Abstract
Western equine encephalitis virus (WEEV), Highlands J virus (HJV), and Fort Morgan virus (FMV) are the sole representatives of the WEE antigenic complex of the genus Alphavirus, family Togaviridae, that are endemic to North America. All three viruses have their ancestry in a recombination event involving eastern equine encephalitis virus (EEEV) and a Sindbis (SIN)-like virus that gave rise to a chimeric alphavirus that subsequently diversified into the present-day WEEV, HJV, and FMV. Here, we present a comparative analysis of the genetic, ecological, and evolutionary relationships among these recombinant-origin viruses, including the description of a nsP4 polymerase mutation in FMV that allows it to circumvent the host range barrier to Asian tiger mosquito cells, a vector species that is normally refractory to infection. Notably, we also provide evidence that the recombination event that gave rise to these three WEEV antigenic complex viruses may have occurred in North America.
Keywords: Fort Morgan virus, Highlands J virus, Western equine encephalitis virus, Western equine encephalitis antigenic complex, Alphavirus, Togavirus, Recombination, Host range, Virus emergence
Introduction
The genus Alphavirus within the family Togaviridae is comprised of 31 arthropod-borne viruses with a worldwide distribution (Powers et al., 2011; Nasar et al., 2012). Of these viruses, four are endemic to North America: eastern equine encephalitis virus (EEEV), western equine encephalitis virus (WEEV), Highlands J virus (HJV), and Fort Morgan virus (FMV) (Weaver et al., 1997). Two additional alphaviruses found in the western United States, Buggy Creek virus and the recently described Stone Lakes virus, are generally regarded as variants of FMV (Hopla et al. 1993; Powers et al., 2001; Brault et al., 2009). WEEV, HJV, and FMV are notable in that they are descendants of a recombination event between a Sindbis (SIN)-like virus and EEEV that is believed to have occurred in the neotropics of South America (Hahn et al., 1988; Strauss and Strauss, 1997). As the major surface glycoproteins (E1 and E2) of the ancestral recombinant were obtained from the SIN-like virus, WEEV, HJV, and FMV are antigenically related (i.e., in neutralization tests) to SINV rather than to EEEV (Calisher et al., 1988). Along with Aura virus (AURAV) and Whataroa virus (WHAV), WEEV, HJV, FMV, and SINV collectively constitute the WEE antigenic complex (Weaver et al., 1997).
Other than the three recombinants, AURAV is the only WEE antigenic complex member found in the New World, having been isolated from Culex and Aedes species of mosquitoes in Brazil and Argentina (Causey et al., 1963; Rümenapf et al., 1994; Rümenapf et al., 1995). As AURAV is endemic to South America and is related to SINV, it initially provided an attractive candidate as the putative SIN-like parental virus of the recombination event. However, as WEEV is more closely related to SINV than it is to AURAV (Rumenapf et al., 1995), and AURAV and SINV are believed to have diverged prior to the recombination event (Weaver et al., 1997), it is likely that EEEV recombined with a virus more closely related to the present-day SINV. SINV (or a SIN-like virus other than AURAV) has not been detected in North or South America, suggesting that after its introduction into the New World (presuming a non-New World origin of SINV), this virus either went extinct or that it may possibly still be circulating endemically (e.g., in the neotropics) but has not been detected. Conversely, EEEV is found only in the New World and represents the sole species constituting the EEE antigenic complex, although South American lineages (lineages II–IV) of EEEV have recently been reclassified as Madaraiga virus (MADV) based on genetic, ecological, epidemiological, and pathogenic differences between North and South American strains (Arrigo et al., 2010).
In North America, EEEV, WEEV, HJV, and FMV all circulate in transmission cycles involving passerine birds as amplifying hosts and hematophagous arthropods as vectors. However, FMV is unique among these viruses in that the normal invertebrate vector is the cimicid swallow bug (Oeciacus vicarius), rather than a mosquito species [i.e., Culiseta (Cs.) melanura for EEEV and HJV; Culex (Cx.) tarsalis for WEEV] (Calisher et al., 1980). The distribution of each of the recombinant alphaviruses in the United States is, for the most part, spatially discrete, and is essentially defined by the geographical range of their respective enzootic vectors. HJV is primarily confined to eastern states along the Gulf and Atlantic seaboard and some inland foci around the Great Lakes region (Cilnis et al., 1996), while WEEV and FMV are endemic throughout most of the western United States (Reisen and Monath, 1988; Pfeffer et al., 2006), although WEEV appears to be declining in North America (Forrester et al., 2008; Bergren et al., 2014). Additionally, FMV is more focally distributed than WEEV due to its unique transmission cycle (outlined below). Interestingly, HJV and EEEV share apparently identical transmission cycles in North America and thus nearly identical geographical ranges (Scott and Weaver, 1989). Although lineages of WEEV and EEEV exist in Central and South America (Srihongse and Galindo, 1967; Mitchell et al., 1987; Weaver et al., 1994; Weaver et al., 1997; Brault et al., 1999), FMV and HJV have not been isolated outside of the United States. Additionally, unlike EEEV and WEEV, neither FMV nor HJV are normally associated with disease in mammals (Hayes and Wallis, 1977; Calisher et al., 1980; Englund et al., 1986; Karabatsos et al., 1988; Przelomski et al., 1988), although all four viruses are avian pathogens to varying degrees (Scott et al., 1984; Ficken et al., 1993; Randolph et al., 1994; Huyvaert et al., 2008).
Fort Morgan virus was first isolated by Hayes et al. (1977) from O. vicarius in eastern Colorado. Since its initial description, FMV (along with Buggy Creek virus and Stone Lakes virus) has been reported from a number of additional western and central states including Nebraska, Oklahoma, Texas, North Dakota, South Dakota, Washington, and California (Calisher et al., 1980; Hopla et al., 1993; Pfeffer et al., 2006; Padhi et al., 2008; Brault et al., 2009; Brown et al., 2009). The primary vertebrate amplifying hosts for FMV are cliff swallows (Petrochelidon pyrrhonata), and to a lesser extent, house sparrows (Passer domesticus), with the latter being inadvertently involved in transmission as they opportunistically occupy cliff swallow nests (Calisher et al., 1980; Scott et al., 1984; O’Brien et al., 2011). O. vicarius is a sedentary ectoparasite that strictly blood-feeds on cliff swallows (and house sparrows which parasitize cliff swallow nests) (Brown et al., 2010a, 2010b). This, in part, likely restricts the geographical range of FMV to more discrete endemic foci compared to that of other North American alphaviruses that are vectored endemically by motile mosquito species that may feed on multiple avian species (Cs. melanura) or are more catholic in host preference (Cx. tarsalis) (Loye, 1985). That Cx. tarsalis and Cx. pipiens have been demonstrated to be refractory to FMV infection following intra-thoracic inoculation, and Aedes (Ae.) albopictus cell cultures do not support replication of FMV (Calisher et al., 1980), suggests that FMV is exclusively adapted to O. vicarius. However, it was also demonstrated that, in vivo, FMV could infect Cs. melanura, the vector for EEEV and HJV (Calisher et al., 1980).
The genomes of two of the recombinants, WEEV and HJV, along with representative progenitors of the parental viruses, EEEV and SINV, have been described previously (Shirako et al., 1991; Weaver et al., 1993; Netolitzky et al., 2000; Allison and Stallknecht, 2009). However, the lack of the FMV genome has precluded a comprehensive comparative evolutionary analysis of the three recombinant viruses, although the genome of the variant Buggy Creek virus was recently sequenced (Forrester et al., 2012). Additionally, the mechanism(s) underlying the inability of FMV to infect mosquito cells remains unknown and represents a novel host range barrier, as most other alphaviruses (including HJV and WEEV) are vectored by mosquitoes. Herein, we present the genome of FMV and analyze the genetic, ecological, and evolutionary relationships of the WEE antigenic complex viruses, including the origin of the ancestral recombinant and other possible recombination events, as well as the in vitro host range adaptation of FMV to mosquito cells.
Results
Genetic relationships of the recombinant WEEV antigenic complex viruses
Genome sequencing of FMV was undertaken to perform genetic and phylogenetic comparisons of the full-length genomes among the three viruses (FMV, WEEV, HJV) known to be derived from the recombination event between EEEV and a SIN-like virus. As the structural polyprotein (C-E3-E2-6K-E1) and 3′ UTR of FMV CM4-146, accounting for ~3.9 kB, have previously been sequenced (Pfeffer et al., 1998), these genomic regions will not be discussed in detail here. Excluding the 5′ cap nucleotide and the 3′ poly(A) tail, the genome of FMV is 11,381 nt in length. The FMV genome is the shortest among the recombinants, being 127 or 145 nt less than either WEEV or HJV, respectively, primarily due to the short 3′ UTR. Whereas HJV and WEEV share a 75% nucleotide and 87% amino acid identity over their entire genomes, FMV is more divergent, with 69% nucleotide and 78% amino acid identity to both HJV and WEEV (Fig. S1). nsP4 was the most conserved (87–88%) protein among the recombinants and the parental EEEV, followed by the capsid (84–88%), nsP1 (80–86%), nsP2 (81–84%), and nsP3 (61–69%) (Fig. S1).
All of the nonstructural proteins of FMV were the same length as in HJV, WEEV, and EEEV, with the exception that the nsP3 protein of FMV (522aa) was of intermediate length between HJV (515aa) and WEEV (532aa), with the nsP3 protein of all the recombinants shorter than that of the parental EEEV (559aa). Although the C-terminal region of nsP3 is the most highly variable coding region of the alphavirus genome, a number of short, conserved amino acid motifs were observed upstream of the nsP3/nsP4 cleavage site sequence in FMV, HJV, WEEV, and EEEV (349-IPSP-352, 401-WSIPS-405, 447-QFLS-450, 456-PAPR-459; numbering based on FMV) (Fig. S2), suggesting that these motifs may have some structural and/or functional role in the C-terminal domain. Similar to HJV, WEEV, and EEEV, the nsP3 gene of FMV contains an opal termination codon (UGA) at genomic positions 5573-5575 followed by a C nucleotide at position 5576 (Strauss et al., 1983). The nsP1/nsP2, nsP2/nsP3, and nsP3/nsP4 cleavage site sequences for FMV were EAGA/GSVE, EAGR/APAY, and RYEAGA/YIFS, respectively, which were identical to that of EEEV, WEEV, and HJV (Strauss and Strauss, 1994; Allison and Stallknecht, 2009). Based on the cleavage site motifs, the theoretical isoelectric point and molecular weight of each of the FMV nonstructural proteins were: nsP1 (6.00; 59.8 kDa), nsP2 (8.80; 89.0 kDa), nsP3 (5.54; 57.4 kDa), and nsP4 (6.32; 68.1 kDa).
Host range relationships of the recombinant WEEV antigenic complex viruses
In vitro growth comparisons between FMV, HJV, and WEEV demonstrated that each virus had similar growth properties and host range, with all three viruses replicating in avian (107.04-108.72 PFU/mL maximum titers observed during a 6-day growth period), mammalian (107.83-108.78 PFU/mL), reptilian (106.62-108.11 PFU/mL) and fish (103.71-104.10 PFU/mL) cells. The finding that the WEEV antigenic complex recombinant viruses replicated in fish cells (derived from fathead minnow), albeit to lower titers than in mammalian, avian, or mosquito cells, is consistent with the hypothesis that alphaviruses may have a marine origin (Forrester et al., 2012) and that the fish alphaviruses [Salmon pancreatic disease virus (SPDV) and its variant, Sleeping disease virus (SDV)] appear to be ancient viruses of the genus (Powers et al., 2001). However, one noticeable difference among the three recombinants was the inability of FMV to infect cells of mosquito origin (C6/36 cells derived from Ae. albopictus), as has previously been documented both in vivo (Cx. tarsalis and Cx. pipiens) and in vitro (Ae. albopictus) (Calisher et al., 1980; Brault et al., 2009).
To assess if FMV could be adapted to replicate in mosquito cells, cultures of C6/36 cells were inoculated with an m.o.i. of 1 of stock CM4-146, followed by four passages at weekly intervals and assaying the fourth passage supernatant for live virus. Recovery of seven C6/36-adapted viruses – designated as CM4-146-C6-1 to 7 (or C6-1 to 7) – followed by growth curve experiments versus the original parental virus (CM4-146) demonstrated that all seven isolates had acquired the ability to replicate efficiently in C6/36 cells (Fig. S3). To further confirm this finding and determine how the mosquito-adapted viruses replicated in other cell lines, we chose the prototype C6/36-adapted virus (C6-1) for multi-step growth curve analysis in different host cells. Growth curves of C6-1 in C6/36 cells demonstrated a maximum mean titer of 106.47 PFU/mL by day six post-infection, while the prototype CM4-146 did not replicate (Fig. 1C). However, adaptation to mosquito cells in C6-1 was accompanied by a slight loss of fitness in other cell lines. This finding is compatible with an adaptive trade-off (Greene et al., 2005b), as C6-1 titers were reduced in both Vero cells (Fig. 1A) and Pekin duck embryo fibroblasts (Fig. 1B) in comparison to CM4-146 at 28 °C, as well as 37 °C (not shown). Comparative analysis between the genomes of the mosquito-adapted variants and parental CM4-146 indicated there were eight nucleotide changes (Fig. 2). Of these changes, three resulted in amino acid substitutions (CM4-146 → C6-1): (i) nsP3 nt position 5575 (X→C), (ii) nsP4 nt position 6168 (Q→R), and (iii) E1 nt position 10920 (I → F). Of the remaining five changes, four were silent third base transitions, and one substitution occurred in the 5′ UTR of the subgenomic 26S RNA, which may potentially influence replication through changes in secondary structure and/or promoter recognition.
Fig. 1.
Multi-step single growth curve analysis of FMV at 28°C in (A) mammalian [African green monkey kidney (Vero E6)], (B) avian [Pekin duck embryo; (PDE)] and (C) mosquito [Aedes albopictus larvae (C6/36)] cell lines. Titration curves of the prototype CM4-146 strain of FMV are shown with an open circle (○), while a C6/36-adapted variant of CM4-146 (C6-1) is indicated with a solid triangle (▲). Data shown are from experiments performed in triplicate with error bars indicating standard deviations.
Fig. 2.
Nucleotide and amino acid changes observed between the parental FMV (CM4-146) and C6/36-adapted variants (C6-1 to -7). Unique changes found in the C6/36-adapted variants (relative to parental CM4-146) are shaded in grey. For each of the eight mutations, the genetic location (gene/UTR), genomic position in nucleotides, and amino acid position in that particular protein (where applicable) is shown. If the mutation is located within a protein-encoding gene, the codon (with the mutation underlined) and resulting amino acid residue are indicated. Note that the nsP4 192 Glu to Arg change is conserved in all C6/36-adapted variants.
Evolutionary history of recombinant WEEV antigenic complex viruses and other alphaviruses
To reveal the evolutionary history of alphaviruses in more detail, we performed a phylogenetic analysis of all known alphaviruses, inferring trees on a gene-by-gene basis (Fig. 3). It was notable that proteome sequences across the genus as a whole were highly divergent (despite the Gblocks pruning), suggesting that the divergences between many of these viruses are ancient, and hence inter-species phylogenies should not be inferred from nucleotide sequences because of the likelihood of widespread site-saturation. Indeed, many deep nodes on these phylogenies lack strong bootstrap support such that aspects of the early evolutionary history of the alphaviruses remain uncertain, even with complete proteome sequence data.
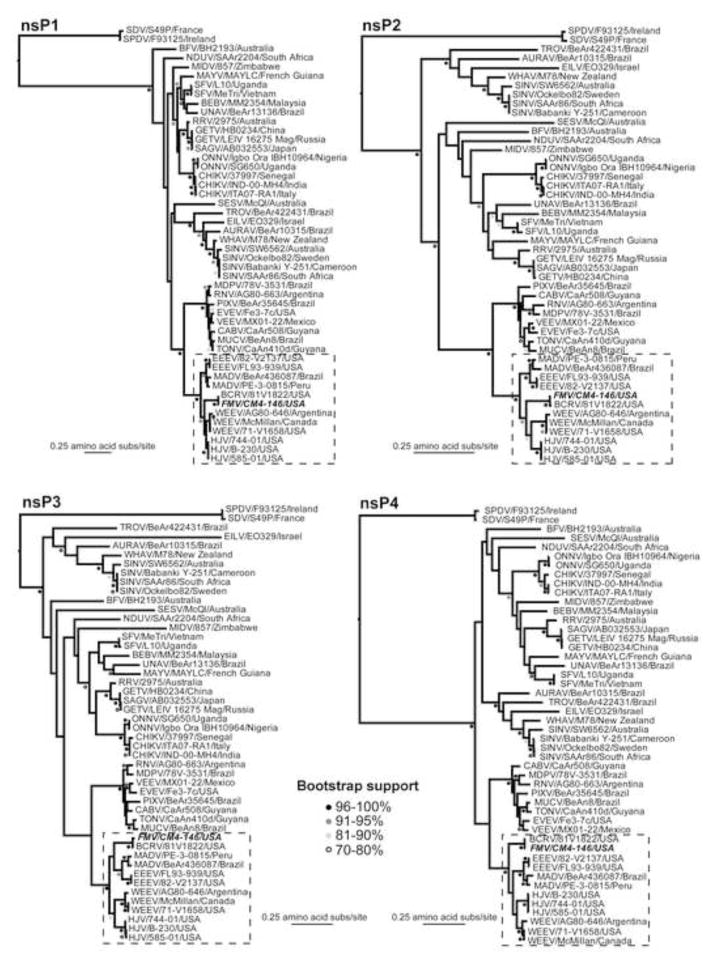
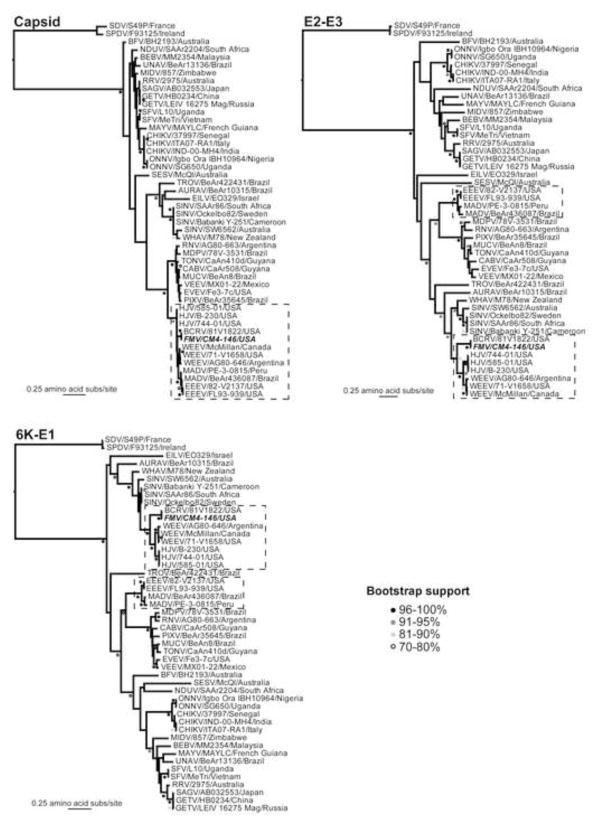
Fig. 3.
Gene-specific maximum likelihood phylogenetic trees of the nonstructural and structural proteins of all currently classified 31 alphavirus species (48 total sequences in the analysis). In some cases, certain proteins (i.e., E3 and 6K) have been combined with their C-terminal adjacent larger protein (i.e., E2 and E1, respectively) due to the small size of the former. All bootstrap values >70% are shown and shaded appropriately. All trees were rooted using the most divergent pair of sequences in the data set (the fish alphaviruses, SPDV and its variant SDV) and which are therefore assumed to represent an outgroup. The sequences of the WEEV antigenic complex recombinants and EEEV/MADV are boxed. Sequences used in the analysis were: Aura virus (AURAV) BeAr 10315 (AF126284); Barmah Forest virus (BFV) BH2193 (BFU73745); Bebaru virus (BEBV) MM2345 (NC_016962); Buggy Creek virus (BCRV; FMV subtype) 81V1822 (HM147986); Cabassou virus (CABV) CaAr508 (AF075259); Chikungunya virus (CHIKV) ITA07-RA1 (EU244823), CHIKV IND-00-MH4 (EF027139), and CHIKV 37997 (AY726732); Eastern equine encephalitis virus (EEEV) FL93-939 (EF151502) and EEEV 82V2137 (U01034); Eilat virus (EILV) EO329 (NC_018615); Everglades virus (EVEV) Fe3-7c (AF075251); Fort Morgan virus (FMV) CM4-146 (NC_013528); Getah virus (GETV) LEIV 16275 Mag (EF631998) and GETV HB0234 (EU015062); Highlands J virus (HJV) 585-01 (NC_012561), HJV 744-01 (GU167952), and HJV B-230 (GQ227789); Madariaga virus (MADV) PE-3.0815 (DQ241303) and MADV BeAr 436087 (EF151503); Mayaro virus (MAYV) MAYLC (DQ001069); Middelburg virus (MIDV) 857 (EF536323); Mosso das Pedras virus (MDPV) 78V-3531(AF075257); Mucambo virus (MUCV) BeAn 8 (AF075253); Ndumu virus (NDUV) SAAr2204 (NC_016959); O’nyong-nyong virus (ONNV) SG650 (AF079456) and ONNV Igbo Ora strain IBH10964 (AF079457); Pixuna virus (PIXV) BeAr35645 (AF075256); Rio Negro virus (RNV) AG80-663 (AF075258); Ross River virus (RRV) 2975 (GQ433360); Sagiyama virus (SAGV; RRV subtype) (AB032553); Salmon pancreas disease virus (SPDV) F93125 (AJ316244); Semliki forest virus (SFV) L10 (AY112987) and SFV Me Tri (EU350586); Sindbis virus (SINV) SW6562 (AF429428), SINV Ockelbo strain Edsbyn 82-5 (M69205), and SIN-like virus S.A.AR86 (U38305); Sleeping disease virus (SDV; SPDV subtype) S49P (AJ316246); Southern elephant seal virus (SESV) McQI (NC_016960); Tonate virus (TONV) CaAn 410d (AF075254); Trocara virus (TROV) BeAr422431 (HM147991); Una virus (UNAV) BeAr 13136 (HM147992); Venezuelan equine encephalitis virus (VEEV) MX01-22 (AY823299); Western equine encephalitis virus (WEEV) McMillan (GQ287640), WEEV 71-V1658 (GQ287645), and WEEV AG80-646 (GQ287646). The FMV sequence is highlighted in bold italic.
While the recombination event involving EEEV and SINV is clear (boxed regions of the phylogenies in Fig. 3), our phylogenetic analysis gave tentative support for a number of additional recombination events involving WEEV antigenic complex viruses that occurred subsequent to the emergence of the ancestral chimeric progenitor. For instance, although the FMV-BCRV cluster is strongly supported as falling basal to a grouping of WEEV and HJV in the nsP1, nsP2, E3-E2, and 6K-E1 phylogenies, a variety of different topological arrangements are observed for the other proteins. In the case of nsP3, FMV-BCRV group with EEEV, and in nsP4, FMV falls as basal to the entire WEEV antigenic complex, while phylogenetic positions are ambiguous in the capsid protein with little resolution in key regions of the phylogeny. Similarly, there are a variety of other differences in tree topology among the individual alphavirus proteins. For example, Trocara virus (TROV) is usually basal to AURAV, WHAV and SINV, yet occupies a more divergent position in 6K-E1. In addition, Mayaro virus (MAYV) forms a sister-group with Una virus (UNAV) in nsP3, E2-E3 and 6K-E1, yet a variety of positions in other genes. While these phylogenetic incongruences are compatible with recombination, a lack of significant bootstrap support (i.e., <70%) at key nodes on the tree means that this process cannot be resolved with certainty.
Discussion
Here we present the first comparative analysis of the three alphaviruses (WEEV, HJV, and FMV) that originated from a recombination event between EEEV and a SIN-like virus. This particular recombination event is an informative case study in viral evolution as it resulted in the emergence of three new viral species that occupy distinct ecological niches. One of the most important ecological differences between WEEV, HJV, and FMV is the host range barrier that exists for FMV in mosquito cells, which is in stark contrast to the vast majority of alphaviruses (including WEEV and HJV), which are normally vectored by mosquitoes. In turn, this suggests that one possible consequence of the adaptation of FMV to an aberrant transmission cycle involving O. vicarious (cimicid swallow bug) may have been the loss of the ability to infect mosquitoes, which likely contributes to the more focal nature of FMV transmission (i.e., cliff swallow nesting areas) in nature relative to that of either WEEV or HJV.
Analysis of the FMV prototype genome that was adapted to replicate in mosquito cells (C6/36) identified two nucleotide changes of interest at positions 5575 and 6168 (Fig. 2). The change at position 5575 (A → U) resulted in the replacement of the nsP3 opal termination codon (UGA) with a cysteine residue (UGU), identical to that previously observed in SINV S.A.AR86, a South African SIN-like virus isolated from Culex spp. in 1954 (Weinbren et al., 1956; Simpson et al., 1996). Loss of the opal termination codon would presumably result in the translation of a single nonstructural polyprotein (nsP1234) at 100% efficiency, which, under normal circumstances, is only produced ~10–20% of the time in alphaviruses containing the nsP3 termination codon due to intermittent readthrough (Strauss and Strauss, 1994). Previous studies involving mutating the opal termination codon to a sense codon in SINV resulted in similar overall yields of infectious particles in comparison to wild-type virus, although RNA synthesis was impaired early in infection at a low m.o.i. (Li and Rice, 1989). In O’nyong’nyong virus (ONNV), an increase in infectivity and dissemination in Anopheles gambiae was associated with the presence of the opal codon over a sense codon (Myles et al., 2006). Although the biological significance of the loss of the nsP3 opal termination in the C6/36-adapted viruses is unknown, it appears unlikely to be directly involved in the adaptation to mosquito cells as the abolishment of the nsP3 opal termination codon was not conserved among the different viruses (Fig. 2).
The second nucleotide change of interest was an A→G transition at genomic position 6168, which resulted in an Arg substitution for a Gln at nsP4 residue 192. Comparative analysis of the regions of difference between CM4-146 and seven C6/36-adapted viruses revealed that, in contrast to other mutations, the nsP4 192 Gln to Arg change was conserved in all C6/36-adapted viruses (Fig. 2). This single mutation in FMV may therefore be essential for overcoming the host range barrier in mosquito cells (Fig. 1C). Interestingly, another basic amino acid substitution (Lys) at the same Gln residue in SINV (nsP4 191) has been shown to be critical in promoter recognition and/or initiation factor binding (Sawicki et al., 1990). For SINV, the Lys substitution for Gln resulted in continued minus-strand synthesis, which is normally terminated around 4 hr post-infection at 37 °C (Sawicki et al., 2003). This inhibition of the cessation of antigenome synthesis was suggested to be either a direct effect of nsP4 binding to a promoter sequence that determines minus/plus strand template specificity, or possibly that the Lys substitution altered the binding of nsP4 to a host initiation factor (Sawicki et al., 1990; also see Lemm et al., 1990). Additionally, an Arg at a nearby residue (SINV nsP4 183) is believed to be important in the function of the minus-strand replicase specifically in mosquito cells through interaction with an Aedes albopictus host factor (Fata et al., 2002a, 2002b). The homology (location and charge) of the FMV Q192R substitution to the SINV Q191K change, coupled with the absence of a conserved substitution in the envelope glycoproteins of all the C6/36-adapted viruses, suggests that the host range barrier to mosquito cells is not at the level of receptor binding and entry, but rather occurs at some stage of replication (e.g., inefficient binding of nsP4 to a host protein required for replication and/or the viral template). This host range barrier appears to be overcome with the positively-charged Arg substitution at nsP4 192. Further analysis such as the development of an infectious clone of CM4-146 to verify the role of nsP4 residue 192 in dictating host range, and whether these results correlate to infection and transmissibility in an in vivo mosquito model, will aid in corroborating these findings. Nevertheless, the apparent acquisition of a new host species through a single nucleotide change – in this case, in the nsP4 polymerase gene – potentially demonstrates the ease by which alphaviruses can jump species and enter into new transmission cycles. Such plasticity can have far reaching biological effects, as epidemic emergence or re-emergence of alphaviruses, such as Chikungunya virus (CHIKV) and Venezuelan equine encephalitis virus (VEEV), has been associated with single mutations that influence host range and/or pathogenicity, which may ultimately result in the dispersal of alphaviruses into regions previously unoccupied (Brault et al., 2004; Greene et al., 2005a; Anishchenko et al., 2006; Tsetsarkin et al., 2007).
Our evolutionary analysis on a gene-by-gene basis resulted in phylogenetic trees that were similar to those published previously (Powers et al., 2001; Forrester et al., 2012), particularly with respect to the recombinant history of the WEEV antigenic complex viruses (Fig. 3). Although this phylogenetic analysis tentatively provided evidence for additional recombination events, for example involving FMV and EEEV in nsP3 (Fig. 3), because these variable tree topologies are not consistently supported by high bootstrap values, whether they truly reflect a past history of recombination is difficult to assess. The reported double recombination event for WEEV (and hence the ancestral recombinant), whereby both the 5′ and 3′ ends would be derived from EEEV (Hahn et al., 1988), suggests that the cis-acting elements contained within both the terminal ends of EEEV may have been a necessary prerequisite for the efficient replication, and, hence, survival of the original recombinant (Strauss and Strauss, 1997). Additionally, as EEEV is unique among the alphaviruses in that it contains five 16nt conserved sequence elements (CSE) in its 3′ UTR (i.e., GYRGYGYAUAAKGCYG; Fig. S4) that encompass nt 27–40 of the 40nt SINV CSE (Strauss and Strauss, 1994), these sequence and predicted structural (stem-loop) similarities between the parental viruses (EEEV and SINV) may have not only facilitated the replication/translation of the ancestral virus, but may also have enabled the initial recombination event itself through pausing, detachment, and reannealing of the polymerase complex at these conserved terminal sequences/structures. Other than additional members of the WEEV antigenic complex, no alphavirus other that EEEV appears to have these similar SINV-like CSE motifs (Strauss and Strauss, 1994).
The geographical origin of the alphaviruses has been a matter of debate (Weaver et al., 1997; Powers et al., 2001; Gould et al., 2009; Forrester et al., 2012). In particular, it has been suggested that the recombination event between EEEV and the SINV-like virus occurred in the tropics of South America (Weaver et al., 1997), or possibly in the Old World (Gould et al., 2009). However, we suggest that this recombination event may have occurred in North America after the establishment of the North American lineage of EEEV. Since both EEEV and HJV use Cs. melanura solely as an enzootic vector and, hence, occupy nearly identical geographical ranges, while other North American WEEV serocomplex recombinant viruses occupy disparate geographical regions (i.e., western United States) and utilize alternative vectors (i.e., Cx. tarsalis, WEEV; O. vicarius, FMV), it is therefore possible that the recombination event between EEEV and the SIN-like virus occurred in Cs. melanura. Notably, Cs. melanura is largely restricted to coastal acid-water swamps from New Brunswick south to eastern Texas (Darsie and Ward, 2005), and is absent from South America, compatible with a recombination event in the (eastern) United States. Further, if the recombination event occurred in South America, then both EEEV and HJV must have been independently transported (or dually transported in a single infected avian host) to North America with both viruses subsequently adapting to an enzootic cycle involving Cs. melanura. However, as Tonate virus (TONV) was apparently recovered from the original pool of the FMV isolate sequenced in this study (Monath et al., 1980), it is possible that alphaviruses may be intermittently transported from South America to the United States and that dual infections in a single host may occur.
The possible recombination of EEEV and the SIN-like virus after the adaptation of EEEV to Cs. melanura in North America is consistent with the concomitant adaptation of HJV to the same habitat-specialized enzootic vector. The lack of reports regarding the isolation of HJV and FMV from South America supports this hypothesis, although it is clear that wider surveillance is required. Additionally, the normal transmission cycle of SINV in the Old World involves passerine birds and Culiseta/Culex species of mosquitoes, and therefore may be more ‘ecologically’ likely to recombine with a North American lineage of EEEV that is maintained in a cycle involving passerine birds and a strictly ornithophilic mosquito species (Cs. melanura). On the other hand, South American lineages of EEEV (i.e., MADV) appear to be maintained in more diverse cycle(s) involving a number of catholic Cx. (Melanoconion) species and, based on the independent evolution of multiple geographically-defined lineages, may be maintained enzootically in ground-dwelling mammalian hosts (Srihongse and Galindo, 1967; Walder et al., 1984; Arrigo et al., 2010). However, additional work is required to determine the exact geographic origin of this important virus recombination event and the role that recombination has, in general, played in shaping alphavirus evolution.
Materials and Methods
Viruses
FMV isolate CM4-146, originally isolated from O. vicarius in Colorado in 1973, was obtained from the American Type Culture Collection (ATCC; Manassas, VA) (lot no. 1907186). CM4-146 was additionally passaged once in Vero E6 cells for generation of stock virus used for genomic sequencing and cell culture assays. Other recombinant WEE serocomplex viruses analyzed for the in vitro studies included WEEV isolate SW-99, obtained from a pool of Cx. tarsalis from Arizona in 1999 and passaged three times in Vero E6 cells, and HJV isolate 585-01, recovered from the brain of a red-tailed hawk (Buteo jamaicensis) from Georgia in 2001 (GenBank accession FJ827631) and passaged twice in Vero E6 cells.
Host ranges of the recombinant WEEV antigenic complex viruses
The in vitro host ranges of FMV, HJV, and WEEV were assessed in a number of mosquito, avian, mammalian, reptilian, and fish cell lines including C6/36 [Asian tiger mosquito (Aedes albopictus) larvae], PDE [Pekin duck (Anas platyrhynchus domesticus) embryo fibroblasts], Vero E6 [African green monkey (Cercopithecus aethiops) kidney epithelium], TH-1 [Eastern box turtle (Terrapene carolina) heart fibroblasts], VH-2 [Russell’s viper (Vipera russelli) heart fibroblasts], and FHM [Fathead minnow (Pimephales promelas) connective tissue and muscle epithelium]. All cell lines were obtained from the ATCC and were grown in minimum essential medium (MEM) supplemented with 2.2g/L NaHCO3, 5–10% fetal bovine serum (FBS), 400 units/mL penicillin, 400μg/mL streptomycin, and 1μg/mL amphotericin (Sigma-Aldrich, St. Louis, MO). Cells were maintained at 28 °C (C6/36, FHM, TH-1, VH-2) or 37 °C (Vero, PDE) in a humidified atmosphere containing 5% CO2. For growth curve analysis of WEEV, HJV, and FMV in each cell line, confluent monolayers in a 3.8cm2 well format were infected at a m.o.i. of ~0.0005–0.001. Wells were harvested daily for six days and viral titers were assessed by 10-fold titrations in Vero E6 cells overlaid with 1% gum tragacanth/1X MEM supplemented with 3% FBS. Cultures were inactivated on day 2–3 post-adsorption with 10% buffered formalin and stained with 0.25% crystal violet for plaque visualization. Dilutions in which 20–100 plaques could be counted (when applicable) were used in determining titers (log10 PFU/mL).
To assess if FMV could be adapted to replicate in C6/36 mosquito cells, 12 aliquots of stock CM4-146 were inoculated into C6/36 cells in a 3.8cm2 well format at an m.o.i. of 1.0 and then wells were passaged (100 μL) at weekly intervals for one month (four passages total). Supernatant of the fourth passage was then assayed for the presence of live virus (as CPE is not observed in C6/36 cells) by plaque assay in Vero E6 cells as stated earlier. Seven of the 12 inoculated wells at passage 4 were shown to contain plaque-forming viruses. Initially, one of these viruses (CM4-146-C6-1, referred to as C6-1) was chosen for additional genomic analysis to potentially determine the changes involved in adaptation to mosquito cells. Additionally, C6-1 stock virus was compared against the parental stock CM4-146 by multiple-step growth curve analysis in Vero, PDE, and C6/36 cells. Cells in a 3.8cm2 well format were inoculated with a m.o.i. of ~0.0005–0.001 of CM4-146 or C6-1 stock viruses and incubated at 28 °C (Vero, PDE, C6/36) and/or 37 °C (Vero, PDE) with 5% CO2 and titrated as mentioned above. To determine which genetic changes in C6-1 were potentially involved in the adaptation to replicate in mosquito cells, the additional six C6/36-adapted viruses (C6-2 through 7) were analyzed to determine if there were any conserved mutations amongst the C6/36-adapted viruses.
RNA and protein sequence analysis
Viral RNA was extracted from stock FMV using a QIAamp Viral RNA Mini kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. To amplify the portion of the FMV genome encompassing the nonstructural protein genes, degenerate primers were designed from conserved regions of a CLUSTALW alignment (Chenna et al., 2003) of genomic sequences of HJV (GenBank ID FJ827631), EEEV (GenBank ID AY705240), and WEEV (GenBank ID NC003908). Gaps in the genome were subsequently filled using FMV-specific primers based on sequence obtained from the degenerate primers. To verify the full-length nucleotide sequence of the structural genes, primers were developed based on the previously available structural polyprotein gene sequence of FMV CM4-146 (GenBank ID AF339475). A list of all primers used in sequencing of the FMV genome may be obtained from the authors. The 5′ and 3′ ends of the FMV genome were sequenced by rapid amplification of cDNA ends (RACE) using a First Choice RLM-RACE Kit (Life Technologies, Carlsbad, CA). RACE amplicons were cloned using a PCR Cloning Kit (Qiagen) and purified using a QIAprep Spin Miniprep Kit (Qiagen) according to the manufacturer’s instructions. The complete genome of FMV CM4-146, along with the C6/36-adapted variant of CM4-146 (C6-1), was constructed from multiple overlapping contigs using Sequencher 4.1.4. (Gene Codes Corporation, Ann Arbor, MI). The genome of FMV CM4-146 was deposited in GenBank under the accession GQ281603. The molecular weights and isoelectric points of the FMV nonstructural proteins were determined using the Compute pI/Mw tool on the ExPASy server (http://www.expasy.ch/tools/pi_tool.html) (Gasteiger et al., 2003).
Phylogenetic analysis
A total of 48 complete genome sequences of alphaviruses were used in a phylogenetic analysis, encompassing all 31 currently identified alphavirus species (Fig. 3). Because of the highly divergent nature of these sequences, they were aligned at the amino acid level using the MUSCLE program (Edgar, 2004), with alignments constructed for the whole proteome and each protein individually. Visual inspection of the alignments revealed regions of highly ambiguous alignment and multiple insertion-deletion (indel) events, which are likely to have an adverse effect on phylogenetic analysis. Consequently, all such highly divergent regions (including all indels) were removed prior to the phylogenetic analysis using the Gblocks package (Talavera and Castresana, 2007). Following Gblocks pruning, phylogenetic analyses were conducted on individual proteins, or groups of proteins, as follows (all data sets comprising 48 sequences): (i) nsP1, 472 amino acids; (ii) nsP2, 653 amino acids; (iii) nsP3, 257 amino acids; (iv) nsP4, 601 amino acids; (v) capsid, 174 amino acids; (vi) E3-E2, 318 amino acids (these proteins were combined because of the short length of E3); and (vii) 6K-E1, 386 amino acids (these proteins were also combined because of the short length of 6K) (see Fig. S1 for genome organization).
Phylogenetic trees were inferred using the maximum likelihood (ML) method available in the PhyML package version 3 (Guindon et al., 2010) and utilized the WAG+Γ model of amino acid substitution (with the shape parameter of the Γ distribution of among-site rate variation estimated from the empirical data) with subtree pruning and regrafting (SPR) branch swapping. To assess the level of support for individual nodes on the trees, a second PhyML analysis was undertaken, this time utilizing 1000 bootstrap pseudo-replicate trees estimated using nearest-neighbor interchange (NNI) branch swapping. Potential reassortment events in the data were identified manually through patterns of phylogenetic incongruence.
Supplementary Material
Nucleotide and amino acid comparisons of the individual genes and genomes of the recombinant alphaviruses (FMV, HJV, and WEEV), along with the parental viruses of the recombination event (EEEV and SINV). Nucleotide and amino acid identities are shown in the upper and lower portions of each panel, respectively. A schematic of the FMV genome is shown to highlight the alphavirus genetic organization, with the approximate portion of the coding region of the recombinant viruses obtained from EEEV highlighted in grey.
Partial amino acid alignment of nsP3 between FMV, HJV, WEEV, and EEEV, showing the lack of conservation in the C-terminal region. The degree of truncation of the three recombinant WEE complex viruses relative to the parental EEEV is shown to the right of the alignment. The read-through opal termination codon adjacent to the RYEAGA/YIFS nsP3/nsP4 cleavage site sequence is highlighted in grey and denoted as an “X”. Discontinuous areas of strict identity within the C-terminal region are boxed.
Multi-step single growth curve analysis of the seven C6/36-adapted variants (C6-1 to 7) of FMV at 28°C in Aedes albopictus larvae (C6/36) cells over 72 hours versus the prototype parental strain from which they were derived (CM4-146). Note that the prototype CM4-146 strain of FMV does not replicate in C6/36 cells.
Nucleotide alignment of the 3′ UTRs of EEEV and SINV demonstrating similar conserved repeat sequence elements (RSEs) that may have aided in the survival (replication/translation) of the ancestral recombinant. Asterisks denote nucleotide identity. The three conserved 40nt RSEs of SINV [structurally predicted to form double stem loop (DSL) structures; not shown] are shaded in grey, with the interim three nucleotides (GAA) between each SL shown in black. The five 16nt RSEs of EEEV (GYRGYGYAUAAKGCYG) are shaded in royal blue. Note that the 2nd, 4th, and 5th EEEV RSEs encompasses nucleotides 27–40 of the SINV 40nt RSE. Also note that EEEV contains a predicted DSL structure highlighted in dark blue (which encompasses the 2nd 16nt CSE) that is very similar to the 1st SINV 40nt RSE. The italicized area covering the 3′ terminal 19 nucleotides represents the 19-nt CSE.
Host range barrier of FMV to mosquito cells is overcome by a nsP4 mutation
New theory on the geographical origin of the EEEV-SINV recombination event
First comprehensive comparative analysis of the recombinant alphaviruses
First gene-specific phylogenetic analysis of the genus Alphavirus
Acknowledgments
Funds for this research were provided through sponsorship from the fish and wildlife agencies of the SCWDS; through the Federal Aid to Wildlife Restoration Act (50 Stat. 917) and Grant Agreement 06ERAG0005, Biological Resources Division, U.S. Geological Survey, U.S. Department of the Interior; and through Cooperative Agreements 0596130032CA and 0696130032CA, Veterinary Services, Animal and Plant Health Inspection Service, U.S. Department of Agriculture. We thank Daniel Mead, University of Georgia, for providing WEEV SW-99. E.C.H. is supported by a National Health and Medical Research Council Australia Fellowship. A.B.A. is supported by a NRSA fellowship (F32AI100545) from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allison AB, Stallknecht DE. Genomic sequencing of Highlands J virus: a comparison to western and eastern equine encephalitis viruses. Virus Res. 2009;145:334–340. doi: 10.1016/j.virusres.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Anishchenko M, Bowen RA, Paessler S, Austgen L, Greene IP, Weaver SC. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc Natl Acad Sci U S A. 2006;103:4994–4999. doi: 10.1073/pnas.0509961103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigo NC, Adams AP, Weaver SC. Evolutionary patterns of eastern equine encephalitis virus in North versus South America suggest ecological differences and taxonomic revision. J Virol. 2010;84:1014–1025. doi: 10.1128/JVI.01586-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergren NA, Auguste AJ, Forrester NL, Negi SS, Braun WA, Weaver SC. Western equine encephalitis virus: evolutionary analysis of a declining alphavirus based on complete genome sequences. J Virol. 2014;88:9260–9267. doi: 10.1128/JVI.01463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault AC, Powers AM, Chavez CL, Lopez RN, Cachón MF, Gutierrez LF, Kang W, Tesh RB, Shope RE, Weaver SC. Genetic and antigenic diversity among eastern equine encephalitis viruses from North, Central, and South America. Am J Trop Med Hyg. 1999;61:579–586. doi: 10.4269/ajtmh.1999.61.579. [DOI] [PubMed] [Google Scholar]
- Brault AC, Powers AM, Ortiz D, Estrada-Franco JG, Navarro-Lopez R, Weaver SC. Venezuelan equine encephalitis emergence: enhanced vector infection from a single amino acid substitution in the envelope glycoprotein. Proc Natl Acad Sci U S A. 2004;101:11344–11349. doi: 10.1073/pnas.0402905101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault AC, Armijos MV, Wheeler S, Wright S, Fang Y, Langevin S, Reisen WK. Stone Lakes virus (family Togaviridae, genus Alphavirus), a variant of Fort Morgan virus isolated from swallow bugs (Hemiptera: Cimicidae) west of the Continental Divide. J Med Entomol. 2009;46:1203–1209. doi: 10.1603/033.046.0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Moore AT, Knutie SA, Komar N. Overwintering of infectious Buggy Creek virus (Togaviridae: Alphavirus) in Oeciacus vicarius (Hemiptera: Cimicidae) in North Dakota. J Med Entomol. 2009;46:391–394. doi: 10.1603/033.046.0227. [DOI] [PubMed] [Google Scholar]
- Brown CR, Moore AT, Young GR, Komar N. Persistence of Buggy Creek virus (Togaviridae, Alphavirus) for two years in unfed swallow bugs (Hemiptera: Cimicidae: Oeciacus vicarius) J Med Entomol. 2010a;47:436–441. doi: 10.1603/me09288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Strickler SA, Moore AT, Knutie SA, Padhi A, Brown MB, Young GR, O’Brien VA, Foster JE, Komar N. Winter ecology of Buggy Creek virus (Togaviridae, Alphavirus) in the Central Great Plains. Vector Borne Zoonotic Dis. 2010b;10:355–363. doi: 10.1089/vbz.2009.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calisher CH, Monath TP, Muth DJ, Lazuick JS, Trent DW, Francy DB, Kemp GE, Chandler FW. Characterization of Fort Morgan virus, an alphavirus of the western equine encephalitis virus complex in an unusual ecosystem. Am J Trop Med Hyg. 1980;29:1428–1440. doi: 10.4269/ajtmh.1980.29.1428. [DOI] [PubMed] [Google Scholar]
- Calisher CH, Karabatsos N, Lazuick JS, Monath TP, Wolff KL. Reevaluation of the western equine encephalitis antigenic complex of alphaviruses (family Togaviridae) as determined by neutralization tests. Am J Trop Med Hyg. 1988;38:447–452. doi: 10.4269/ajtmh.1988.38.447. [DOI] [PubMed] [Google Scholar]
- Causey OR, Casals J, Shope RE, Udomsakdi S. Aura and Una, two new Group A arthropod-borne viruses. Am J Trop Med Hyg. 1963;12:777–781. doi: 10.4269/ajtmh.1963.12.777. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the CLUSTAL series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cilnis MJ, Kang W, Weaver SC. Genetic conservation of Highlands J viruses. Virology. 1996;218:343–351. doi: 10.1006/viro.1996.0203. [DOI] [PubMed] [Google Scholar]
- Darsie RF, Jr, Ward RA. Identification and geographical distribution of the mosquitoes of North America, north of Mexico. University Press of Florida; Gainesville, Florida: 2005. [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund JA, Breningstall GN, Heck LJ, Lazuick JS, Karabatsos N, Calisher CH, Tsai TF. Diagnosis of western equine encephalitis in an infant by brain biopsy. Pediatr Infect Dis. 1986;5:382–384. doi: 10.1097/00006454-198605000-00026. [DOI] [PubMed] [Google Scholar]
- Fata C, Sawicki SG, Sawicki DL. Alphavirus minus strand RNA synthesis: Identification of a role for Arg183 of the nsP4 polymerase. J Virol. 2002a;76:8632–8640. doi: 10.1128/JVI.76.17.8632-8640.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata C, Sawicki SG, Sawicki DL. Modification of Asn374 of nsP1 suppresses a Sindbis nsP4 minus strand polymerase mutant. J Virol. 2002b;76:8641–8649. doi: 10.1128/JVI.76.17.8641-8649.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficken MD, Wages DP, Guy JS, Quinn JA, Emory WH. High mortality of domestic turkeys associated with Highlands J virus and eastern equine encephalitis virus infections. Avian Dis. 1993;37:585–590. [PubMed] [Google Scholar]
- Forrester NL, Kenney JL, Deardorff E, Wang E, Weaver SC. Western equine encephalitis submergence: lack of evidence for a decline in virus virulence. Virology. 2008;380:170–172. doi: 10.1016/j.virol.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester NL, Palacios G, Tesh RB, Savji N, Guzman H, Sherman M, Weaver SC, Lipkin WI. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J Virol. 2012;86:2729–2738. doi: 10.1128/JVI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould EA, Coutard B, Malet H, Morin B, Jamal S, Weaver S, Gorbalenya A, Moureau G, Baronti C, Delogu I, Forrester N, Khasnatinov M, Gritsun T, de Lamballerie X, Canard B. Understanding the alphaviruses: Recent research on important emerging pathogens and progress towards their control. Antiviral Res. 2010;87:111–124. doi: 10.1016/j.antiviral.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene IP, Paessler S, Austgen L, Anishchenko M, Brault AC, Bowen RA, Weaver SC. Envelope glycoprotein mutations mediate equine amplification and virulence of epizootic venezuelan equine encephalitis virus. J Virol. 2005a;79:9128–9133. doi: 10.1128/JVI.79.14.9128-9133.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene IP, Wang E, Deardorff ER, Milleron R, Domingo E, Weaver SC. Effect of alternating passage on adaptation of sindbis virus to vertebrate and invertebrate cells. J Virol. 2005b;79:14253–14260. doi: 10.1128/JVI.79.22.14253-14260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hahn CS, Lustig S, Strauss EG, Strauss JH. Western equine encephalitis virus is a recombinant virus. Proc Natl Acad Sci U S A. 1988;85:5997–6001. doi: 10.1073/pnas.85.16.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CG, Wallis RC. Ecology of western equine encephalomyelitis in the eastern United States. Adv Virus Res. 1977;21:37–83. doi: 10.1016/s0065-3527(08)60761-7. [DOI] [PubMed] [Google Scholar]
- Hayes RO, Francy DB, Lazuick JS, Smith GC, Gibbs EPJ. Role of the cliff swallow bug (Oeciacus vicarius) in the natural cycle of a western equine encephalitis-related alphavirus. J Med Entomol. 1977;14:257–262. [Google Scholar]
- Hopla CE, Francy DB, Calisher CH, Lazuick JS. Relationship of cliff swallows, ectoparasites, and an alphavirus in west-central Oklahoma. J Med Entomol. 1993;30:267–272. doi: 10.1093/jmedent/30.1.267. [DOI] [PubMed] [Google Scholar]
- Huyvaert KP, Moore AT, Panella NA, Edwards EA, Brown MB, Komar N, Brown CR. Experimental inoculation of house sparrows (Passer domesticus) with buggy creek virus. J Wildl Dis. 2008;44:331–340. doi: 10.7589/0090-3558-44.2.331. [DOI] [PubMed] [Google Scholar]
- Karabatsos N, Lewis AL, Calisher CH, Hunt AR, Roehrig JT. Identification of Highlands J virus from a Florida horse. Am J Trop Med Hyg. 1988;39:603–606. doi: 10.4269/ajtmh.1988.39.603. [DOI] [PubMed] [Google Scholar]
- Lemm JA, Durbin RK, Stollar V, Rice CM. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J Virol. 1990;64:3001–3011. doi: 10.1128/jvi.64.6.3001-3011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GP, Rice CM. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J Virol. 1989;63:1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loye JE. The life history and ecology of the cliff swallow bug, Oeciacus vicarious (Hemiptera: Cimicidae) Ent Med et Parasitol. 1985;23:133–139. [Google Scholar]
- Mitchell CJ, Monath TP, Sabattini MS, Daffner JF, Cropp CB, Calisher CH, Darsie RF, Jr, Jakob WL. Arbovirus isolations from mosquitoes collected during and after the 1982–1983 epizootic of western equine encephalitis in Argentina. Am J Trop Med Hyg. 1987;36:107–113. doi: 10.4269/ajtmh.1987.36.107. [DOI] [PubMed] [Google Scholar]
- Monath TP, Lazuick JS, Cropp CB, Rush WA, Calisher CH, Kinney RM, Trent DW, Kemp GE, Bowen GS, Francy DB. Recovery of Tonate virus (“Bijou Bridge” strain), a member of the Venezuelan equine encephalomyelitis virus complex, from Cliff Swallow nest bugs (Oeciacus vicarius) and nestling birds in North America. Am J Trop Med Hyg. 1980;29:969–983. doi: 10.4269/ajtmh.1980.29.969. [DOI] [PubMed] [Google Scholar]
- Myles KM, Kelly CL, Ledermann JP, Powers AM. Effects of an opal termination codon preceding the nsP4 gene sequence in the O’Nyong-Nyong virus genome on Anopheles gambiae infectivity. J Virol. 2006;80:4992–4997. doi: 10.1128/JVI.80.10.4992-4997.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasar F, Palacios G, Gorchakov RV, Guzman H, Da Rosa AP, Savji N, Popov VL, Sherman MB, Lipkin WI, Tesh RB, Weaver SC. Eilat virus, a unique alphavirus with host range restricted to insects by RNA replication. Proc Natl Acad Sci U S A. 2012;109:14622–14627. doi: 10.1073/pnas.1204787109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netolitzky DJ, Schmaltz FL, Parker MD, Rayner GA, Fisher GR, Trent DW, Bader DE, Nagata LP. Complete genomic RNA sequence of western equine encephalitis virus and expression of the structural genes. J Gen Virol. 2000;81:151–159. doi: 10.1099/0022-1317-81-1-151. [DOI] [PubMed] [Google Scholar]
- O’Brien VA, Moore AT, Young GR, Komar N, Reisen WK, Brown CR. An enzootic vector-borne virus is amplified at epizootic levels by an invasive avian host. Proc Biol Sci. 2011;278:239–246. doi: 10.1098/rspb.2010.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi A, Moore AT, Brown MB, Foster JE, Pfeffer M, Gaines KP, O’Brien VA, Strickler SA, Johnson AE, Brown CR. Phylogeographical structure and evolutionary history of two Buggy Creek virus lineages in the western Great Plains of North America. J Gen Virol. 2008;89:2122–2131. doi: 10.1099/vir.0.2008/001719-0. [DOI] [PubMed] [Google Scholar]
- Pfeffer M, Kinney RM, Kaaden OR. The alphavirus 3′-nontranslated region: size heterogeneity and arrangement of repeated sequence elements. Virology. 1998;240:100–108. doi: 10.1006/viro.1997.8907. [DOI] [PubMed] [Google Scholar]
- Feffer M, Foster JE, Edwards EA, Brown MB, Komar N, Brown CR. Phylogenetic analysis of Buggy Creek virus: evidence for multiple clades in the Western Great Plains, United States of America. Appl Environ Microbiol. 2006;72:6886–6893. doi: 10.1128/AEM.00868-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, Weaver SC. Evolutionary relationships and systematics of the alphaviruses. J Virol. 2001;75:10118–101131. doi: 10.1128/JVI.75.21.10118-10131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers A, Huang H, Roehrig J, Strauss E, Weaver S. Family Togaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy: Classification and Nomenclature of Viruses, Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; San Diego, California: 2012. pp. 1103–1110. [Google Scholar]
- Przelomski MM, O’Rourke E, Grady GF, Berardi VP, Markley HG. Eastern equine encephalitis in Massachusetts: a report of 16 cases, 1970–1984. Neurology. 1988;38:736–739. doi: 10.1212/wnl.38.5.736. [DOI] [PubMed] [Google Scholar]
- Randolph KD, Vanhooser SL, Hoffman M. Western equine encephalitis virus in emus in Oklahoma. J Vet Diagn Invest. 1994;6:492–493. doi: 10.1177/104063879400600417. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Monath TP. Western equine encephalomyelitis. In: Monath TP, editor. The Arboviruses: Epidemiology and Ecology. V. CRC Press, Inc; Boca Raton, Florida: 1988. pp. 89–137. [Google Scholar]
- Rümenapf T, Strauss EG, Strauss JH. Subgenomic mRNA of Aura alphavirus is packaged into virions. J Virol. 1994;68:56–62. doi: 10.1128/jvi.68.1.56-62.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rümenapf T, Strauss EG, Strauss JH. Aura virus is a New World representative of Sindbis-like viruses. Virology. 1995;208:621–633. doi: 10.1006/viro.1995.1193. [DOI] [PubMed] [Google Scholar]
- Sawicki DL, Barkhimer DB, Sawicki SG, Rice CM, Schlesinger S. Temperature sensitive shut-off of alphavirus minus strand RNA synthesis maps to a nonstructural protein, nsP4. Virology. 1990;174:43–52. doi: 10.1016/0042-6822(90)90052-s. [DOI] [PubMed] [Google Scholar]
- Sawicki DL, Silverman RH, Williams BR, Sawicki SG. Alphavirus minus-strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R. J Virol. 2003;77:1801–1811. doi: 10.1128/JVI.77.3.1801-1811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott TW, Bowen GS, Monath TP. A field study on the effects of Fort Morgan virus, an arbovirus transmitted by swallow bugs, on the reproductive success of cliff swallows and symbiotic house sparrows in Morgan County, Colorado, 1976. Am J Trop Med Hyg. 1984;33:981–991. doi: 10.4269/ajtmh.1984.33.981. [DOI] [PubMed] [Google Scholar]
- Scott TW, Weaver SC. Eastern equine encephalomyelitis virus: epidemiology and evolution of mosquito transmission. Adv Virus Res. 1989;37:277–328. doi: 10.1016/s0065-3527(08)60838-6. [DOI] [PubMed] [Google Scholar]
- Shirako Y, Niklasson B, Dalrymple JM, Strauss EG, Strauss JH. Structure of the Ockelbo virus genome and its relationship to other Sindbis viruses. Virology. 1991;182:753–764. doi: 10.1016/0042-6822(91)90616-j. [DOI] [PubMed] [Google Scholar]
- Simpson DA, Davis NL, Lin SC, Russell D, Johnston RE. Complete nucleotide sequence and full-length cDNA clone of S.A.AR86, a South African alphavirus related to Sindbis. Virology. 1996;222:464–469. doi: 10.1006/viro.1996.0445. [DOI] [PubMed] [Google Scholar]
- Srihongse S, Galindo P. The isolation of eastern equine encephalitis virus from Culex (Melanoconion) taeniopus Dyar and Knab in Panama. Mosq News. 1967;27:74–76. [Google Scholar]
- Strauss EG, Rice CM, Strauss JH. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A. 1983;80:5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss JH, Strauss EG. Recombination in alphaviruses. Semin Virol. 1997;8:85–94. [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3(12):e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walder R, Suarez OM, Calisher CH. Arbovirus studies in southwestern Venezuela during 1973–1981. II Isolation and further studies of Venezuelan and Eastern Equine Encephalitis, Una, Itaqui and Moju viruses. Am J Trop Med Hyg. 1984;33:483–491. [PubMed] [Google Scholar]
- Weaver SC, Hagenbaugh A, Bellew LA, Netesov SV, Volchkov VE, Chang GJ, Clarke DK, Gousset L, Scott TW, Trent DW, Holland JJ. A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology. 1993;197:375–390. doi: 10.1006/viro.1993.1599. [DOI] [PubMed] [Google Scholar]
- Weaver SC, Hagenbaugh A, Bellew LA, Gousset L, Mallampalli V, Holland JJ, Scott TW. Evolution of alphaviruses in the eastern equine encephalomyelitis complex. J Virol. 1994;68:158–169. doi: 10.1128/jvi.68.1.158-169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Kang W, Shirako Y, Rumenapf T, Strauss EG, Strauss JH. Recombinational history and molecular evolution of western equine encephalomyelitis complex alphaviruses. J Virol. 1997;71:613–623. doi: 10.1128/jvi.71.1.613-623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbren MP, Kokernot RH, Smithburn KC. Strains of Sindbis-like virus isolated from culicine mosquitoes in the Union of South Africa. I Isolation and properties. S Afr Med J. 1956;30:631–636. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide and amino acid comparisons of the individual genes and genomes of the recombinant alphaviruses (FMV, HJV, and WEEV), along with the parental viruses of the recombination event (EEEV and SINV). Nucleotide and amino acid identities are shown in the upper and lower portions of each panel, respectively. A schematic of the FMV genome is shown to highlight the alphavirus genetic organization, with the approximate portion of the coding region of the recombinant viruses obtained from EEEV highlighted in grey.
Partial amino acid alignment of nsP3 between FMV, HJV, WEEV, and EEEV, showing the lack of conservation in the C-terminal region. The degree of truncation of the three recombinant WEE complex viruses relative to the parental EEEV is shown to the right of the alignment. The read-through opal termination codon adjacent to the RYEAGA/YIFS nsP3/nsP4 cleavage site sequence is highlighted in grey and denoted as an “X”. Discontinuous areas of strict identity within the C-terminal region are boxed.
Multi-step single growth curve analysis of the seven C6/36-adapted variants (C6-1 to 7) of FMV at 28°C in Aedes albopictus larvae (C6/36) cells over 72 hours versus the prototype parental strain from which they were derived (CM4-146). Note that the prototype CM4-146 strain of FMV does not replicate in C6/36 cells.
Nucleotide alignment of the 3′ UTRs of EEEV and SINV demonstrating similar conserved repeat sequence elements (RSEs) that may have aided in the survival (replication/translation) of the ancestral recombinant. Asterisks denote nucleotide identity. The three conserved 40nt RSEs of SINV [structurally predicted to form double stem loop (DSL) structures; not shown] are shaded in grey, with the interim three nucleotides (GAA) between each SL shown in black. The five 16nt RSEs of EEEV (GYRGYGYAUAAKGCYG) are shaded in royal blue. Note that the 2nd, 4th, and 5th EEEV RSEs encompasses nucleotides 27–40 of the SINV 40nt RSE. Also note that EEEV contains a predicted DSL structure highlighted in dark blue (which encompasses the 2nd 16nt CSE) that is very similar to the 1st SINV 40nt RSE. The italicized area covering the 3′ terminal 19 nucleotides represents the 19-nt CSE.