Abstract
Background
Protein kinase D (PKD or PKD1) is a serine/threonine protein kinase that has been shown to play a role in a variety of cellular processes; however, the function of PKD1 in the skin has not been fully investigated. The balance between proliferation and differentiation processes in the predominant cells of the epidermis, the keratinocytes, is essential for normal skin function.
Objective
To investigate the effect of PKD1 deficiency on proliferation and differentiation of epidermal keratinocytes.
Methods
We utilized a floxed PKD1 mouse model such that infecting epidermal keratinocytes derived from these mice with an adenovirus expressing Cre-recombinase allowed us to determine the effect of PKD1 gene loss in vitro. Proliferation and differentiation were monitored using qRT-PCR, Western blot, transglutaminase activity assays, [3H]thymidine incorporation into DNA and cell cycle analysis.
Results
A significant decrease in PKD1 mRNA and protein levels was achieved in adenoviral Cre-recombinase-infected cells. Deficiency of PKD1 resulted in significant increases in the mRNA and protein expression of various differentiation markers such as loricrin, involucrin, and keratin 10 either basally and/or upon stimulation of differentiation. PKD1-deficient keratinocytes also showed an increase in transglutaminase expression and activity, indicating an anti-differentiative role of PKD1. Furthermore, the PKD1-deficient keratinocytes exhibited decreased proliferation. However, PKD1 loss had no effect on stem cell marker expression.
Conclusions
Cre-recombinase-mediated knockdown represents an additional approach demonstrating that PKD1 is an anti-differentiative, pro-proliferative signal in mouse keratinocytes.
1. Introduction
Keratinocytes are the predominant cell type present in the outermost layer of the skin, the epidermis. Keratinocytes undergo a distinct pattern of proliferation and differentiation. The proliferating keratinocytes in the basal layer move upward upon growth arrest and initially express mature keratins 1 and 10 in the first differentiated epidermal layer, the spinous layer, followed by intermediate markers such as involucrin. The upper differentiated granular layer is marked by expression of late (e.g., loricrin) differentiation markers. Late differentiation is also accompanied by expression/activation of proteins, like transglutaminase, that are essential for the formation of the cornified envelope and corneocytes at the outer layer of the epidermis, the stratum corneum. Although the epidermis is similar in mice and humans, differences exist. Thus, in humans, the stratum spinosum is composed of multiple layers of keratinocytes, whereas in mouse epidermis the stratum spinosum is usually a single cell layer [1]. Also, average epidermal thickness (30μM) in mouse skin is less than that of human epidermis (50μM) [2]. In addition, human hair follicles cycle asynchronously through growth (anagen), involution (catagen) and resting (telogen) phases, while those of mice are synchronized [3].
The equilibrium between keratinocyte proliferation and differentiation is essential for maintaining the protective barrier and to prevent skin diseases (reviewed in [4–7]). For example, in squamous and basal cell carcinoma, the malignant cells fail to complete the keratinocyte differentiation program. Abnormal keratinocyte differentiation also characterizes another human skin disease, psoriasis, treatment costs of which are estimated at over $1 billion annually (www.aad.org). Therefore, to develop better treatments, it is important to understand the regulation of keratinocyte proliferation and differentiation.
Protein kinase D or protein kinase D1 (PKD or PKD1) is a serine/threonine protein kinase that has been shown to play a role in a variety of cell processes (reviewed in [8–10]). PKD has been classified as the first member, PKD1, of a new family that includes also PKD2 and PKD3, with sequence homology to calcium/calmodulin-dependent kinases in the catalytic domain. PKD can be activated by transphosphorylation mediated by protein kinase C and a Src/Abl tyrosine kinase cascade in response to a variety of growth factors, hormones and other extracellular signals (reviewed in [10, 11]). PKD is also activated by oxidative stress [12–16] and DNA damage [17]. Importantly, we have recently shown activation of PKD in keratinocytes exposed to ultraviolet B (UVB) irradiation, the primary risk factor for the development of non-melanoma skin cancer, providing a link between these cancers, sun exposure (i.e., UVB), and PKD [18].
Ours and others’ results suggest that PKD plays a role in regulating keratinocyte proliferation and differentiation. Increased PKD levels in mouse epidermal carcinomas [19], decreased DNA synthesis [19, 20] and increased differentiation upon inhibition of its activity [20], and increased DNA synthesis [18] and promoter activity of keratin 5, a basal layer marker, [21] upon PKD overexpression all suggest the pro-proliferative role of PKD in keratinocytes. Reversal of calcium-differentiated keratinocytes to a proliferative basal-like phenotype upon switching of the cells to a low calcium medium has also been described [22], and this low calcium switch-induced mitogenic response is dependent upon PKD signaling. Recently, in vivo studies involving targeted deletion of PKD1 in epidermal keratinocytes suggested a pro-proliferative role of PKD1 during wounding and upon challenge with tumor-promoting phorbol esters [23]. These authors also suggested that PKD1 is dispensable, under normal conditions, for skin development and homeostasis [23]. However, this report did not show cumulative quantified results of multiple animals for proliferative or differentiative markers under basal conditions nor was epidermal PKD1 deletion verified in the skin in situ. In addition, possible compensatory effects related to the loss of PKD1 in utero could not be excluded. On the other hand, Ivanova and coworkers have shown that siRNA-mediated down-regulation of PKD1 decreases human keratinocyte proliferation, although these authors failed to demonstrate PKD1 knockdown at the protein level [24]. Thus, all of the strategies used to date have both benefits and the potential for artifacts, and the results from multiple approaches can only strengthen the evidence for PKD1’s role in keratinocytes.
To determine PKD’s role in keratinocyte function, we have used a recently generated transgenic mouse model [25] in which the PKD1 gene is flanked by loxP sites (i.e., a floxed PKD1 mouse model). Infecting epidermal keratinocytes derived from these mice with an adenovirus possessing Cre-recombinase allowed us to determine the effect of PKD1 loss in vitro. Our hypothesis was that PKD1 exerts anti-differentiative effects on epidermal keratinocytes such that its loss will result in increased differentiation.
2. Materials and methods
2.1. Materials
Antibodies were purchased as described in Table 1 of the supplementary materials. PVDF membrane was from Millipore (Billerica, MA), and iScript cDNA synthesis kits were purchased from Bio-Rad (Hercules, CA.). The keratinocyte serum-free media (K-SFM) kit was purchased from Life Technologies (Carlsbad, CA). [3H]Thymidine and [3H]putrescine were purchased from Dupont/NEN (Boston, MA).
2.2. Adenoviral constructs amplification
The GFP-tagged adenovirus construct for Cre-recombinase was purchased from Vector Biolabs (Philadelphia, PA). Adenoviruses containing wild type-PKD (WT-PKD) and tyrosine-463-to-phenylalanine PKD mutant (mutant-PKD) constructs were made previously in our laboratory using the AdEasy adenoviral system as described elsewhere [18, 26]. The amplification and purification of viruses was performed as described earlier [18, 27].
2.3. Cell culture and experimental design
Primary epidermal mouse keratinocytes were prepared from 1–3 day old neonatal floxed-PKD1 [25] or outbred ICR CD-1 mice (Harlan Laboratories, Indianapolis, IN) and cultured in K-SFM containing 50μM CaCl2. Isolated keratinocytes were seeded on 6-well plates using dialyzed fetal bovine serum-containing plating media as described earlier [28]. For PKD1 depletion experiments, the day after plating, when the floxed-PKD1 keratinocytes were approximately 40% confluent, the keratinocytes were infected with Cre-recombinase (also expressing GFP) adenovirus or GFP adenovirus [at a multiplicity of infection (MOI) of 0.5 or 5] in K-SFM. The virus-containing media was removed 24 hours post-infection and replaced with K-SFM and every 24 hours thereafter. 48 hours after infection, the cells were stimulated to differentiate with K-SFM containing a moderately elevated calcium concentration (125μM) [29, 30]. After 24 hours of calcium treatment (for a total of 72 hours after viral infection), the cells were harvested either for quantitative RT-PCR (qRT-PCR) or Western analysis. For time course experiments, 125μM calcium-containing medium was added 24 hours prior to termination of the experiments and harvesting of the cells. For over-expression studies, CD-1 mouse keratinocytes were incubated with K-SFM and allowed to grow for 1–2 days. Sub-confluent cells were infected with adenoviral constructs over-expressing wild-type PKD (WT-PKD) or tyrosine-463-to-phenylalanine PKD (mutant-PKD) or with GFP alone (Vector) using an MOI of 50. After 24 hours of adenoviral infection, the cells were harvested for qRT-PCR or Western analysis.
2.4. RNA extraction, cDNA synthesis, and quantitative RT-PCR
After treatment the keratinocytes were processed for total RNA extraction using PerfectPure RNA tissue kits (5 PRIME, Inc, Gaithersburg, MD, USA) as per the manufacturer’s protocol. Spectroscopic quality testing and quantitation of total RNA was performed using a Nanodrop instrument (NanoDrop Technologies, Wilmington, DE). Equal quantities of total RNA (1μg) were reverse transcribed using iScript cDNA synthesis kits (Bio-Rad Laboratories, Hercules, CA, USA) following the manufacturer’s instructions. Equal volumes of 5-times diluted cDNA were used in qRT-PCR reactions with Taqman probes purchased from Applied Biosystems (Applied Biosystems, Grand Island, NY), as indicated in Supplementary Table 2. The qRT-PCR reaction was performed using the Fast Reagent PCR Master Mix (Applied Biosystems) and the StepOnePlus Real-Time PCR System (Applied Biosystems) as per the manufacturer’s protocol. The relative gene expression was calculated by the delta-delta Ct method using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and ribosomal protein, large, P0 (Rplp0) as endogenous control genes (average value of the two genes) and normalized to the GFP (Vector)-infected group.
2.5. Western blot analysis
For Western analysis, primary keratinocytes were solubilized using hot lysis buffer [0.1875M Tris-HCl (pH 8.5), 3% SDS, and 1.5mM EDTA], scraped and homogenized by repeated pipetting. After removal of an aliquot for determination of protein, the lysates were then diluted with 3x sample buffer (30% glycerol, 15% beta-mercaptoethanol, 1% bromophenol blue, 54% water) to constitute Laemmli buffer [31]. Equal amounts of protein were subjected to SDS-PAGE and transferred to PVDF membranes, which were incubated with the appropriate antibodies after blocking. Immunoreactivity was visualized using an Odyssey imaging system (LI-COR, Biosciences, Lincoln, NE) or using an enhanced chemiluminescence plus detection system (GE Healthcare, Biosciences, Pittsburgh, PA) and Hyperfilm.
2.6. Transglutaminase activity
Transglutaminase activity was monitored by the cross-linking of [3H]putrescine to casein as described previously [32].
2.7. Measurement of DNA synthesis and content
For measurement of cell proliferation, [3H]thymidine incorporation into DNA was assayed as described previously [33]. Additionally, the percentage of cells in S-phase was analyzed using flow cytometric analysis as described earlier [34].
2.8. Statistical analysis
Data from at least three independent experiments are presented as means ± SEM. Group mean values were defined using one-way analysis of variance with a Newman-Keuls post-hoc test (GraphPad Prism, La Jolla, CA). Significant differences were defined at ***, P≤0.001; **, P≤0.01; and *, P≤0.05 compared to control group or ††† or ƒƒƒ or §§§, P≤0.001; †† or ƒƒ, P≤0.01; and † or ƒ, P≤0.05 as compared between the indicated groups.
3. Results
3.1. Adenoviral expression of Cre-recombinase in floxed PKD1 keratinocytes results in PKD1 deficiency
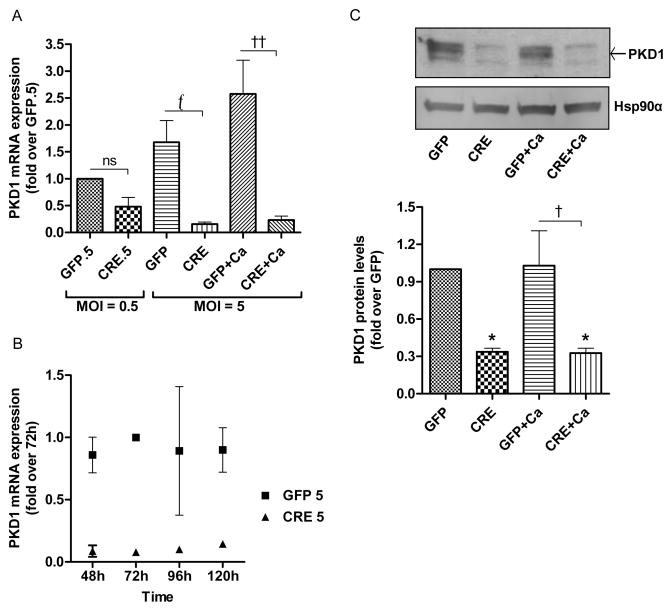
Using a floxed PKD1 mouse model and infecting epidermal keratinocytes derived from these mice with an adenovirus expressing Cre-recombinase (CRE), versus a green fluorescent protein (GFP) control vector, we determined the effect of PKD1 gene loss in vitro. First, we compared the infection efficiency of the two adenoviruses by examining GFP protein expression in the infected keratinocytes after 72 hours of adenoviral infection. Western analysis showed that comparable amounts of GFP protein were expressed in vector- and CRE-infected keratinocytes (Supplementary Fig. 1A). Further, Cre-recombinase was expressed in keratinocytes that were infected with Cre-adenovirus and not in GFP-infected keratinocytes (Supplementary Fig. 1B). After 72 hours of adenoviral infection, a significant reduction in PKD1 mRNA expression was achieved in adenoviral Cre-recombinase-infected keratinocytes depending on the dose of adenovirus used (Fig. 1A). Thus, although no significant decrease in PKD1 mRNA levels was observed with a lower dose (0.5 multiplicity of infection; MOI) of adenovirus expressing Cre-recombinase, the higher dose (5 MOI) produced a significant reduction in PKD1 mRNA levels (Fig. 1A). We also investigated the effect of Cre-recombinase adenoviral infection on PKD1 mRNA expression at different time points. PKD1 mRNA levels decreased in a time-dependent manner, with a significant reduction in PKD1 expression observed at the 72-hour time point after infection (Fig. 1B). Therefore, this time point was used for subsequent experiments. We also analyzed mRNA expression of PKD2 to confirm that Cre-recombinase acted only on the PKD1 gene and not on PKD2. The qRT-PCR analysis showed that PKD2 mRNA expression was not significantly altered upon Cre infection (Supplementary Fig. 2).
Fig 1. PKD1 depletion is achieved with adenoviral Cre-recombinase in floxed PKD1 keratinocytes.
(A–C) Keratinocytes from floxed PKD1 neonatal mice were infected with Cre-recombinase (CRE)- or GFP vector (GFP)-expressing adenoviruses for 24h. The medium was replaced with 50μM calcium-containing control medium or elevated calcium (125μM calcium; Ca)-containing medium at 48h (post-infection) for 24h, and then cells were harvested for qRT-PCR or Western analysis. (A–B) Quantitative RT-PCR was performed to determine the expression of the PKD1 gene (A) for the two indicated multiplicities of infection (MOI) and (B) for different time points of infection. (C) Western blot (upper panel) and densitometric analysis of multiple Western blots (lower panel) of PKD1 protein, with Hsp90α as the loading control, are shown. Please note that based on previous studies, the middle band represents unphosphorylated PKD1 as indicated by the arrow, the lower band represents PKD2 and the upper band(s) represent phosphorylated, activated PKD1 and PKD2. Data represent the means ± SEM from at least 3 independent experiments.
In parallel with mRNA results, PKD1 protein expression was also significantly reduced upon infection with adenovirus expressing Cre-recombinase (Fig. 1C, upper and lower panels). There is no PKD1-specific antibody available; the antibody we used recognizes both PKD1 and PKD2. Based on our previous results in keratinocytes [18, 35], the uppermost band represents phosphorylated, activated forms of PKD1 and PKD2, while the middle and lowermost band represents unphosphorylated PKD1 and PKD2, respectively [18, 21].
3.2. PKD1 deficiency increased the expression of loricrin, a late differentiation marker, upon induction of differentiation
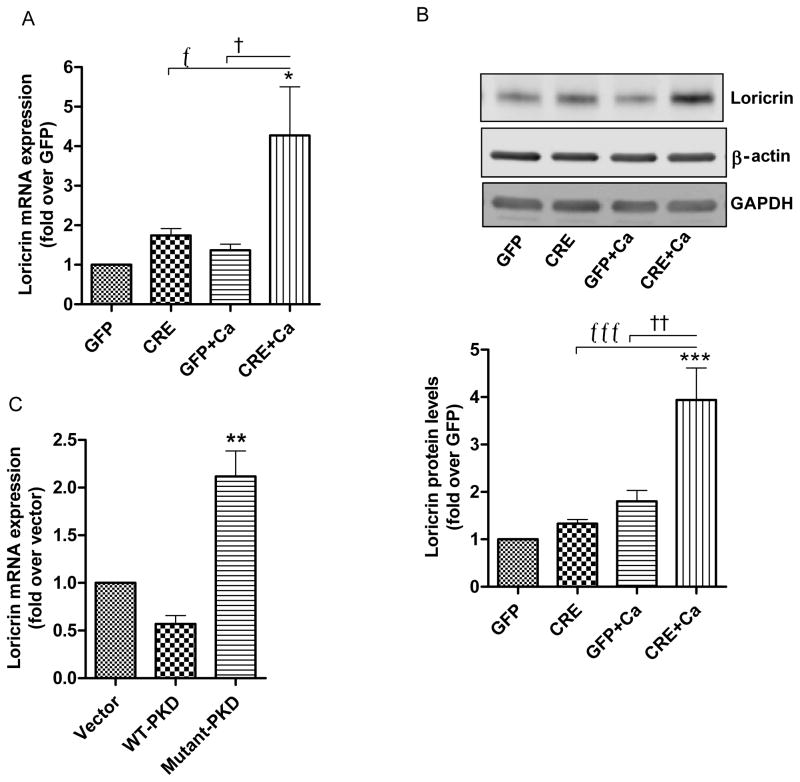
To investigate the effect of PKD1 deficiency on keratinocyte differentiation, we first studied the expression of the late differentiation marker, loricrin. Upon induction of differentiation by increasing the extracellular calcium level (to 125μM), the PKD1-deficient cells showed significantly increased mRNA expression (Fig. 2A) and protein levels (Fig. 2B, upper and lower panels) of loricrin. We also investigated the effect of PKD1 on expression of loricrin following adenovirus-mediated overexpression of wild-type PKD (WT-PKD) and a dominant-negative tyrosine-463-to-phenylalanine PKD mutant (mutant-PKD) [18] adenoviral construct into wild-type (CD-1) keratinocytes. Western blot results confirm the overexpression of PKD (Supplementary Fig. 3). Loricrin mRNA expression was significantly increased under basal conditions in keratinocytes overexpressing mutant-PKD as compared to vector- or WT-PKD-overexpressing cells (Fig. 2C).
Fig 2. Effect of PKD1 depletion on a late differentiation marker, loricrin.
(A–B) Keratinocytes from floxed PKD1 neonatal mice were infected with Cre-recombinase (CRE)- or GFP vector (GFP)-expressing adenoviruses for 24h. The medium was replaced with 50μM calcium-containing control medium or elevated calcium (125μM calcium; Ca)-containing medium at 48h (post-infection) for 24h. (A) Quantitative RT-PCR was performed to determine the expression of the loricrin gene. (B) A representative Western blot (upper panel) and a densitometric analysis (N=4) (lower panel) of loricrin protein, with β-actin and GAPDH as loading controls, are shown. (C) Sub-confluent keratinocytes from CD-1 neonates were infected with the indicated adenoviral constructs for 24h and analyzed for loricrin mRNA expression using qRT-PCR. Data represent the means ± SEM from at least 3 independent experiments.
3.3. PKD1 deficiency increased the expression of involucrin, an intermediate differentiation marker
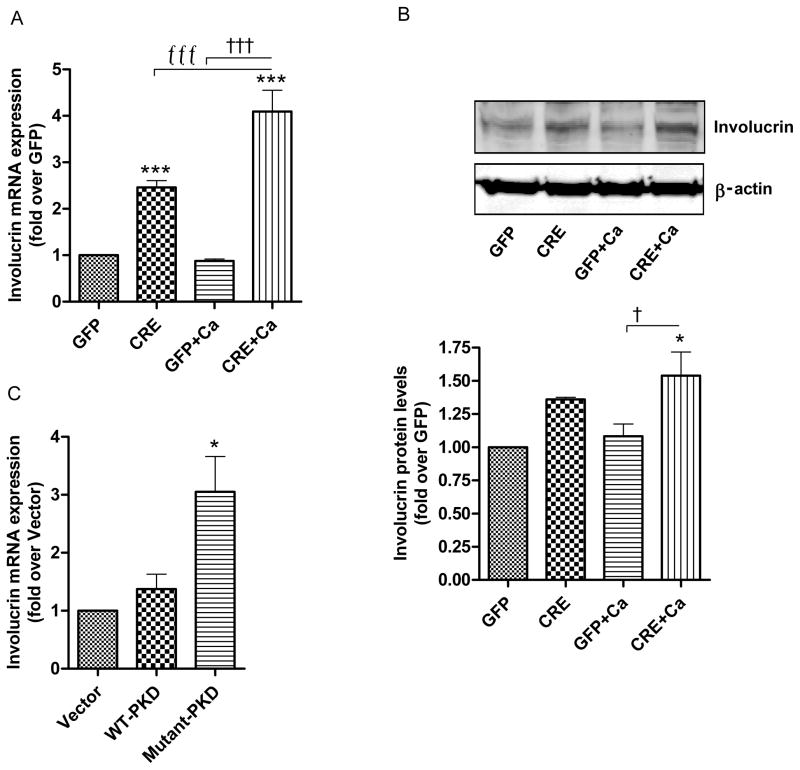
In contrast to loricrin, which required calcium-induced differentiation in order to observe a significant effect of PKD deficiency, the mRNA expression of the intermediate differentiation marker involucrin was significantly increased upon PKD1 ablation under basal conditions (Fig. 3A). Induction of differentiation by increasing the extracellular calcium level resulted in a further increase in involucrin mRNA (Fig. 3A) as well as protein expression (Fig. 3B, upper and lower panels). In line with the results observed with PKD loss, overexpression of the mutant PKD resulted in significantly increased involucrin mRNA expression as compared to overexpression of the vector or WT-PKD (Fig. 3C).
Fig 3. Effect of PKD1 depletion on an intermediate differentiation marker, involucrin.
(AB) Keratinocytes from floxed PKD1 neonates were infected with Cre-recombinase (CRE)- or GFP vector (GFP)-expressing adenoviruses for 24h. The medium was replaced with 50μM calcium-containing control medium or elevated calcium (125μM calcium; Ca)-containing medium at 48h (post-infection) for 24h. (A) Quantitative RT-PCR was performed to determine the expression of the involucrin gene. (B) A representative Western blot (upper panel) and densitometric analysis (N=3) (lower panel) of involucrin protein, with β-actin as the loading control, are shown. (C) Sub-confluent keratinocytes from CD-1 neonatal mice were infected with the indicated adenoviral constructs for 24h and analyzed for involucrin mRNA expression using qRT-PCR. Data represent the means ± SEM from at least 3 independent experiments.
3.4. PKD1 deficiency increased the protein expression of keratin 10, an early differentiation marker
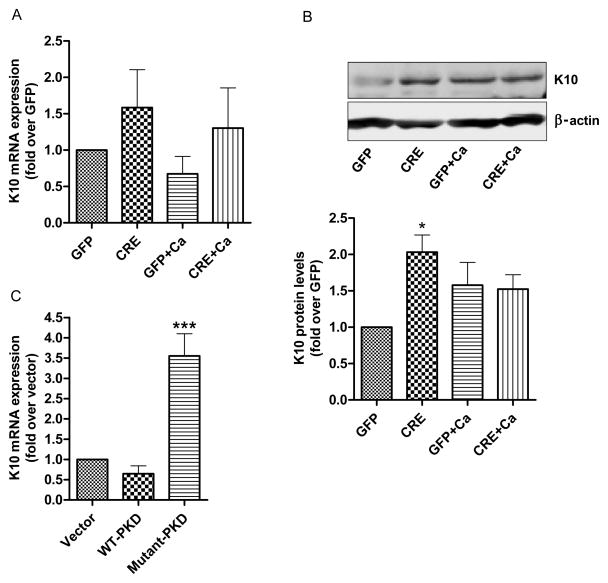
We then investigated the effect of PKD1 deletion on the expression of keratin 10 (K10), an early differentiation marker. Although no significant difference in mRNA expression of K10 was observed, there was a trend toward increased K10 expression with PKD1 depletion (Fig. 4A). Indeed, a significant increase in K10 protein levels was detected in keratinocytes in which the PKD1 gene was deleted with Cre-recombinase (Fig. 4B, upper and lower panels). Also, the mRNA expression of K10 in keratinocytes overexpressing mutant-PKD was significantly greater than in vector- or WT-PKD-overexpressing cells (Fig. 4C).
Fig 4. Effect of PKD1 depletion on an early differentiation marker, keratin 10 (K10).
(AB) Keratinocytes from floxed PKD1 neonates were infected with Cre-recombinase (CRE)- or GFP vector (GFP)-expressing adenoviruses for 24h. The medium was replaced with 50μM calcium-containing control medium or elevated calcium (125μM calcium; Ca)-containing medium at 48h (post-infection) for 24h. (A) Quantitative RT-PCR was performed to determine the expression of the K10 gene. (B) A representative Western blot (upper panel) and a densitometric analysis (N=3) (lower panel) of K10 protein, with β-actin as the loading control, are shown. (C) Sub-confluent keratinocytes from CD-1 neonatal mice were infected with the indicated adenoviral constructs for 24h and analyzed for K10 mRNA expression using qRT-PCR. Data represent the means ± SEM from at least 3 independent experiments.
3.5. PKD1 deficiency increased the expression and activity of transglutaminase
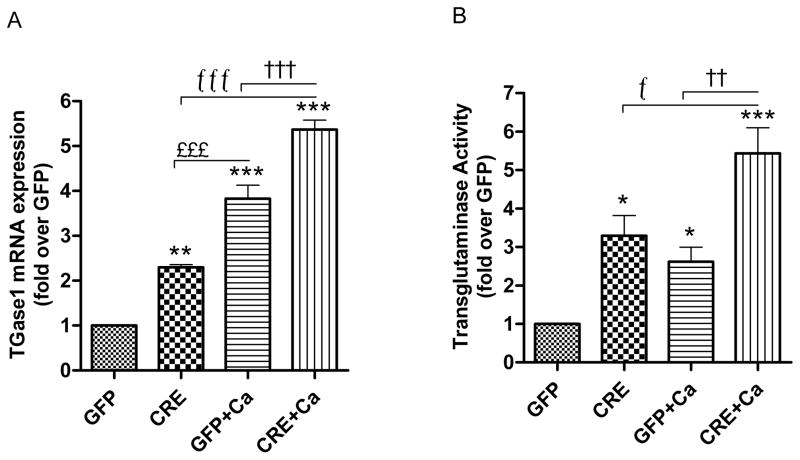
The transglutaminase 1 (TGase 1) enzyme is expressed during the terminal differentiation of keratinized squamous epithelium and is involved in the formation of cornified cell envelopes in terminally differentiating epidermal keratinocytes [36, 37]. Therefore, we investigated the effect of PKD1 loss on TGase1 expression and TGase activity. There was a significant increase in TGase1 mRNA expression in PKD1-deficient keratinocytes both under basal and differentiative (elevated calcium) conditions (Fig. 5A). As expected, an increase in TGase1 expression was observed with calcium treatment in GFP- as well as CRE-infected groups. Because TGase activity is regulated not only at the transcriptional level but also post-transcriptionally and post-translationally [38], to determine if this increase in TGase1 expression resulted in increased activity, we measured TGase activity. TGase activity was significantly increased in PKD1-deficient keratinocytes under both basal and differentiative conditions (Fig. 5B).
Fig.5. Effect of PKD1 depletion on expression and activity of transglutaminase.
(A–B) Keratinocytes from floxed PKD1 neonates were infected with Cre-recombinase (CRE)- or GFP vector (GFP)-expressing adenoviruses for 24h. The medium was replaced with 50μM calcium-containing control medium or elevated calcium (125μM calcium; Ca)-containing medium at 48h (post-infection) for 24h. (A) Quantitative RT-PCR was performed to determine the expression of the transglutaminase (TGase) 1 gene. (B) TGase activity is shown as fold over the GFP-infected control. Data represent the means ± SEM from at least 3 independent experiments.
3.6. Role of PKD1 in regulating keratinocyte proliferation
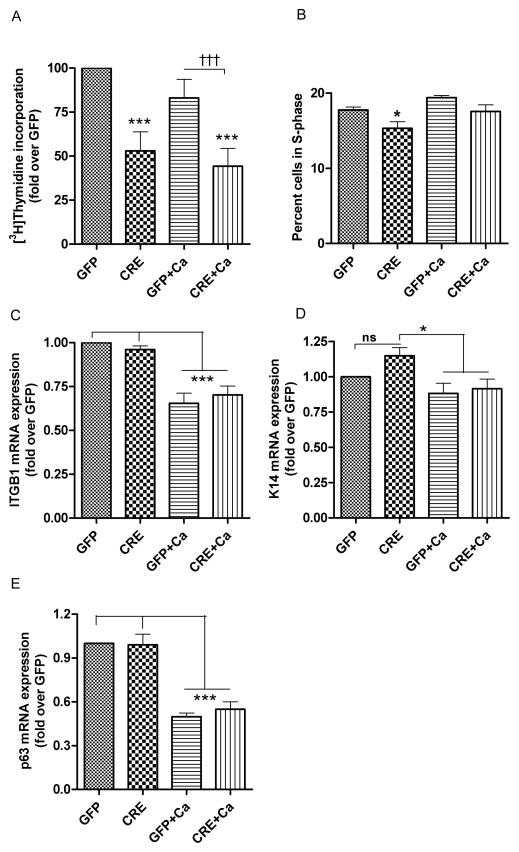
To study the effect of PKD1 loss on keratinocyte proliferation, we measured [3H]thymidine incorporation into DNA. [3H]Thymidine incorporation into DNA (i.e., proliferation) was reduced in keratinocytes with PKD1 deficiency compared to keratinocytes with intact PKD1 under basal conditions or upon stimulation of differentiation (Fig. 6A). We also analyzed the cell cycle using propidium iodide staining and flow cytometry. The number of cells in S-phase was reduced in keratinocytes with PKD1 deficiency compared to keratinocytes with intact PKD1 under basal conditions (Fig. 6B). Since PKD1 is localized predominantly in the basal layer of the epidermis (where stem cells are usually located), we wished to determine if PKD1 depletion can reduce the “stemness” of keratinocytes. Therefore, we examined mRNA expression of several stem cell markers (integrin β1 encoded by ITGB1, keratin 14 or K14 and p63) known to characterize epidermal stem cells [39]. Our results showed that PKD1 depletion had no significant effect on the mRNA levels of these markers (Figure 6C, D, E). However, an elevated calcium concentration inhibited the expression of these stem cell markers (Figure 6C, D, E), as expected since increased extracellular calcium levels induce keratinocyte differentiation. These results suggest that PKD1 has no effect on the “stemness” of the keratinocytes.
Fig 6. Effect of PKD1 depletion on keratinocyte proliferation and “stemness”.
(A–B) Keratinocytes from floxed PKD1 neonatal mice were infected with Cre-recombinase (CRE)- or GFP vector (GFP)-expressing adenoviruses for 24h. Cells were refed with 50μM calcium-containing control medium or elevated calcium (125μM calcium; Ca)-containing medium at 48h (post-infection) for 24h. (A) At the end of the treatment cells were analyzed for [3H]thymidine incorporation into DNA as described in the Methods section. (B) Cells were processed for propidium iodide staining and flow cytometry to analyze the percentage of cells in S-phase. Data represent the means ± SEM from at least 3 independent experiments. (C, D, E) Quantitative RT-PCR was performed to determine the expression of the stem cell markers ITGB1 (C), K14 (D) and p63 (E). Data represent the means ± SEM from at least 3 independent experiments. ns: non-significant.
4. Discussion
The major findings of this study are that Cre-recombinase-mediated PKD1 deficiency in PKD1 floxed keratinocytes resulted in increased keratinocyte differentiation as shown by increased expression, levels and/or activity of various differentiation markers. These results confirm that PKD1 is an anti-differentiative and pro-proliferative signaling enzyme in mouse keratinocytes, similar to human keratinocytes [24], but are unique in that they were obtained with the use of the floxed PKD1 mouse model rather than overexpression [40] or RNA interference [24], with the attendant caveats of these methods. Thus, in this study, we used PKD1-floxed mice that possess loxP sites flanking exons 12 through 14 of the PKD1 gene, which encodes part of the catalytic domain. This catalytic domain of PKD1 can then be deleted using Cre-recombinase enzyme in primary cultures of mouse keratinocytes obtained from floxed-PKD1 mice, to render keratinocytes PKD1-deficient.
Although Rashel et al. [23] generated epidermal-specific conditional PKD1 knockout mice using the PKD1 floxed mice, they reported (as data not shown) no difference in skin phenotype under normal conditions. However, in this model, PKD1 was deleted in utero and changes in the levels or activity of one or more proteins could have compensated for the lack of PKD1 in the unstressed epidermis. Although these authors showed no change in the protein expression of PKD2, they overlooked the possibility of increased activity of PKD2. Moreover, they did not directly show the levels of PKD1 in skin samples; rather, they showed Western analysis of in vitro cultured keratinocytes derived from PKD1 floxed versus knockout animals [23]. Although PKD1 levels were reduced in the knockout keratinocytes, PKD1 was nevertheless detected in these cells but its presence was dismissed as arising from contaminating (non-keratinocyte) cells (with no data to support this contention). It seems possible that the protein purported to be PKD1 in these blots is instead active PKD2, which migrates more slowly when activated and autophosphorylated [18]. Also, it is noteworthy that keratinocyte proliferation and differentiation (as well as the proteins that regulate these processes) differ among preparations of keratinocytes and with the confluence of the cells. Therefore, the different keratinocyte cultures derived from floxed PKD1 versus PKD1 knockout animals may not be entirely comparable. On the other hand, in a previous study using siRNA to knock down PKD1 in human keratinocytes, Ivanova and coworkers were unable to show an effect on PKD1 at the protein level [24]. In addition, siRNA technology has a risk of off-target effects [41, 42]. Therefore, we attempted to investigate the effect of deletion of PKD1 in post-natal keratinocytes in vitro by another method that allowed us to use the same keratinocyte preparation to generate PKD1 knockout and non-knockout keratinocytes for valid comparisons, without the potential artifacts of RNA interference technologies. This technique resulted in decreased PKD1 mRNA and protein expression in hard-to-transfect primary cultures of mouse keratinocytes (Fig. 1A and 1C).
We suspected that PKD1 deficiency alone might not be sufficient for observing changes in various differentiation markers, particularly late markers, under basal growth conditions. Therefore, to maximize the effect of PKD1 loss, we examined differentiation under both basal conditions and in keratinocytes stimulated to differentiate by raising extracellular calcium levels. Previous reports implicate a role of increased calcium in the induction of differentiation, as an increasing gradient of calcium concentration from the proliferating basal layer to the outermost living differentiated (granular) layer is observed in the epidermis in situ [43–45]. Also, ablation of the G-protein-coupled receptor for extracellular calcium, the calcium-sensing receptor, impairs epidermal differentiation in knockout mice in vivo [46]. In vitro, low medium calcium levels promote a basal-like state of the keratinocyte, and raising the medium calcium concentration triggers keratinocyte differentiation [47]. Moreover, differentiation marker expression is reportedly maximally induced in mouse keratinocytes by calcium concentrations in the range of 90 to about 120μM [29, 30]. Therefore, we stimulated keratinocytes to differentiate using moderately elevated calcium levels (125μM), and this strategy allowed us to identify additional differentiation markers that appear to be repressed by PKD1. Upon stimulation of differentiation, the late differentiation marker, loricrin was significantly increased in PKD1-deficient keratinocytes at both the mRNA (Fig. 2A) and protein (Fig. 2B) levels. These results are in line with the fact that in the epidermis loricrin is expressed in the upper granular layer where the calcium concentration is high [48] and where PKD1 levels are low [21, 49]. However, basally loricrin expression was not affected by the presence or absence of PKD1. The expression of the intermediate differentiation marker, involucrin, was also increased in PKD1-deficient keratinocytes (Fig. 3). This increase was observed in PKD1-deficient keratinocytes under basal as well as differentiating conditions. Similar to our results, siRNA-mediated PKD1 knockdown in human keratinocytes resulted in increased involucrin mRNA expression [24]. Also, we have previously reported that PKD overexpression with an involucrin promoter reporter constructs results in decreased promoter activity [21].
In human keratinocytes an increase in K10 mRNA was observed with siRNA-mediated PKD1 knockdown [24]. However, we did not observe any significant differences in mRNA expression of the early differentiation marker, K10, in keratinocytes with or without PKD1 deficiency (Fig. 4A). This might be due to down-regulation of K10 expression as keratinocytes mature beyond the early differentiation stage. Nevertheless, there was a significant difference in the protein levels in these groups. Keratinocytes with PKD1 loss expressed significantly higher K10 protein levels as compared to keratinocytes with intact PKD1 (Fig. 4B). In addition, using PKD wild-type and mutant adenoviral overexpression constructs, we supported the results obtained by Cre-recombinase-mediated floxed PKD1 loss. Mutant-PKD expression led to significant increases in loricrin, involucrin and K10 mRNA levels (Fig. 2C, 3C, 4C). However, WT-PKD overexpression showed no significant effect on expression of these differentiation markers. This may be due to the fact that under basal growth conditions keratinocytes exhibit partial activation of PKD, as observed by its detectable autophosphorylation under such conditions [21]; thus, PKD may already be suppressing the expression of these markers basally. Consistent with this idea, mutant PKD overexpression likely released endogenous PKD-mediated suppression of these differentiation markers to result in their increased expression in wild-type keratinocytes.
The functionality of PKD1 depletion on keratinocyte differentiation was assessed by investigating the expression and activity of transglutaminase. This enzyme is necessary for proper cornification of keratinoctyes during terminal differentiation [36, 37]. In line with the results obtained for other differentiation markers, TGase1 expression and TGase activity was significantly increased in PKD1-deficient keratinocytes under basal and differentiative conditions (Fig. 5). These results suggest that endogenous PKD1 has anti-differentiative effects on mouse epidermal keratinocytes such that its loss promotes differentiation.
We and others have shown a pro-proliferative role for PKD in keratinocytes [19, 20]. Consistent with these reports, [3H]thymidine incorporation into DNA, as well as the percentage of cells in the S-phase of the cell cycle, was reduced in PKD1-deficient keratinocytes (Fig. 6A and B), indicating decreased proliferation in these cells. These results suggest the pro-proliferative role of PKD1 in keratinocytes; however, we did not observe any significant effect of loss of PKD1 on other epidermal proliferation markers like keratin 14 and keratin 5 or on stem cell markers, suggesting that PKD1 likely promotes a transit amplifying cell phenotype. However, co-expressing PKD with reporter constructs increased the promoter activity of keratin 5 [21].
Together, our data provide genetic evidence that PKD1 is an anti-differentiative and pro-proliferative enzyme in mouse keratinocytes in vitro. While Rashel et al. [23] reported no effect of PKD1 loss on keratinocyte proliferation or differentiation in epidermis in vivo under basal conditions, these authors did observe reduced proliferation in healing wounds as well as a resistance to tumor formation. The possible reasons underlying these somewhat disparate results in vivo and in vitro are many and include: the techniques used (immunohistochemistry and immunofluorescence) may be insufficiently sensitive to observe effects on proliferation and differentiation without perturbation in the in vivo experiments, the in vivo loss of PKD1 in utero could be compensated for by alterations in the levels or activity or other signaling proteins under basal conditions or in vitro culture may artificially accentuate effects on these parameters. These results support the idea that PKD plays an important role in initiating or promoting hyperproliferative skin diseases like basal cell carcinoma and psoriasis, as suggested by the dysregulation of epidermal PKD levels in these diseases [49]. Thus, PKD1 can potentially be therapeutically targeted in these hyperproliferative disorders to improve skin function.
Supplementary Material
Highlights.
Effect of protein kinase D1 (PKD1) deficiency was examined in keratinocytes in vitro.
PKD1 was deleted by infection of floxed keratinocytes with Cre-recombinase adenovirus.
PKD1 gene deletion decreased PKD1 mRNA and protein levels in keratinocytes.
PKD1 deletion stimulated keratinocyte differentiation marker expression.
PKD1 deletion inhibited keratinocyte proliferation.
Acknowledgments
We appreciate Purnima Merai for assisting with the preparation of primary mouse keratinocytes. The authors are grateful to Dr. Eric N. Olson (University of Texas Southwestern Medical Center, Dallas, TX) for kindly providing us the floxed PKD1 transgenic mouse model and Dr. Alex Toker (Harvard Medical School, Boston, MA) for providing the PKD constructs used for adenoviral constructs. This study was supported in part by NIH award #R21AR057321 and by VA Merit Award #01CX000590. WBB is supported by a VA Research Career Scientist Award. The contents of this article do not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
The authors have no conflict of interest to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Menon GK. New insights into skin structure: scratching the surface. Adv Drug Deliv Rev. 2002;54 (Suppl 1):S3–17. doi: 10.1016/s0169-409x(02)00121-7. [DOI] [PubMed] [Google Scholar]
- 2.So P, Kim H, Kochevar I. Two-Photon deep tissue ex vivo imaging of mouse dermal and subcutaneous structures. Opt Express. 1998;3:339–350. doi: 10.1364/oe.3.000339. [DOI] [PubMed] [Google Scholar]
- 3.Slominski AT, Zmijewski MA, Zbytek B, Tobin DJ, Theoharides TC, Rivier J. Key role of CRF in the skin stress response system. Endocr Rev. 2013;34:827–884. doi: 10.1210/er.2012-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol. 2007;23:93–113. doi: 10.1146/annurev.cellbio.23.090506.123357. [DOI] [PubMed] [Google Scholar]
- 6.Proksch E, Brandner JM, Jensen JM. The skin: an indispensable barrier. Exp Dermatol. 2008;17:1063–1072. doi: 10.1111/j.1600-0625.2008.00786.x. [DOI] [PubMed] [Google Scholar]
- 7.Bikle DD, Ng D, Tu CL, Oda Y, Xie Z. Calcium- and vitamin D-regulated keratinocyte differentiation. Mol Cell Endocrinol. 2001;177:161–171. doi: 10.1016/s0303-7207(01)00452-x. [DOI] [PubMed] [Google Scholar]
- 8.Guha S, Tanasanvimon S, Sinnett-Smith J, Rozengurt E. Role of protein kinase D signaling in pancreatic cancer. Biochem Pharmacol. 2010;80:1946–1954. doi: 10.1016/j.bcp.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaggi M, Du C, Zhang W, Balaji KC. Protein kinase D1: a protein of emerging translational interest. Front Biosci. 2007;12:3757–3767. doi: 10.2741/2349. [DOI] [PubMed] [Google Scholar]
- 10.Steinberg SF. Regulation of protein kinase D1 activity. Mol Pharmacol. 2012;81:284–291. doi: 10.1124/mol.111.075986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozengurt E. Protein kinase D signaling: multiple biological functions in health and disease. Physiology (Bethesda) 2011;26:23–33. doi: 10.1152/physiol.00037.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storz P, Doppler H, Johannes FJ, Toker A. Tyrosine phosphorylation of protein kinase D in the pleckstrin homology domain leads to activation. J Biol Chem. 2003;278:17969–17976. doi: 10.1074/jbc.M213224200. [DOI] [PubMed] [Google Scholar]
- 13.Storz P, Doppler H, Toker A. Protein kinase Cdelta selectively regulates protein kinase D-dependent activation of NF-kappaB in oxidative stress signaling. Mol Cell Biol. 2004;24:2614–2626. doi: 10.1128/MCB.24.7.2614-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Storz P, Doppler H, Toker A. Activation loop phosphorylation controls protein kinase D-dependent activation of nuclear factor kappaB. Mol Pharmacol. 2004;66:870–879. doi: 10.1124/mol.104.000687. [DOI] [PubMed] [Google Scholar]
- 15.Storz P, Doppler H, Toker A. Protein kinase D mediates mitochondrion-to-nucleus signaling and detoxification from mitochondrial reactive oxygen species. Mol Cell Biol. 2005;25:8520–8530. doi: 10.1128/MCB.25.19.8520-8530.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storz P, Toker A. Protein kinase D mediates a stress-induced NF-kappaB activation and survival pathway. EMBO J. 2003;22:109–120. doi: 10.1093/emboj/cdg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Besirli CG, Johnson EM., Jr The activation loop phosphorylation of protein kinase D is an early marker of neuronal DNA damage. J Neurochem. 2006;99:218–225. doi: 10.1111/j.1471-4159.2006.04116.x. [DOI] [PubMed] [Google Scholar]
- 18.Arun SN, Kaddour-Djebbar I, Shapiro BA, Bollag WB. Ultraviolet B irradiation and activation of protein kinase D in primary mouse epidermal keratinocytes. Oncogene. 2011;30:1586–1596. doi: 10.1038/onc.2010.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rennecke J, Rehberger PA, Furstenberger G, Johannes FJ, Stohr M, Marks F, et al. Protein-kinase-Cmu expression correlates with enhanced keratinocyte proliferation in normal and neoplastic mouse epidermis and in cell culture. Int J Cancer. 1999;80:98–103. doi: 10.1002/(sici)1097-0215(19990105)80:1<98::aid-ijc19>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 20.Shapiro BA, Ray S, Jung E, Allred WT, Bollag WB. Putative conventional protein kinase C inhibitor Godecke 6976 [12-(2-cyanoethyl)-6,7,12,13-tetrahydro-13-methyl-5-oxo-5H-indolo(2,3-a)pyrrolo(3 ,4-c)-carbazole] stimulates transglutaminase activity in primary mouse epidermal keratinocytes. J Pharmacol Exp Ther. 2002;302:352–358. doi: 10.1124/jpet.302.1.352. [DOI] [PubMed] [Google Scholar]
- 21.Dodd EM, Ristich VL, Ray S, Lober RM, Bollag WB. Regulation of protein kinase D during differentiation and proliferation of primary mouse keratinocytes. J Invest Dermatol. 2005;125:294–306. doi: 10.1111/j.0022-202X.2005.23780.x. [DOI] [PubMed] [Google Scholar]
- 22.Jadali A, Ghazizadeh S. Protein kinase D is implicated in the reversible commitment to differentiation in primary cultures of mouse keratinocytes. J Biol Chem. 2010;285:23387–23397. doi: 10.1074/jbc.M110.105619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rashel M, Alston N, Ghazizadeh S. Protein kinase d1 has a key role in wound healing and skin carcinogenesis. J Invest Dermatol. 2014;134:902–909. doi: 10.1038/jid.2013.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanova P, Atanasova G, Poumay Y, Mitev V. Knockdown of PKD1 in normal human epidermal keratinocytes increases mRNA expression of keratin 10 and involucrin: early markers of keratinocyte differentiation. Arch Dermatol Res. 2008;300:139–145. doi: 10.1007/s00403-008-0832-7. [DOI] [PubMed] [Google Scholar]
- 25.Fielitz J, Kim MS, Shelton JM, Qi X, Hill JA, Richardson JA, et al. Requirement of protein kinase D1 for pathological cardiac remodeling. Proc Natl Acad Sci U S A. 2008;105:3059–3063. doi: 10.1073/pnas.0712265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shapiro BA, Olala L, Arun SN, Parker PM, George MV, Bollag WB. Angiotensin II-activated protein kinase D mediates acute aldosterone secretion. Mol Cell Endocrinol. 2010;317:99–105. doi: 10.1016/j.mce.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jung EM, Betancourt-Calle S, Mann-Blakeney R, Griner RD, Bollinger Bollag W. Sustained phospholipase D activation is associated with keratinocyte differentiation. Carcinogenesis. 1999;20:569–576. doi: 10.1093/carcin/20.4.569. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Tucker RW, Hennings H, Yuspa SH. Chelation of intracellular Ca2+ inhibits murine keratinocyte differentiation in vitro. J Cell Physiol. 1995;163:105–114. doi: 10.1002/jcp.1041630112. [DOI] [PubMed] [Google Scholar]
- 30.Yuspa SH, Kilkenny AE, Steinert PM, Roop DR. Expression of murine epidermal differentiation markers is tightly regulated by restricted extracellular calcium concentrations in vitro. The Journal of cell biology. 1989;109:1207–1217. doi: 10.1083/jcb.109.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Griner RD, Qin F, Jung E, Sue-Ling CK, Crawford KB, Mann-Blakeney R, et al. 1,25-dihydroxyvitamin D3 induces phospholipase D-1 expression in primary mouse epidermal keratinocytes. J Biol Chem. 1999;274:4663–4670. doi: 10.1074/jbc.274.8.4663. [DOI] [PubMed] [Google Scholar]
- 33.Griner RD, Bollag WB. Inhibition of [(3)H]thymidine transport is a nonspecific effect of PDMP in primary cultures of mouse epidermal keratinocytes. J Pharmacol Exp Ther. 2000;294:1219–1224. [PubMed] [Google Scholar]
- 34.Dransfield DT, Griner RD, Ray S, Keskintepe M, Bollag WB. 8-Cl-adenosine induces growth arrest without differentiation of primary mouse epidermal keratinocytes. J Invest Dermatol. 2001;117:1588–1593. doi: 10.1046/j.0022-202x.2001.01572.x. [DOI] [PubMed] [Google Scholar]
- 35.Dodd MEBS, Arun S, Ray S, Bollag RJ, Bollag WB. Vitamin D Signaling in the Epidermis. In: Jimenez ML, editor. Vitamins and Antioxidant Research. Nova Science Publishers, Inc; Hauppauge, New York: 2008. pp. 3–49. [Google Scholar]
- 36.Kim SY, Chung SI, Yoneda K, Steinert PM. Expression of transglutaminase 1 in human epidermis. J Invest Dermatol. 1995;104:211–217. doi: 10.1111/1523-1747.ep12612769. [DOI] [PubMed] [Google Scholar]
- 37.Michel S, Bernerd F, Jetten AM, Floyd EE, Shroot B, Reichert U. Expression of keratinocyte transglutamine mRNA revealed by in situ hybridization. J Invest Dermatol. 1992;98:364–368. doi: 10.1111/1523-1747.ep12499806. [DOI] [PubMed] [Google Scholar]
- 38.Eckert RL, Sturniolo MT, Jans R, Kraft CA, Jiang H, Rorke EA. TIG3: a regulator of type I transglutaminase activity in epidermis. Amino Acids. 2009;36:739–746. doi: 10.1007/s00726-008-0123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Poojan S, Verma V, Verma MK, Lohani M. Rapid isolation of integrin rich multipotent stem cell pool and reconstruction of mouse epidermis equivalent. Am J Stem Cells. 2014;3:27–36. [PMC free article] [PubMed] [Google Scholar]
- 40.Arun SN, Xie D, Dodd ME, Zhong X, Bollag WB. The potential use of protein kinase D inhibitors for prevention/treatment of epidermal tumors. J Dermatol Sci. 2010;60:29–39. doi: 10.1016/j.jdermsci.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Liu P, Chou HH. Whole-genome thermodynamic analysis reduces siRNA off-target effects. PLoS One. 2013;8:e58326. doi: 10.1371/journal.pone.0058326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naito Y, Ui-Tei K. Designing functional siRNA with reduced off-target effects. Methods Mol Biol. 2013;942:57–68. doi: 10.1007/978-1-62703-119-6_3. [DOI] [PubMed] [Google Scholar]
- 43.Elias P, Ahn S, Brown B, Crumrine D, Feingold KR. Origin of the epidermal calcium gradient: regulation by barrier status and role of active vs passive mechanisms. J Invest Dermatol. 2002;119:1269–1274. doi: 10.1046/j.1523-1747.2002.19622.x. [DOI] [PubMed] [Google Scholar]
- 44.Lee SH, Elias PM, Proksch E, Menon GK, Mao-Quiang M, Feingold KR. Calcium and potassium are important regulators of barrier homeostasis in murine epidermis. J Clin Invest. 1992;89:530–538. doi: 10.1172/JCI115617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menon GK, Elias PM, Lee SH, Feingold KR. Localization of calcium in murine epidermis following disruption and repair of the permeability barrier. Cell Tissue Res. 1992;270:503–512. doi: 10.1007/BF00645052. [DOI] [PubMed] [Google Scholar]
- 46.Tu CL, Crumrine DA, Man MQ, Chang W, Elalieh H, You M, et al. Ablation of the calcium-sensing receptor in keratinocytes impairs epidermal differentiation and barrier function. J Invest Dermatol. 2012;132:2350–2359. doi: 10.1038/jid.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pillai S, Bikle DD, Mancianti ML, Cline P, Hincenbergs M. Calcium regulation of growth and differentiation of normal human keratinocytes: modulation of differentiation competence by stages of growth and extracellular calcium. J Cell Physiol. 1990;143:294–302. doi: 10.1002/jcp.1041430213. [DOI] [PubMed] [Google Scholar]
- 48.Ishida-Yamamoto A, Hohl D, Roop DR, Iizuka H, Eady RA. Loricrin immunoreactivity in human skin: localization to specific granules (L-granules) in acrosyringia. Arch Dermatol Res. 1993;285:491–498. doi: 10.1007/BF00376822. [DOI] [PubMed] [Google Scholar]
- 49.Ristich VL, Bowman PH, Dodd ME, Bollag WB. Protein kinase D distribution in normal human epidermis, basal cell carcinoma and psoriasis. Br J Dermatol. 2006;154:586–593. doi: 10.1111/j.1365-2133.2005.07073.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.