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Didierlaurent A, Ferrero I, Otten LA, Dubois B, Reinhardt M, Carlsen H, et al. Flagellin promotes myeloid differentiation factor 88-dependent development of Th2-type response. J Immunol. 2004;172:6922–30. doi: 10.4049/jimmunol.172.11.6922. [DOI] [PubMed] [Google Scholar]
- 37.Letran SE, Lee S-J, Atif SM, Uematsu S, Akira S, McSorley SJ. TLR5 functions as an endocytic receptor to enhance flagellin-specific adaptive immunity. Eur J Immunol. 2010;41:29–38. doi: 10.1002/eji.201040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunn-Walters DK, Isaacson PG, Spencer J. Sequence analysis of human IgVH genes indicates that ileal lamina propria plasma cells are derived from Peyer’s patches. Eur J Immunol. 1997;27:463–7. doi: 10.1002/eji.1830270217. [DOI] [PubMed] [Google Scholar]
- 39.Sanders CJ, Franchi L, Yarovinsky F, Uematsu S, Akira S, Núñez G, et al. Induction of adaptive immunity by flagellin does not require robust activation of innate immunity. Eur J Immunol. 2009;39:359–71. doi: 10.1002/eji.200838804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lopez-Yglesias AH, Zhao X, Quarles EK, Lai MA, VandenBos T, Strong RK, et al. Flagellin Induces Antibody Responses through a TLR5- and Inflammasome-Independent Pathway. J Immunol. 2014;192:1587–96. doi: 10.4049/jimmunol.1301893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marshak-Rothstein A. Toll-like receptors in systemic autoimmune disease. Nature Reviews Immunology. 2006;6:823–35. doi: 10.1038/nri1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gururajan M, Jacob J, Pulendran B. Toll-Like Receptor Expression and Responsiveness of Distinct Murine Splenic and Mucosal B-Cell Subsets. PLoS ONE. 2007:2. doi: 10.1371/journal.pone.0000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dorner M, Brandt S, Tinguely M, Zucol F, Bourquin J-P, Zauner L, et al. Plasma cell toll-like receptor (TLR) expression differs from that of B cells, and plasma cell TLR triggering enhances immunoglobulin production. Immunology. 2009;128:573–9. doi: 10.1111/j.1365-2567.2009.03143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flores-Langarica A, Marshall JL, Hitchcock J, Cook C, Jobanputra J, Bobat S, et al. Systemic Flagellin Immunization Stimulates Mucosal CD103(+) Dendritic Cells and Drives Foxp3(+) Regulatory T Cell and IgA Responses in the Mesenteric Lymph Node. J Immunol. 2012;189:5745–54. doi: 10.4049/jimmunol.1202283. [DOI] [PubMed] [Google Scholar]
- 45.Hooper LV, Littman DR, Macpherson AJ. Interactions Between the Microbiota and the Immune System. Science. 2012;336:1268–73. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, et al. Innate and Adaptive Immunity Interact to Quench Microbiome Flagellar Motility in the Gut. Cell Host & Microbe. 2013;14:571–81. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gewirtz AT. Dominant-negative TLR5 polymorphism reduces adaptive immune response to flagellin and negatively associates with Crohn’s disease. AJP: Gastrointestinal and Liver Physiology. 2006;290:G1157–63. doi: 10.1152/ajpgi.00544.2005. [DOI] [PubMed] [Google Scholar]
- 48.Pasare C, Medzhitov R. Control of B-cell responses by Toll-like receptors. Nature. 2005;438:364–8. doi: 10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 49.Vijay-Kumar M, Carvalho FA, Aitken JD, Fifadara NH, Gewirtz AT. TLR5 or NLRC4 is necessary and sufficient for promotion of humoral immunity by flagellin. Eur J Immunol. 2010;40:3528–34. doi: 10.1002/eji.201040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2014;63:1069–80. doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Relman DA. The human microbiome: ecosystem resilience and health. Nutrition Reviews. 2012;70:S2–S9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proceedings of the National Academy of Sciences. 2007;104:13780–5. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lepage P, Häsler R, Spehlmann ME, Rehman A, Zvirbliene A, Begun A, et al. Twin Study Indicates Loss of Interaction Between Microbiota and Mucosa of Patients With Ulcerative Colitis. Gastroenterology. 2011;141:227–36. doi: 10.1053/j.gastro.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 55.Xenoulis PG, Palculict B, Allenspach K, Steiner JRM, Van House AM, Suchodolski JS. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiology Ecology. n.d;66:579–89. doi: 10.1111/j.1574-6941.2008.00556.x. [DOI] [PubMed] [Google Scholar]
- 56.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, et al. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn’s disease involving the ileum. Isme J. 2007;1:403–18. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 57.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2003 doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 58.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology. 1997 doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 59.Forbes SJ, Martinelli D, Hsieh C, Ault JG, Marko M, Mannella CA, et al. Association of a Protective Monoclonal IgA with the O Antigen of Salmonella enterica Serovar Typhimurium Impacts Type 3 Secretion and Outer Membrane Integrity. Infect Immun. 2012;80:2454–63. doi: 10.1128/IAI.00018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suo Z, Yang X, Avci R, Deliorman M, Rugheimer P, Pascual DW, et al. Antibody Selection for Immobilizing Living Bacteria. Anal Chem. 2009;81:7571–8. doi: 10.1021/ac9014484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stanton TB, Savage DC. Motility as a factor in bowel colonization by Roseburia cecicola, an obligately anaerobic bacterium from the mouse caecum. J Gen Microbiol. 1983;130:173–83. doi: 10.1099/00221287-130-1-173. [DOI] [PubMed] [Google Scholar]
- 62.Scott KP, Martin JC, Chassard C, Clerget M, Potrykus J, Campbell G, et al. Substrate-driven gene expression in Roseburia inulinivorans: Importance of inducible enzymes in the utilization of inulin and starch. Proceedings of the National Academy of Sciences. 2011;108:4672–9. doi: 10.1073/pnas.1000091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2:328–39. doi: 10.1016/j.chom.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 64.Lönnermark E, Nowrouzinan F, Adlerberth I, Ahrné S, Wold A, Friman V. Oral and faecal lactobacilli and their expression of mannose-specific adhesins in individuals with and without IgA deficiency. International Journal of Medical Microbiology. 2012;302:53–60. doi: 10.1016/j.ijmm.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 65.Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–11. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, et al. Shotgun metaproteomics of the human distal gut microbiota. Isme J. 2008;3:179–89. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 67.Gewirtz AT, Yu Y, Krishna US, Israel DA, Lyons SL, Peek RM., Jr Helicobacter pyloriFlagellin Evades Toll-Like Receptor 5–Mediated Innate Immunity. J Infect Dis. 2004;189:1914–20. doi: 10.1086/386289. [DOI] [PubMed] [Google Scholar]
- 68.Andersen-Nissen E, Smith KD, Bonneau R, Strong RK, Aderem A. A conserved surface on Toll-like receptor 5 recognizes bacterial flagellin. Journal of Experimental Medicine. 2007;204:393–403. doi: 10.1084/jem.20061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wlasiuk G, Khan S, Switzer WM, Nachman MW. A History of Recurrent Positive Selection at the Toll-Like Receptor 5 in Primates. Molecular Biology and Evolution. 2009;26:937–49. doi: 10.1093/molbev/msp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ, et al. A Common Dominant TLR5 Stop Codon Polymorphism Abolishes Flagellin Signaling and Is Associated with Susceptibility to Legionnaires’ Disease. Journal of Experimental Medicine. 2003;198:1563–72. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al-Daghri NM, Clerici M, Al-Attas O, Forni D, Alokail MS, Alkharfy KM, et al. A Nonsense Polymorphism (R392X) in TLR5 Protects from Obesity but Predisposes to Diabetes. J Immunol. 2013;190:3716–20. doi: 10.4049/jimmunol.1202936. [DOI] [PubMed] [Google Scholar]
- 72.Klimosch SN, Forsti A, Eckert J, Knezevic J, Bevier M, Schonfels von W, et al. Functional TLR5 Genetic Variants Affect Human Colorectal Cancer Survival. Cancer Research. 2013;73:7232–42. doi: 10.1158/0008-5472.CAN-13-1746. [DOI] [PubMed] [Google Scholar]