Abstract
BK Polyomavirus (BKPyV) is a ubiquitous nonenveloped human virus that can cause severe disease in immunocompromised populations. After internalization into renal proximal tubule epithelial cells, BKPyV traffics through the ER and enters the cytosol. However, it is unclear how the virus enters the nucleus. In this study, we elucidate a role for the nuclear localization signal located on the minor capsid proteins VP2 and VP3 during infection. Site-directed mutagenesis of a single lysine in the basic region of the C-terminus of the minor capsid proteins abrogated their nuclear localization, and the analogous genomic mutation reduced infectivity. Additionally, through use of the inhibitor ivermectin and knockdown of importin β1, we found that the importin α/β pathway is involved during infection. Overall these data are the first to show the significance of the NLS of the BKPyV minor capsid proteins during infection in a natural host cell.
Keywords: polyomavirus, nuclear entry, intracellular virus trafficking
Introduction
Polyomaviruses are ubiquitous in the human population. The polyomavirus BKPyV usually infects in early childhood, establishing a subclinical persistent infection in the host. However, reactivation in immunosuppressed individuals, particularly bone marrow and kidney transplant patients, can lead to severe disease (Jiang et al., 2009a). Currently there are no BKPyV-specific antivirals and the only treatment is palliative therapy or reduction of immunosuppression, with the latter treatment leaving renal transplant patients open to organ rejection. In addition to the intrinsic value of uncovering aspects of viral and host biology, understanding the life cycle of the virus within the host cell will provide much needed information for future development of targeted therapies to lessen BKPyV reactivation and disease.
The BKPyV particle is nonenveloped and consists of three structural proteins, including the major capsid protein VP1 and minor capsid proteins VP2 and VP3. The structural proteins encapsidate the 5.2 Kb circular double stranded DNA genome, which takes the form of a minichromosome associated with cellular histones (Meneguzzi et al., 1978). The outer layer of the viral particle is made up of 72 VP1 pentamers stabilized by intermolecular disulfide bonds (Nilsson et al., 2005). Associated with each pentamer is one molecule of either VP2 or VP3, which are not exposed to the surface of the particle until disassembly begins within the endoplasmic reticulum (ER) of the host cell (Bennett et al., 2013). VP2 and VP3 are expressed from the same reading frame and share an identical C-terminus, but VP2 contains a unique N-terminal sequence that can be myristoylated (Krauzewicz et al., 1990). Our lab has previously shown in renal proximal tubule epithelial (RPTE) cells that BKPyV reaches the ER at 8 – 10 hpi (Jiang et al., 2009b), then traffics through the cytosol at 16 hpi (Bennett et al., 2013), followed by nuclear entry and early gene expression beginning at 24 hpi (Low et al., 2004). Data suggest that BKPyV and other polyomaviruses exit the ER and enter the cytosol using components of the ER-associated degradation pathway (Bennett et al., 2013; Geiger et al., 2011; Goodwin et al., 2011; Jiang et al., 2009b; Walczak et al., 2014).
Previous studies have provided conflicting evidence for the nuclear entry mechanism of polyomaviruses. The virus could enter the nucleus through the nuclear pore complex, since partial uncoating would make the virus particle small enough for active transport through the pore (Pante and Kann, 2002). Studies from Nakanishi et al. have shown an interaction between importins α and β and the SV40 chromosome in the cytosol, suggesting the use of the canonical nuclear import pathway during SV40 nuclear entry (Nakanishi et al., 2007; Nakanishi et al., 2002). Importins α and β are adaptor proteins that recognize and bind a nuclear transport signal made up of basic amino acids called a nuclear localization signal (NLS). Certain basic amino acids in the C-terminus of SV40 VP2 and VP3 (VP2/3) were found to be necessary for the interaction with the importins, suggesting that entry of SV40 through the nuclear pore complex is mediated by interactions between an NLS on the minor capsid proteins VP2/3 and the canonical nuclear import machinery (Nakanishi et al., 2007; Nakanishi et al., 2002). However, studies by Butin-Israeli et al. provided evidence that SV40 infection leads to breakdown of the nuclear lamina and disruption of the nuclear envelope, allowing the virus to pass from the ER lumen directly into the nucleus (Butin-Israeli et al., 2011).
In this study we have evaluated the role of the nuclear localization signal of the minor capsid proteins VP2 and VP3 during entry of BKPyV in RPTE cells. We found that a specific lysine in VP2 and VP3 was important for localization to the nucleus, and that mutation of this lysine attenuated infection and hindered viral entry at a point after ER trafficking. These findings further describe the trafficking route of BKPyV, and provide a model where polyomavirus can enter the nucleus from the cytosol through the nuclear pore, using the minor capsid protein NLS as the import signal.
Results
Lysine 319 on VP2 and Lysine 200 on VP3 are critical for nuclear localization of the minor capsid proteins
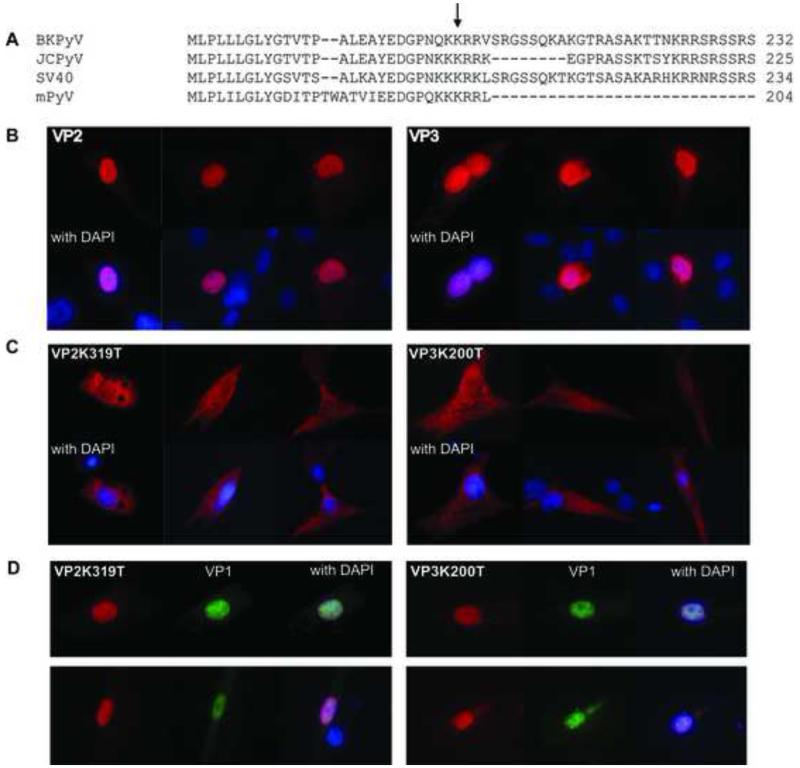
Because BKPyV VP2 and VP3 share homology with SV40, especially in the basic C-terminal amino acids important for nuclear localization of the SV40 proteins (Gharakhanian and Kasamatsu, 1990; Gharakhanian et al., 1987) (Fig. 1A), we hypothesized that VP2 and VP3 of BKPyV would also possess a nuclear localization signal that mapped to these residues. To test this, we expressed VP2 or VP3 alone in RPTE cells by transfection of expression plasmids. In RPTE cells, both VP2 and VP3 showed nuclear localization (Fig. 1B). We next determined the amino acids necessary to confer this nuclear localization. It has been previously shown that lysine 202 on SV40 VP3 and the same amino acid at residue 320 of VP2 was vital for nuclear localization of the SV40 minor capsid proteins (Gharakhanian and Kasamatsu, 1990). Therefore, we targeted the homologous lysine in BKPyV, K319 of VP2 and K200 of VP3, for mutagenesis (Fig. 1A, arrow). Site-directed mutagenesis was used to convert the lysine to a threonine, and the mutant constructs were transfected into RPTE cells to test localization of the proteins. Indeed, the mutants VP2K319T and VP3K200T no longer localized to the nucleus (Fig. 1C).
Figure 1. Subcellular localization of NLS mutant VP2 and VP3 proteins.
(A) VP3 Lysine 200 is conserved in a number of polyomavirus species. (B) Wild type VP2 and VP3 expression plasmids were transfected in RPTE cells, fixed at 24 h, and stained with anti-VP2/3 (red). Nuclei were stained with DAPI (blue). Both VP2 and VP3 are nuclear, as demonstrated by co-staining (bright purple) with DAPI. (C) RPTE cells were transfected with mutant VP3 (VP3K200R) or VP2 (VP2K319R) in the same manner. (D) RPTE cells were co-transfected with VP1 and either VP2 or VP3, and stained for VP2/3, VP1, and DAPI.
For SV40, co-expression of VP1 allows for nuclear import of nuclear localization-defective minor capsid proteins (Ishii et al., 1994; Kasamatsu and Nehorayan, 1979). To test whether this was also the case for BKPyV, we co-transfected a VP1 expression vector with each of the VP2K319T and VP3K200T expression vectors. The presence of VP1 led to nuclear localization of both VP2K319T and VP3K200T (Fig. 1D). This observation suggests that the nuclear localization signal on VP1 enables import of VP1-VP2 or VP1-VP3 complexes. These results also suggest that assembly of capsids, which takes place in the nucleus, should not be affected by the presence of a mutation in the minor capsid protein NLS.
The VP2/3 NLS is important for entry in RPTE cells
To evaluate the role of the minor capsid protein NLS during infection, a point mutation was created in the viral genome to convert lysine 319 of VP2 and 200 of VP3 to a threonine as was done in the expression plasmids. Mutant and wild type genomes were transfected into RPTE cells to produce virus. Lysates from the transfected cells were blind passaged to infect a new set of RPTE cells. Those infected RPTE cells were then harvested after two weeks and the quantity of viral genomes in the lysates was measured using qPCR. This crude stock was then used to grow a large scale stock of purified virus. Of note, similar numbers of viral genomes were measured for both purified and mutant viruses. PCR and DNA sequencing was performed to ensure that the mutation was still present in the final purified virus (data not shown). Additionally, we wanted to confirm that VP2 and VP3 were incorporated into virions, even though our transfections showed that the NLS-mutant VP2/3 could still be imported into the nucleus by VP1. Equal genomes were resolved by SDS-PAGE and the resulting western blot was probed for VP2/3, showing the presence of VP2 and VP3 in both wild type and mutant viruses (Fig. 2A). While the VP2:VP3 ratio appears slightly different, this is normal variation that we detect even among different batches of wild type virus (data not shown). To test infectivity of the mutant compared to wild type, equal numbers of genomes were used to infect RPTE cells. Whole cell lysates were collected at 24 hpi, resolved by SDS-PAGE, and probed for the early protein large T Antigen (TAg). Compared to wild type virus (Dun), Dunlop containing the VP2/VP3 lysine mutation (DunM) expressed less TAg (Fig. 2B), suggesting a possible defect in entry.
Figure 2. Mutation in VP2/3 NLS attenuates infectivity of BKPyV.
(A) Equal genome numbers of wild type (Dun) and NLS mutant (DunM) viruses were separated by SDS-PAGE and analyzed by Western blotting for the presence of VP2 and VP3. (B) RPTE cells were infected with Dun and DunM virus at an MOI of 104 genomes per cell for 24 h. Whole cell lysates were resolved by SDS-PAGE and probed for TAg and β actin. Quantification was performed using the Li-Cor Odyssey system, with mock and mutant TAg levels normalized to Dun, set to a value of 1. Results represent at least three independent experiments. (C) RPTE cells were infected with equal genomes and harvested immediately or 24 h after adsorption under alkylating conditions, then whole cell lysates were resolved under reducing or non-reducing conditions by SDS-PAGE and probed for VP1 and GAPDH. (D) RPTE cells were infected with equal genomes and left untreated or treated with BFA as a control for non-specific staining, then fixed at 24 hpi and stained with anti-VP2/3 and DAPI.
To assess whether early entry events prior to nuclear import were disrupted by the introduction of the mutation, we evaluated disassembly and ER trafficking. The disulfide bonds between the VP1 pentamers become reduced as the virus traffics through the ER; this disassembly process can be detected beginning around 12 hpi, and corresponds with increasing quantities of VP1 monomers and oligomers in lysates prepared under non-reducing conditions (Jiang et al., 2009b). To compare mutant and wild type virus, RPTE cells were infected and protein lysates were harvested at 0 hpi and 24 hpi under alkylating conditions. The lysates were analyzed by SDS-PAGE under non-reducing conditions to detect VP1 disassembly events that had occurred in the cell. At 0 hpi, there was an absence of VP1 that entered the gel because the virus particles were still fully assembled (Fig. 2C). Similar levels of VP1 disassembly intermediates were seen for both the wild type and mutant virus at 24 hpi (Fig. 2C), suggesting that the mutant is able to disassemble in the ER in a similar manner to wild type virus. As another way of testing trafficking of the mutant virus through the ER, the exposure of the minor capsid proteins VP2 and VP3 was assessed by immunofluorescent staining. VP2 and VP3 become accessible to antibody recognition as the virus particle becomes partially disassembled in the ER, and thus immunofluorescent detection of the minor capsid proteins can be used as a readout for disassembly (Norkin et al., 2002). For both mutant and wild type BKPyV, staining for VP2/3 showed similar levels of puncta throughout the cells (Fig. 2D). In cells treated with brefeldin A (BFA), which inhibits trafficking to the ER, no puncta were seen, indicating the specificity of the assay. Together these data show that disassembly and ER trafficking still occurred during entry of the mutant virus. Therefore, the defect leading to reduced TAg expression in mutant virus infected cells is likely occurring at a step between initial disassembly and nuclear entry.
Importin β - mediated nuclear import is involved during infection
Since a classical NLS usually leads to nuclear import through interaction with the transporter proteins importin α/β, we first evaluated the role of this pathway during infection through the use of the inhibitor ivermectin, which specifically inhibits the importin α/β nuclear import pathway without affecting other nuclear import pathways (Wagstaff et al., 2012). RPTE cells were infected with wild type BKPyV and treated with 10 μM ivermectin. Early gene expression of wild type BKPyV was evaluated by measuring TAg protein by western blot and mRNA levels by RT-qPCR. TAg protein levels were much lower in the treated lysates compared to those treated with the vehicle butanol (Fig. 3A). Since the TAg protein itself normally undergoes importin α/β-mediated nuclear import, TAg mRNA levels were also measured to verify that the inhibitor was not simply causing instability of the TAg protein by preventing its nuclear localization. Quantitative RT-PCR demonstrated that TAg mRNA levels were also decreased in the inhibitor-treated samples (Fig. 3B), suggesting that the decrease in TAg protein was due to less viral gene expression from inhibition of BKPyV entry into the nucleus. Interestingly, the mutant virus was also sensitive to ivermectin treatment, as there was a decrease in TAg mRNA expression from the mutant, suggesting that the mutant virus retains the ability to use the nuclear import pathway, albeit at a reduced level.
Figure 3. The canonical nuclear import α/β 1 pathway is important for infection.
(A) RPTE cells were infected at an MOI of 104 genomes/cell and treated with ivermectin (+I) or butanol (untreated, UT). Whole cell lysates were harvested at 24 hpi and resolved by SDS-PAGE, then probed for TAg and β actin. (B) RPTE cells were infected with 104 genomes/cell of wild-type (Dun) or mutant (DunM) and treated with ivermectin (+I) at 10 μM at the beginning of infection or left untreated. RNA was harvested at 24 hpi and reverse-transcription (RT) was performed followed by qPCR to measure TAg cDNA relative to GAPDH. (C) RPTE cells were transfected with Importin β1 siRNA (Imp β) or non-targeting siRNA (N.T.), and then infected 2 d after transfection with wild type virus. Whole cell lysates were harvested at 24 hpi, resolved by SDS-PAGE, and probed for TAg, B actin, and importin β1. (D) Transfection and infection was performed as in C, and RNA was harvested at 24 hpi. RT and qPCR was performed as before. Results represent three independent experiments.
To further address a potential role for an importin-mediated nuclear entry pathway in BKPyV infection, we knocked down importin 1 protein levels through siRNA silencing. One day after siRNA transfection, RPTE cells were infected with wild type BKPyV, and protein or RNA was harvested at 24 hpi. Western blotting for importin 1 protein showed an approximately 50% knockdown (Fig. 3C, middle row). TAg levels measured by western blotting were decreased in the knock-down samples compared to those treated with a non-targeting siRNA, and TAg mRNA was also decreased (Fig. 3D). These data provide additional support for the role of the importin α/β canonical nuclear import pathway.
Discussion
The cellular entry pathway of polyomaviruses is only partially understood, and many details remain to be elucidated. Previous studies have provided evidence that after BKPyV or SV40 is internalized, it travels through endocytosis to the ER, where disassembly begins (Engel et al., 2011; Goodwin et al., 2011; Jiang et al., 2009b; Schelhaas et al., 2007). After ER trafficking, the virus must enter the nucleus, and we and others have provided data supporting a model in which polyomaviruses exit the ER into the cytosol before nuclear entry, using the host pathway of ER-associated degradation (ERAD) (Bennett et al., 2013; Geiger et al., 2011; Goodwin et al., 2011; Jiang et al., 2009b; Walczak et al., 2014). However, the mechanism of polyomavirus nuclear entry, and specifically BKPyV nuclear entry, has so far remained ill-defined. In this report, we have provided evidence that an NLS on the minor capsid proteins of BKPyV plays a role during entry, suggesting that after the virus particle exits the ER, the minor capsid proteins facilitate the process of nuclear import.
Previously, both of the minor capsid proteins of SV40 were shown to localize to the nucleus, and the necessary amino acid sequences were identified to include a QPNKKRR sequence (Nakanishi et al., 2002). BKPyV capsid proteins have extensive homology with SV40 proteins, but the localization properties of BKPyV VP2 and VP3 have not been assessed previously, particularly in the context of infection and in a natural host cell such as RPTE cells. Here we have demonstrated nuclear localization of both BKPyV VP2 and VP3 during expression in RPTE cells and identified a lysine in the basic C-terminal sequence that was important for this localization. The requirement for this lysine seems to be specific as a mutation to similarly charged arginine also abrogated nuclear localization of VP2/3 (data not shown). Of note, after transfection into RPTE cells of the VP2 and VP3 expression plasmids, VP2 and VP3-positive cell numbers were relatively low, perhaps due to the lytic properties of the minor capsid proteins (Daniels et al., 2006).
After we identified a C-terminal VP2/3 residue required for nuclear localization, we created the corresponding mutation in the genome, simultaneously mutating both VP2 and VP3. We confirmed incorporation of VP2 and VP3 into the DunM virus by western blot. Because the presence of wild type VP1 enables nuclear import of nuclear localization-deficient VP2 or VP3 (Fig. 1B), we could not assess differences in wild type and mutant VP2/3 localization after expression from the viral genome, since VP1 is expressed concurrently with VP2 and VP3. However, decreased TAg protein and mRNA levels from the mutant at 24 h after infection with equal genomes implies a defect during viral entry. Since VP2/3 exposure in the ER still occurred, and the capsid became reduced to a similar extent as wild type, it is likely that the mutation does not cause a defect in the early, pre-nuclear entry trafficking steps. Interestingly, we observed an increased amount of VP1 disassembly products during entry of the DunM virus. This could be due to an increased particle to infectious genome ratio, or a non-productive pathway may be favored for a nuclear entry-defective virus, leading to increased disassembly products.
The importance of an NLS implies the involvement of the classical importin α/β 1 import pathway during nuclear entry. Indeed, knockdown of importin 1 and treatment with ivermectin both inhibited infection, supporting a model where BKPyV co-opts this endogenous nuclear entry mechanism using the NLS of the minor capsid proteins. The nuclear import inhibitor ivermectin specifically targets the importin α/β1-mediated import pathway without affecting the other import pathways including importin β-mediated and transportin-mediated import (Wagstaff et al., 2012). The NLS is recognized first by the adapter importin α, then importin β1 interacts with the substrate-NLS-importin α complex to allow transport through the nuclear pore (Yoneda et al., 1999). The inhibitory effect of ivermectin on BKPyV infection further supports the use of active transport through the nuclear pore complex by polyomaviruses for gaining access to the nucleus.
A number of observations point to an alternative nuclear entry pathway existing along with import through the nuclear pore. The BKPyV NLS mutation only led to attenuated infectivity, and both ivermectin treatment and importin β1 knockdown led to about a 50% decrease in early gene expression. One alternative way for a cytosolic viral particle to enter the nucleus is during mitosis, when nuclear envelope breakdown occurs (Butin-Israeli et al., 2011). Additionally, it is possible that BKPyV can use an alternative NLS from the C-terminal one identified here. VP1 contains an NLS (Ishii et al., 1996; Moreland and Garcea, 1991), but this motif is thought to be buried within the partially disassembled particle, such that entry would rely on the minor capsid proteins. However, it is possible that this VP1 NLS is available during entry in RPTE cells, or that another VP2 and/or VP3 NLS functions in the context of the virus structure that does not function when the proteins are expressed individually. Since ivermectin treatment inhibited the mutant to a similar extent as the wild type virus, this suggests that the mutant can somehow still use the canonical import pathway, supporting the use of another viral NLS.
Recent data implicating a possible connection between the nuclear import and ERAD machinery might provide insight into a model for BKPyV nuclear entry in RPTE cells. Importin β1 has been shown to be necessary for the retrotranslocation of certain ERAD substrates (Zhong et al., 2011), making it reasonable to suspect that the nuclear import pathway provides a way for the movement of the virus out of the ER to be energetically favorable, as it crosses into the cytosol, interacts with nuclear import machinery on the cytosolic side of the ER membrane, and proceeds into the nucleus through the nuclear pore.
In summary, we have demonstrated that a basic lysine in the C-terminus of the minor capsid proteins is required for nuclear import of the individually expressed minor capsid proteins. Additionally, we have shown the involvement of the importin 1 canonical import pathway during entry. Our data suggest a model where partially disassembled BKPyV exits the ER into the cytosol, where the exposed NLS of VP2/VP3 is used to bring the viral genome into the nucleus via the nuclear pore complex; but another route of entry into the nucleus may exist as well.
Materials and Methods
Cell Culture
RPTE cells were grown in renal epithelial basal growth medium (REGM) with SingleQuots Bulletkit from Lonza at 37°C and 5% CO2 and passaged up to six times as previously described (Abend et al., 2007).
Transfection
The VP2 and VP3 expression plasmids were obtained from Addgene, Cambridge, MA (plasmid numbers 32109, 32110). The VP1 expression plasmid was obtained from Christopher Buck, NCI, Bethesda, MD. RPTE cells in a 12-well were transfected using 3 μl Mirus LT-1 transfection reagent, and 1 μg plasmid DNA following manufacturer’s instructions (or 0.5 μg of each plasmid for the co-transfection). LT-1 was mixed with 100 μl Opti-mem and incubated 15 min before adding the DNA and then 25 min at room temperature.
Infections
Purified wild-type and mutant BKPyV (Dunlop genetic background) was propagated and purified as previously described (Abend et al., 2007). Titers were based on genomes/μl after virus purification, measured by qPCR as described below. RPTE cells were infected at 70% confluence with a multiplicity of infection of 104 genomes per cell. Cells were pre-chilled 15 min and inoculated with virus for one hour at 4°C, then washed once with cold media before adding warm media and incubating at 37°C until the desired time post infection. Ivermectin (Sigma) was dissolved in ethanol according to manufacturer’s instructions and used at a final concentration of 10 μM.
Quantitative PCR (qPCR) titering of genomes
To calculate genomes, 5 μl of virus stock was treated with 2 μl DNAse for 1 h at 37°C, then treated with 50 μg proteinase K for 1 h at 37, followed by isolation of the viral genomes by the QIAquick PCR purification kit (Qiagen). Dilutions of the isolated DNA were used for qPCR and final concentrations were calculated from a standard curvein genomes/μl. QPCR was done using primers 5′ TGTGATTGGGATTCAGTGCT 3′ and 5′ AAGGAAAGGCTGGATTCTGA 3’in the TAg open reading frame.
Preparation of cell lysates
Whole cell protein lysates were harvested in E1A buffer (50 mM HEPES pH 7, 250 mM NaCl, 0.1% NP40, with protease inhibitors 5 μg/ml PMSF, 5 μg/ml aprotinin, 50 mM NaF, 0.2 mM Na-orthovanadate, and 5 μg/ml leupeptin added just before use). For alkylating conditions, 20 mM N-ethyl maleimide (Sigma) was added to the E1A buffer. Protein concentrations were determined using a Bradford assay after clarifying the lysate.
Western Blots
Equal amounts of protein were resolved by SDS-PAGE on a 10% gel (8% for TAg), under reducing or non-reducing conditions and transferred overnight onto a nitrocellulose membrane at 60 V by wet transfer. Membranes were blocked in 2% nonfat dry milk in PBS containing 0.1% Tween 20 (PBS-T). Antibodies were used at the following concentrations diluted in PBS-T: anti-TAg PAb416 1:3000, anti-VP1P5G6 1:10,000, anti-VP2/3 1:3000, anti-GFP 1:2000, anti-β-Actin (Cell Signaling) 1:5000, HRP-conjugated anti-mouse and anti-rabbit (GE) 1:5000. For western blots analyzed by the Odyssey system, membranes were blocked in Odyssey blocking buffer (LI-COR) and probed with the indicated primary antibodies or anti-mouse or anti-rabbit secondary antibodies diluted in Odyssey blocking buffer with 0.1% Tween 20.
Immunofluorescence microscopy
At 24 hours post transfection, cells were fixed with cold 95% EtOH/5% acetic acid. Cells were blocked with 5% goat serum in PBS, and then coverslips were co-stained for indirect immunofluorescence with 1:500 dilution of rabbit anti-VP2/3 and/or 1:200 mouse anti-VP1 in goat serum. Secondary antibodies Alexa Flour® 594 goat anti-rabbit IgG or Alexa Fluor® 488 goat anti-mouse (Invitrogen) were used at a 1:200 dilution. Cells were imaged with an inverted fluorescence microscope.
Reverse transcription and qPCR
At 24 hours post-infection, RNA was harvested in Trizol and isolated using the Zymo Research Direct-zol RNA mini-prep kit. RNA (150ng) was treated with DNase I then reverse transcription was performed on RNA using the Qiangen Omniscript RT kit. QPCR was performed on cDNA using primers for TAg (same as above), or GAPDH 5′ GCCTCAAGATCATCAGCAAT 3′ and 5′ CTGTGGTCATGAGTCCTTCC 3′. Fold change in expression was determined using the ΔΔCt method (Schmittgen and Livak, 2008).
Highlights.
polyomaviruses must deliver their genome to the nucleus to replicate
the minor capsid proteins have a well-conserved nuclear localization signal
mutation of this NLS diminishes, but does not completely inhibit, infection
Acknowledgments
This work was supported in part by a University of Michigan Rackham research grant to S.M.B., by NIH grant AI060584 to M.J.I., and in part by NIH grant CA046592 to the University of Michigan Cancer Center. We thank Mengxi Jiang and Katherine R. Spindler for their helpful comments about this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abend JR, Low JA, Imperiale MJ. Inhibitory effect of gamma interferon on BK virus gene expression and replication. J Virol. 2007;81:272–279. doi: 10.1128/JVI.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett SM, Jiang M, Imperiale MJ. Role of cell-type-specific endoplasmic reticulum-associated degradation in polyomavirus trafficking. J Virol. 2013;87:8843–8852. doi: 10.1128/JVI.00664-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butin-Israeli V, Ben-nun-Shaul O, Kopatz I, Adam SA, Shimi T, Goldman RD, Oppenheim A. Simian virus 40 induces lamin A/C fluctuations and nuclear envelope deformation during cell entry. Nucleus. 2011;2:320–330. doi: 10.4161/nucl.2.4.16371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Rusan NM, Wilbuer AK, Norkin LC, Wadsworth P, Hebert DN. Simian virus 40 late proteins possess lytic properties that render them capable of permeabilizing cellular membranes. J Virol. 2006;80:6575–6587. doi: 10.1128/JVI.00347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel S, Heger T, Mancini R, Herzog F, Kartenbeck J, Hayer A, Helenius A. Role of endosomes in simian virus 40 entry and infection. J Virol. 2011;85:4198–4211. doi: 10.1128/JVI.02179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R, Andritschke D, Friebe S, Herzog F, Luisoni S, Heger T, Helenius A. BAP31 and BiP are essential for dislocation of SV40 from the endoplasmic reticulum to the cytosol. Nat Cell Biol. 2011;13:1305–1314. doi: 10.1038/ncb2339. [DOI] [PubMed] [Google Scholar]
- Gharakhanian E, Kasamatsu H. Two independent signals, a nuclear localization signal and a Vp1-interactive signal, reside within the carboxy-35 amino acids of SV40 Vp3. Virology. 1990;178:62–71. doi: 10.1016/0042-6822(90)90379-6. [DOI] [PubMed] [Google Scholar]
- Gharakhanian E, Takahashi J, Kasamatsu H. The carboxyl 35 amino acids of SV40 Vp3 are essential for its nuclear accumulation. Virology. 1987;157:440–448. doi: 10.1016/0042-6822(87)90286-8. [DOI] [PubMed] [Google Scholar]
- Goodwin EC, Lipovsky A, Inoue T, Magaldi TG, Edwards AP, Van Goor KE, Paton AW, Paton JC, Atwood WJ, Tsai B, DiMaio D. BiP and multiple DNAJ molecular chaperones in the endoplasmic reticulum are required for efficient simian virus 40 infection. MBio. 2011;2:e00101–00111. doi: 10.1128/mBio.00101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Minami N, Chen EY, Medina AL, Chico MM, Kasamatsu H. Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. Journal of virology. 1996;70:1317–1322. doi: 10.1128/jvi.70.2.1317-1322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii N, Nakanishi A, Yamada M, Macalalad MH, Kasamatsu H. Functional complementation of nuclear targeting-defective mutants of simian virus 40 structural proteins. Journal of virology. 1994;68:8209–8216. doi: 10.1128/jvi.68.12.8209-8216.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Johnson SF, Imperiale MJ. The role of polyomaviruses in human disease. Virology. 2009a;384:266–273. doi: 10.1016/j.virol.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Tsai B, Imperiale MJ. Early events during BK virus entry and disassembly. J Virol. 2009b;83:1350–1358. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu H, Nehorayan A. Vp1 affects intracellular localization of Vp3 polypeptide during simian virus 40 infection. Proceedings of the National Academy of Sciences of the United States of America. 1979;76:2808–2812. doi: 10.1073/pnas.76.6.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauzewicz N, Streuli CH, Stuart-Smith N, Jones MD, Wallace S, Griffin BE. Myristylated polyomavirus VP2: role in the life cycle of the virus. Journal of virology. 1990;64:4414–4420. doi: 10.1128/jvi.64.9.4414-4420.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J, Humes HD, Szczypka M, Imperiale M. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology. 2004;323:182–188. doi: 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Meneguzzi G, Pignatti PF, Barbanti-Brodano G, Milanesi G. Minichromosome from BK virus as a template for transcription in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:1126–1130. doi: 10.1073/pnas.75.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland RB, Garcea RL. Characterization of a nuclear localization sequence in the polyomavirus capsid protein VP1. Virology. 1991;185:513–518. doi: 10.1016/0042-6822(91)90811-o. [DOI] [PubMed] [Google Scholar]
- Nakanishi A, Li PP, Qu Q, Jafri QH, Kasamatsu H. Molecular dissection of nuclear entry-competent SV40 during infection. Virus research. 2007;124:226–230. doi: 10.1016/j.virusres.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi A, Shum D, Morioka H, Otsuka E, Kasamatsu H. Interaction of the Vp3 nuclear localization signal with the importin alpha 2/beta heterodimer directs nuclear entry of infecting simian virus 40. Journal of virology. 2002;76:9368–9377. doi: 10.1128/JVI.76.18.9368-9377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Miyazaki N, Xing L, Wu B, Hammar L, Li TC, Takeda N, Miyamura T, Cheng RH. Structure and assembly of a T=1 virus-like particle in BK polyomavirus. J Virol. 2005;79:5337–5345. doi: 10.1128/JVI.79.9.5337-5345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norkin LC, Anderson HA, Wolfrom SA, Oppenheim A. Caveolar endocytosis of simian virus 40 is followed by brefeldin A-sensitive transport to the endoplasmic reticulum, where the virus disassembles. J Virol. 2002;76:5156–5166. doi: 10.1128/JVI.76.10.5156-5166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelhaas M, Malmstrom J, Pelkmans L, Haugstetter J, Ellgaard L, Grunewald K, Helenius A. Simian Virus 40 depends on ER protein folding and quality control factors for entry into host cells. Cell. 2007;131:516–529. doi: 10.1016/j.cell.2007.09.038. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Wagstaff KM, Sivakumaran H, Heaton SM, Harrich D, Jans DA. Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem J. 2012;443:851–856. doi: 10.1042/BJ20120150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CP, Ravindran MS, Inoue T, Tsai B. A cytosolic chaperone complexes with dynamic membrane J-proteins and mobilizes a nonenveloped virus out of the endoplasmic reticulum. PLoS Pathog. 2014;10:e1004007. doi: 10.1371/journal.ppat.1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda Y, Hieda M, Nagoshi E, Miyamoto Y. Nucleocytoplasmic protein transport and recycling of Ran. Cell Struct Funct. 1999;24:425–433. doi: 10.1247/csf.24.425. [DOI] [PubMed] [Google Scholar]
- Zhong Y, Wang Y, Yang H, Ballar P, Lee JG, Ye Y, Monteiro MJ, Fang S. Importin beta interacts with the endoplasmic reticulum-associated degradation machinery and promotes ubiquitination and degradation of mutant alpha1-antitrypsin. J Biol Chem. 2011;286:33921–33930. doi: 10.1074/jbc.M111.272906. [DOI] [PMC free article] [PubMed] [Google Scholar]