Abstract
Fuchs endothelial corneal dystrophy (FECD) is a genetically heterogeneous disease. Hypothesizing that cellular senescence may be relevant in FECD pathogenesis, genetically undifferentiated late-onset FECD endothelial samples were analyzed to identify common changes of specific senescence-related transcripts. Total RNA was extracted from 21 FECD endothelial samples retrieved from patients undergoing lamellar keratoplasty due to clinically diagnosed end-stage FECD and from 12 endothelial samples retrieved from normal autopsy eyes. Taqman low density array (TLDA) cards were used to analyze differential expression of 89 cellular senescence-related transcripts. Result validation was performed using individual real-time PCR assays. TLDA-analysis demonstrated differential expression of 31 transcripts (fold-change >1.5; p<0.05). Thereof, 27 showed significant up-regulation and 4 significant down-regulation. Markedly elevated mRNA-levels of the constitutively active and reactive oxygen species-generating enzyme NOX4 were found in all evaluable FECD samples. In addition, increased expression of CDKN2A and its transcriptional activators ETS1 and ARHGAP18 (SENEX) along with decreased expression of CDKN2A inhibitor ID1 were detected in FECD samples. Consistent over-expression of NOX4 in FECD endothelial samples suggests a role as pathogenic factor and as a potential new treatment target in FECD. Transcriptional up-regulation of the CDKN2A-pathway provides further evidence for increased cellular senescence in FECD endothelium.
Keywords: Fuchs endothelial corneal dystrophy, pathogenesis, corneal endothelium, cellular senescence, NOX4, CDKN2A, oxidative stress
Fuchs endothelial corneal dystrophy (FECD) is a bilateral disorder of the corneal endothelium. Morphologically, it is characterized by an accelerated decrease of corneal endothelial cell (CEC) density over several decades with abnormal subendothelial deposition of extracellular matrix (ECM) and formation of posterior excrescences (guttae) of Descemet’s membrane.
Previous studies have demonstrated an imbalance of oxidant and antioxidant factors contributing to FECD pathogenesis.(Jurkunas et al., 2010) Particularly, nuclear factor erythroid 2-related factor 2 (Nrf2), a transcription factor binding the antioxidant response element and activating antioxidant defense, is deregulated, and decreased Nrf2-levels may be involved in the pathogenic cascade eventually leading to oxidative DNA-damage and apoptosis induction in FECD endothelium.(Jurkunas et al., 2010)
CECs do not normally proliferate in vivo, and are arrested in G1 phase. However, proliferation of CECs may be induced under optimized in vitro conditions.(Engelmann et al., 1988) Joyce and colleagues found that prolonged exposure of normal CECs to oxidative stress causes a loss of proliferative capacity and reduced cellular function.(Joyce et al., 2009) This state is referred to as stress-induced premature senescence (SIPS). Cells undergoing SIPS and concomitantly altered gene expression may considerably disrupt the cellular microenvironment and thus tissue homeostasis. Due to their non-dividing nature, their positioning within the light path, and their high metabolic activity, CECs from the corneal center are particularly prone to oxidative stress.(Joyce, 2012) Normal CECs from the central cornea of old donors exhibit increased protein expression of senescence-related markers CDKN1A and CDKN2A when compared to CECs from younger individuals.(Enomoto et al., 2006) In a similar fashion, we have recently demonstrated that increased protein expression of CDKN1A and CDKN2A is also evident in FECD endothelium when compared to normal tissues from donors of the same age.(Matthaei et al., 2013b) This suggests that cellular senescence affects at least a certain proportion of CECs in FECD.
In the present study we generated an expression profile of senescence-related transcripts in an extended set of FECD endothelial samples in order to analyze CEC senescence in FECD in more detail. Here we show for the first time that the constitutively active and reactive oxygen species (ROS)-generating enzyme NOX4 [NADPH (reduced form of nicotinamide adenine dinucleotide phosphate) oxidase 4] is highly over-expressed in FECD endothelium. An imbalance of decreased Nrf2-expression and increased NOX4 expression could significantly contribute to the emergence of oxidative stress and consecutive senescence induction in CECs from FECD patients. In addition, our results suggest a highlighted role of increased expression of CDKN2A and its transcriptional activators ETS1 and ARHGAP18 (SENEX) along with decreased expression of the CDKN2A inhibitor ID1 in FECD pathogenesis.
Endothelial samples (stripped Descemet’s membrane with attached endothelial cells) from FECD corneas and normal eyes were included in the present study. FECD endothelial samples were directly retrieved from patients undergoing endothelial keratoplasty due to clinically diagnosed end-stage FECD and no history of ocular surgery. Normal samples were retrieved from autopsy eyes without corneal pathology, glaucoma, or history of ocular surgery. Briefly, corneoscleral buttons were excised from autopsy eyes. Comparable to lamellar transplant surgery, the endothelium was circularly scored in the mid-peripheral area (about 9mm diameter) using a scalpel blade. The Descemet membrane and endothelial cells within this scored area were subsequently stripped and preserved. For the array experiment endothelial samples from n = 15 FECD patients (mean age ± standard error of the mean (SEM) 62.6 ± 1.6 years; male-to-female ratio 5:10) and n=8 autopsy eyes (donor mean age ± SEM 67.3 ± 3.7 years; male-to-female ratio 3:5, death-to-preservation time ± SEM 12.25 ± 2.36 hours) were used. For the validation of differentially expressed transcripts endothelial samples from n = 8 FECD patients (including n=6 independent samples and n=2 samples form the initial array experiment, mean age ± SEM 67.5 ± 4.8 years; male-to-female ratio 2:6) and n=8 autopsy eyes (including n=2 independent samples, n=2 samples from fellow eyes of the initial array experiment and n=4 samples from the initial array experiment, donor mean age ± SEM 65.5 ± 6.5 years; male-to-female ratio 3:5, death-to-preservation time ± SEM 13.31 ± 2.53 hours) were used. Studies using human tissues were approved by The Johns Hopkins Institutional Review Board and adhered to the tenets of the Declaration of Helsinki. Written informed consent was obtained from all patients.
For total RNA extraction, a combined TRIzol/spin-column protocol was used (modified from http://www.med.upenn.edu/kaestnerlab). Briefly, stripped Descemet’s membrane with attached CECs were placed directly into 1.3ml TRIzol (Invitrogen, Carlsbad, CA). Endothelial cells were lysed by gentle pipetting through a standard 1000μl pipet tip and incubated for five minutes at room temperature. A volume of 260μl chloroform was added and the sample was vigorously shaken for 15 seconds. Following another incubation at room temperature, the samples were centrifuged for 5 minutes at 12,000g and 4°C. The a queous phase was gently removed and mixed in a 1:1 ratio with 70% ethanol and centrifuged for 30 seconds at 12,000g and 4°C through an RNeasy mini column (Qiagen, Valencia, CA) placed in a 2ml collection tube. This was followed by a centrifugation step for 30 seconds at 12,000g at 4°C with 700μl buffer RW1 and two centrifugation steps for 30 seconds and 2 minutes, respectively, at 12,000g and 4°C with 500 μl buffer RPE.
After an empty centrifugation step to completely dry the RNeasy silica-gel membrane, the RNA was eluted in 30μl RNase-free water by centrifugation at 12,000g for 1 minute. The RNA content of each sample was measured using NanoDrop 2000 spectrophotometry (Thermo Scientific, Waltham, MA). RNA was reverse transcribed to cDNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems (ABI), Foster City, CA, USA) according to manufacturer’s instructions
Custom TaqMan Low Density Array Cards (TLDA, ABI) were used for the expression analysis of 89 senescence related transcripts. For preamplification the individual cDNA samples were mixed with custom PreAmp Pool (prepared by ABI) and TaqMan PreAmp Master Mix (ABI) in a 1:1:2 ratio. Preamplification cycles were run on a Veriti 96-Well Thermal Cycler (ABI) according to manufacturer’s instructions. Subsequently, 14μl of pre-amplification-product of each sample was mixed with 206μl nuclease-free water and 220μl Taqman Universal Master Mix, no UNG (ABI) (reaction mix). 400 μl of the reaction mix per sample was loaded onto a custom TLDA card (ABI) and the cards were run on a 7900HT real-time PCR system (ABI) under standard conditions according to manufacturer’s instructions. The Sequence Detection System software package version 2.3 (sds software, ABI) was used for data collection. The resulting files were exported and analyzed using the Expression Suite v1.0 software package (ABI).
Datasets were analyzed using the Expression Suite v1.0 software package (ABI) as previously described.(Matthaei et al., 2013a) The experimental data were grouped into files from diseased (FECD) and normal biological replicates using the normal group as reference. A cycle threshold (Ct) value of 40 was set for detectable RNA expression. Endothelial samples showing Ct values above this threshold or presenting uneven amplification for a given target were considered as being non-evaluable and omitted from further analyses for the given target. GUSB (GUSB-Hs99999908_m1), HPRT1 (HPRT1-Hs99999909_m1), ACTB (ACTB-Hs99999903_m1) and 18S (18S-Hs99999901_s1) were used as housekeeping genes. Relative expression of >1.5-fold (up- or down-regulated) and a p-value of <0.05 were set as thresholds for significant differential expression of individual transcripts.
Differential expression of individual transcripts was validated using individual Taqman assays (ABI). Individual assays used were: NOX4-Hs00418356_m1, CDKN2A-Hs00923894_m1, ETS1-Hs00428293_m1, ID1-Hs03676575_s1, FN1-Hs00365052_m1, ARHGAP18-Hs00364379_m1, NFKB1-Hs00765730_m1, IGF1-Hs01547656_m1, IGFBP7-Hs00266026_m1, IGFBP5-Hs00181213_m1, ACTB-Hs99999903_m1.
RNA extraction and reverse transcription was performed as described above. The pre-amplification of cDNA samples was performed using a self-prepared preamplification pool (individual assays diluted in TE buffer at a ratio of 1:100). CDNA samples were mixed with the preamplification pool and Taqman PreAmp Master Mix (ABI) in a ratio of 1:1:2 and preamplified on a Veriti 96-Well Thermal Cycler (ABI) according to manufacturer’s protocol. Preamplified cDNA samples were diluted 1:20 in TE buffer. For each real-time PCR reaction, 5μl diluted preamplification product were mixed with 1μl Taqman Gene Expression Assay (ABI), 10μl TaqMan Universal Master Mix, no UNG (ABI) and 4μl nuclease-free water and run on a StepOne Plus thermal cycler (ABI).
The cycling conditions were 50°C for 2 minutes, 95° C for 10 minutes, 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute. Data sets were expo rted and analyzed using the Expression Suite v1.0 software package (ABI). ACTB was used as housekeeping gene.
The Expression Suite v1.0 software package (ABI) and PRISM4 software package (GraphPad, La Jolla, CA) were used for statistical analyses applying Student’s t test. Pooled FECD (FECD1 + FECD2) vs control (Control1 + Control2) cohorts were analyzed in the validation experiment.
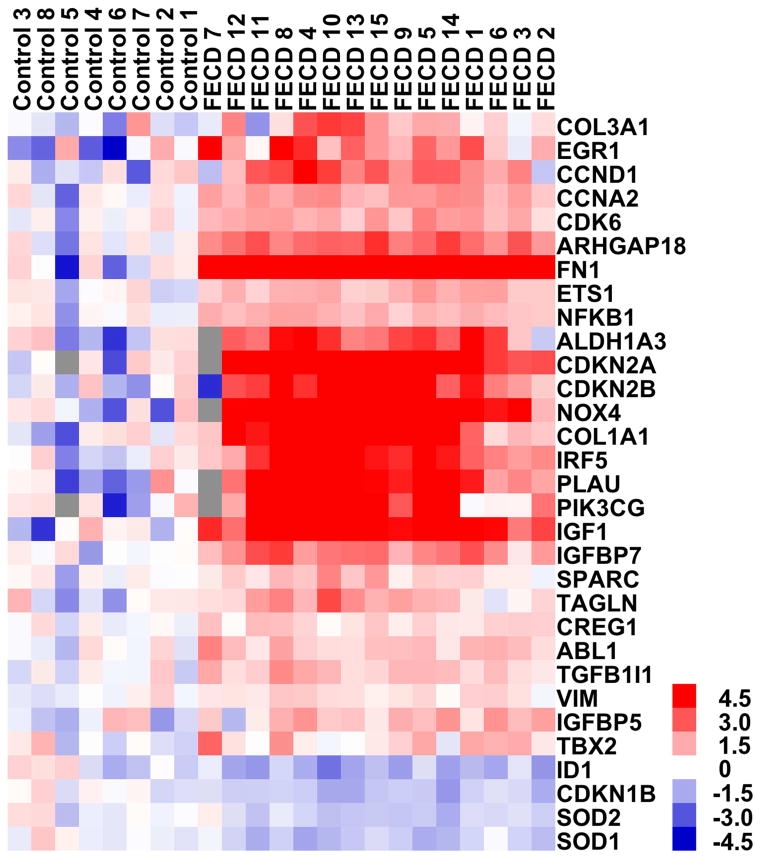
A total of 87 of the examined transcripts showed detectable expression in the TLDA experiment. Sixty-two genes showed a relative quantity (RQ)-value of >1 and 25 genes showed a RQ-value of <1 in FECD compared to normal samples. Using a fold-change of 1.5-fold and a p-value <0.05 as threshold for the detection of significant changes, 27 genes were found to be up-regulated and 4 genes were found to be down-regulated. A detailed list of differentially regulated transcripts is presented in Table 1. Using hierarchical clustering (here shown in the form of a heat map, see Figure 1) all samples segregated into the respective FECD or normal control group.
Table 1.
Differentially expressed transcripts in Fuchs endothelial corneal dystrophy (FECD) compared to normal control samples
| Gene (Taqman Assay) | RQa | P-Value |
|---|---|---|
| FN1 (Hs00365052_m1) | 1073.26 | 0.000 |
| CDKN2A (Hs00923894_m1) | 152.64 | 0.001 |
| NOX4 (Hs00418356_m1) | 62.06 | 0.000 |
| IGF1 (Hs01547656_m1) | 37.01 | 0.000 |
| PLAU (Hs01547054_m1) | 36.76 | 0.007 |
| COL1A1 (Hs00164004_m1) | 23.23 | 0.003 |
| PIK3CG (Hs00277090_m1) | 16.13 | 0.002 |
| CDKN2B (Hs00793225_m1) | 13.68 | 0.001 |
| IRF5 (Hs00158114_m1) | 13.37 | 0.001 |
| ALDH1A3 (Hs00167476_m1) | 11.94 | 0.002 |
| EGR1 (Hs00152928_m1) | 10.53 | 0.042 |
| ARHGAP18 (Hs00364379_m1) | 7.25 | 0.000 |
| CCND1 (Hs00765553_m1) | 5.70 | 0.002 |
| IGFBP7 (Hs00266026_m1) | 4.78 | 0.000 |
| CCNA2 (Hs00996788_m1) | 3.65 | 0.000 |
| COL3A1 (Hs00943809_m1) | 3.14 | 0.018 |
| CDK6 (Hs01026371_m1) | 3.10 | 0.000 |
| TAGLN (Hs01038777_g1) | 2.99 | 0.007 |
| ETS1 (Hs00428293_m1) | 2.69 | 0.000 |
| IGFBP5 (Hs00181213_m1) | 2.69 | 0.004 |
| NFKB1 (Hs00765730_m1) | 2.66 | 0.000 |
| TGFB1I1 (Hs00210887_m1) | 2.29 | 0.000 |
| ABL1 (Hs01104728_m1) | 2.29 | 0.000 |
| TBX2 (Hs00172983_m1) | 2.09 | 0.021 |
| SPARC (Hs00234160_m1) | 1.90 | 0.003 |
| CREG1 (Hs00171585_m1) | 1.68 | 0.000 |
| VIM (Hs00185584_m1) | 1.53 | 0.009 |
| SOD2 (Hs00167309_m1) | 0.63 | 0.027 |
| SOD1 (Hs00533490_m1) | 0.55 | 0.021 |
| CDKN1B (Hs00153277_m1) | 0.53 | 0.009 |
| ID1 (Hs03676575_s1) | 0.46 | 0.029 |
RQ=relative quantity
Figure 1. Analysis of corneal endothelial expression of cellular senescence-related transcripts in Fuchs endothelial corneal dystrophy (FECD, n=15) and normal control samples (n=8) using Taqman low density arrays (TLDAs).
The heat-map summarizes the expression levels and the unsupervised hierarchical clustering of the differentially expressed (fold-change >1.5, p<0.05) transcripts in FECD samples and normal control samples and shows clear distribution of each sample in the FECD or normal control group (grey square: non-evaluable amplification).
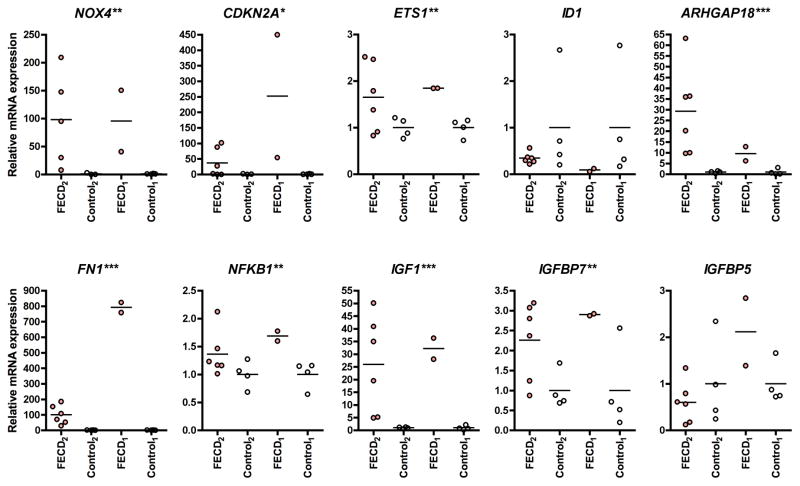
Ten out of 31 transcripts that had been found to be differentially regulated in the initial TLDA experiment were re-analyzed in a validation experiment using individual Taqman real-time PCR assays (Figure 2). Two cohorts of samples were analyzed: The first cohort (FECD1 n=2, Control1 n=4) consisted of endothelial samples from the initial array experiment. The second cohort (FECD2 n=6, Control2 n=4) consisted of new samples (due to reduced availability of autopsy tissues suitable for our study we included two autopsy control samples that had been retrieved from fellow eyes of samples used in the TLDA experiment in this group). The mean RQ-values of FECD samples from the initial array experiment/from the validation-experiment in old samples (FECD1 vs. Control1)/from the validation-experiment in new samples (FECD2 vs. Control2) (p-value FECD1+2 vs. Control1+2) were: NOX4 62.06/95.6/97.99 (p=0.003); CDKN2A 152.64/252.0/36.58 (p=0.035); ETS1 2.69/1.85/1.65 (p=0.005); ID1 0.46/0.09/0.34 (p=0.051); ARHGAP18 7.25/9.5/29.22 (p<0.001); FN1 1073.26/791.3/100.3 (p<0.001); NFKB1 2.66/1.69/1.36 (p=0.004); IGF1 37.01/32.18/25.95 (p<0.001); IGFBP7 4.78/2.9/2.26 (p=0.003); IGFBP5 2.69/2.11/0.6 (p=0.46).
Figure 2. Validation of differentially expressed transcripts in Fuchs endothelial corneal dystrophy (FECD) and normal control samples.
Differential expression of n=10 transcripts was validated using individual real-time PCR assays in representative samples from the initial array experiment (FECD1: n=2, Controls1: n=4) and in a second set of independent samples/fellow eyes (FECD2: n=6, Controls2: n=4). Horizontal bars represent means. ACTB was used as housekeeping gene. Asterisks next to the gene names on individual graphs indicate that *p<0.05, **p<0.01, ***p<0.001 for combined cohorts 1 and 2, FECD vs. Controls.
A variety of genes and chromosomal loci affected in FECD have recently been identified. They include COL8A2(Biswas et al., 2001; Gottsch et al., 2005), SLC4A11(Vithana et al., 2008), TCF4(Baratz et al., 2010), TCF8(Riazuddin et al., 2010), LOXHD1(Riazuddin et al., 2012), AGBL1(Riazuddin et al., 2013). However, the molecular pathomechanisms unifying these genetic modifications and leading to the common FECD phenotype are still poorly understood. Based on the genetic heterogeneity underlying this disease and hypothesizing that cellular senescence may be relevant in FECD pathogenesis, the present study was designed as a transcriptional analysis to identify common changes of distinct senescence-related transcripts in genetically undifferentiated FECD endothelium. CECs from 21 FECD patients and 12 normal autopsy eyes were analyzed. Real-time PCR representing the gold standard for quantitative gene expression analysis was used due to its high specificity and sensitivity. Our results allow for two important conclusions to be drawn:
Constant and high transcriptional over-expression of the constitutively active and ROS-generating enzyme NOX4 in all evaluable FECD samples suggests that NOX4 may be an important contributor to the oxidant—antioxidant imbalance and to cellular senescence induction in FECD endothelium.(Jurkunas et al., 2010; Matthaei et al., 2013b) This introduces NOX4 as a new candidate target for the treatment of FECD using readily available NOX4-inhibitors or NOX4-targeted siRNA.
Increased expression of cellular senescence marker CDKN2A and its transcriptional activators ETS1 and ARHGAP18 (SENEX) along with reduced expression of transcriptional CDKN2A-repressor ID1 provides further evidence for a prominent role of this signaling pathway and the presence of senescent endothelial cells in FECD.
All evaluable endothelial samples from FECD patients showed marked over-expression of NOX4. NOX4 is a member of the NAPDH oxidase (NOX) family, which is one of the most important sources for the generation of ROS in organs such as the kidney.(Gorin and Block, 2013) It is considered to be a constitutively active enzyme. Thus, NOX4 activity and concomitant output of ROS are primarily regulated by its cellular expression level.(Gorin and Block, 2013) NOX4 may be induced by several factors including TGF-β and IGF-1 and plays a critical role in mediating the pro-fibrotic effect of TGF-β.(Jiang et al., 2014) Its inhibition provides a new therapeutic approach for a variety of fibrotic diseases including idiopathic pulmonary fibrosis, atherosclerosis, diabetic kidney disease and liver fibrosis.(Gorin and Block, 2013) In this regard, the two small-molecule inhibitors GKT136901 and GKT137831 (Genkyotex, Geneva, Switzerland) have recently attracted much interest.(Gorin and Block, 2013)
Previous broadbased comparative gene expression analyses showed proteomic overexpression of clusterin and TGFBIP in FECD endothelium, which was confirmed by a more recent study (Weller et al., 2014), as well as the decreased expression of antioxidant peroxiredoxins.(Jurkunas et al., 2009; Jurkunas et al., 2008a; Jurkunas et al., 2008b). Transcriptional gene analysis in FECD endothelium demonstrated reduced expression of mitochondrial genes and differential expression of genes involved in energy metabolism, pump function and antiapoptotic or antioxidant cell defense.(Gottsch et al., 2003)
There is profound evidence supporting the role of oxidative stress in the pathogenesis of FECD.(Jurkunas et al., 2010) Endothelial oxidative DNA damage in FECD is attributed to an imbalance of oxidant and antioxidant factors in CECs based on an abnormal response of the transcription factor Nrf2 and its antioxidant targets (including superoxide dismutases).(Jurkunas et al., 2010) Our results showing decreased transcriptional levels of SOD1 and SOD2 (as shown previously by others (Jurkunas et al., 2010)) in FECD endothelial samples support these results. In addition, they suggest that augmented NOX4 expression/activity could exacerbate this oxidant-antioxidant imbalance. A similar imbalance between Nrf2 and NOX4 has recently been studied in a mouse model for lung injury.(Hecker et al., 2014) The study describes that myofibroblasts from young mice exhibit transient senescence and susceptibility to apoptosis, which allow fibrosis resolution.(Hecker et al., 2014) In contrast, myofibroblasts in injured tissue from old mice show a Nrf2 – Nox4 imbalance sustaining a senescent and apoptosis-resistant phenotype, which prevents the resolution of fibrosis and makes the tissues from old animals susceptible to fibrotic disorders.(Hecker et al., 2014)
Considering these investigations, our additional results in this aging disease seem to be of particular interest: Our analysis confirms increased expression of CDKN2A and of its transcriptional activators ETS1 and ARHGAP18 (SENEX) along with decreased expression of its transcriptional repressor ID1 in CECs from FECD patients.(Krishnamurthy et al., 2004; Ohtani et al., 2001) CDKN2A is an important biomarker and effector of aging.(Krishnamurthy et al., 2004) The idea of studying increased CDKN2A expression as an indicator for biologically older tissue before transplantation is not new.(McGlynn et al., 2009) CDKN2A maintains hypophosphorylation of the retinoblastoma protein (Rb) by binding to CDK4, thus preventing its dissociation from the transcription factor E2F1. This suppresses the entry of E2F1 into the nucleus with subsequent transcription of genes that promote transition from the G1 to the S phase. The transcriptional activator ETS1 is directly antagonized by ID1: ID1 prevents interaction of ETS1 with the CDKN2A promoter. ID1-null mouse embryo fibroblasts undergo premature senescence and CDKN2A expression with increasing age is strongly correlated with ETS1 expression in mice and rats.(Alani et al., 2001; Krishnamurthy et al., 2004) Similarly, ARHGAP18 (SENEX) is a regulator of stress-induced premature senescence by increasing both CDKN2A mRNA and protein levels together with Rb.(Coleman et al., 2010) Therefore, our results suggest that cellular senescence may affect CECs in FECD pathogenesis when undergoing oxidative stress.
Finally and in addition to the results described above, the significant over-expression of FN1 and (as described earlier (Matthaei et al., 2013a)) of COL1A1 supports the hypothesis of other groups, that a transformation of CECs to fibroblastic cells in the context of endothelial-to-mesenchymal-transition (EnMT) occurs in FECD.(Okumura et al., 2013) Moreover, the over-expression of FN1, COL1A1, COL3A1 is in line with a recently published study investigating ECM alterations in FECD in more detail.(Weller et al., 2014) However, further analyses at the protein level and functional studies are necessary with respect to all gene expression differences described in this study in order to establish the role of cellular senescence and its relation to EnMT and abnormal ECM deposition, as well as the role of potential new therapeutic approaches in the future.
We conclude that this study of senescence-associated transcripts demonstrates for the first time a marked over-expression of NOX4 in CECs from FECD patients and introduces the constitutively active enzyme NOX4 as a new candidate therapeutic target in FECD. In addition, our results support the prominent role of the CDKN2A pathway and warrant further proteomic and functional analyses of NOX4 and CDKN2A in FECD pathogenesis.
Highlights.
Expression analysis of cellular senescence-related transcripts in FECD endothelium
Increased expression of NOX4 was found in FECD endothelium
Increased CDKN2A, ETS1 and ARHGAP18 with decreased ID1 was found in FECD endothelium
This suggests a role of NOX4 and of CDKN2A/senescence as pathogenic factors in FECD
Acknowledgments
Grant Information: National Institutes of Health (NIH EY019874 to ASJ), Medical Illness Counseling Center (to ASJ), Research to Prevent Blindness (to Wilmer Eye Institute), Deutsche Forschungsgemeinschaft (DFG MA 5110/2-1 to MM), Fuchs Dystrophy Research Grant (Wilmer Eye Institute to MM), Richard Lindstrom/Eye Bank Association of America Research Grant (to MM), and grants from the J. Willard and Alice S. Marriott Foundation, Edward Colburn, Richard Dianich, Barbara Freeman, Mary Finegan, Stanley Friedler, MD, Herbert Kasoff, Diane Kemker, Jean Mattison, Florenz Ourisman, Lee Silverman, and Norman Tunkel, PhD (all to ASJ).
The authors thank Shannath Merbs, MD, PhD, Ray Enke, PhD, Verity Oliver, PhD, and Jin Song, MD, PhD, for providing human corneal control samples.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. P Natl Acad Sci USA. 2001;98:7812–7816. doi: 10.1073/pnas.141235398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratz KH, Tosakulwong N, Ryu E, Brown WL, Branham K, Chen W, Tran KD, Schmid-Kubista KE, Heckenlively JR, Swaroop A, Abecasis G, Bailey KR, Edwards AO. E2-2 protein and Fuchs’s corneal dystrophy. N Engl J Med. 2010;363:1016–1024. doi: 10.1056/NEJMoa1007064. [DOI] [PubMed] [Google Scholar]
- Biswas S, Munier FL, Yardley J, Hart-Holden N, Perveen R, Cousin P, Sutphin JE, Noble B, Batterbury M, Kielty C, Hackett A, Bonshek R, Ridgway A, McLeod D, Sheffield VC, Stone EM, Schorderet DF, Black GC. Missense mutations in COL8A2, the gene encoding the alpha2 chain of type VIII collagen, cause two forms of corneal endothelial dystrophy. Hum Mol Genet. 2001;10:2415–2423. doi: 10.1093/hmg/10.21.2415. [DOI] [PubMed] [Google Scholar]
- Coleman PR, Hahn CN, Grimshaw M, Lu Y, Li X, Brautigan PJ, Beck K, Stocker R, Vadas MA, Gamble JR. Stress-induced premature senescence mediated by a novel gene, SENEX, results in an anti-inflammatory phenotype in endothelial cells. Blood. 2010;116:4016–4024. doi: 10.1182/blood-2009-11-252700. [DOI] [PubMed] [Google Scholar]
- Engelmann K, Bohnke M, Friedl P. Isolation and Long-Term Cultivation of Human Corneal Endothelial-Cells. Invest Ophth Vis Sci. 1988;29:1656–1662. [PubMed] [Google Scholar]
- Enomoto K, Mimura T, Harris DL, Joyce NC. Age differences in cyclin-dependent kinase inhibitor expression and rb hyperphosphorylation in human corneal endothelial cells. Invest Ophthalmol Vis Sci. 2006;47:4330–4340. doi: 10.1167/iovs.05-1581. [DOI] [PubMed] [Google Scholar]
- Gorin Y, Block K. Nox4 and diabetic nephropathy: With a friend like this, who needs enemies? Free Radic Biol Med. 2013;61C:130–142. doi: 10.1016/j.freeradbiomed.2013.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottsch JD, Bowers AL, Margulies EH, Seitzman GD, Kim SW, Saha S, Jun AS, Stark WJ, Liu SH. Serial analysis of gene expression in the corneal endothelium of Fuchs’ dystrophy. Invest Ophthalmol Vis Sci. 2003;44:594–599. doi: 10.1167/iovs.02-0300. [DOI] [PubMed] [Google Scholar]
- Gottsch JD, Sundin OH, Liu SH, Jun AS, Broman KW, Stark WJ, Vito EC, Narang AK, Thompson JM, Magovern M. Inheritance of a novel COL8A2 mutation defines a distinct early-onset subtype of fuchs corneal dystrophy. Invest Ophthalmol Vis Sci. 2005;46:1934–1939. doi: 10.1167/iovs.04-0937. [DOI] [PubMed] [Google Scholar]
- Hecker L, Logsdon NJ, Kurundkar D, Kurundkar A, Bernard K, Hock T, Meldrum E, Sanders YY, Thannickal VJ. Reversal of persistent fibrosis in aging by targeting nox4-nrf2 redox imbalance. Sci Transl Med. 2014;6:231ra247. doi: 10.1126/scitranslmed.3008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Liu GS, Dusting GJ, Chan EC. NADPH oxidase-dependent redox signaling in TGF-beta-mediated fibrotic responses. Redox Biol. 2014;2:267–272. doi: 10.1016/j.redox.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce NC. Proliferative capacity of corneal endothelial cells. Experimental Eye Research. 2012;95:16–23. doi: 10.1016/j.exer.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce NC, Zhu CC, Harris DL. Relationship among oxidative stress, DNA damage, and proliferative capacity in human corneal endothelium. Invest Ophthalmol Vis Sci. 2009;50:2116–2122. doi: 10.1167/iovs.08-3007. [DOI] [PubMed] [Google Scholar]
- Jurkunas UV, Bitar M, Rawe I. Colocalization of increased transforming growth factor-beta-induced protein (TGFBIp) and Clusterin in Fuchs endothelial corneal dystrophy. Invest Ophthalmol Vis Sci. 2009;50:1129–1136. doi: 10.1167/iovs.08-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkunas UV, Bitar MS, Funaki T, Azizi B. Evidence of oxidative stress in the pathogenesis of fuchs endothelial corneal dystrophy. Am J Pathol. 2010;177:2278–2289. doi: 10.2353/ajpath.2010.100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkunas UV, Bitar MS, Rawe I, Harris DL, Colby K, Joyce NC. Increased clusterin expression in Fuchs’ endothelial dystrophy. Invest Ophthalmol Vis Sci. 2008a;49:2946–2955. doi: 10.1167/iovs.07-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkunas UV, Rawe I, Bitar MS, Zhu C, Harris DL, Colby K, Joyce NC. Decreased expression of peroxiredoxins in Fuchs’ endothelial dystrophy. Invest Ophthalmol Vis Sci. 2008b;49:2956–2963. doi: 10.1167/iovs.07-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthaei M, Hu J, Kallay L, Eberhart CG, Cursiefen C, Qian J, Lackner EM, Jun AS. Endothelial Cell MicroRNA Expression in Human Late-Onset Fuchs Dystrophy. Invest Ophthalmol Vis Sci. 2013a doi: 10.1167/iovs.13-12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthaei M, Lackner EM, Meng H, Hicks JL, Meeker AK, Eberhart CG, Jun AS. Tissue Microarray Analysis of Cyclin-Dependent Kinase Inhibitors p21 and p16 in Fuchs Dystrophy. Cornea. 2013b;32:473–478. doi: 10.1097/ICO.0b013e31826f324e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn LM, Stevenson K, Lamb K, Zino S, Brown M, Prina A, Kingsmore D, Shiels PG. Cellular senescence in pretransplant renal biopsies predicts postoperative organ function. Aging Cell. 2009;8:45–51. doi: 10.1111/j.1474-9726.2008.00447.x. [DOI] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- Okumura N, Kay EP, Nakahara M, Hamuro J, Kinoshita S, Koizumi N. Inhibition of TGF-beta signaling enables human corneal endothelial cell expansion in vitro for use in regenerative medicine. Plos One. 2013;8:e58000. doi: 10.1371/journal.pone.0058000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin SA, Parker DS, McGlumphy EJ, Oh EC, Iliff BW, Schmedt T, Jurkunas U, Schleif R, Katsanis N, Gottsch JD. Mutations in LOXHD1, a recessive-deafness locus, cause dominant late-onset Fuchs corneal dystrophy. Am J Hum Genet. 2012;90:533–539. doi: 10.1016/j.ajhg.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin SA, Vasanth S, Katsanis N, Gottsch JD. Mutations in AGBL1 Cause Dominant Late-Onset Fuchs Corneal Dystrophy and Alter Protein-Protein Interaction with TCF4. American Journal of Human Genetics. 2013;93:758–764. doi: 10.1016/j.ajhg.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riazuddin SA, Zaghloul NA, Al-Saif A, Davey L, Diplas BH, Meadows DN, Eghrari AO, Minear MA, Li YJ, Klintworth GK, Afshari N, Gregory SG, Gottsch JD, Katsanis N. Missense mutations in TCF8 cause late-onset Fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am J Hum Genet. 2010;86:45–53. doi: 10.1016/j.ajhg.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vithana EN, Morgan PE, Ramprasad V, Tan DT, Yong VH, Venkataraman D, Venkatraman A, Yam GH, Nagasamy S, Law RW, Rajagopal R, Pang CP, Kumaramanickevel G, Casey JR, Aung T. SLC4A11 mutations in Fuchs endothelial corneal dystrophy. Hum Mol Genet. 2008;17:656–666. doi: 10.1093/hmg/ddm337. [DOI] [PubMed] [Google Scholar]
- Weller JM, Zenkel M, Schlotzer-Schrehardt U, Bachmann BO, Tourtas T, Kruse FE. Extracellular Matrix Alterations in Late-Onset Fuchs’ Corneal Dystrophy. Invest Ophth Vis Sci. 2014;55:3700–3708. doi: 10.1167/iovs.14-14154. [DOI] [PubMed] [Google Scholar]