Abstract
Humans have developed the perception, production and processing of sounds into the art of music. A genetic contribution to these skills of musical aptitude has long been suggested. We performed a genome-wide scan in 76 pedigrees (767 individuals) characterized for the ability to discriminate pitch (SP), duration (ST) and sound patterns (KMT), which are primary capacities for music perception. Using the Bayesian linkage and association approach implemented in program package KELVIN, especially designed for complex pedigrees, several SNPs near genes affecting the functions of the auditory pathway and neurocognitive processes were identified. The strongest association was found at 3q21.3 (rs9854612) with combined SP, ST and KMT test scores (COMB). This region is located a few dozen kilobases upstream of the GATA binding protein 2 (GATA2) gene. GATA2 regulates the development of cochlear hair cells and the inferior colliculus (IC), which are important in tonotopic mapping. The highest probability of linkage was obtained for phenotype SP at 4p14, located next to the region harboring the protocadherin 7 gene, PCDH7. Two SNPs rs13146789 and rs13109270 of PCDH7 showed strong association. PCDH7 has been suggested to play a role in cochlear and amygdaloid complexes. Functional class analysis showed that inner ear and schizophrenia related genes were enriched inside the linked regions. This study is the first to show the importance of auditory pathway genes in musical aptitude.
Keywords: musical aptitude, family study, linkage, association, auditory pathway, cognition, SNP
Introduction
Music is universal to humankind and is present even in the animal kingdom; for example birds and whales also sing. The ability to recognize complex musical sounds is already present in newborns1. In addition to simple sensory perception, the processing of music includes numerous cognitive tasks, emotion, learning, and memory2. Extensive research has been performed with brain imaging methods to understand the neural events involved in music processing: What does music activate in the brain? How do the brains of musicians and non-musicians differ? Music and other sounds, such as speech, activate multiple regions in the brain. For example, Broca's area, the prefrontal cortex, and the amygdala are activated as a response to any auditory stimulation2. Moreover, brain lesion studies have shown that separate brain areas handle temporal and melodic differences in music3.
The perception of sounds occurs in the auditory pathway (Fig. 1). Sounds are recognized by cochlear cells in the inner ear and transmitted as electronic signals through the auditory nerve into the brain through the superior olivary nucleus and inferior colliculus to the thalamus and auditory cortex, where sounds are primarily recognized. In addition to the auditory pathway, neuroimaging studies have confirmed that the actual music perception also affects multiple other regions in the brain2, 3.
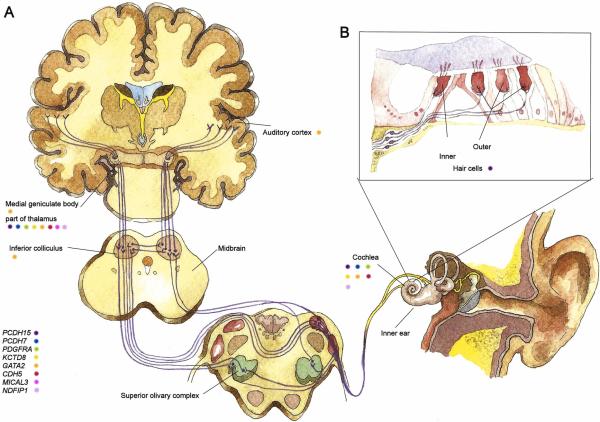
Figure 1. Auditory pathway with our best-associated genes localized according to their known expression patterns.
The perception of sounds occurs in the auditory pathway (A). Sounds are recognized by cochlear hair cells in the inner ear (B) and transmitted as electronic signals through the auditory nerve into the brain. The pathway goes through the superior olivary complex and inferior colliculus to the medial geniculate body in the thalamus and further to the auditory cortex where sounds are primarily recognized. On the bottom left, genes associated with the musical aptitude features in this study are listed. The known expressions patterns of these genes are shown with colored dots next to the text indicating the corresponding part of the auditory pathway. Only expressions within the auditory pathway are considered herein. Note that the brain sections are not to scale.
Based on family and twin studies, musical aptitude is at least partially inherited4-9. Heritability estimates of 0.21-0.68 for a set of three tests of musical aptitude were obtained7. In musical ability studies, attention has been paid to extreme phenotypes such as absolute pitch (AP; the ability to identify notes without reference)4 and amusia (“tone deafness”)6. For both of these traits, the recurrence risk for siblings is increased4, 6, indicating familial enrichment. Chromosome 8q24.21 has been linked to AP in a genome-wide study8 but no results have yet been published for amusia. Concerning the normal variation of music perception skills, the chromosome 4q22-23 region has been implicated as a candidate locus by two independent studies: our pilot study7 and another study in Mongolian families9.
The aim of this study was to identify genetic variants associated with sensory perception, given herein as the detection and discrimination of sounds, and auditory structuring capacities in humans. In order to reach this goal we performed a genome-wide linkage and association scan in 76 families using over 660 000 SNPs. Multiple regions were identified that contained genes affecting auditory pathway and cognitive functions.
Materials and methods
Family material
The collected sample included 99 extended Finnish families comprising 915 individuals. Each family included 2 – 50 individuals spanning 1 to 4 generations. The participant ages ranged from 7 – 94 years and 41% of them were male (Supplementary Table 1). Subjects older than 7 years old were tested with the musical aptitude tests, and DNA was collected from individuals older than 12 years old. The Ethical Committee of Helsinki University Central Hospital approved the study, and informed consent was obtained from all participants or their parents.
The first 15 families were selected based on having several professional musicians7. The rest of the families were collected with no prior knowledge about their interest in music via advertisements in magazines, webpages and mailing lists. There were only a few professional musicians in these families. The difference in the ascertainment criteria was taken into consideration in the statistical analyses (see below).
Phenotype
In this study, musical aptitude was assessed using three music tests: the auditory structuring ability test (Karma Music Test, KMT)10 and Carl Seashore's subtests of pitch (SP) and time discrimination (ST)11. These tests measure music perception skills, not music production.
The KMT measures the recognition of melodic contour, grouping, relational pitch processing, and gestalt principles, the same potentially innate musical cognitive operations reported by Justus and Hutsler12. In this test, the subject's task is to detect abstract sound patterns and compare them to find structural changes. In each item (N=40), a pattern is played with varying instruments three times and then a test sequence is played once (Supplementary Fig. 1). The participant is asked if the test sequence is same or different from the first pattern played. Three sample items from the KMT are available at http://www.hi.helsinki.fi/music/english/samples.htm.
SP and ST measure the ability to detect small differences between two sequentially presented tones. SP measures the ability to detect differences in tone pitch, and ST in tone duration. ST and SP consist of 50 items each. The reliabilities of the music test scores are 0.88 for the KMT, 0.91 for the SP and 0.78 for the ST7. Testing was performed as a group test that lasted approximately one hour. The tests were played through loud speakers.
To analyze the measured musical aptitude as one variable, a combined test score (COMB) was created. The COMB was computed as the sum of the three individual test scores after the KMT music score was scaled to the same range as the other music scores (from 25 to 50 pts.) (for the original ranges see Supplementary Fig. 2). Notably, there is some overlap among the test scores: the correlations between the tests are 0.38 between SP and ST, 0.42 between KMT and ST, and 0.61 between KMT and SP13.
The heritabilities were estimated with SOLAR14 as 0.46, 0.68, 0.21 and 0.60 for the KMT, SP, ST and COMB, respectively. Because the heritability of ST was low, it was not included in the test score-specific linkage analyses. However, it was included in the COMB scores to have more complete perspective of musical aptitude (see Supplementary Fig. 3).
Genotyping
DNA was isolated from peripheral blood using the phenol-chloroform method and was available from 799 participants. The samples were genotyped with Illumina HumanOmniExpress 12 1.0V SNP chip (Illumina Inc., San Diego, California, USA). Genotyping was conducted at the Wellcome Trust Centre for Human Genetics, Oxford University. The chip includes 733 202 SNPs. Genotype calls and genotyping quality control were performed with GenomeStudio. Genotyping failed in two samples, which were removed from the following analyses. The call rates for the included samples were higher than 99% (minimum 99.18%, mean 99.74%).
Quality control
Quality control was performed with PLINK 1.0715. Relatedness was evaluated with the identity by descent (IBD) calculations. In total, three subjects were removed due to incorrect relatedness. In the remaining data, the Mendelian error rate was further tested with PedCheck16, and it was less than 0.1% per individual. No individuals needed to be removed for missing data (5%). Uninformative families were excluded from the further analyses. Thus, a total of 767 individuals (632 genotyped, 699 phenotyped) from 76 families remained in the analysis.
Markers were removed for missingness (>5%), Mendelian inconsistencies and low minor allele frequency (<5%). A total of 664 177 markers remained, which were all used for the association analysis. For the linkage analysis, the SNP data had to be thinned. First, the minor allele frequency was required to be >0.25 to maximize the information per selected SNP. Next, SNPs in high linkage disequilibrium (LD) with each other were removed. LD pruning was performed with the variant inflation factor (VIF) method in PLINK with a VIF limit of 1.25 (corresponding to an LD of 0.2 as measured by R2). Additionally, the remaining SNPs were pruned for a map distance of less than 0.2cM (Rutgers map v.217). Finally, the remaining SNPs were rerun for Mendelian errors with PedCheck16. In total, 10 742 SNPs were included in the linkage analyses.
Linkage and association analysis
The software package KELVIN v2.4.118 was used for the genetic analyses. KELVIN supports combined linkage (posterior probability of linkage, PPL) and linkage disequilibrium (posterior probability of linkage disequilibrium, PPLD) analyses. KELVIN is also capable of handling large pedigrees, so we were able to keep the very large families intact. Multipoint linkage analysis was done using a unique “hybrid” approach using Markov chain Monte Carlo (MCMC) for the marker data19 and exact likelihood calculations for the trait data20.
The three musical aptitude phenotypes (KMT, SP and COMB) were analyzed under a quantitative trait (QT) model. This model is parameterized in terms of the disease allele frequency, three genotypic means and three genotypic variances. Normality is assumed at the genotypic level, but not at the population level, and there is no inflation of scores under violations of normality21. Also included in the likelihood is the admixture parameter of Smith22, allowing for the possibility of locus heterogeneity. All trait parameters are integrated out of the final statistic, using essentially uniform prior distributions (ordering constraints are imposed on the means and unbounded parameters are integrated over a finite range18, 21), implicitly allowing for dominant, recessive and additive models. This provides a robust approximation for mapping complex traits in terms of the marginal model at each locus and because the parameters are integrated out, no specific assumptions regarding their values are required. The likelihood also contains two location parameters: the recombination fraction θ and the standardized LD parameter D', representing trait-marker association due to physical proximity. This enables us to carry out combined linkage and association analysis using the same likelihood framework.
The PPL and PPLD statistics are calculated on a probability scale from 0 to 1, which is interpreted directly as the probability of a trait gene being linked to/associated with the location/marker examined. Based on earlier calculations23, the prior probability at each location is set to 2%, so that PPLs >2% indicate (some degree) of evidence in favor of a trait gene at that locus, while PPLs <2% represent evidence against the location. The prior probability of LD given linkage (L) is also set to 0.02, so that in the absence of prior linkage information P(L&LD) = 0.0004. (See also WTCC Consortium 200724 for justification of a comparable figure). The PPL (or PPLD) accumulates evidence across datasets through Bayesian sequential updating, a mathematically rigorous approach, which allows heterogeneity across datasets, here, between the two differently ascertained batches (Subset 1: 14 families, N=195; Subset 2: 62 families, N=572). Another important distinguishing feature of the sequentially updated PPL (PPLD) is that it accumulates evidence both for and against linkage (or association) as the data accrue.
The PPL is a measure of statistical evidence, not a decision-making procedure. There are, therefore, no “significance levels” associated with it (i.e., no specific cutoffs beyond which we declare significance) and it is not interpreted in terms of associated error probabilities25, 26.
By the same token, no multiple testing corrections are applied to the PPL, just as one would not “correct” a measure of the temperature made in one location for temperature readings taken at different locations27.
The PPLD analyses are robust to population substructure by virtue of being family-based; in particular, in these medium- to large-sized pedigrees, the association evidence is driven largely by transmission patterns and not just by underlying allele frequencies in founders.
The association results outside of linked regions were pruned to exclude the most probable false positives: only signal clusters with at least 2 SNPs with PPLD≥20% were included as true positive signals. The signal clusters were identified as SNPs with R2≥0.5 with the reference SNP.
Finally, we performed pedigree-wise association and linkage analyses with the 12 largest families (Supplementary Fig. 4). Each family was analyzed separately by both linkage and association (KELVIN byPed) analyses.
Gene information
The nearest genes to the associated SNPs were identified according to UCSC28. LD analysis (with PLINK 1.07) was used to study if the associated SNPs were genetically near the genes. LD was evaluated between the associated SNP and SNPs within 500kb region. Unrelated individuals (N=192) were used for the evaluation. This pairwise LD information is shown in the regional association plots. Additionally, the recombination rate for overall LD structure was retrieved from the HapMap CEU (Utah residents with ancestry from northern and western Europe) data29.
Putative regulatory regions close to GATA2 at 3q21.3 were predicted using UCSC (hg19), conservation from the Placental Chain/Net track28 and DNase I hypersensitivity from ENCODE Digital DNase I Hypersensitivity Cluster in 125 cell types30. The known enhancers for GATA2 were gathered from literature.
Haplotype analysis
Haplotyping and haplotype association analysis of the best-associated region at 3q21.3 was carried out with PLINK 1.07 haplotype-based quantitative trait association. The best-associated haplotype spanned over ten markers, from rs3803 to rs4613470 (TGGGTGCCGA), with p=0.001923. Note however that the PLINK haplotyping procedure handles only partial family structures.
Functional class analysis
The linkage results were further studied to evaluate the biological functions of the genes within the linked regions. Genes were included within 2cM regions around the linkage peaks with PPL≥0.2 (Supplementary Table 2). Narrow linkage regions (where 2cM region is equivalent to less than 2Mb region) were widened to altogether span 2Mb. In total, 286 genes were included in the analysis. The genes from the best-linked regions (PPL≥0.5) are listed in Supplementary Table 3.
Functional class analysis of the genes was performed with IPA (www.ingenuity.com). The identified functional classes with less than 5 genes linked in our data, and classes with overlapping functions were removed. We did not carry out the functional analysis with association results due to the small number of genes pinpointed by association.
Figures and Tables
Regional association plots were produced with R 2.15.2 (www.r-project.org). These figures were generated using a modified version of a code from Broad institute (http://www.broadinstitute.org/diabetes/scandinavs/figures.html). For the position information, hg18 was used in all figures and tables.
Results
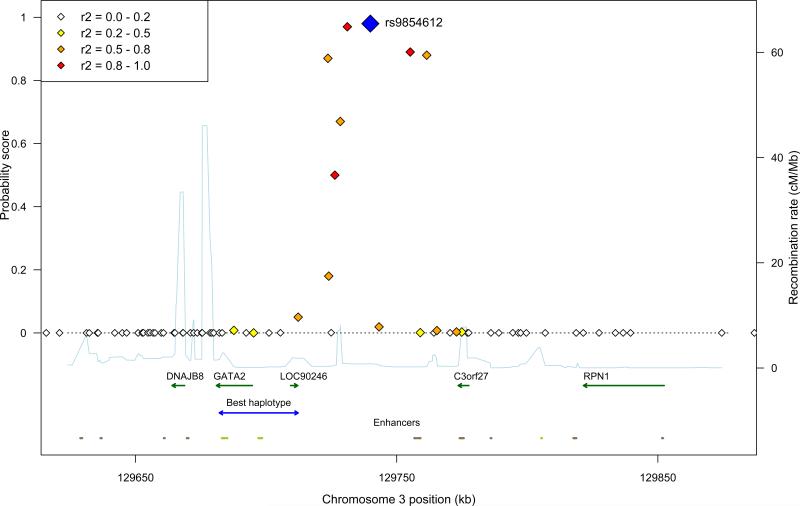
The strongest association was found at 3q21.3 (the highest probability of marker-trait LD was 0.98 at rs9854612 with COMB) (Fig. 2-3; Table 1). This region is located a few dozen kilobases upstream of the GATA binding protein 2 (GATA2) gene. Notably, the best-associating haplotype spans the entire gene (Fig. 3, Supplementary Table 4). Other genes in the region include LOC90246 and C3orf27 of unknown function. The region was further studied to identify possible regulatory information concerning the associated SNPs. Although no known regulatory sites were found within the region, GATA2 had known regulatory sites on both sides of the SNPs31-36. Also, we could identify a putative regulatory site within the associated region (Fig. 3).
Figure 2. Genome-wide association and linkage results for musical aptitude: A) COMB, B) KMT and C) SP.
The best linkage was obtained on chromosome 4 for SP. Evidence against linkage was found across 85.1%, 86.6% and 87.5% of the genome, and PPL≥0.10 occurred over only 2.3%, 2.4% and 0.9% of the genome for COMB, KMT and SP, respectively. D) Enlargement of the 4p15-q26 region of chromosome 4. The studied SNPs are marked with open diamonds. The colors indicate different phenotypes: COMB, purple; SP, green and KMT, blue. The location of the best linkage peak from Park et al.9 (grey line) is also shown. For the sake of clarity, only the genes mentioned in this article have been depicted. White circles denote the centromeres. The Y-axes show the strength of the association and linkage as probability scores ranging from 0 to 1.
Figure 3. Regional association plot from chromosome 3.
The posterior probability of LD (trait-marker association) is given on the Y-axis. The best associated SNP and the region with the best haplotype are marked in blue. All the genes in the region are shown. The linkage disequilibrium between the best associated SNP, rs9854612 (blue diamond), and the other SNPs are color-coded according to their pairwise R-squared values (see figure legend). The light blue curve shows the background recombination rate. Predicted enhancers (grey) and the known GATA2 enhancers (light green) are shown at the bottom.
Table 1. The strongest associations for each of the phenotypes.
A PPLD>0.5 was required for the location to be listed. For each association, the nearest genes in the regions are named, with their distance to the best-associated SNP given.
| Location | Genes | Association (PPLD) | ||||||
|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Other SNPs with PPLD>0.5* | Gene | Distance (kb) | KMT | SP | COMB | |
| 1p31.1 | rs4630083 | rs4291455 |
LRRIQ3
**
BC041341 ** |
Leucine-rich repeats and IQ motif containing 3 uncharacterized |
316 371 |
0.70 | 0.03 | 0.56 |
| 3q21.3 | rs9854612 | rs7433900 rs6769565 rs9819395 rs7629705 rs7635061 rs2335050 |
LOC90246
C3orf27 GATA2 |
uncharacterized Chromosome 3 open reading frame 27 GATA binding protein 2 |
28 34 45 |
0.52 | 0.78 | 0.98 |
| 4p15.1 | rs13146789 | rs13109270 | PCDH7 | Protocadherin 7 | 53 | 0.12 | 0.34 | 0.81 |
| 5q31.3 | rs2961694 | rs7729341 | NDFIP1 | Nedd4 family interacting protein 1 | 9 | 0.61 | <0.01 | 0.03 |
| 11q21 | rs2186739 | - |
FAM76B
**
SESN3 ** |
Family with sequence similarity 76, member B Sestrin 3 | 175 363 |
0.58 | 0.27 | 0.40 |
SNPs nearby the best-associated SNP.
Gene not in the same LD cluster with the given SNPs.
Note. While effect size estimates can be difficult to interpret, it is possible to calculate the degree of genetic determination from Kelvin output; see Supplementary Table 8.
Chromosome 4 harbors several regions of interest, shown by linkage covering a vast region from 4p15-q24 (Fig. 2, Table 2). Interestingly, all our phenotypes are linked to this region. The highest probability of linkage was obtained for phenotype SP at 4p14 (PPL=0.86, Fig. 2, Table 2). The linked region is located next to the region harboring the protocadherin 7 gene, PCDH7, where was found the best association on the whole chromosome 4 (PPLD=0.81). The associated SNPs (rs13146789 and rs13109270) were located 53kb upstream of the PCDH7 gene at 4p15.1, (Table 1, Supplementary Fig. 5).
Table 2. The best linkages.
The regions with a multipoint linkage score PPL>0.5 with at least one of the studied phenotypes are listed. The strongest PPL score is bolded. Additionally, the SNPs showing the highest associations inside each linkage region are given accompanied by their PPLD values and the nearest gene(s). The strength of the association is depicted as the posterior probability of linkage disequilibrium between the SNP and the trait (PPLD).
| Location | Linkage (PPL) | The best association (PPLD) inside the linked region | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Chr | cM | KMT | SP | COMB | SNP | Trait | PPLD | Gene | |
| 4p14-13 | 62 | 0.05 | 0.86 | 0.18 | rs4349633 | KMT | 0.07 | LIMCH1 | LIM and calponin homology domains 1 |
| 4p12-q12 | 66 -72 | 0.24 | 0.08 | 0.50 | rs11732997 rs2412501 |
COMB COMB |
0.28 0.34 |
KCTD8
PDGFRA |
Potassium channel tetramerization domain containing 8
Platelet-derived growth factor receptor, alpha polypeptide |
| 16q21-22.1 | 84 - 86 | 0.57 | 0.01 | 0.18 | rs7188225 | KMT | 0.08 |
LOC283867
CDH5 |
- Cadherin 5, type 2 (vascular endothelium) |
| 18q12.3-21.1 | 66 - 70 | 0.02 | 0.04 | 0.55 | rs617459 | SP | 0.07 | SETBP1 | SET binding protein 1 |
| 22q11.1-.21 | 0 - 16 | 0.30 | 0.68 | 0.48 | rs4819640 | KMT | 0.11 | MICAL3 | Microtubule associated monooxygenase, calponin and LIM domain containing 3 |
The KMT and COMB were linked to 4p12-q12. Within this region, the best-associated SNPs were located near the platelet-derived growth factor receptor, alpha polypeptide (PDGFRA) (PPLD=0.34) and potassium channel tetramerization domain containing 8 (KCTD8) (PPLD=0.28) genes.
Nedd4 family interacting protein 1 (NDFIP1) gene was associated at 5p31.3 (PPLD=0.61) (Supplementary Fig. 6). NDFIP1 has a role in immune response37.
At chromosomes 1p31.1 (PPLD 0.70 with KMT) and 11q21 (PPLD 0.58 with KMT) association could not be linked to any known gene (Table 1, Supplementary Figs. 7 and 8). However, the associated SNPs at 1p31.1 were inside long non-coding RNA (Ensembl ENSG00000223479), which may have expression in human brain (based on microarray data from Sestan Lab at Yale University, UCSC).
We also found some evidence for 4q21.23-22.1 and 4q24 being linked to the KMT scores (PPL=0.35 and PPL=0.30, respectively). A single pedigree association analysis supported the scavenger receptor class B, member 2 (SCARB2) gene at 4q21.1 (rs17001659; PPLD=0.17 with COMB; Family #10; Supplementary Table 5), which is connected to epilepsy in humans and to deafness in mice38.
There were also other loci linked to musical aptitude, especially on chromosomes 16, 18 and 22 (Fig. 2, Table 2). On chromosome 22, SP was linked to 22q11.21, which is a well-known DiGeorge syndrome (DGS) region (MIM#188400). On chromosome 16, the KMT scores were linked to 16q21-22.1 with a PPL of 0.57 (Table 2). The highest PPLD score (0.08) in the region was obtained at rs7188225 between the cadherin 5, type 2 (CDH5) gene and LOC28386. Interestingly, the region has also been pinpointed in autosomal recessive nonsyndromic hearing impairment39. Furthermore, the COMB scores were linked to 18q12.3-21.1 (PPL=0.55, Table 2). This region includes the lipoxygenase homology domains 1 (LOXHD1) gene, which is expressed in the mechanosensory hair cells in the inner ear (Fig. 1). Mutations in this gene lead to auditory defects40.
The associations were pruned according to SNP clusters to exclude possible false positive signals. However, there were 7 associated SNPs (PPLD≥0.2) that had no neighboring SNPs with R2≥0.5. These singleton results could also be true positive signals and the results for these SNPs are listed on Supplementary Table 6.
A functional class analysis for the linked genes showed some interesting functions: schizophrenia (p-value 1.0×10-7) and inner ear development (p-value 4.7×10-4) were among the best-associated functions (Supplementary Table 7).
Discussion
This report describes the results of a GWAS of music perception skills using large pedigrees with over 660 000 SNPs. The strongest association was found at 3q21.3 near GATA2. GATA2 is an important transcription factor involved in the development of several organs, such as the inner ear41 and inferior colliculus (IC)42, which are both involved in the auditory pathway (Fig. 1). Importantly, tonotopic mapping occurs in the IC: different pitches map to different cells43. In the midbrain in general, GATA2 determines the identity of GABAergic neurons (neurons that secrete gamma-Aminobutyric acid)42. In this respect, it is noteworthy that a decrease in the number of GABAergic neurons in the IC is linked to age-related hearing loss44. Furthermore, a closely related factor, GATA3, regulates GATA2 during inner ear development. Mutations in GATA3 are known to cause deafness45, but to the best of our knowledge, mutations in GATA2 have not been shown to cause major hearing defects. Notably, our best-associated SNPs are located in the regulatory region of GATA2 and include several highly conserved DNase I hypersensitivity sites28 (Fig. 3), suggesting that the differences affecting musical aptitude are regulatory rather than structural.
Near the linked region at 4p14, the best association was found near PCDH7 gene. PCDH7 is a very plausible candidate because it is expressed in, for example, the developing cochlea46 (chicken) and the mouse amygdaloid complex both during the postnatal period and in adult animals47. In neuroscientific studies, amygdala has been linked to emotional interpretation of musical sounds and may not have other impact on perceptual processing48. Additionally, PCDH15 (10q21.1), which in our data is associated with a probability of 0.39 (to KMT), plays a crucial role in hair cell sensory transduction49 and is a known deafness gene50. Inside that linked region at 4p14, there were also interesting genes. Although associations remained low within this region (Table 2), it contains genes such as the cholinergic receptor, nicotinic, alpha 9 (neuronal) (CHRNA9)51 and the paired-like homeobox 2b (PHOX2B) gene52, which both impact inner ear development.
Inside another linked region on chromosome 4, 4p12-q12, associations were found near genes PDGFRA and KCTD8. KCTD8 is expressed in the spiral ganglion of the cochlea53 (Fig. 1). KCTD8 also interacts with GABA receptors GABRB1 and GABRB2, whose genes are both found within same linked region. PDGFRA is expressed in the hippocampus54 and cochlea55 in mice.
Our result at 4q21.23-22.1 and 4q24 are in agreement with those of our preliminary study7 (chromosome 4q22) and a study by Park et al.9 (chromosome 4q23) (Fig. 2D). All these findings on chromosome 4 might indicate a common locus.
In many of the linked regions, the associations remained quite low. The linkage analysis and association analysis have power to detect very different types of genetic effects: association involves allelic effects, which need to be quite homogeneous across pedigrees in order to be detectable but need not confer large relatives risks; linkage analysis targets loci containing genes of large effect, and it can find such genes even when they are causally relevant only in a subset of families. Under even modest levels of allelic heterogeneity, it is possible to find a strong (true) linkage peak in a region with no allelic associations; conversely, for homogeneous allelic effects involving low relative risks, an allelic association may not generate a linkage signal. Thus the two types of analysis are complementary, and may or may not yield concordant findings at any given locus.
Notably, the regions pinpointed for absolute pitch8 were not replicated in our materials. Also, we did not replicate our previous associations near the AVPR1A gene56.
According to the model of auditory theory, traits that we usually understand as musical abilities are secondary skills, whereas primary capacities are a prerequisite for the development of these skills57 (Supplementary Fig. 3). The secondary musical skills are culture-dependent and modified by the environment, and the primary capacities should reflect innate musical aptitude. In principle, the tests we used mainly measure these primary capacities. However, especially the KMT is considered as a test of cognitive functions instead of mere sensory capacities58.
Overall, the best-linked and associated regions show several genes mostly related to the auditory pathway, not only specifically to inner ear function but also to neurocognitive processes (Supplementary Table 3; Fig. 1). In particular, GATA2, PCDH7 and PCDH15, all of which are known to affect inner ear development, were found to associate strongly with musical aptitude. The functional class analysis showed enrichment of genes related to the development of inner ear. A bit surprisingly, the strongest enriched class was schizophrenia, which might refer to the importance of genes related to cognition in general. Naturally, the allelic architecture contributing to neurocognitive disorders need not be the same as those we observe for musical aptitude. Notably, our best associations were found in the 5’ regions of the candidate genes, thereby emphasizing the possible importance of regulatory variations instead of any actual structural differences.
Obviously, genes that function in the development of the auditory pathway affect its efficiency and resolution ability in processing the auditory signals necessary for identifying differences between pitches or short melodies. Meanwhile, the neurocognitive functions of the genes pinpointed (e.g. schizophrenia related genes) might relate to the overall cognitive skills needed to remember and interpret a series of sounds. Several neuroscientific studies2, 59, 60 have suggested that music perception induces neuroplasticity and enhances neurogenesis, which might also be important in musical aptitude. The neuroplasticity might be the connection between the schizophrenia related genes and musical aptitude.
We acknowledge that musical aptitude is a complex behavioral trait and that our tests account for only a part of the phenotype. Environmental factors, such as the childhood musical environment, the example set by parents and siblings, and music education affect musical abilities. Currently, we aim to analyze these effects in more detail, based on wide questionnaire data collected from the families and from sporadic individuals genotyped in this study. We argue that our findings provide a valuable background for molecular studies and research on the interplay of genes and the environment with respect to musical ability.
Supplementary Material
Acknowledgements
We are grateful to the families for their participation. We thank the High-Throughput Genomics Group at the Wellcome Trust Centre for Human Genetics (funded by Wellcome Trust grant reference 090532/Z/09/Z and MRC Hub grant G0900747 91070) for the generation of the genotype data. Daniel Borshagovski is thanked for information about the regulatory elements. Johanna Lumme is thanked for the artwork and Minna Varhala for her expert laboratory work. Funding for this study was provided by the Academy of Finland #13371, the Finnish Cultural Foundation to IJ. LUV is funded by the Helsinki University Research Foundation and the Paulo Foundation. VJV is funded by the National Institutes of Health (grant R01 MH086117 and U24 MH068457). This work was also supported in part by an allocation of computing time from the Ohio Supercomputer Center Grant PCCR0002 to VJV.
Footnotes
Author contributions
Study concept and design: PO, VJV, IJ. Sample collection and interpretation: JO, LUV, PR, KK. Genotyping: LUV. Data cleaning and management of the sample data: JO. Analysis of the genotype data: JO, YH. Drafting and critical revision of the manuscript: JO, PO, YH, VJV, IJ. All authors gave approval to the final manuscript.
Conflict of interest
The authors declare no conflict of interest.
Supplementary information
Supplementary information is available at Molecular Psychiatry's website.
References and Notes
- 1.Perani D, Saccuman MC, Scifo P, Spada D, Andreolli G, Rovelli R, et al. Functional specializations for music processing in the human newborn brain. Proc Natl Acad Sci U S A. 2010;107:4758–4763. doi: 10.1073/pnas.0909074107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koelsch S. Toward a neural basis of music perception – a review and updated model. Front Psychol. 2011;2:1–20. doi: 10.3389/fpsyg.2011.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peretz I, Zatorre RJ. Brain organization for music processing. Annu Rev Psychol. 2005;56:89–114. doi: 10.1146/annurev.psych.56.091103.070225. [DOI] [PubMed] [Google Scholar]
- 4.Baharloo S, Johnston PA, Service SK, Gitschier J, Freimer NB. Absolute pitch: an approach for identification of genetic and nongenetic components. Am J Hum Genet. 1998;62:224–231. doi: 10.1086/301704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drayna D, Manichaikul A, de Lange M, Snieder H, Spector T. Genetic correlates of musical pitch recognition in humans. Science. 2001;291:1969–1972. doi: 10.1126/science.291.5510.1969. [DOI] [PubMed] [Google Scholar]
- 6.Peretz I, Cummings S, Dubé M. The genetics of congenital amusia (tone deafness): a family- aggregation study. Am J Hum Genet. 2007;81:582–588. doi: 10.1086/521337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pulli K, Karma K, Norio R, Sistonen P, Göring HHH, Järvelä I. Genome-wide linkage scan for loci of musical aptitude in Finnish families: evidence for a major locus at 4q22. J Med Genet. 2008;45:451–456. doi: 10.1136/jmg.2007.056366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theusch E, Basu A, Gitschier J. Genome-wide study of families with absolute pitch reveals linkage to 8q24.21 and locus heterogeneity. Am J Hum Genet. 2009;85:112–119. doi: 10.1016/j.ajhg.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park H, Lee S, Kim H, Ju YS, Shin J, Hong D, et al. Comprehensive genomic analyses associate UGT8 variants with musical ability in a Mongolian population. J Med Genet. 2012;49:747–752. doi: 10.1136/jmedgenet-2012-101209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karma K. Musical aptitude definition and measure validation: ecological validity can endanger the construct validity of musical aptitude tests. Psychomusicology. 2007;19:79–90. [Google Scholar]
- 11.Seashore C, Lewis D, Saetveit J. Seashore Measures of Musical Talents (Manual) The Psychological Corp; New York, USA: 1960. [Google Scholar]
- 12.Justus T, Hustler JJ. Fundamental issues in the evolutionary psychology of music: Assessing innateness and domain specificity. Music Perception. 2005;23:1–27. [Google Scholar]
- 13.Ukkola-Vuoti L, Oikkonen J, Onkamo P, Karma K, Raijas P, Järvelä I. Association of the arginine vasopressin receptor 1A (AVPR1A) haplotypes with listening to music. J Hum Genet. 2011;56:324–329. doi: 10.1038/jhg.2011.13. [DOI] [PubMed] [Google Scholar]
- 14.Almasy L, Blangero J. Multipoint Quantitative-Trait Linkage Analysis in General Pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vieland VJ, Huang Y, Seok S, Burian J, Catalyurek U, O'Connell J, et al. Kelvin: a Software Package for Rigorous Measurement of Statistical Evidence in Human Genetics. Hum Hered. 2011;72:276–288. doi: 10.1159/000330634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas A, Gutin A, Abkevich V, Bansal A. Multilocus linkage analysis by blocked Gibbs sampling. Statistics and computing. 2000;10:259–269. [Google Scholar]
- 20.Huang Y, Thomas A, Vieland VJ. Employing MCMC under the PPL framework to analyze sequence data in large pedigrees. Front Genet. 2013;4:59. doi: 10.3389/fgene.2013.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartlett CW, Vieland VJ. Accumulating quantitative trait linkage evidence across multiple datasets using the posterior probability of linkage. Genet Epidemiol. 2007;31:91–102. doi: 10.1002/gepi.20193. [DOI] [PubMed] [Google Scholar]
- 22.Smith CAB. Testing for heterogeneity of recombination fraction values in Human Genetics. Ann Hum Genet. 1963;27:175–182. doi: 10.1111/j.1469-1809.1963.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 23.Elston RC, Lange K. The prior probability of autosomal linkage. Ann Hum Genet. 1975;38:341–350. doi: 10.1111/j.1469-1809.1975.tb00619.x. [DOI] [PubMed] [Google Scholar]
- 24.WTCC Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Royall R. Statistical evidence: a likelihood paradigm. Vol. 71. CRC press; 1997. [Google Scholar]
- 26.Vieland JV, Hodge SE. Review of Statistical Evidence: A Likelihood Paradigm. Am J Hum Genet. 1998;63:283–289. [Google Scholar]
- 27.Vieland VJ. Thermometers: something for statistical geneticists to think about. Hum Hered. 2006;61:144–156. doi: 10.1159/000093775. [DOI] [PubMed] [Google Scholar]
- 28.Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, et al. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res. 2013;41:D64–69. doi: 10.1093/nar/gks1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The International HapMap Consortium The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 30.ENCODE Project Consortium. Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Y, Yamamoto M, Engel JD. GATA2 is required for the generation of V2 interneurons. Development. 2000;127:3829–3838. doi: 10.1242/dev.127.17.3829. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi-Osaki M, Ohneda O, Suzuki N, Minegishi N, Yokomizo T, Takahashi S, et al. GATA motifs regulate early hematopoietic lineage-specific expression of the Gata2 gene. Mol Cell Biol. 2005;25:7005–7020. doi: 10.1128/MCB.25.16.7005-7020.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grass JA, Jing H, Kim S, Martowicz ML, Pal S, Blobel GA, et al. Distinct functions of dispersed GATA factor complexes at an endogenous gene locus. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khandekar M, Brandt W, Zhou Y, Dagenais S, Glover TW, Suzuki N, et al. A Gata2 intronic enhancer confers its pan-endothelia-specific regulation. Development. 2007;134:1703–1712. doi: 10.1242/dev.001297. [DOI] [PubMed] [Google Scholar]
- 35.Wozniak RJ, Keles S, Lugus JJ, Young KH, Boyer ME, Tran TM, et al. Molecular hallmarks of endogenous chromatin complexes containing master regulators of hematopoiesis. Mol Cell Biol. 2008;28:6681–6694. doi: 10.1128/MCB.01061-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nozawa D, Suzuki N, Kobayashi-Osaki M, Pan X, Engel JD, Yamamoto M. GATA2-dependent and region-specific regulation of Gata2 transcription in the mouse midbrain. Genes Cells. 2009;14:569–582. doi: 10.1111/j.1365-2443.2009.01289.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Tong X, Ye X. Ndfip1 negatively regulates RIG-I-dependent immune signaling by enhancing E3 ligase Smurf1-mediated MAVS degradation. J Immunol. 2012;189:5304–5313. doi: 10.4049/jimmunol.1201445. [DOI] [PubMed] [Google Scholar]
- 38.Berkovic SF, Dibbens LM, Oshlack A, Silver JD, Katerelos M, Vears DF, et al. Array-based gene discovery with three unrelated subjects shows SCARB2/LIMP-2 deficiency causes myoclonus epilepsy and glomerulosclerosis. Am J Hum Genet. 2008;82:673–684. doi: 10.1016/j.ajhg.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Basit S, Lee K, Habib R, Chen L, Umm-e-Kalsoom, Santos-Cortez RLP, et al. DFNB89, a novel autosomal recessive nonsyndromic hearing impairment locus on chromosome 16q21-q23.2. Hum Genet. 2011;129:379–385. doi: 10.1007/s00439-010-0934-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grillet N, Schwander M, Hildebrand MS, Sczaniecka A, Kolatkar A, Velasco J, et al. Mutations in LOXHD1, an evolutionarily conserved stereociliary protein, disrupt hair cell function in mice and cause progressive hearing loss in humans. Am J Hum Genet. 2009;85:328–337. doi: 10.1016/j.ajhg.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haugas M, Lilleväli K, Hakanen J, Salminen M. Gata2 is required for the development of inner ear semicircular ducts and the surrounding perilymphatic space. Dev Dyn. 2010;239:2452–2469. doi: 10.1002/dvdy.22373. [DOI] [PubMed] [Google Scholar]
- 42.Lahti L, Achim K, Partanen J. Molecular regulation of GABAergic neuron differentiation and diversity in the developing midbrain. Acta Physiol (Oxf) 2013;207:616–627. doi: 10.1111/apha.12062. [DOI] [PubMed] [Google Scholar]
- 43.De Martino F, Moerel M, van de Moortele P, Ugurbil K, Goebel R, Yacoub E, et al. Spatial organization of frequency preference and selectivity in the human inferior colliculus. Nat Commun. 2013;4:1386. doi: 10.1038/ncomms2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caspary D, Milbrandt JC, Helfert RH. Central auditory aging: gaba changes in the inferior colliculus. Exp Gerontol. 1995;30:349–360. doi: 10.1016/0531-5565(94)00052-5. [DOI] [PubMed] [Google Scholar]
- 45.Van Esch H, Groenen P, Nesbit MA, Schuffenhauer S, Lichtner P, Vanderlinden G, et al. GATA3 haplo-insufficiency causes human HDR syndrome. Nature. 2000;406:419–422. doi: 10.1038/35019088. [DOI] [PubMed] [Google Scholar]
- 46.Lin J, Yan X, Wang C, Guo Z, Rolfs A, Luo J. Anatomical expression patterns of delta-protocadherins in developing chicken cochlea. J Anat. 2012;221:598–608. doi: 10.1111/j.1469-7580.2012.01568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hertel N, Redies C, Medina L. Cadherin expression delineates the divisions of the postnatal and adult mouse amygdala. J Comp Neurol. 2012;520:3982–4012. doi: 10.1002/cne.23140. [DOI] [PubMed] [Google Scholar]
- 48.Gosselin N, Peretz I, Johnsen E, Adolphs R. Amygdala damage impairs emotion recognition from music. Neuropsychologia. 2007;45:236–244. doi: 10.1016/j.neuropsychologia.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Sotomayor M, Weihofen WA, Gaudet R, Corey DP. Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction. Nature. 2012;492:128–132. doi: 10.1038/nature11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, et al. Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet. 2001;69:25–34. doi: 10.1086/321277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katz E, Elgoyhen AB, Gómez-Casati ME, Knipper M, Vetter DE, Fuchs PA, et al. Developmental regulation of nicotinic synapses on cochlear inner hair cells. J Neurosci. 2004;24:7814–7820. doi: 10.1523/JNEUROSCI.2102-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ousdal OT, Anand Brown A, Jensen J, Nakstad PH, Melle I, Agartz I, et al. Associations between variants near a monoaminergic pathways gene (PHOX2B) and amygdala reactivity: a genome-wide functional imaging study. Twin Res Hum Genet. 2012;15:273–285. doi: 10.1017/thg.2012.5. [DOI] [PubMed] [Google Scholar]
- 53.Metz M, Gassmann M, Fakler B, Schaeren-Wiemers N, Bettler B. Distribution of the auxiliary GABAB receptor subunits KCTD8, 12, 12b, and 16 in the mouse brain. J Comp Neurol. 2011;519:1435–1454. doi: 10.1002/cne.22610. [DOI] [PubMed] [Google Scholar]
- 54.Di Pasquale G, Davidson BL, Stein CS, Martins I, Scudiero D, Monks A, et al. Identification of PDGFR as a receptor for AAV-5 transduction. Nat Med. 2003;9:1306–1312. doi: 10.1038/nm929. [DOI] [PubMed] [Google Scholar]
- 55.Ballana E, Wang J, Venail F, Estivill X, Puel J, Arbonès ML, et al. Efficient and specific transduction of cochlear supporting cells by adeno-associated virus serotype 5. Neurosci Lett. 2008;442:134–139. doi: 10.1016/j.neulet.2008.06.060. [DOI] [PubMed] [Google Scholar]
- 56.Ukkola LT, Onkamo P, Raijas P, Karma K, Järvelä I. Musical aptitude is associated with AVPR1A-haplotypes. PLoS One. 2009;4:e5534. doi: 10.1371/journal.pone.0005534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karma K. Auditory and Visual Temporal Structuring: How Important is Sound to Musical Thinking? Psychology of Music. 1994;22:20–30. [Google Scholar]
- 58.Tervaniemi M, Ilvonen T, Karma K, Alho K, Näätänen R. The musical brain: brain waves reveal the neuropsychological basis of musicality in human subjects. Neuroscience letters. 1997;226:1–4. doi: 10.1016/s0304-3940(97)00217-6. [DOI] [PubMed] [Google Scholar]
- 59.Salimpoor VN, van den Bosch I, Kovacevic N, McIntosh AR, Dagher A, Zatorre RJ. Interactions between the nucleus accumbens and auditory cortices predict music reward value. Science. 2013;340:216–219. doi: 10.1126/science.1231059. [DOI] [PubMed] [Google Scholar]
- 60.Herholz SC, Zatorre RJ. Musical Training as a Framework for Brain Plasticity: Behavior, Function, and Structure. Neuron. 2012;76:486–502. doi: 10.1016/j.neuron.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 61.Mäkinen V, Parkkonen M, Wessman M, Groop P, Kanninen T, Kaski K. High-throughput pedigree drawing. Eur J Hum Genet. 2005;13:987–989. doi: 10.1038/sj.ejhg.5201430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.